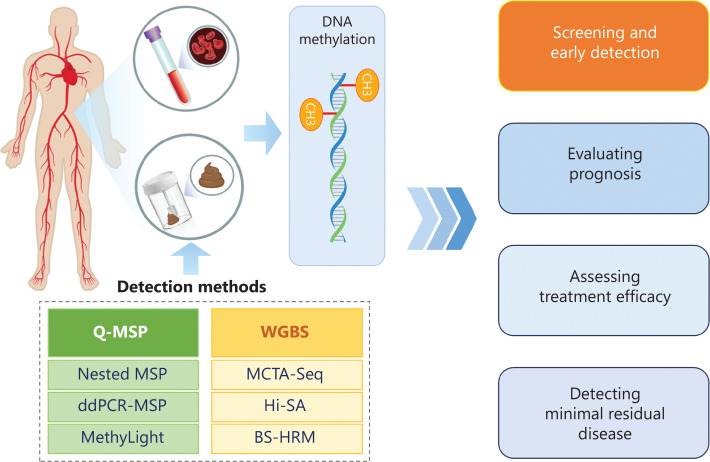
Cancer is one of the leading causes of death worldwide. The early diagnosis of cancer remains one of the greatest cancer research challenges. Epigenetic alterations, such as altered DNA methylation, that occur during the early stages of carcinogenesis have been proposed as candidate cancer biomarkers. In recent years detection of small amounts of methylated DNA in samples, including blood and stool, has demonstrated the feasibility of DNA methylation as a molecular cancer biomarker. The translational promise of aberrant DNA methylation includes screening and detecting cancer, evaluating prognosis, assessing treatment efficacy, and detecting minimal residual disease (Figure 1). The application of DNA methylation biomarkers for cancer detection has been studied most intensively. Alterations in DNA methylation patterns in the genome have been observed across malignancies and usually occur before other detectable genetic changes1. Therefore, biomarker mining for the early diagnosis of cancer based on DNA methylation has emerged as a promising field and has become a focus of research globally. Although hundreds of DNA methylation biomarkers have displayed great potential for early cancer detection, only a few methylation biomarkers have been used in the clinical setting to date. The National Medical Products Administration (NMPA) in China has approved 20 methylation-based commercial kits for cancer diagnosis. More than one-half of these kits are used for colorectal cancer (CRC) diagnosis (11); one kit is used for gastric cancer, three for cervical cancer, two for lung cancer, and the remaining three are used for the diagnosis of gliomas, and liver and bladder cancers. In the US, seven DNA methylation-based assays are available commercially to help clinicians make better treatment decisions in patients with cancer2. Two assays can be used to detect CRC and one can be used to detect > 50 types of tumors.
Figure 1.
Main technologies for DNA methylation detection and clinical applications.
Unlike Western countries, gastric cancer and CRC are highly prevalent in China with > 480,000 patient-related deaths, accounting for 20.1% of all cancer-related deaths3. The incidence of CRC in China has rapidly increased. CRC currently ranks second with respect to morbidity among all malignancies3. The incidence of gastric cancer in China is among the highest worldwide, accounting for > 45% of all new gastric cancer cases3. Gastric cancer and CRC have a poor prognosis and are difficult to diagnose in the early stages due to a lack of characteristic clinical manifestations. In high-risk groups, endoscopy with tissue biopsies is the gold standard for diagnosing gastric cancer and CRC; however, endoscopy is invasive and highly dependent on the judgment and experience of the endoscopic specialist. Unfortunately, the currently available protein markers, such as CEA, CA19-9 and CA72-4, are ineffective in detecting early-stage gastrointestinal cancer owing to a low sensitivity. There is an ongoing quest for reliable non-invasive biomarkers with better sensitivity and specificity for the detection of gastrointestinal cancer to complement the currently available screening methods. Gastric cancer and CRC share many biological features. For example, both stomach and colorectum epithelia are derived from endoderm. Normal cells undergo a hyperplasia-neoplasia-cancerous process during tumorigenesis to become cancerous. Notably, gastric cancer and CRC share many aberrant DNA methylations, including SEPT9, MGMT, and SDC2. Therefore, in this perspective we focused on the progress in research involving DNA methylation-based diagnostics for gastric cancer and CRC screening and early detection.
Clinical applications of DNA methylation biomarkers for detecting early gastrointestinal cancers
Colorectal cancer
Screening for CRC using a fecal immunochemical test (FIT) has been shown to reduce CRC-related mortality; however, a FIT is limited by relatively low specificity and sensitivity for early CRC detection. Recently, several methylated genes have been studied epigenetically as alternative biomarkers to FIT.
Blood-based DNA methylation biomarkers for screening and early detection of CRC
To date, several potential blood-based DNA methylation biomarkers have been identified for CRC detection, including BCAT1, BMP3, C9orf50, CDKN2A, CLIP4, KCNQ5, MLH1, NDRG4, PRIMA1, SDC2, SEPT9, SFRP2, and VIM2,4 (Table 1). In fact, the best-known blood epigenetic marker for CRC is SEPT9. Methylated SEPT9 is the only single-gene methylation biomarker approved by the U.S. Food & Drug Administration (FDA) for CRC detection, as well as the first methylation biomarker approved by the NMPA in China. Methylation changes in SEPT9, a member of the septin family, which is involved in cytokinesis and cytoskeletal organization, have been linked to multiple cancers. In case-control and opportunistic screening studies, plasma methylated SEPT9 demonstrated approximately 70% sensitivity and 90% specificity for detecting CRC4. In a large prospective CRC screening cohort, the sensitivity and specificity of methylated SEPT9 were estimated to be 48.2% and 91.5%, respectively5. Furthermore, among patients with TNM and Duke stage progression, the positive methylated SEPT9 rates gradually increase6. Of note, the criteria for determining methylated SEPT9 positivity vary across studies. For example, in some studies a positive reaction was indicated by a methylated SEPT9 curve exceeding the prespecified threshold of 50 polymerase chain reaction (PCR) cycles5, whereas a predetermined threshold of 45 PCR cycles was applied in other studies7. In addition, there is inconsistency in the PCR repeat systems used across different studies; most studies use triplicate PCR reactions, while other studies use double replicates5. Therefore, the methylated SEPT9 test performance across studies may reflect differences in the study populations, different interpretation thresholds among commercially available kits, and differences between study settings (retrospective case-control study vs. opportunistic vs. population-based screening).
Table 1.
Overview of promising DNA methylation biomarkers used in the diagnosis of CRC and adenomas
| Gene | Test | Sample type | Cohort size |
Sensitivity (%) | Specificity (%) | Method | NMPA approval (date) | Reference PMID | |
|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | ||||||||
| Single methylated gene | |||||||||
| BMP3 | CRC | Blood | 50 | 45 | 40 | 94 | BS-HRM | No | 29892846 |
| Stool | 40 | 35 | 40 | 85 | MSP | No | 29142517 | ||
| AA | Stool | 40 | 36 | 33.3 | 85 | MSP | No | 29142517 | |
| CDKN2A | CRC | Blood | 10 | 52 | 38 | 100 | MSP | No | 11801557 |
| Stool | 31 | 30 | 40 | 96.8 | MSP | No | 21033217 | ||
| Adenoma | Stool | 31 | 25 | 24 | 96.8 | MSP | No | 21033217 | |
| MGMT | CRC | Stool | 24 | 52 | 48.1 | 100 | MSP | No | 17352030 |
| Adenoma | Stool | 24 | 21 | 28.6 | 100 | MSP | No | 17352030 | |
| MLH1 | CRC | Blood | 19 | - | 33 | 100 | PCR | No | 11221878 |
| NDRG4 | CRC | Blood | 16 | 84 | 54.8 | 78.1 | Nested MSP | No | 25663916 |
| Stool | 16 | 84 | 76.2 | 89.1 | Nested MSP | No | 25663916 | ||
| AA | Stool | 40 | 36 | 27.8 | 80 | MSP | No | 29142517 | |
| PRIMA1 | CRC | Blood | 37 | 47 | 80.9 | 73 | MSP | No | 28753106 |
| Adenoma | Blood | 37 | 37 | 70.3 | 73 | MSP | No | 28753106 | |
| SDC2 | CRC | Blood | 125 | 131 | 87.0 | 95.2 | MSP | No | 23747112 |
| Stool | 713 | 359 | 83.8 | 98.0 | MSP | Yes | 33126908 | ||
| Adenoma | Blood | 37 | 37 | 81.1 | 97.3 | MSP | No | 28753106 | |
| AA | Stool | 713 | 38 | 42.1 | 98.0 | MSP | Yes | 33126908 | |
| SEPT9 | CRC | Blood | 295 | 291 | 76.6 | 95.9 | MSP | Yes (2015) | 27133379 |
| Stool | 76 | 72 | 83.3 | 92.1 | qMSP | No | 32373158 | ||
| Adenoma | Blood | 295 | 214 | 9.8 | 95.9 | MSP | No | 27133379 | |
| AA | Blood | 81 | 13 | 30.8 | 90.1 | qMSP | No | 32373158 | |
| AA | Stool | 76 | 12 | 66.7 | 92.1 | qMSP | No | 32373158 | |
| SFRP2 | CRC | Blood | 37 | 47 | 72.3 | 89.2 | MSP | No | 28753106 |
| Stool | 40 | 35 | 60.0 | 87.5 | MSP | No | 29142517 | ||
| Adenoma | Blood | 37 | 37 | 83.8 | 89.2 | MSP | No | 28753106 | |
| AA | Stool | 40 | 36 | 27.8 | 87.5 | MSP | No | 29142517 | |
| TFPI2 | CRC | Stool | 53 | 61 | 93.4 | 94.3 | qMSP | No | 33958894 |
| Adenoma | Stool | 53 | 16 | 81.3 | 94.3 | qMSP | No | 33958894 | |
| Vimentin | CRC | Blood | 110 | 81 | 59 | 93 | qMSP | No | 19684580 |
| Stool | 38 | 22 | 41 | 95 | qMSP | No | 19684580 | ||
| AA | Stool | 38 | 20 | 45 | 95 | qMSP | No | 19684580 | |
| Methylated gene panel | |||||||||
| NDRG4, BMP3, mutation KRAS, hemoglobin | CRC | Stool | 9167 | 65 | 92.3 | 86.6 | Multitarget assay | Yes (2020) | 24645800 |
| AA | Stool | 9167 | 757 | 42.4 | 86.6 | ||||
| C9orf50, KCNQ5, CLIP4 | CRC | Blood | 91 | 143 | 85 | 99 | ddPCR | No | 31727158 |
| MGMT, hMLH1, Vimentin | CRC | Stool | 37 | 60 | 75.0 | 86.5 | MSP | No | 19617759 |
| Adenoma | Stool | 37 | 52 | 59.6 | 86.5 | No | |||
| SFRP2, TFPI2, NDRG4, BMP3 | CRC | Stool | 40 | 35 | 94.3 | 55.0 | MSP | No | 29142517 |
| AA | Stool | 40 | 36 | 72.2 | 55.0 | No | |||
| SDC2, TFPI2 | CRC | Stool | 217 | 289 | 96.6 | 96.4 | MSP | Yes (2022) | 35004840 |
| Adenoma | Stool | 217 | 190 | 80.0 | 95.7 | ||||
| SDC2, SFRP2 | CRC | Stool | 1345 | 42 | 92.9 | 93.3 | MSP | Yes (2022) | 34933958 |
| AA | Stool | 1345 | 302 | 35.1 | 93.3 | ||||
| SEPT9, SDC2, BCAT1 | CRC | Blood | 60 | 104 | 82.7 | 96.9 | MSP | Yes (2022) | 34382948 |
| SDC2, NPY, FGF5, PDX1 | CRC | Stool | 856 | 419 | 91.2 | 91.1 | MSP | Yes (2023) | NA& |
| AA | Stool | 856 | 124 | 75.8 | 91.1 | ||||
&Retrieved from https://www.nmpa.gov.cn. AA, advanced adenomas; BS-HRM, bisulfite-specific high-resolution melting analysis; CRC, colorectal cancer; MSP, methylation-specific PCR; qMSP, quantitative methylation-specific PCR.
Stool-based DNA methylation biomarkers for screening and early detection of CRC
In addition to blood, stool is another promising sample source for CRC detection. Cancer cells released from tumor tissues accumulate in the stool, forming the basis for stool testing to identify tumor-specific hypermethylation changes and gene mutations. Numerous hypermethylated genes, including BMP3, CDKN2A, FGF5, hMLH1, MGMT, NDRG4, NPY, PDX1, SDC2, SEPT9, SFRP2, TFPI2, and VIM, have been analyzed in fecal DNA for CRC early detection2,4 (Table 1). Among these methylation-based CRC diagnostic biomarkers, methylated VIM, BMP3, NDRG4, and SDC2 have demonstrated robustness for clinical use. Methylated VIM was the first stool-based methylation biomarker approved for CRC detection8; however, a meta-analysis involving 8 studies concluded unsatisfactory diagnostic performance of methylated VIM, with a sensitivity of 54.6% and a specificity of 88.5%9. Methylated SDC2 was the first stool-based methylation assay for CRC detection approved by the NMPA in China. The sensitivity of methylated SDC2 in fecal DNA for CRC was 83.8%, 42.1% for advanced adenomas, and 87.0% for early-stage CRC (stage I-II)10. Methylated SDC2 appears to be the most accurate single gene among stool DNA methylation tests for detecting CRC based on a meta-analysis9, albeit large-sample clinical trials are needed for further validation.
Combined detection of multiple targets
Although single-gene methylation biomarkers have demonstrated promising specificity for CRC, the sensitivity is insufficient. Therefore, multigene combined testing, which has attracted much attention in recent years, may improve the sensitivity of CRC detection. Imperiale et al.11 proposed the use of FIT in addition to assessing KRAS mutations, aberrant NDRG4, and BMP3 methylation for the early detection of CRC in stool samples. FIT demonstrated a 73.8% sensitivity and 94.9% specificity when used independently in CRC detection, and a 92.3% sensitivity and 86.6% specificity when combined with DNA testing11. Although the sensitivity of the multitarget stool DNA test did not vary significantly according to cancer stage or location within the colon, the sensitivity was relatively higher in distal advanced precancerous lesions than in proximal lesions (54.5% vs. 33.2%)11. This panel of multitarget stool DNA tests has been approved by the U.S. FDA and the NMPA in China for CRC diagnosis. In addition, the NMPA in China has approved several novel multigene methylation stool test kits for the detection of CRC, including SDC2/TFPI2, SDC2/SFRP2, SEPT9/SDC2/BCAT1, and SDC2/NPY/FGF5/PDX1 (Table 1). The specificity of multigene combined testing is slightly lower than single-gene methylation testing, but the sensitivity is significantly better, which implies that multitarget combination testing is a promising future research domain.
Strengths and weaknesses between blood- and stool-based DNA methylation biomarkers for CRC detection
No head-to-head studies have compared the efficacy of these commercially available methylated gene detection kits in the same patient cohort. Based on studies with small sample sizes, methylated gene detection in stool samples did not demonstrate superiority over the detection of the same genes in plasma samples (Table 1). Of note, a blood sample can be obtained safely and objectively at any time, while a stool sample may not be collectible on demand. It is difficult to control feces quality and the characteristics of feces, such as loose or watery stools, may affect the test results. Moreover, fecal methylation testing cannot be used to monitor recurrence after surgical resection. Notably, the methylation biomarker detection rate in advanced adenomas was relatively low whether serum, plasma, or feces was analyzed. Although several methylation detection kits have been approved by the NMPA in China, it is important to note that the kits are a supplement to colonoscopy, not a replacement.
Gastric cancer
Although early screening for gastric cancer via gastroscopy may improve overall survival12, the availability of reliable, simple, and non-invasive screening tests is more limited than for CRC. Several studies have recently been conducted to identify DNA methylation-based biomarkers in the plasma, serum, gastric juice, and fecal samples for gastric cancer diagnosis, albeit with varying specificity and sensitivity13. Early detection and in vitro diagnostics for gastric cancer have yet to reach clinics en masse.
Blood-based DNA methylation biomarkers for screening and early detection of gastric cancer
Several potential blood-based diagnostic methylation biomarkers have been identified for gastric cancer detection, including C13orf18, DLEC1, FLNC, HODX10, MGMT, PCDH10, RNF180, RPRM, RPRML, RUNX3, SEPT9, SFRP2, SOX17, THBS1, UCHL1, and ZNF56913,14 (Table 2). RNF180 is one of the ring finger protein genes involved in the degradation of its substrates as an E3 ubiquitin ligase. Genes belonging to this family have been implicated in various biological processes, including cell growth, differentiation, and tumorigenesis15. Our previous study showed that the average methylation rate and methylated CpG sites within the RNF180 promoter region in tissues increased with the severity of gastric mucosal lesions16,17. Therefore, methylated RNF180 may serve as a candidate biomarker for gastric cancer. As mentioned earlier, methylated SEPT9 has been identified as a non-invasive diagnostic biomarker for CRC; however, methylated SEPT9 is not CRC-specific. Elevated levels of methylated SEPT9 have been observed in various cancers, with 48%–56% of gastric cancer patients also testing positive for methylated SEPT918. One study reported that the RS19 test is a new blood-based methylation assay for early gastric cancer detection that combines two methylated genes (RNF180 and SEPT9) in a single reaction to improve the rate for early-stage gastric cancer and gastric dysplasia detection14. The RS19 test is an effective approach with good sensitivity (62.2%) and high specificity (84.8%) for detecting gastric cancer14. The plasma RS19 test has higher sensitivity than methylated SEPT9 or RNF180 alone in detecting gastric cancer and gastric dysplasia14. This study had the largest reported sample size, exceeding 1000 cases14. The RS19 test is the first epigenetic biomarker approved by the NMPA in China for detecting gastric cancer and is commercially available. Currently, the authors are conducting a multicenter community-based gastrointestinal cancer screening program using methylated RNF180, SEPT9, FIT, and Helicobacter pylori stool antigen (NCT05996458). In addition, another retrospective study presented a DNA methylation-based panel (ELMO1, ZNF569, and C13orf18) for distinguishing gastric cancer19. The study was limited by a relatively small sample size (36 patients with gastric cancer and 38 controls). It is anticipated that results from a larger study on screening, surveillance, or other intended-use populations will provide additional confirmation. Ongoing clinical trials are currently exploring the performance of novel blood DNA methylation-based panels for gastric cancer diagnosis (clinical trials.gov: NCT04511559, NCT04947995, NCT05224596, NCT05336058, NCT05347524, and NCT05668910; https://www.chictr.org.cn/: ChiCTR2300075157). Additional methylation kits for gastric cancer screening may become available for clinical use in the future.
Table 2.
Overview of promising DNA methylation biomarkers used in the diagnosis of gastric cancer
| Methylated sites | Sample type | Cohort size |
Sensitivity (%) | Specificity (%) | Method | NMPA approval (date) | Reference PMID | |
|---|---|---|---|---|---|---|---|---|
| Controls | Cases | |||||||
| Single methylated gene | ||||||||
| DLEC1 | Blood | 40 | 82 | 80.5 | 93 | Q-MCP | No | 26550574 |
| FLNC | Blood | 40 | 82 | 67.1 | 93 | Q-MCP | No | 26550574 |
| HODX10 | Blood | 34 | 131 | 48.1 | 80 | MSP | No | 28529617 |
| PCDH10 | Blood | 202 | 101 | 94.1 | 97.03 | MSP | No | 27330867 |
| RNF180 | Blood | 527 | 650 | 46.2 | 87.3 | MSP | No | 37584087 |
| RPRM | Blood | 88 | 96 | 47 | 93 | MSP | No | 32431794 |
| RPRML | Blood | 25 | 25 | 56 | 88 | MethyLight | No | 33322837 |
| RUNX3 | Blood | 34 | 131 | 42.7 | 79.2 | MSP | No | 28529617 |
| SDC2 | Stool | 90 | 66 | 40.9 | 93.3 | PCR | No | 33765723 |
| SEPT9 | Blood | 527 | 650 | 40.0 | 96.0 | MSP | No | 37584087 |
| SFRP2 | Blood | 50 | 92 | 60.9 | 86 | Q-PCR | No | 32379490 |
| SOX17 | Blood | 20 | 73 | 58.9 | 100 | MSP | No | 23403728 |
| THBS1 | Blood | 40 | 82 | 63.4 | 94.2 | Q-MCP | No | 26550574 |
| TERT | Stool | 90 | 66 | 36.4 | 90 | PCR | No | 33765723 |
| RASSF2 | Stool | 90 | 66 | 31.8 | 93.3 | PCR | No | 33765723 |
| SFRP2 | Stool | 90 | 66 | 22.7 | 90 | PCR | No | 33765723 |
| UCHL1 | Blood | 40 | 82 | 56.1 | 89.5 | Q-MCP | No | 26550574 |
| ZIC1 | Blood | 34 | 131 | 69.5 | 69.2 | MSP | No | 28529617 |
| Methylated gene panel | ||||||||
| CABIN1, DOCK10, KCNQ5 | Blood | 82 | 89 | 64 | 93 | MCTA-Seq | No | 34791072 |
| ELMO1, ZNF569, C13orf18 | Blood | 38 | 36 | 86 | 95 | MSP | No | 29844130 |
| HODX10, RUNX3 | Blood | 34 | 131 | 72.5 | 65 | MSP | No | 28529617 |
| MGMT, p15, hMLH1 | Blood | 22 | 20 | 75 | 54 | MSP | No | 18837952 |
| RASSF2, SFRP2 | Stool | 101 | 21 | 57.1 | 89.4 | Hi-SA | No | 19700653 |
| RNF180, SEPT9 | Blood | 527 | 650 | 62.2 | 84.8 | MSP | Yes (2020) | 37584087 |
| RPRM, RUNX3 | Blood | 88 | 96 | 82 | 89 | MSP | No | 32431794 |
| SDC2, TERT, hemoglobin | Stool | 90 | 66 | 66.7 | 78.9 | PCR | No | 33765723 |
| WIF1, SDC2, TFPI2, NDRG4 | Stool | 107 | 35 | 67.5 | 97.81 | ColoCaller | No | 35419280 |
| ZIC1, RUNX3, HODX10 | Blood | 34 | 131 | 91.6 | 50 | MSP | No | 28529617 |
MSP, methylation-specific PCR; MCTA-seq, methylated CpG tandem amplification and sequencing; Q-PCR, quantitative real-time PCR; q-MSP, quantitative methylation-specific PCR; Hi-SA, high-sensitivity assay for bisulfite DNA.
Stool-based DNA methylation biomarkers for screening and early detection of gastric cancer
Unlike CRC, only a few studies have investigated stool-based DNA methylation biomarkers for gastric cancer diagnosis (Table 2). Because the shedding of gastric tumor cells occurs in the upper gastrointestinal tract, tumor DNA passes through the intestines and is expelled from the body with feces after exposure to gastric acid, bile, and digestive enzymes. As a result, there is a minimal amount of tumor DNA available for testing in the stool. Existing studies have also shown that stool DNA methylation-based biomarkers do not exhibit good performance in detecting gastric cancer.
Future developments and perspective
Although DNA methylation biomarkers outperform traditional markers, such as CEA, CA19-9, and CA125, in diagnosing early-stage gastric cancer and CRC, the overall sensitivity and specificity remain insufficient to fully meet the needs of cancer screening, especially for gastric cancer. Importantly, the impact of DNA methylation biomarker-based screening on reducing the incidence and mortality of gastrointestinal cancer remains unclear. Another potential limitation of DNA methylation biomarkers for routine cancer screening is the higher cost. To address these challenges and needs, several considerations are essential. First, specific combination algorithms are needed to better consolidate existing DNA methylation biomarkers and traditional tumor markers to improve the sensitivity of early cancer detection. Second, the genome has approximately 28 million CpG sites, which have enormous potential for mining. Therefore, it is necessary to mine and integrate novel methylation biomarkers as diagnostic targets using genome-wide profiling. Third, large randomized controlled trials are needed to verify whether DNA methylation marker-based cancer screening can reduce the incidence and mortality of gastrointestinal cancer. Concurrent health and economic evaluations during such trials are necessary to assess cost-effectiveness. Fourth, there is a stepwise accumulation of DNA methylation of tumor suppressor genes from precancers-to-cancers. Understanding whether patients without neoplastic lesions who test positive for DNA methylation biomarkers have a higher risk of developing cancer than the general population is also crucial. Therefore, quantitative detection and dynamic observation of DNA methylation levels may be helpful for these patients to determine whether or not the lesion is malignant.
Presently, all DNA methylation kits approved by the NMPA in China are used for the diagnosis of a single cancer type. The advantage of biomarkers for single cancer screening is the relatively clear identification of the corresponding target lesion in patients with positive detection. Moreover, the sensitivity and specificity of a single cancer methylation gene for cancer detection are high and the cost is relatively low, which warrants further development. However, a drawback is that for whole-body screening, multiple markers need testing with a substantial increase in costs. Therefore, pan-cancer DNA methylation biomarkers are more suitable for individuals undergoing whole-body cancer screening. PATHFINDER evaluated a pan-cancer early-detection blood test based on DNA methylation signatures20. The latest study supports the feasibility of this blood test for multicancer early detection20. Unfortunately, this pan-cancer screening technique overlooks 80% of early-stage tumors (stage I-II), indicating substantial room for improvement in sensitivity20. The U.S. FDA has approved this test as a groundbreaking advance, marking the commencement of a new era of global early cancer screening. Given the rapid advances in sequencing and analytical and computational technologies, DNA methylation biomarkers are emerging as a significant advance in optimizing cancer screening.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant Nos 82202837, 81730016, and 81972761), and the National Key R & D Program of China (Grant Nos. 2016YFC1303200, 2022YFC2505100, and 2017YFC0908300).
Conflict of interest statement
No potential conflicts of interest are disclosed.
Author contributions
Conceived and designed the paper: Xianchun Gao and Yongzhan Nie.
Drafted the paper: Xianchun Gao, Hui Liu, Jun Yu.
Revised the paper: Yongzhan Nie.
References
- 1.Qian C, Zou X, Li W, Li Y, Yu W. The outpost against cancer: universal cancer only markers. Cancer Biol Med. 2023;20:806–15. doi: 10.20892/j.issn.2095-3941.2023.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davalos V, Esteller M. Cancer epigenetics in clinical practice. CA Cancer J Clin. 2023;73:376–424. doi: 10.3322/caac.21765. [DOI] [PubMed] [Google Scholar]
- 3.Maomao C, He L, Dianqin S, Siyi H, Xinxin Y, Fan Y, et al. Current cancer burden in China: epidemiology, etiology, and prevention. Cancer Biol Med. 2022;19:1121–38. doi: 10.20892/j.issn.2095-3941.2022.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatemi N, Tierling S, Es HA, Varkiani M, Mojarad EN, Aghdaei HA, et al. DNA methylation biomarkers in colorectal cancer: clinical applications for precision medicine. Int J Cancer. 2022;151:2068–81. doi: 10.1002/ijc.34186. [DOI] [PubMed] [Google Scholar]
- 5.Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–25. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun J, Fei F, Zhang M, Li Y, Zhang X, Zhu S, et al. The role of (m)SEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC cancer. 2019;19:450. doi: 10.1186/s12885-019-5663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu D, Zhou G, Jin P, Zhu J, Li S, Wu Q, et al. Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn. 2016;18:535–45. doi: 10.1016/j.jmoldx.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Grady WM, Yu M, Markowitz SD. Epigenetic alterations in the gastrointestinal tract: current and emerging use for biomarkers of cancer. Gastroenterology. 2021;160:690–709. doi: 10.1053/j.gastro.2020.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gachabayov M, Lebovics E, Rojas A, Felsenreich DM, Latifi R, Bergamaschi R. Performance evaluation of stool DNA methylation tests in colorectal cancer screening: a systematic review and meta-analysis. Colorectal Dis. 2021;23:1030–42. doi: 10.1111/codi.15521. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Liu S, Wang H, Zheng L, Zhou C, Li G, et al. Robust performance of a novel stool DNA test of methylated SDC2 for colorectal cancer detection: a multicenter clinical study. Clin Epigenetics. 2020;12:162. doi: 10.1186/s13148-020-00954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–97. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 12.Xu H, Li W. Early detection of gastric cancer in China: progress and opportunities. Cancer Biol Med. 2022;19:1622–8. doi: 10.20892/j.issn.2095-3941.2022.0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Dong Y, Zhang H, Zhao Y, Miao T, Mohseni G, et al. DNA methylation drives a new path in gastric cancer early detection: current impact and prospects. Genes Dis. 2024;11:847–60. doi: 10.1016/j.gendis.2023.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nie Y, Gao X, Cai X, Wu Z, Liang Q, Xu G, et al. Combining methylated SEPTIN9 and RNF180 plasma markers for diagnosis and early detection of gastric cancer. Cancer Commun (Lond) 2023;43:1275–9. doi: 10.1002/cac2.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung KF, Lam CN, Wu K, Ng EK, Chong WW, Cheng AS, et al. Characterization of the gene structure, functional significance, and clinical application of RNF180, a novel gene in gastric cancer. Cancer. 2012;118:947–59. doi: 10.1002/cncr.26189. [DOI] [PubMed] [Google Scholar]
- 16.Han F, Sun LP, Liu S, Xu Q, Liang QY, Zhang Z, et al. Promoter methylation of RNF180 is associated with h.Pylori infection and serves as a marker for gastric cancer and atrophic gastritis. Oncotarget. 2016;7:24800–9. doi: 10.18632/oncotarget.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang N, Gao X, Yuan Q, Fu X, Wang P, Cai F, et al. E3 ubiquitin ligase RNF180 prevents excessive PCDH10 methylation to suppress the proliferation and metastasis of gastric cancer cells by promoting ubiquitination of DNMT1. Clin Epigenetics. 2023;15:77. doi: 10.1186/s13148-023-01492-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song L, Chen Y, Gong Y, Wan J, Guo S, Liu H, et al. Opportunistic screening and survival prediction of digestive cancers by the combination of blood mSEPT9 with protein markers. Ther Adv Med Oncol. 2020;12:1758835920962966. doi: 10.1177/1758835920962966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson BW, Suh YS, Choi B, Lee HJ, Yab TC, Taylor WR, et al. Detection of gastric cancer with novel methylated DNA markers: discovery, tissue validation, and pilot testing in plasma. Clin Cancer Res. 2018;24:5724–34. doi: 10.1158/1078-0432.CCR-17-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schrag D, Beer TM, McDonnell CH, 3rd, Nadauld L, Dilaveri CA, Reid R, et al. Blood-based tests for multicancer early detection (PATHFINDER): a prospective cohort study. Lancet. 2023;402:1251–60. doi: 10.1016/S0140-6736(23)01700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]