ABSTRACT
Candida auris is an emerging yeast pathogen of major concern because of its ability to cause hospital outbreaks of invasive candidiasis and to develop resistance to antifungal drugs. A majority of C. auris isolates are resistant to fluconazole, an azole drug used for the treatment of invasive candidiasis. Mechanisms of azole resistance are multiple, including mutations in the target gene ERG11 and activation of the transcription factors Tac1b and Mrr1, which control the drug transporters Cdr1 and Mdr1, respectively. We investigated the role of the transcription factor Upc2, which is known to regulate the ergosterol biosynthesis pathway and azole resistance in other Candida spp. Genetic deletion and hyperactivation of Upc2 by epitope tagging in C. auris resulted in drastic increases and decreases in susceptibility to azoles, respectively. This effect was conserved in strains with genetic hyperactivation of Tac1b or Mrr1. Reverse transcription PCR analyses showed that Upc2 regulates ERG11 expression and also activates the Mrr1/Mdr1 pathway. We showed that upregulation of MDR1 by Upc2 could occur independently from Mrr1. The impact of UPC2 deletion on MDR1 expression and azole susceptibility in a hyperactive Mrr1 background was stronger than that of MRR1 deletion in a hyperactive Upc2 background. While Upc2 hyperactivation resulted in a significant increase in the expression of TAC1b, CDR1 expression remained unchanged. Taken together, our results showed that Upc2 is crucial for azole resistance in C. auris, via regulation of the ergosterol biosynthesis pathway and activation of the Mrr1/Mdr1 pathway. Notably, Upc2 is a very potent and direct activator of Mdr1.
IMPORTANCE
Candida auris is a yeast of major medical importance causing nosocomial outbreaks of invasive candidiasis. Its ability to develop resistance to antifungal drugs, in particular to azoles (e.g., fluconazole), is concerning. Understanding the mechanisms of azole resistance in C. auris is important and may help in identifying novel antifungal targets. This study shows the key role of the transcription factor Upc2 in azole resistance of C. auris and shows that this effect is mediated via different pathways, including the regulation of ergosterol biosynthesis and also the direct upregulation of the drug transporter Mdr1.
KEYWORDS: ergosterol, efflux pumps, antifungal resistance, zinc cluster transcription factor, Candida albicans, Candida glabrata
INTRODUCTION
Candida auris is an emerging yeast pathogen that has spread in all continents since 2009 (1). Five genotypically distinct clades have been identified in different geographical areas: Clade I (South Asia), Clade II (East Asia), Clade III (South Africa), Clade IV (South America), and Clade V (Iran) (2). Similarly to other Candida spp., C. auris can cause severe infections such as candidemia or other forms of invasive candidiasis, accounting now for a substantial and variable proportion (5%–30%) of these infections in some areas (2). Some particular features make C. auris a unique and distinct human fungal pathogen, such as its ability to cause nosocomial outbreaks and its potential for the rapid development of resistance to all antifungal drug classes (2). In particular, acquired resistance to fluconazole, an azole drug used for the treatment of invasive candidiasis, is common in C. auris, affecting most isolates (90%–100%) of Clades I and III and variable proportions (15%–60%) of Clades II and IV isolates (2–6).
Mechanisms of azole resistance in C. auris are multiple and close to those reported in other Candida spp., including mutations in the azole target gene (ERG11) and activation of the drug transporters Cdr1 and Mdr1 under the control of their respective transcription factors Tac1b and Mrr1 (7–10). Some gain-of-function (GOF) mutations of TAC1b and MRR1 conferring azole resistance in C. auris have been identified (8, 9, 11).
The transcription factor Upc2 is another key regulator of azole resistance in Candida spp., which has not yet been investigated in C. auris (12, 13). In Candida albicans and Candida glabrata, Upc2 controls ERG11 and other genes of the ergosterol biosynthesis pathway (13–17). Moreover, Upc2 was shown to have a possible regulatory function for the drug transporters Cdr1 and Mdr1 (18–21).
The aim of the present study was to assess the role of Upc2 in azole resistance of C. auris and to decipher its mechanisms and connections with other pathways of azole resistance.
RESULTS
Upc2 is crucial for azole resistance of C. auris
To assess the role of Upc2 in the antifungal resistance of C. auris, we first deleted UPC2 in a C. auris strain of Clade IV (IV.1) to generate the upc2Δ strain (Table 1). Deletion of UPC2 resulted in a significant decrease in minimal inhibitory concentration (MIC) for both fluconazole and voriconazole compared to the background strain (0.5 vs 4 µg/mL and 0.008 vs 0.03 µg/mL, respectively) (Table 2). We also observed a slight (one dilution) increase in amphotericin B MIC in the upc2Δ strain, while susceptibility to micafungin was not affected (Table 2).
TABLE 1.
Description of the C. auris strains in this study
| Isolate ID | Name | Description | Reference |
|---|---|---|---|
| 17 | IV.1 | Clinical isolate | (22) |
| JLY0133 | upc2Δ | IV.1 upc2Δ::NatR | This study |
| JLY0095 | UPC2HA | IV.1 PAHD1-UPC2-3XHA Tag-TACT1 ::SAT1 | This study |
| JLY0094 | TAC1bHA | IV.1 PAHD1-TAC1b-3XHA Tag-TACT1 ::SAT1 | This study |
| JLY0051 | MRR1HA | IV.1 PAHD1-MRR1-3XHA Tag-TACT1 ::SAT1 | This study |
| JLY0136 | TAC1bHA/upc2Δ | IV.1 PAHD1-TAC1b-3XHA Tag-TACT1 ::SAT1/upc2Δ::HygR | This study |
| JLY0137 | MRR1HA/upc2Δ | IV.1 PAHD1-MRR1-3XHA Tag-TACT1 ::SAT1/upc2Δ::HygR | This study |
| JLY0129 | UPC2HA/mdr1Δ | IV.1 PAHD1-UPC2-3XHA Tag-TACT1 ::SAT1/mdr1Δ::HygR | This study |
| JLY0130 | UPC2HA/mrr1Δ | IV.1 PAHD1-UPC2-3XHA Tag-TACT1 ::SAT1/mrr1Δ::HygR | This study |
TABLE 2.
MIC values to antifungal drugs of the different strains of this study
| Strain | Fluconazole MIC (µg/mL) |
Voriconazole MIC (µg/mL) |
Amphotericin B MIC (µg/mL) |
Micafungin MIC (µg/mL) |
|---|---|---|---|---|
| IV.1 | 4 | 0.03 | 2 | 0.125 |
| upc2∆ | 0.5 | 0.008 | 4 | 0.125 |
| UPC2HA | 32 | 0.125 | 1 | 0.125 |
| TAC1bHA | 128 | 2 | 2 | 0.125 |
| TAC1bHA/upc2∆ | 2 | 0.015 | 4 | 0.125 |
| MRR1HA | 32 | 0.125 | 2 | 0.125 |
| MRR1HA/upc2∆ | 1 | 0.015 | 4 | 0.125 |
| UPC2HA/mdr1∆ | 32 | 0.125 | 1 | 0.125 |
| UPC2HA/mrr1∆ | 16 | 0.06 | 1 | 0.125 |
We then generated a C. auris strain overexpressing UPC2 (UPC2HA) using the hyperactivation system for zinc cluster transcription factors by fusion of a 3xHA Tag at its C-terminal locus, as previously described (Table 1) (18, 23). HA tagging of Upc2p in the UPC2HA strain was confirmed by western blot analysis (Fig. S5). The UPC2HA strain exhibited higher MICs for both fluconazole and voriconazole compared to the IV.1 strain (32 vs 4 µg/mL and 0.12 vs 0.03 µg/mL, respectively) (Table 2). Of note, we also observed a slight (one dilution) decrease in amphotericin B MIC in the UPC2HA strain, while micafungin MIC was unchanged (Table 2).
Because overexpression of the drug transporters Cdr1 and Mdr1, under the control of the transcription factors Tac1b and Mrr1, respectively, represents important mechanisms of azole resistance in C. auris (8–11), we assessed the impact of UPC2 deletion in the background of Tac1b or Mrr1 hyperactivation. For this purpose, we generated strains with a C-terminal 3xHA Tag of TAC1b or MRR1 in the wild-type IV.1 strain (TAC1bHA and MRR1HA, respectively) and in the upc2Δ strain (TAC1bHA/upc2Δ and MRR1HA/upc2Δ, respectively; Table 1). Western blot analysis demonstrated HA tagging of Tac1bp and Mrr1p in the TAC1bHA and MRR1HA strains, respectively (Fig. S5). While the TAC1bHA and MRR1HA strains exhibited fluconazole and voriconazole MICs that were significantly higher compared to the IV.1 strain (i.e., considered fluconazole-resistant according to the tentative MIC breakpoints) (24), UPC2 deletion could restore azole susceptibility in the TAC1bHA/upc2Δ and MRR1HA/upc2Δ strains (Table 2).
These results show that Upc2 is required for azole resistance of C. auris.
As a next step, we investigated the mechanisms by which Upc2 controls azole resistance in C. auris and its connections with other pathways of azole resistance, such as the ergosterol biosynthesis pathway and the drug transporter systems Tac1b/Cdr1 and Mrr1/Mdr1. For this purpose, we performed real-time reverse transcription PCR (RT-PCR) analyses to measure UPC2, ERG11, TAC1b, CDR1, MRR1, and MDR1 expressions in the different mutant strains of this study.
Upc2 is independent of Tac1b and Mrr1
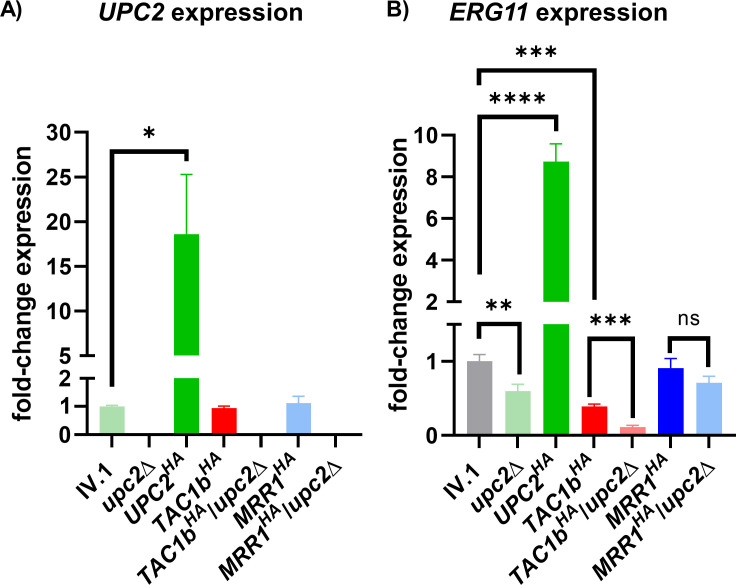
We first performed RT-PCR analyses of UPC2 expression (Fig. 1A). As expected, we observed a complete loss of UPC2 expression in the upc2Δ strain and a significant overexpression of UPC2 (18.61-fold) in the UPC2HA strain compared to the IV.1 strain. UPC2 expression was not affected by hyperactivation of Tac1b or Mrr1 in the TAC1bHA and MRR1HA strains, respectively. These results show that Upc2 is not regulated by Tac1b or Mrr1.
Fig 1.
Relative expression of UPC2 (A) and ERG11 (B) in the different strains of this study. Results are expressed as fold-change compared to the wild-type IV.1 strain. Bars represent means with standard deviations of three biological replicates. The brackets show the conditions that were compared using the t-test method and interpreted as statistically significant: * P-value ≤ 0.05, ** P-value ≤ 0.01, *** P-value ≤ 0.001, and **** P-value ≤ 0.0001; not statistically significant: ns.
Upc2 regulates Erg11
Because Upc2 is known to control the ergosterol biosynthesis pathway via regulation of Erg11 in other Candida spp. (13, 15, 16, 18), we also measured ERG11 expression (Fig. 1B). As expected, ERG11 transcript levels were significantly decreased in the upc2Δ strain and increased in the UPC2HA strain compared to the IV.1 strain. Hyperactivation of Tac1b induced a significant decrease in ERG11 expression, which was not the case following Mrr1 hyperactivation.
These results confirm that Upc2 controls azole resistance of C. auris via regulation of Erg11. We also observed that upregulation of Tac1b was associated with downregulation of the ergosterol biosynthesis pathway, possibly as a compensatory effect.
Upc2 induces azole resistance independently of Cdr1
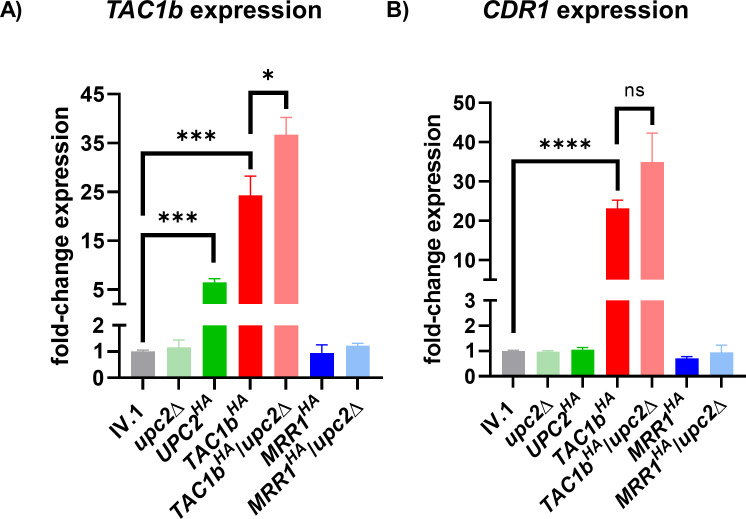
Because previous studies in C. glabrata suggest a link between Upc2, the transcription factor Pdr1 (ortholog of C. auris Tac1b), and its target the drug transporter Cdr1 (13, 21), we then measured TAC1b and CDR1 expressions in the different generated strains (Fig. 2A and B). While deletion of UPC2 did not result in significant changes in TAC1b and CDR1 expressions, we observed a significant increase in TAC1b expression but not CDR1, in the UPC2HA strain. As expected, the TAC1bHA strain displayed strong overexpression of both TAC1b and CDR1, which could not be reduced by UPC2 deletion. On the contrary, we observed a slight overexpression of TAC1b in the TAC1bHA/upc2Δ compared to the TAC1bHA strain, which may correspond to some compensatory effect.
Fig 2.
Relative expression of TAC1b (A) and CDR1 (B) in the different strains of this study. Results are expressed as fold-change compared to the wild-type IV.1 strain. Bars represent means with standard deviations of three biological replicates. The brackets show the conditions that were compared using the t-test method and interpreted as statistically significant: * P-value ≤ 0.05, ** P-value ≤ 0.01, *** P-value ≤ 0.001, and **** P-value ≤ 0.0001; not statistically significant: ns.
Expression of TAC1b and CDR1 was unchanged in the MRR1HA and MRR1HA/upc2Δ strains compared to the IV.1 strain, which suggests that the Tac1b/Cdr1 pathway is independent of Mrr1.
Taken together, these results show that Upc2 activation does not significantly impact CDR1 expression despite some upregulation of TAC1b.
Upc2 regulates Mrr1 and Mdr1
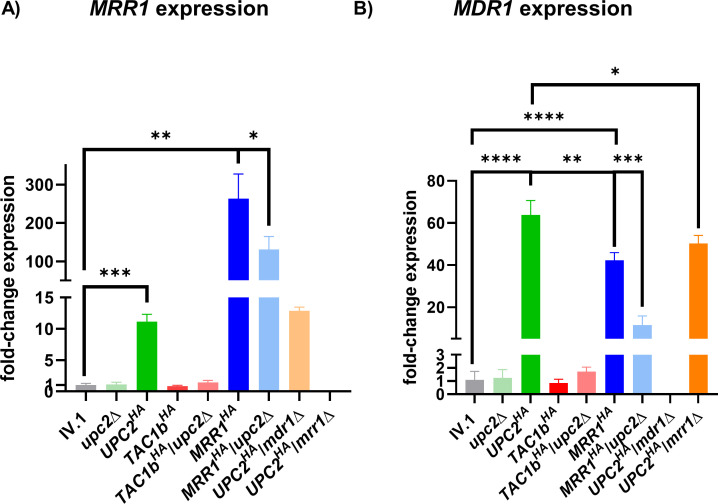
Because previous works in C. albicans have suggested a possible link between Upc2 and the Mrr1/Mdr1 pathway (14, 18), we then performed RT-PCR analyses of MRR1 and MDR1 expressions in the different mutant strains (Fig. 3A and B). No significant changes in MDR1 expression were found in the upc2Δ strain compared to the IV.1 strain. However, we observed a significant increase in MRR1 and MDR1 expressions in the UPC2HA strain. Notably, MDR1 expression resulting from Upc2 hyperactivation was particularly high (>60-fold compared to that of the wild-type IV.1 strain). As expected, hyperactivation of Mrr1 in the IV.1 strain resulted in significant upregulation of both MRR1 and MDR1. Of note, expression of MDR1 was significantly higher in the UPC2HA strain compared to the MRR1HA strain. UPC2 deletion in the MRR1HA background resulted in a significant decrease of both MRR1 and MDR1 expressions.
Fig 3.
Relative expression of MRR1 (A) and MDR1 (B) in the different strains of this study. Results are expressed as fold-change compared to the wild-type IV.1 strain. Bars represent means with standard deviations of three biological replicates. The brackets show the conditions that were compared using the t-test method and interpreted as statistically significant: * P-value ≤ 0.05, ** P-value ≤ 0.01, *** P-value ≤ 0.001, and **** P-value ≤ 0.0001.
No significant changes in MRR1 and MDR1 expressions were observed in the TAC1bHA and TAC1bHA/upc2Δ strains compared to the IV.1 strain, which further suggests that the Tac1b/Cdr1 and Mrr1/Mdr1 pathways are independent of each other. Globally, these results show that Upc2 can upregulate both Mrr1 and Mdr1.
Upc2 can control Mdr1 via mechanisms that are independent of Mrr1
To further investigate the link between Upc2 and the Mrr1/Mdr1 pathway, we generated two strains with deletions of MRR1 and MDR1 in the UPC2HA strain (UPC2HA/mrr1Δ and UPC2HA/mdr1Δ, respectively; Table 1). Fluconazole and voriconazole MICs were slightly decreased in the UPC2HA/mrr1Δ, but not in the UPC2HA/mdr1Δ strain, when compared to the UPC2HA strain (Table 2). RT-PCR analyses showed that MDR1 deletion had no impact on MRR1 expression in the UPC2HA strain background (Fig. 3A). Surprisingly, high level of MDR1 expression could be maintained in the Upc2-hyperactivated strain despite the loss of MRR1 (Fig. 3B). MDR1 expression in the UPC2HA/mrr1Δ strain was only slightly decreased compared to the UPC2HA strain and similar to that observed in the MRR1HA strain. On the contrary, a significant decrease in MDR1 expression was observed with the loss of UPC2 in the MRR1HA background.
These results demonstrate that Upc2 can directly regulate Mdr1 independently from Mrr1. Moreover, the role of Upc2 in controlling Mdr1 appears to be stronger than that of Mrr1. However, hyperactivated Upc2 can maintain azole resistance even in the absence of Mdr1, probably via its effect on the ergosterol biosynthesis pathway.
DISCUSSION
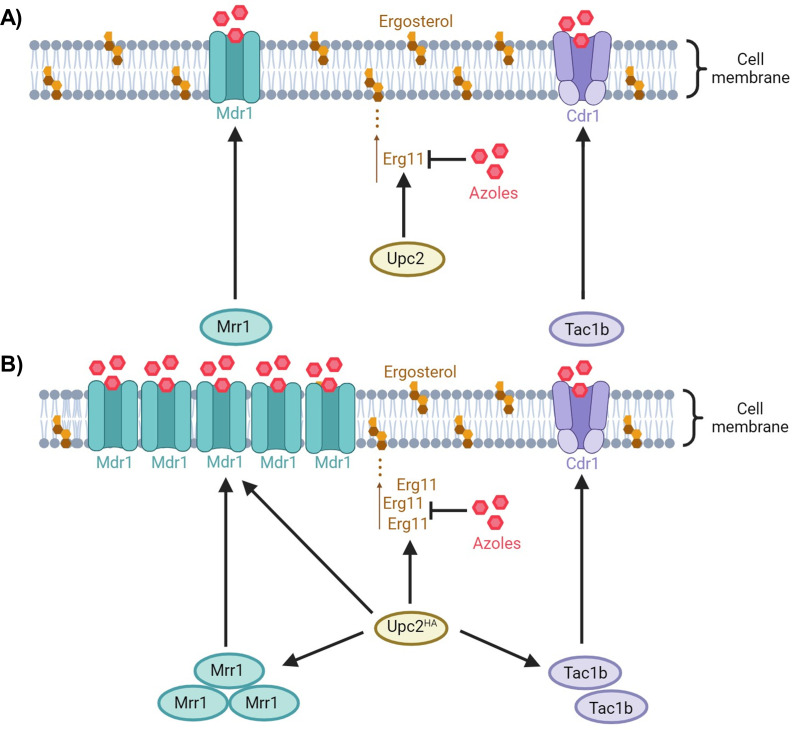
The transcription factor Upc2 was shown to be a key regulator of azole resistance in Candida spp. (12, 20). In this study, we analyzed the Upc2-dependent pathways of azole resistance in C. auris. Our results, summarized in the schematic representation of Fig. 4, show some similar and distinct mechanisms of Upc2-mediated azole resistance in C. auris compared to C. albicans or C. glabrata.
Fig 4.
Schematic representation of the role of Upc2 in azole resistance of C. auris. In standard conditions (A), Upc2 regulates Erg11 (target of the azole drugs). Upc2 hyperactivation [Upc2HA (B)] results in upregulation of Erg11. Moreover, Upc2 induces high level of Mdr1 by direct upregulation and indirect control via Mrr1. Although Upc2 hyperactivation results in some upregulation of Tac1b, Cdr1 levels remain unchanged. Black arrows = positive regulation.
First, Upc2 is known to regulate Erg11 and the ergosterol biosynthesis pathway in C. albicans and C. glabrata (12, 13, 15, 16, 18). In the present study, we demonstrated that Upc2 controls the same pathway in C. auris. Deletion of UPC2 resulted in decreased ERG11 expression and reduced resistance to fluconazole and voriconazole, while its hyperactivation led to the opposite effect.
Previous analyses in C. albicans showed that Upc2 regulates other genes involved in azole resistance, such as drug transporters of the major facilitator superfamily (MFS) or ATP-binding cassette (ABC) family (14, 17). Two of these drug transporters, Cdr1 (ABC) and Mdr1 (MFS), which are mainly controlled by the transcription factors Tac1 and Mrr1, respectively, are key regulators of azole resistance in Candida spp. (25, 26). Genome-wide location profiling analyses found that Upc2 binds to the promoters of MDR1 and CDR1 in C. albicans (17). However, the actual role of Upc2 in the regulation of these genes is unclear. Some GOF mutations in UPC2 were found to be associated with increased MDR1 expression in C. albicans (14). However, Mdr1 upregulation via Mrr1 hyperactivation or chemical compounds occurred independently of Upc2, while activation of Upc2 only resulted in a slight upregulation of Mdr1, which was dependent on Mrr1 (18). Among ABC transporters, only Cdr11, but not Cdr1, was found to be activated by Upc2 in C. albicans (14). In C. glabrata, deletion of UPC2A (UPC2 ortholog) in a strain harboring a GOF mutation of PDR1 (TAC1 ortholog) could drastically reduce azole resistance despite conserved high expression levels of CDR1 (13). However, its deletion in an azole-susceptible strain could partially reduce the fluconazole-induced expression of both PDR1 and CDR1 (13). Indeed, other studies showed that UPC2A could directly bind to the PDR1 and CDR1 promoters (19, 21). Interestingly, inhibition of the ergosterol biosynthesis pathway by different approaches led to increased expression of PDR1 and CDR1, which could be reduced following UPC2 deletion (21). Transcriptomic analyses in C. glabrata showed that Upc2 could induce expression of some membrane protein genes that are also upregulated by Pdr1, which further supports the link between Upc2 and the Pdr1/Cdr1 pathway in this yeast pathogen (20).
We investigated the link between Upc2 and the Tac1b/Cdr1 and Mrr1/Mdr1 pathways in C. auris. We found that strong hyperactivation of Tac1b or Mrr1 was not sufficient to maintain azole resistance in the absence of Upc2. These results are consistent with previous observations in C. albicans and C. glabrata (12, 13). We observed that hyperactivation of Upc2 resulted in a significant overexpression of TAC1b, which was, however, much inferior to that obtained in the Tac1b-hyperactivated strain and possibly therefore not sufficient to induce an increase in CDR1 expression. We concluded that Cdr1 upregulation was not sufficient to induce azole resistance in the absence of Upc2, as previously observed in C. glabrata (13). Our results also support a possible regulation of Tac1b by Upc2 in C. auris, which has been demonstrated in C. glabrata (21). However, this effect seems marginal in C. auris, as it did not result in any impact on CDR1 expression, despite high overexpression of UPC2.
The link between Upc2 and the Mrr1/Mdr1 pathway seems more relevant in C. auris according to our results. Indeed, we observed that Upc2 hyperactivation could induce significant overexpression of both MRR1 and MDR1. Interestingly, Upc2 hyperactivation reached a level of MRR1 expression that was lower compared to that observed in the Mrr1-hyperactivated strain but level of MDR1 expression that was higher. This suggested that Upc2 can control Mdr1 by mechanisms that are mediated by Mrr1 but also independent of Mrr1. This was further confirmed by demonstrating the ability of Upc2 to induce overexpression of MDR1 even in the absence of Mrr1. MDR1 expression was actually more impacted by UPC2 deletion in the hyperactive Mrr1 background than by MRR1 deletion in the hyperactive Upc2 background, which suggests that Upc2 may be a predominant transcription factor controlling Mdr1. These results differ from those previously reported in C. albicans, where the impact of Upc2 activation on MDR1 expression was modest and remained dependent on Mrr1 (18). The regulatory mechanisms behind the control of MDR1 by UPC2 remain unexplored, and future studies should address the occupancy of Upc2 in the MDR1 promoter of C. auris. Moreover, the actual relevance of this mechanism of azole resistance mediated by Mdr1 via Upc2 remains unclear. Indeed, we also observed that hyperactivation of Upc2 could maintain high level of azole resistance even in the absence of Mdr1, which suggests that the major impact of Upc2 on azole resistance is probably mediated via upregulation of Erg11.
Interestingly, our data also show that UPC2 deletion or hyperactivation had distinct effects on amphotericin B MIC (i.e., slight MIC increase and decrease, respectively), which may result from the impact of Upc2 on the ergosterol biosynthesis pathway.
In conclusion, this study shows that Upc2 is a major and universal regulator of azole resistance in C. auris, cross-talking with other important resistance pathways, as it has been previously demonstrated in other Candida spp. However, our results highlighted distinct features about the connections of Upc2 with other pathways of azole resistance in C. auris compared to C. albicans and C. glabrata. Indeed, in C. auris, the link between Upc2 and the Mrr1/Mdr1 pathway seems predominant when compared to the two other species, while the link with the Tac1b/Cdr1 pathway appears to be less relevant than that observed in C. glabrata.
GOF mutations in transcription factors have often been linked to drug resistance of fungal pathogens (27). In C. auris, such mutations have been identified in Tac1b and Mrr1 (8, 9, 11). Whether acquired GOF mutations of Upc2 may be responsible for azole resistance in C. auris clinical isolates is still unknown. Further analyses of clinical isolates are warranted to assess the actual relevance of Upc2-mediated azole resistance mechanisms in C. auris.
MATERIALS AND METHODS
Strains, plasmids, and media
The C. auris strains and the plasmids used in this study are listed in Table 1 and Table S1, respectively. Plasmids pJK795 containing the NatR cassette and pYM70 containing the HygR cassette were used as sources of nourseothricin and hygromycin resistances, respectively (28, 29). The plasmid Clp-pACT1-3xFLAG-MNase-SV40-CYC-SAT1 (a gift from Adnane Sellam, Institute of Cardiology of Montreal, Canada), which contains the nourseothricin resistance cassette SAT1 and C. auris neutral site CauNI, was used for constructs of the hyperactivated zinc cluster transcription factors (30). Isolates were grown in yeast extract-peptone-dextrose (YEPD) medium. Cultures were incubated for 16–20 h at 37°C on solid YEPD agar plates or in liquid YEPD under constant agitation (220 rpm).
The antifungal drugs fluconazole, voriconazole, amphotericin B (Sigma-Aldrich, St. Louis, MO), and micafungin (Selleck Chemicals, Houston, TX) were obtained as powder suspensions and dissolved in dimethyl sulfoxide (DMSO) as stocks of 10 mg/mL for fluconazole and 1 mg/mL for other drugs.
Escherichia coli DH5α was used as a host for plasmid constructions and propagation. DH5α was grown in Luria-Bertani broth or agar plates supplemented with ampicillin (100 µg/mL, AppliChem, Darmstadt, Germany) when required for 16–20 h at 37°C. Plasmids were extracted with the Plasmid Mini Kit (Qiagen, Hilden, Germany). All primers used in this study are listed in Table S2. All DNA sequences used for plasmid constructions were amplified from the genome of C. auris strain IV.1 unless otherwise specified.
Construction of plasmids for hyperactivation of zinc finger transcription factors
For hyperactivation of zinc cluster transcription factors, we used a model where the target gene is put under the control of the ADH1 promoter and tagged at its C-terminal locus by a 3xHA sequence, as previously described (Fig. S1) (18, 23). The ADH1 promotor was amplified by the primers ADH1p_PF_KpnI and ADH1p_PR_KasI_NheI. The PCR product digested by KpnI and NheI was cloned at the NheI/KpnI sites of the plasmid Clp-pACT1-3xFLAG-MNase-SV40-CYC-SAT1 to generate pjli6.
For the hyperactivation of Mrr1, the MRR1 nucleotide sequence was amplified by the primers MRR1_PF_KasI and MRR1_PR_BsrGI_Tag_CS_NruI_NheI (containing the 3xHA Tag sequence). The PCR product was digested by the primers KasI and NheI and cloned at KasI/NheI sites of pjli6 to generate pjli7. The ACT1 terminator was amplified by the primers ACT1t_PF_NheI and ACT1t_PR_NheI, and the PCR product was then cloned at NheI site of pjli7 to generate pjli8.
For the hyperactivation of Tac1b, the nucleotide sequence of TAC1b was amplified by the primers TAC1b_PF_KasI and TAC1b_PR_BsrGI. The PCR product was digested by KasI and BsrGI and then cloned at KasI/BsrGI sites of pjli8 to generate pjli11.
For the hyperactivation of Upc2, the nucleotide sequence of UPC2 was amplified by the primers UPC2_PF_KasI and UPC2_PR_BsrGI. The PCR product was digested by KasI and BsrGI and then cloned at KasI/BsrGI sites of pjli8 to generate pjli12. Absence of mutations resulting from PCR amplification was verified by sequencing. Plasmids pjli8, pjli11, and pjli12 were linearized by StuI for integration in the CauNI locus of the IV.1 strain to generate the strains MRR1HA, TAC1bHA, and UPC2HA (for hyperactivation of Mrr1, Tac1b, and Upc2, respectively).
Constructs for UPC2 deletion
For UPC2 deletion in the IV.1 strain, the construct was made by fusion PCR of three PCR products (Fig. S2A). The first PCR product consisted of the 5′ flanking region (580 bp) of UPC2 and was amplified with primers UPC2_del_PF1 and UPC2_del_PR1. The second PCR product consisted of the NatR cassette, which was amplified from pJK795 with primers UPC2_del_PF2 and UPC2_del_PR2. The third PCR product consisted of the 3′ flanking region (520 bp) of UPC2 and was amplified with primers UPC2_del_PF3 and UPC2_del_PR3. The three PCR products were purified with the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). The fusion PCR was performed with the nested primers UPC2_del_PF4 and UPC2_del_PR4 in the presence of 1.3 M of betaine.
For UPC2 deletion in the TAC1bHA and MRR1HA strains, the selection marker HygR (hygromycin resistance) was used in place of NatR (nourseothricin resistance), which was already used for the generation of these mutant strains (Fig. S2B). The same flanking regions of UPC2 were amplified using primers UPC2_del_PF1 and UPC2_del_PR1(hygro) for the 5′ flanking region and UPC2_del_PF3(hygro) and UPC2_del_PR3 for the 3′ flanking region. The HygR gene was amplified from the plasmid pYM70 with primers UPC2_del_PF2(hygro) and UPC2_del_PR2(hygro). The fusion PCR was performed with the nested primers UPC2_del_PF4 and UPC2_del_PR4.
The fusion PCR products were used for transformation in IV.1, TAC1bHA, and MRR1HA strains to generate strains upc2Δ, TAC1bHA/upc2Δ, and MRR1HA/upc2Δ, respectively (Fig. S3).
Constructs for MRR1 and MDR1 deletion in the UPC2HA strain
Construction of MDR1 and MRR1 deletion cassettes was performed as described in our previous publication, using the HygR cassette as a selection marker to substitute the target gene (8). The fusion PCR products were used for transformation in strain UPC2HA to generate strains UPC2HA/mrr1Δ and UPC2HA/mdr1Δ, with deletion of MRR1 and MDR1, respectively.
Transformations in C. auris
Transformations in C. auris were performed by a CRISPR-Cas9 approach. RNA-protein complexes (RNPs) reconstituted with purified Cas9 protein combined with scaffold- and gene-specific guide RNAs were used as previously described (31). Gene-specific RNA guides were designed to contain 20 bp homologous sequences of the upstream and downstream regions of the target region for integration. Sequences of RNA guides are shown in Table S2. The mix of the guide RNAs, the Cas9 endonuclease 3NLS (Integrated DNA Technologies Inc., Coralville, IA), and tracrRNA (universal transactivating CRISPR RNA) were prepared according to a previously described protocol (32).
Transformation of C. auris was performed by electroporation with about 1 µg of the constructs as previously described (32). Transformants were selected at 37°C on YEPD containing 200 µg/mL of nourseothricin (Werner BioAgents, Jena, Germany) for transformations in the IV.1 strain or 600 µg/mL of hygromycin B (Corning, Corning, NY) for transformations in the TAC1bHA and MRR1HA strains. Correct integration of the constructs was verified by PCR screening for all the HA-tagged strains (Fig. S4A and B), the UPC2 deletion strains (Fig. S6A through C), the MDR1 deletion strains (Fig. S7A and B), and the MRR1 deletion strains (Fig. S8A and B).
Protein extraction and western blot analysis
Protein extracts were obtained from whole cell extracts of the IV.1, MRR1HA, TAC1bHA, and UPC2HA strains after precipitation with trichloroacetic acid as previously described (33). Proteins were separated through electrophoresis using Mini-PROTEAN TGX gel (Bio-Rad Laboratories, Hercules, CA) and blotted onto nitrocellulose membranes. Mrr1p, Tac1bp, and Upc2p were detected by primary HA Tag Monoclonal Antibody (Invitrogen, ThermoFisher Scientific, Waltham, MA) with 1/2,500 dilution in 5% bovine serum albumin in phosphate-buffered saline with 1% Tween (BSA-PBS-T) and secondary Goat anti-Mouse IgG (H + L) antibody (Invitrogen, ThermoFisher Scientific, Waltham, MA) with 1/500 dilution in BSA-PBS-T.
Antifungal susceptibility testing
MIC of fluconazole, voriconazole, amphotericin B, and micafungin were determined for the C. auris isolates according to the procedure of the Clinical and Laboratory Standards Institute (CLSI, M27, 4th edition) (34).
RT-PCR
Each C. auris isolate was grown overnight in 5 mL of liquid YEPD under constant agitation at 37°C. Cultures were diluted to a density of 0.75 × 107 cells/mL in 5 mL of fresh YEPD and were grown at 37°C under constant agitation for 2–3 h to reach a density of 1.5 × 107 cells/mL. Total RNA was extracted with the Quick-RNA Fungal/Bacterial Miniprep Kit (Zymo Research, Freiburg im Breisgau, Germany), and total RNA extracts were treated with DNase using the DNA-Free Kit (Thermo Fisher Scientific Inc., Waltham, MA). The concentration of purified RNA was measured with a NanoDrop 1000 instrument (Witec AG, Sursee, Switzerland). RNA was stored at −80°C until use. Each isolate was prepared in biological triplicates.
One microgram of RNA of each isolate was converted into cDNA using the Transcriptor high-fidelity cDNA synthesis kit (Roche, Basel, Switzerland). RT-PCR was performed in 96-well plates using the PowerUp SYBR Green Master Mix (Applied Biosystems, Waltham, MA) with primers of the targeted genes (Supplementary Material S1) and supplemented with nuclease-free water up to 20 µL for each reaction. The primers used for ACT1, CDR1, MDR1, ERG11, TAC1b, MRR1, and UPC2 amplification were mentioned in Table S2. Each experiment was performed in biological triplicates and technical duplicates. The QuantStudio Software Program (Thermo Fisher Scientific Inc., Waltham, MA) including a melt curve stage was used for real-time PCR with activation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s, and annealing/extension at 60°C for 1 min. Gene expression was calculated with the threshold cycle (2−ΔΔCT) method and normalized to the ACT1 expression (35). Results were analyzed by the t-test method (GraphPad).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the Swiss National Science Foundation (SNSF, project number 310030_192611) and the Santos-Suarez Foundation.
Contributor Information
Frederic Lamoth, Email: Frederic.Lamoth@chuv.ch.
Dimitrios Kontoyiannis, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Kelly Ishida, Universidade de Sao Paulo, Sao Paulo, Brazil.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.03526-23.
Tables S1 and S2; Figures S1 to S8.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chakrabarti A, Singh S. 2020. Multidrug-resistant Candida auris: an epidemiological review. Expert Rev Anti Infect Ther 18:551–562. doi: 10.1080/14787210.2020.1750368 [DOI] [PubMed] [Google Scholar]
- 3. Escandón P, Chow NA, Caceres DH, Gade L, Berkow EL, Armstrong P, Rivera S, Misas E, Duarte C, Moulton-Meissner H, Welsh RM, Parra C, Pescador LA, Villalobos N, Salcedo S, Berrio I, Varón C, Espinosa-Bode A, Lockhart SR, Jackson BR, Litvintseva AP, Beltran M, Chiller TM. 2019. Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in amphotericin B resistance. Clin Infect Dis 68:15–21. doi: 10.1093/cid/ciy411 [DOI] [PubMed] [Google Scholar]
- 4. Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, Lee SY, Chun S, Lee JH, Choi HJ, Kee SJ, Kim SH, Shin MG. 2019. Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol 57:e01624-18. doi: 10.1128/JCM.01624-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maphanga TG, Naicker SD, Kwenda S, Muñoz JF, van Schalkwyk E, Wadula J, Nana T, Ismail A, Coetzee J, Govind C, Mtshali PS, Mpembe RS, Govender NP, for GERMS-SA . 2021. In vitro antifungal resistance of Candida auris isolates from bloodstream infections, South Africa. Antimicrob Agents Chemother 65:e0051721. doi: 10.1128/AAC.00517-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Szekely A, Borman AM, Johnson EM. 2019. Candida auris isolates of the Southern Asian and South African lineages exhibit different phenotypic and antifungal susceptibility profiles in vitro. J Clin Microbiol 57:e02055-18. doi: 10.1128/JCM.02055-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. doi: 10.1093/jac/dkx480 [DOI] [PubMed] [Google Scholar]
- 8. Li J, Coste AT, Bachmann D, Sanglard D, Lamoth F. 2022. Deciphering the Mrr1/Mdr1 pathway in azole resistance of Candida auris. Antimicrob Agents Chemother 66:e0006722. doi: 10.1128/aac.00067-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li J, Coste AT, Liechti M, Bachmann D, Sanglard D, Lamoth F. 2021. Novel ERG11 and TAC1b mutations associated with azole resistance in Candida auris. Antimicrob Agents Chemother 65:e02663-20. doi: 10.1128/AAC.02663-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rybak JM, Doorley LA, Nishimoto AT, Barker KS, Palmer GE, Rogers PD. 2019. Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob Agents Chemother 63:e00057-19. doi: 10.1128/AAC.00057-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rybak JM, Muñoz JF, Barker KS, Parker JE, Esquivel BD, Berkow EL, Lockhart SR, Gade L, Palmer GE, White TC, Kelly SL, Cuomo CA, Rogers PD, Berman J. 2020. Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio 11:e00365-20. doi: 10.1128/mBio.00365-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vasicek EM, Berkow EL, Flowers SA, Barker KS, Rogers PD. 2014. UPC2 is universally essential for azole antifungal resistance in Candida albicans. Eukaryot Cell 13:933–946. doi: 10.1128/EC.00221-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whaley SG, Caudle KE, Vermitsky JP, Chadwick SG, Toner G, Barker KS, Gygax SE, Rogers PD. 2014. UPC2A is required for high-level azole antifungal resistance in Candida glabrata. Antimicrob Agents Chemother 58:4543–4554. doi: 10.1128/AAC.02217-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flowers SA, Barker KS, Berkow EL, Toner G, Chadwick SG, Gygax SE, Morschhäuser J, Rogers PD. 2012. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell 11:1289–1299. doi: 10.1128/EC.00215-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. MacPherson S, Akache B, Weber S, De Deken X, Raymond M, Turcotte B. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob Agents Chemother 49:1745–1752. doi: 10.1128/AAC.49.5.1745-1752.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nagi M, Nakayama H, Tanabe K, Bard M, Aoyama T, Okano M, Higashi S, Ueno K, Chibana H, Niimi M, Yamagoe S, Umeyama T, Kajiwara S, Ohno H, Miyazaki Y. 2011. Transcription factors CgUPC2A and CgUPC2B regulate ergosterol biosynthetic genes in Candida glabrata. Genes Cells 16:80–89. doi: 10.1111/j.1365-2443.2010.01470.x [DOI] [PubMed] [Google Scholar]
- 17. Znaidi S, Weber S, Al-Abdin OZ, Bomme P, Saidane S, Drouin S, Lemieux S, De Deken X, Robert F, Raymond M. 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot Cell 7:836–847. doi: 10.1128/EC.00070-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schubert S, Barker KS, Znaidi S, Schneider S, Dierolf F, Dunkel N, Aïd M, Boucher G, Rogers PD, Raymond M, Morschhäuser J. 2011. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob Agents Chemother 55:2212–2223. doi: 10.1128/AAC.01343-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vu B.G, Moye-Rowley WS. 2022. Nonidentical function of Upc2A binding sites in the Candida glabrata CDR1 promoter. Genetics 222:iyac135. doi: 10.1093/genetics/iyac135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vu Bao Gia, Stamnes MA, Li Y, Rogers PD, Moye-Rowley WS, Heitman J. 2021. The Candida glabrata Upc2A transcription factor is a global regulator of antifungal drug resistance pathways. PLoS Genet 17:e1009582. doi: 10.1371/journal.pgen.1009582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vu BG, Thomas GH, Moye-Rowley WS, Goldman GH. 2019. Evidence that ergosterol biosynthesis modulates activity of the Pdr1 transcription factor in Candida glabrata. mBio 10:e00934-19. doi: 10.1128/mBio.00934-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Theill L, Dudiuk C, Morales-Lopez S, Berrio I, Rodríguez JY, Marin A, Gamarra S, Garcia-Effron G. 2018. Single-tube classical PCR for Candida auris and Candida haemulonii identification. Rev Iberoam Micol 35:110–112. doi: 10.1016/j.riam.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 23. Schillig R, Morschhäuser J. 2013. Analysis of a fungus-specific transcription factor family, the Candida albicans zinc cluster proteins, by artificial activation. Mol Microbiol 89:1003–1017. doi: 10.1111/mmi.12327 [DOI] [PubMed] [Google Scholar]
- 24. US Centers for Disease Control and Prevention (CDC) . Candida auris. Antifungal susceptibility testing and interpretation. Available from: https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html. Retrieved 27 Sep 2023.
- 25. Nishimoto AT, Sharma C, Rogers PD. 2020. Molecular and genetic basis of azole antifungal resistance in the opportunistic pathogenic fungus Candida albicans. J Antimicrob Chemother 75:257–270. doi: 10.1093/jac/dkz400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whaley SG, Rogers PD. 2016. Azole resistance in Candida glabrata. Curr Infect Dis Rep 18:41. doi: 10.1007/s11908-016-0554-5 [DOI] [PubMed] [Google Scholar]
- 27. Cowen LE, Sanglard D, Howard SJ, Rogers PD, Perlin DS. 2014. Mechanisms of antifungal drug resistance. Cold Spring Harb Perspect Med 5:a019752. doi: 10.1101/cshperspect.a019752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Basso LR Jr, Bartiss A, Mao Y, Gast CE, Coelho PSR, Snyder M, Wong B. 2010. Transformation of Candida albicans with a synthetic hygromycin B resistance gene. Yeast 27:1039–1048. doi: 10.1002/yea.1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shen J, Guo W, Köhler JR. 2005. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect Immun 73:1239–1242. doi: 10.1128/IAI.73.2.1239-1242.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tebbji F, Khemiri I, Sellam A, Mitchell AP. 2020. High-resolution genome-wide occupancy in Candida spp. using ChEC-seq. mSphere 5:e00646-20. doi: 10.1128/mSphere.00646-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grahl N, Demers EG, Crocker AW, Hogan DA, Mitchell AP. 2017. Use of RNA-protein complexes for genome editing in non-albicans Candida species. mSphere 2:e00218-17. doi: 10.1128/mSphere.00218-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kannan A, Asner SA, Trachsel E, Kelly S, Parker J, Sanglard D, Heitman J. 2019. Comparative genomics for the elucidation of multidrug resistance in Candida lusitaniae. mBio 10:e02512-19. doi: 10.1128/mBio.02512-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mamnun YM, Schüller C, Kuchler K. 2004. Expression regulation of the yeast PDR5 ATP-binding cassette (ABC) transporter suggests a role in cellular detoxification during the exponential growth phase. FEBS Lett 559:111–117. doi: 10.1016/S0014-5793(04)00046-8 [DOI] [PubMed] [Google Scholar]
- 34. Reference method for broth dilution antifungal susceptibility testing of yeasts, 4th edition, M27. 2017. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2; Figures S1 to S8.
An accounting of the reviewer comments and feedback.