ABSTRACT
The emergence and international dissemination of multi-drug resistant Staphylococcus aureus (S. aureus) strains challenge current antibiotic-based therapies, representing an urgent threat to public health worldwide. In the U.S. alone, S. aureus infections are responsible for 11,000 deaths and 500,000 hospitalizations annually. Biofilm formation is a major contributor to antibiotic tolerance and resistance-induced delays in empirical therapy with increased infection severity, frequency, treatment failure, and mortality. Developing novel treatment strategies to prevent and disrupt biofilm formation is imperative. In this article, we test the Secretion Modification Region (SMR) peptides for inhibitory effects on resistant S. aureus biofilm-forming capacity by targeting the molecular chaperone DnaK. The dose effect of SMR peptides on biofilm formation was assessed using microtiter plate methods and confocal microscopy. Interaction between the antagonist and DnaK was determined by immune precipitation with anti-Flag M2 Affinity and Western blot analysis. Increasing SMR peptide concentrations exhibited increasing blockade of S. aureus biofilm formation with significant inhibition found at 18 µM, 36 µM, and 72 µM. This work supports the potential therapeutic benefit of SMR peptides in reducing biofilm viability and could improve the susceptibility to antimicrobial agents.
IMPORTANCE
The development of anti-biofilm agents is critical to restoring bacterial sensitivity, directly combating the evolution of resistance, and overall reducing the clinical burden related to pervasive biofilm-mediated infections. Thus, in this study, the SMR peptide, a novel small molecule derived from the HIV Nef protein, was preliminarily explored for anti-biofilm properties. The SMR peptide was shown to effectively target the molecular chaperone DnaK and inhibit biofilm formation in a dose-dependent manner. These results support further investigation into the mechanism of SMR peptide-mediated biofilm formation and inhibition to benefit rational drug design and the identification of therapeutic targets.
KEYWORDS: bacterial biofilm, Staphylococcus aureus, SMR peptide, heat shock protein, DnaK
INTRODUCTION
Global clinical burden of Staphylococcus aureus
Staphylococcus aureus (S. aureus) is a ubiquitous gram-positive bacterium responsible for a plethora of community and nosocomial infections ranging from mild skin and soft tissue infections to life-threatening conditions, including pneumonia, meningitis, osteomyelitis, endocarditis, bacteremia, and toxic shock syndrome (1–3). The high abundance of S. aureus within the environment as well as within the flora of humans and animal species contribute to a large reservoir of virulence factors and antibiotic resistance tactics (4). Over the years, S. aureus has developed resistance to multiple antibiotics and has been designated as an ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogen (5, 6). According to the CDC’s Antibiotic Resistance Threats Report, S. aureus is continuing to readily disseminate dangerous antimicrobial resistance genes through mobile genetic elements (7, 8). The global spread of types and subtypes of methicillin-resistant S. aureus (MRSA) has led to an expanding public health crisis (9). Patients with MRSA frequently have chronic infections with a high risk for bacterial seeding to critical secondary sites resulting in severe acute infections and death (9). In a 2019 global mortality study analyzing 33 bacterial pathogens, S. aureus ranked within the top five causes of death (10).
S. aureus biofilm formation promotes antimicrobial tolerance and resistance
A large degree of the clinical burden due to S. aureus can be attributed, as Reffuveille et al. refer, to the “superstructure” of biofilms (11). Approximately 80% of the S. aureus nosocomial infections are biofilm associated (11). S. aureus biofilms inherently exhibit recalcitrance to treatment, thereby increasing the severity, recurrence, chronicity, and mortality of infections. S. aureus biofilms perpetuate both antimicrobial tolerance and resistance by two primary mechanisms as follows: (i) the biofilm acts as a protective barrier against physical stress, antibiotics, and host immune response. Bacteria living in a biofilm can exhibit a 10- to 1,000-fold increase in antibiotic tolerance compared to similar bacteria living in a planktonic state (12). (ii) The biofilm serves as a platform for complex microbial communal networking and antimicrobial resistance transmission. It is important to distinguish between a biofilm’s capacity for antibiotic tolerance vs resistance. Antibiotic tolerance is a temporary state of reduced susceptibility to antibiotics, often associated with biofilms, whereas antibiotic resistance involves genetic changes that enable bacteria to grow and divide in the presence of antibiotics (13).
During biofilm formation, bacteria produce and surround themselves in an extracellular polymeric substance (EPS) consisting of polysaccharides, lipids, nucleic acids, and protein (14). EPS establishes the functional and structural integrity of a biofilm, acting as a key mediator for physiochemical interactions. Antimicrobial tolerance at the biofilm surface is due in part to this EPS matrix that limits antibiotic diffusion. Furthermore, any antibiotics that penetrate the biofilm are unlikely to maintain functionality. Proteolytic enzymes, metabolic byproducts, and other environmental factors of the biofilm compromise antibiotic structure and action (15). For example, low oxygen levels reduce the bactericidal effects of antibiotics tobramycin and ciprofloxacin, whereas pH changes can negatively impact aminoglycoside action. Persister or dormant bacterial subpopulations within the biofilm enter a “spore-like” state and are tolerant to extreme conditions, such as chemical treatment and antibiotic activity that act by targeting cell division. The survival of these persister cells is not due to any genetic changes; upon release from the biofilm, they begin dividing again and return to their pre-persister susceptibility profiles (16–31). Antimicrobial tolerance and resistance within the biofilm are attributed to the heterogeneity of the bacterial populations and the proximity of the bacterial cells that facilitate a highly active transmission of resistant genetic elements. Conjugation and horizontal gene transfer are the predominant modes that reinforce pervasive phenotypic traits throughout the bacterial populations. For example, efflux pumps are readily disseminated through horizontal gene transfer. Despite the health burden due to S. aureus biofilms, many antibiotic assays for susceptibility and resistance are based on planktonic cells, and there are few therapies that prevent and/or disrupt biofilm formation (32, 33).
Clinical relevance of antimicrobial peptides (AMPs)
AMPs are considered a promising alternative to conventional antibiotics (34). AMPs exhibit broad-spectrum activity and a robust bactericidal effect on multi-drug resistant (MDR) microorganisms (34). However, the use of AMPs requires a comprehensive understanding of the complex interactions between peptides, bacterial resistance mechanisms, and host factors (34, 35). Notable disadvantages that limit the clinical application of AMPs include difficulties in maintaining effective levels (e.g., proteolytic degradation, fast clearance by the kidney and liver, etc.) and toxicity (34). Additionally, AMPs are subject to bacterial resistance development (36). For example, bacteria can alter their membrane composition, charge, or surface molecules to prevent effective targeting by AMPs (36). Synergistic approaches that combine AMPs with therapeutic agents such as antibiotics or other AMPs may offer a more effective strategy to prevent resistance development and enhance antimicrobial activity (36). Continued research efforts into AMPs to improve efficacy and bioavailability, reduce toxicity, and limit the acquisition of resistance will aid in combatting the antimicrobial-resistance health crisis (34).
Rationale of SMR peptide targeting of DnaK for biofilm disruption
DnaK is a molecular chaperone within the heat shock protein 70 family. DnaK acts by binding nascent polypeptide chains and assists in protein assembly, refolding, maintenance, and degradation (37). Although the mechanism is unclear, DnaK has been identified as important to biofilm formation and is predicted to be critical for the appropriate folding of biofilm scaffolding proteins. For example, in Escherichia coli, DnaK contributed to amyloid curli expression. In turn, DnaK represents a potential therapeutic target for inhibition to disrupt and prevent the formation of biofilms (38, 39).
Our lab group has designed and developed a series of SMR peptides to target Mortalin, a heat shock protein with a high degree of homology to DnaK (37, 40, 41). In this study, we aim to explore the effects on biofilms from targeting DnaK with the SMR peptides. The SMRwt and SMRmut peptide sequences are depicted in Fig. 1. SMRmut, a negative control, has an alanine substitution that inactivates the function of the antagonist peptide. Modification of the SMR peptide with a 13-mer cell-penetrating peptide (CPP) derived from the HIV-1 Tat protein in C-terminus enables penetration through bacterial cell membranes. The SMR-FLAG is the same as the SMR-CPPtat with a FLAG sequence replacing the CPP (42, 43).
Fig 1.

Design and development of the SMR peptides. (A) The difference between SMRwt-CPPtat and SMRmut-CPPtat is that SMRwt-CPPtat contains a valine, whereas SMRmut-CPPtat contains an alanine at the first position from the N-terminus. CPPtat stands for Cell-Penetrating Peptide Tat and comprises arginine and lysine residues. CPPtat is a potent transduction agent that can penetrate cell membranes and efficiently transport cargo into cells. (B) The SMRwt-FLAG peptide has an N-terminus histidine and a C-terminus hydroxylamine, followed by the FLAG epitope tag. The SMRmut-FLAG peptide is the same as the wild-type peptide, but with a point mutation at the first amino acid from the N-terminus, changing the valine to an alanine.
In our previous studies, we have found multiple novel effects of the SMR peptides through targeting mortalin in HIV/AIDS and breast cancer (28). Similar to DnaK, mortalin is involved in the translocation and folding of proteins to regulate many processes, including cell growth, apoptosis, and autophagy. Mortalin also interacts with proteins, such as Nef, to elicit the secretion of extracellular vesicles (EV) that mediate intercellular communication and environmental response coordination. Both antibody inhibition and miRNA knockdown experiments confirmed that blocking mortalin blocks Nef-EV secretion in Nef-transfected cells, as well as disrupts mortalin/Nef interactions occurring through the SMR domain (28). Our recent research demonstrated that SMR peptide treatment in breast cancer cells blocked EV secretion, induced cell cycle arrest at the G2/M phase, and reduced cellular proliferation (40, 41). We hypothesized that SMR peptide treatment of resistant S. aureus will interact with DnaK, like mortalin, and block both the folding of scaffolding proteins and EV release, to compromise the structural integrity of the biofilm and the EV-mediated intercellular networking, respectively. We addressed this hypothesis by microtiter plate (MtP) assays, immunoprecipitation assays, and confocal microscopy. This study offers insights into the development of novel treatment strategies to mitigate the impact of this versatile and resilient pathogen.
RESULTS
SMRwt peptide interacts with DnaK in E. coli and S. aureus strains
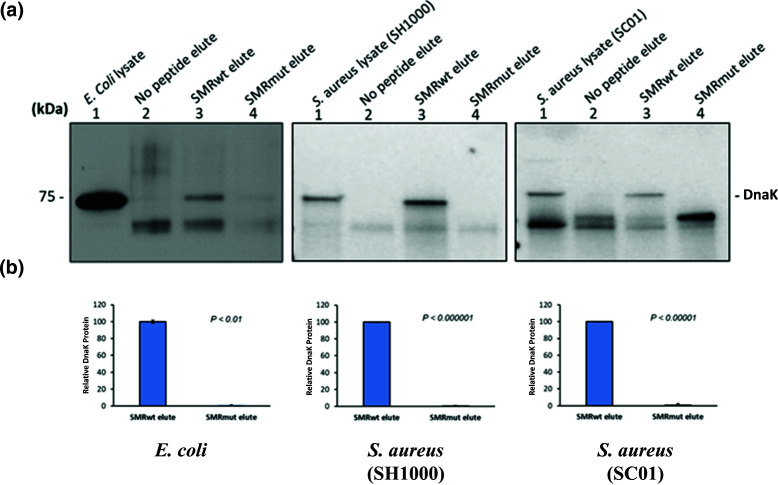
The interaction between the SMRwt peptide and DnaK was studied using an immunoprecipitation assay. In brief, E. coli (Invitrogen MAX Efficiency Stbl2 Competent cells), S. aureus (SH1000), and S. aureus (SC01) cell lysates were screened with an anti-flag M2 affinity gel under three different conditions as follows: SMRwt vs SMRmut vs no peptide. The bound proteins were eluted and subsequently separated using SDS-PAGE. The Western blot analysis with the DnaK antibody revealed a SMRwt band, which was absent under the SMRmut and no peptide conditions (Fig. 2A). A significantly higher elution percentage was observed for SMRwt compared to SMRmut (Fig. 2B). Specifically, the P-values were as follows: E. coli (Invitrogen MAX Efficiency Stbl2 Competent cells) P-value < 0.01, S. aureus (SH1000) P-value < 0.000001, and S. aureus (SC01) P-value < 0.00001. This indicated that the SMRwt peptide was interacting with DnaK in a manner that increased its retention in the affinity gel that was not seen for SMRmut.
Fig 2.
SMR peptides interact with target DnaK in E. coli and S. aureus strains. (A) Western blot analysis of the DnaK antibody depicted the interaction between the SMRwt and DnaK following an immunoprecipitation assay. E. coli (Invitrogen MAX efficiency Stbl2 competent), S. aureus (SH1000), and S. aureus (SC01) were grown in media without antibiotics. The cells were lysed and the resultant lysate was screened by an Anti-Flag M2 affinity under three conditions as follows: SMRwt, SMRmut, and the absence of both peptides. (B) Bar graphs depict the SMRwt and SMRmut elute band percentage intensity. Results are presented as means ± standard deviations (n = 3).
SMRwt peptide significantly inhibits S. aureus biofilm formation
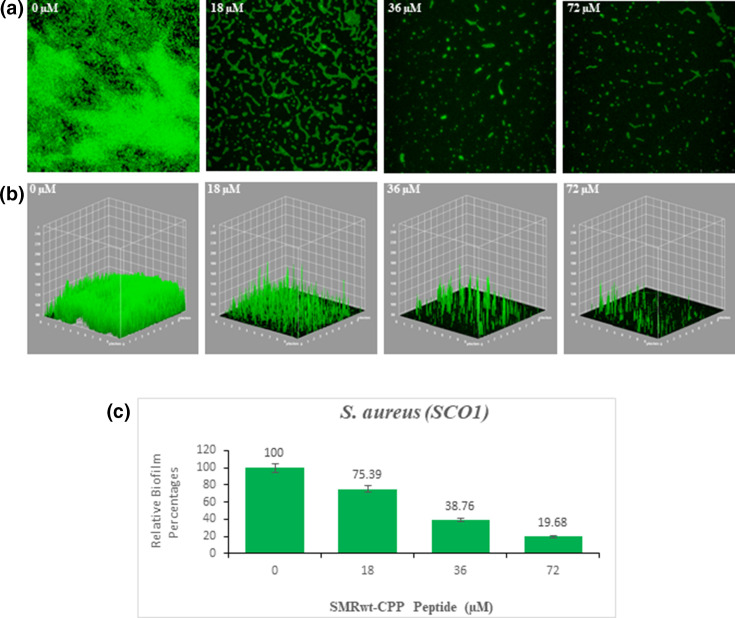
The SMRwt peptide effectively blocked biofilm formation of S. aureus (SC01) as shown through MtP methods and confocal microscopy (Fig. 3 and 4). For the MtP assays, S. aureus (SC01) was seeded with either SMRwt or SMRmut, and biofilm formation was measured using the crystal violet staining method (OD570). The data were normalized for growth (OD600). A clear inhibitory effect was identified with increasing SMRwt concentrations contributing to significantly decreased biofilm formation (Fig. 3). Significant differences relative to the negative control were noted at SMR peptide concentrations of 36 µM with a P-value < 0.022 and 72 µM with a P-value < 0.015. Three-dimensional confocal microscopy findings reinforced this premise of SMR peptide-mediated biofilm formation (Fig. 4B). The dose effect observed through confocal microscopy aligned strongly with the crystal violet staining findings. Specifically, SMR dosages 18 µM, 36 µM, and 72 µM resulted in significantly decreased biofilm formation of 75.39%, 38.76%, and 19.68%, respectively (Fig. 4C).
Fig 3.
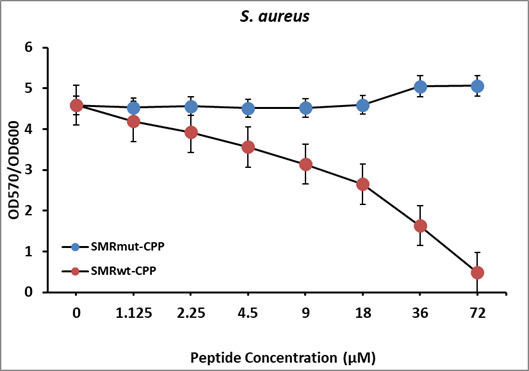
SMR peptides inhibit S. aureus biofilm formation in a dose-dependent manner. S. aureus (SC01) was grown at 37°C to the stationary phase. The cultures were diluted 100-fold in fresh medium and seeded into a 96-well microtiter plate with either SMRwt or SMRmut at the indicated concentrations. After 24 hours, biofilm formation was measured using the crystal violet staining method (OD570). The data were normalized for growth (OD600). Error bars represent the mean ± SD of two independent experiments. Significant differences relative to the negative control were noted at SMR peptide concentrations of 36 µM with a P-value < 0.022 and 72 µM with a P-value < 0.015.
Fig 4.
Confocal microscopy and three-dimensional (3D) views of S. aureus biofilms reveal effective SMR peptide antagonism. S. aureus (SC01) was grown at 37°C to the stationary phase. The cultures were diluted 100-fold in fresh media and seeded onto a MatTek glass bottom plate. After 24 hours, the biofilms were stained with SYTO9, observed by Leica confocal microscopy, and analyzed with ImageJ. (A) Confocal images, (B) 3D images, and (C) bar graphs depict the inhibitory effect of SMR peptides on S. aureus biofilm formation. Error bars represent the mean ± SD of three independent experiments. Significant differences relative to the negative control were noted at SMR peptide concentrations of 18 µM with a P-value < 0.05 as well as 36 µM and 72 µM with a P-value < 0.005.
DISCUSSION
S. aureus biofilms facilitate life-threatening infections recalcitrant to antibiotic therapy, contributing to a substantial burden on healthcare systems worldwide (44–47). The intrinsic and acquired biofilm-specific mechanisms for tolerance and resistance against antimicrobial agents, host immune response, and environmental stressors reinforce persistent and severe nosocomial infections (45). Despite these clinical threats of S. aureus biofilm-associated infections, currently, there are no anti-biofilm treatments available (48). Thus, there have been intensive research efforts on the discovery and development of agents to prevent and/or disrupt biofilms. DnaK has been identified as a promising target with its chemical inhibition resulting in effective biofilm prevention (38, 49). Elimination of DnaK functionality through either gene deletion or direct inhibition has been found to reduce adherence, the production of amyloid curli, as well as the robustness of the biofilm in both E. coli and S. aureus (38). Furthermore, in a murine model, DnaK blockade diminished S. aureus resistance to unfavorable environmental factors and overall survivability (50, 51).
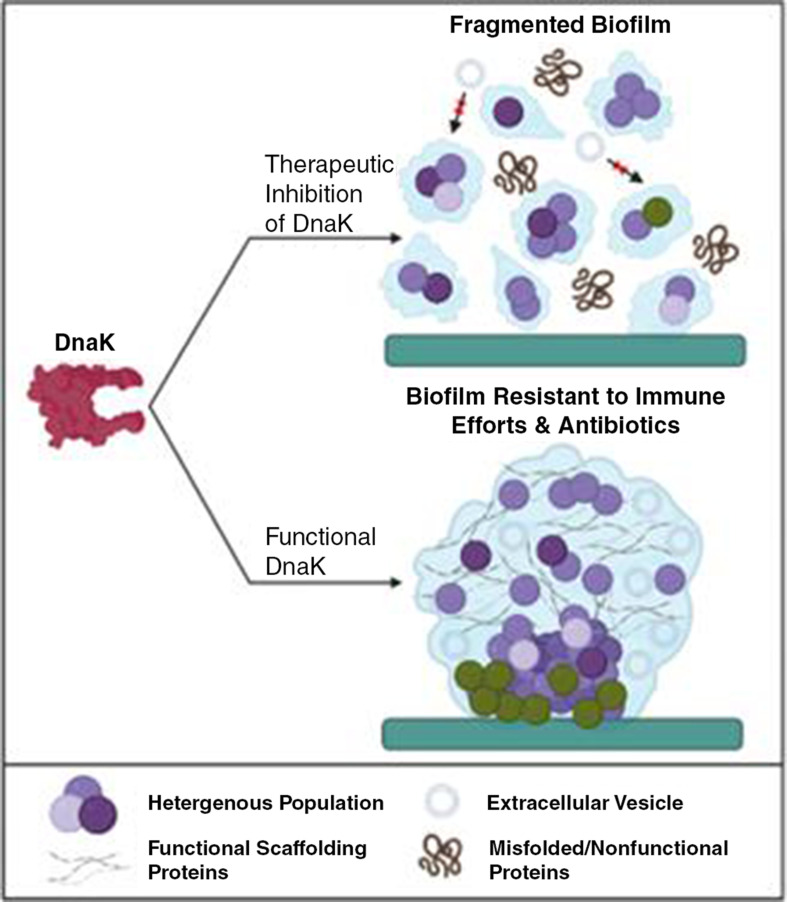
In this study, SMRwt peptide was shown to physically interact with DnaK in E. coli (Invitrogen MAX Efficiency Stbl2 Competent cells), S. aureus (SH1000), and S. aureus (SC01) using an immune-precipitation assay with western blotting (Fig. 2A). There was a significant difference between the band percentage intensity of the SMRwt elute compared with the SMRmut elute (Fig. 2B). As SMRwt was shown to interact with DnaK, it likely could interfere with its activity. This would have implications for the regulation of various cellular processes in the bacterium and catalyzed the proposed model for the effect of SMR peptide-mediated DnaK inhibition on biofilm structure and intercellular communication in S. aureus (Fig. 5). Given previous studies on the potential of DnaK inhibition, our lab theorized that SMRwt could disrupt S. aureus biofilm formation through two primary mechanisms (Fig. 5) as follows: (1) the structural integrity of the biofilm could be compromised through a loss of appropriate folding of scaffolding proteins like amyloid curli. (2) As the SMRwt peptide is capable of EV blockade in both viral infected cells and cancer cells, we postulated that it could similarly affect bacterial intercellular communication networking that is reliant upon EV transmission (52, 53).
Fig 5.
Proposed model for the impact of DnaK inhibition, using SMR peptide, on biofilm structure and intercellular communication. DnaK, a molecular chaperone, is essential to biofilm formation. Although there is a lack of knowledge on the specific mechanism of DnaK, previous studies have identified DnaK as key to amyloid curli expression within the biofilms of E. coli. We propose that DnaK is required to appropriately fold functional scaffolding proteins that contribute to the structural integrity of a biofilm. Therefore, we propose that therapeutic inhibition of DnaK with the SMRwt peptide resulted in an accumulation of misfolded proteins with subsequent biofilm degradation. In addition, we theorize that the intercellular communication transit of extracellular vesicles was blocked, by SMRwt targeting of DnaK, further exacerbating dysfunction within the biofilm. Created with BioRender.
In alignment with the proposed DnaK inhibition model, the data illustrated an inhibitory effect of the SMRwt peptide on the biofilm formation of drug-resistant S. aureus (SC01) in a dose-dependent manner using the crystal violet staining method and confocal microscopy (Fig. 3 and 4). These inhibitory effects on biofilm formation were not observed with the SMRmut peptide. This supports the expectation that the valine at the first position of the SMRwt peptide is essential for its functionality. Other chemical DnaK inhibitors, such as Myr, have shown a similar inhibitory effect on biofilm formation with a corresponding decrease in amyloid curli (38). SMRwt peptide could serve as a possible anti-bacterial therapeutic by DnaK targeting or as a model for rational drug design to inhibit MDR S. aureus biofilm formation (50, 54–70).
In most clinical cases, biofilms are already established (71). Thus, an effective anti-biofilm agent should be capable of disrupting a preformed biofilm (71). We aim to further explore the SMRwt peptide’s impact against preformed biofilms. In preliminary experiments, we observed biofilm disruption following SMRwt-CPP peptide application to a preformed S. aureus biofilm. Although the precise mechanism by which SMRwt-CPP disrupts biofilms and the specific bacterial target(s) involved remains unclear, these results provide evidence that this peptide may be a promising anti-biofilm candidate. This research is significant in the broader context of addressing biofilm-related infections, highlighting targets for inhibition, and offering potential solutions with less selective pressure for antibiotic resistance emergence.
Conclusion
These findings and other data suggest that DnaK antagonism with SMRwt peptide should be investigated further for therapeutic applications. As biofilms act as a platform for promoting antimicrobial resistant tactics, generating anti-biofilm agents will aid in addressing the global threat regarding a dried-up antibiotic discovery pipeline and increasing the prevalence of antibiotic resistance. The SMRwt peptide may be a viable alternative to traditional antibiotics for treating drug-resistant bacteria. However, the exact mechanism of action of the SMRwt peptide is unclear. Determining the mechanism of action for SMRwt peptide biofilm antagonism as well as other clinically relevant parameters, such as in vivo efficacy, optimal dosage, and delivery method, will be critical to future development efforts.
MATERIALS AND METHODS
Strains and growth conditions
The S. aureus SC01 (MRSA) and SH1000 strains were kindly provided by Dr. Jorge Antonio Benitez’s Laboratory and were grown on Mueller–Hinton agar plates with glucose and without antibiotics at 37°C overnight. The E. coli (Invitrogen MAX Efficiency Stbl2 Competent cells) strains cells were grown overnight at 30°C in LB Broth without antibiotics.
Synthesis of SMR-CPPtat and SMR-FLAG peptides
The SMR-CPPtat and SMR-FLAG peptides were custom-made by InnoPep Inc. (San Diego, CA). The amino acid (aa) sequence for the four peptides used in this study is as shown in Fig. 1. SMRwt and SMRmut differ at the first position aa: SMRwt contains a valine, whereas SMRmut contains an alanine. CPPtat is a peptide of arginine and lysine residues that can penetrate the cell membrane. The SMR-FLAG peptides are identical to the SMR-CPP peptides except for the FLAG sequence replacing the CPP sequence.
Evaluation of biofilm-forming capacity
Strains treated with SMR peptides were screened for their biofilm-forming capacity using MtP methods and a crystal violet assay described by Stepanovic et al. (72). In triplicate, one colony was inoculated to 5 mL of LB or 5 mL of BHI medium and incubated at 37°C overnight with agitation (250 rpm). For evaluating the dose effect of SMR peptides, 20 µL of the overnight cultures to 1 mL LB with 0.5% glucose for E. coli or BHI with 2% glucose for S. aureus medium with the appropriate concentration of the SMR peptide were mixed well and transferred (100 µL) to 5 wells of the 96 well of the MtP. Negative controls were the SMRmut peptides as well as medium without strains. Plates were incubated at 37°C for S. aureus 24 hours without shaking. After the incubation time, the OD at 600 nm was determined using the SpectraMax M5 Microplate Reader. The plates were washed three times with phosphate-buffered saline (PBS) pH 7.4, and adherent bacteria were stained with 0.1% crystal violet. After 30 minutes, the plates were washed three times with PBS, the crystal violet-stained biofilm re-dissolved in dimethylsulfoxide, and the plates read at an OD of 570 nm (OD570). Biofilm production was normalized for growth and expressed as the OD570/OD600 ratio for each well.
Confocal microscopy
Confocal microscopy was used to visualize biofilm formation. Samples were grown on glass slides and stained with SYTO 9 (Invitrogen, Carlsbad, CA). The biofilms were prepped for confocal microscopy by first growing them in appropriate media for the respective strains with different concentrations of the SMR peptides. After the biofilm formation, the biofilms were stained with 10 µM SYTO9 and incubated at 30°C for 30 minutes in the dark. The biofilms were then washed to remove any extra dye and immediately observed using a TCS-SP2 microscope (Leica Microsystems, Heidelberg, Germany), equipped with a 63 × oil immersion objective. The SYTO9 dye was excited at 483 nm and the fluorescence emission was detected at 503 nm. 3D images were captured every micrometer throughout the biofilm depth. The images were then visualized and processed using the Leica LAS AF software (Leica Microsystems). Quantitative analyses of the images were performed using the COMSTAT software (http://www.imageanalysis.dk/; Heydorn et al., 2000). At least three images were collected from each of the three independent experiments (nine stacks in total) for analysis.
Immunoprecipitation and Western blot analysis
The interaction between the antagonist and DnaK was determined by immune precipitation with anti-Flag M2 Affinity and Western blot analysis. E. coli (Invitrogen MAX Efficiency Stbl2 Competent cells), S. aureus (SH1000), and S. aureus (SC01) were grown in media without antibiotics and then lysed. The lysate was then screened using anti-flag M2 affinity gel and either the SMRwt or SMRmut peptide, or in the absence of both peptides. The eluted protein samples were separated using SDS-PAGE on 4–20% or 8–16% Tris-HCl Criterion precast gels (Bio-Rad) and transferred to the nitrocellulose membrane, 0.45 µm. The membrane was washed in Tris-Buffered Saline (TBS; Bio-Rad) for 5 minutes, blocked with 5% nonfat milk in TTBS (TBS with 0.1% Tween 20) for 1 hour by shaking at room temperature, processed for immunoblotting using a specific primary mouse DnaK antibody by shaking at 4°C overnight, followed by a secondary HRP-conjugated IgG (H + L) antibody. Protein bands were detected using Western Blotting Luminol Reagent (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) followed by exposure to Image Quant LAS 4000 (FUJIFILM Medical Systems USA, Inc). Densitometry analysis was performed using ImageJ software (National Institutes of Health, Bethesda, MD).
Statistical analysis
Experiments were performed in three biological replicates with the exception of Figure 3 (n=2). Data obtained from the static biofilm assay were log-transformed to stabilize the variance and to make the approximation to the normal distribution. The statistical analysis was performed using the student’s t-test (73–77).
ACKNOWLEDGMENTS
The following grants supported this research: NIH/NIAID 1R21AI095150-01A1, NIH/NIMHD 8G12MD007602, and NIH/NIMHD 8U54MD007588. NIH/RCMI/U54 Pilot Grant.
We thank the Morehouse School of Medicine Facility Study Design, Biostatistics, and Data Management Core Facility’s Dr. Feng-Xia Yan for guidance in statistical analysis. We also thank the Department of Microbiology, Biochemistry, and Immunology's Dr. Yusuf Omosun for reviewing the manuscript.
M.-B.H. and V.C.B. discussed and designed the experiments. M.-B.H., M.S., and J.Y.W. performed the experiments. M.-B.H. wrote and edited the paper and analyzed data through statistical analysis. J.Y.W. and D.B. modified the paper, and M.S., M.-B.H., D.B., and V.C.B. reviewed and revised the paper. All the authors read and approved the final version of the review.
Contributor Information
Ming-Bo Huang, Email: mhuang@msm.edu.
Ilana Kolodkin-Gal, The Hebrew University of Jerusalem, Rehovot, Israel.
REFERENCES
- 1. Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 2. Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhattacharya M, Wozniak DJ, Stoodley P, Hall-Stoodley L. 2015. Prevention and treatment of Staphylococcus aureus biofilms. Expert Rev Anti Infect Ther 13:1499–1516. doi: 10.1586/14787210.2015.1100533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gajdács M. 2019. The continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics (Basel) 8:52. doi: 10.3390/antibiotics8020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. 2019. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol 10:539. doi: 10.3389/fmicb.2019.00539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. CDC . 2022. National estimates for antibiotic resistance. Available from: https://www.cdc.gov/drugresistance/national-estimates.html
- 7. Bauer AW, Kirby WM, Sherris JC, Turck M. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496. [PubMed] [Google Scholar]
- 8. David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. doi: 10.1128/CMR.00081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rai A, Khairnar K. 2021. Overview of the risks of Staphylococcus aureus infections and their control by bacteriophages and bacteriophage-encoded products. Braz J Microbiol 52:2031–2042. doi: 10.1007/s42770-021-00566-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. GBD 2019 Antimicrobial Resistance Collaborators . 2022. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the global burden of disease study 2019. Lancet 400:2221–2248. doi: 10.1016/S0140-6736(22)02185-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fany R, Jérôme J, Quentin V, Céline M, Sophie C G. 2017. Staphylococcus aureus Biofilms and their impact on the medical field. In Shymaa E, Laura ECA (ed), The rise of virulence and antibiotic resistance in Staphylococcus aureus. IntechOpen. [Google Scholar]
- 12. Uruén C, Chopo-Escuin G, Tommassen J, Mainar-Jaime RC, Arenas J. 2020. Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics (Basel) 10:3. doi: 10.3390/antibiotics10010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34 [DOI] [PubMed] [Google Scholar]
- 14. Staudt C, Horn H, Hempel DC, Neu TR. 2004. Volumetric measurements of bacterial cells and extracellular polymeric substance glycoconjugates in biofilms. Biotechnol Bioeng 88:585–592. doi: 10.1002/bit.20241 [DOI] [PubMed] [Google Scholar]
- 15. Thi MTT, Wibowo D, Rehm BHA. 2020. Pseudomonas aeruginosa biofilms. Int J Mol Sci 21:8671. doi: 10.3390/ijms21228671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fatima M, Amin A, Alharbi M, Ishtiaq S, Sajjad W, Ahmad F, Ahmad S, Hanif F, Faheem M, Khalil AAK. 2023. Quorum quenchers from Reynoutria japonica in the battle against methicillin-resistant Staphylococcus aureus (MRSA). Molecules 28:2635. doi: 10.3390/molecules28062635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kang M-G, Khan F, Tabassum N, Cho K-J, Jo D-M, Kim Y-M. 2023. Inhibition of biofilm and virulence properties of pathogenic bacteria by silver and gold nanoparticles synthesized from Lactiplantibacillus sp. Strain C1. ACS Omega 8:9873–9888. doi: 10.1021/acsomega.2c06789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ürer EK, Aslantaş Ö, Tek E, Yılmaz MA, Ergün Y. 2023. Antimicrobial susceptibility and biofilm forming ability of staphylococci from subclinical buffalo mastitis. J Dairy Res:1–4. doi: 10.1017/S0022029923000080 [DOI] [PubMed] [Google Scholar]
- 19. Hamad PA. 2023. Phenotypic and molecular detection of biofilm formation in methicillin-resistant Staphylococcus aureus isolated from different clinical sources in Erbil city. Mediterr J Hematol Infect Dis 15:e2023016. doi: 10.4084/MJHID.2023.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sabino YNV, Cotter PD, Mantovani HC. 2023. Anti-virulence compounds against Staphylococcus aureus associated with bovine mastitis: a new therapeutic option?. Microbiol Res 271:127345. doi: 10.1016/j.micres.2023.127345 [DOI] [PubMed] [Google Scholar]
- 21. Sille IE, Pissinis DE, Fagali NS, Ghilini F, Urrutia MN, Schilardi PL. 2023. Antimicrobial-loaded polyacrylamide hydrogels supported on titanium as reservoir for local drug delivery. Pathogens 12:202. doi: 10.3390/pathogens12020202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shobha B, Ashwini BS, Ghazwani M, Hani U, Atwah B, Alhumaidi MS, Basavaraju S, Chowdappa S, Ravikiran T, Wahab S, Ahmad W, Lakshmeesha TR, Ansari MA. 2023. Trichoderma-mediated Zno nanoparticles and their antibiofilm and antibacterial activities. J Fungi (Basel) 9:133. doi: 10.3390/jof9020133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ismail S, Gaglione R, Masi M, Padhi S, Rai AK, Omar G, Cimmino A, Arciello A. 2023. Ephedra foeminea as a novel source of antimicrobial and anti-biofilm compounds to fight multidrug resistance phenotype. IJMS 24:3284. doi: 10.3390/ijms24043284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boonsilp S, Sikora J, Rupprom K, Acılıoğlu S, Homkaew A, Nutalai D, Phumisantiphong U, Wongsuk T. 2023. Molecular characterization and antibiotic resistance of Staphylococcus aureus isolated from clinical specimens in an urban university hospital in Bangkok, Thailand. Acta Microbiol Immunol Hung 70:61–72. doi: 10.1556/030.2023.01942 [DOI] [PubMed] [Google Scholar]
- 25. Bąchor U, Junka A, Brożyna M, Mączyński M. 2023. The in vitro impact of Isoxazole derivatives on pathogenic biofilm and cytotoxicity of fibroblast cell line. Int J Mol Sci 24:2997. doi: 10.3390/ijms24032997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bottagisio M, Palombella S, Lopa S, Sangalli F, Savadori P, Biagiotti M, Sideratou Z, Tsiourvas D, Lovati AB. 2022. Vancomycin-nanofunctionalized peptide-enriched silk fibroin to prevent methicillin-resistant Staphylococcus epidermidis-induced femoral nonunions in rats. Front Cell Infect Microbiol 12:1056912. doi: 10.3389/fcimb.2022.1056912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sohail M, Muzzammil M, Ahmad M, Rehman S, Garout M, Khojah TM, Al-Eisa KM, Breagesh SA, Hamdan RMA, Alibrahim HI, Alsoliabi ZA, Rabaan AA, Ahmed N. 2023. Molecular characterization of community- and hospital- acquired methicillin-resistant Staphylococcus aureus isolates during COVID-19 pandemic. Antibiotics (Basel) 12:157. doi: 10.3390/antibiotics12010157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saeed SI, Vivian L, Zalati C, Sani NIM, Aklilu E, Mohamad M, Noor AAM, Muthoosamy K, Kamaruzzaman NF. 2023. Antimicrobial activities of graphene oxide against biofilm and intracellular Staphylococcus aureus isolated from bovine mastitis. BMC Vet Res 19:10. doi: 10.1186/s12917-022-03560-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katsipis G, Pantazaki AA. 2023. Serrapeptase impairs biofilm, wall, and phospho-homeostasis of resistant and susceptible Staphylococcus aureus. Appl Microbiol Biotechnol 107:1373–1389. doi: 10.1007/s00253-022-12356-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu D, Xi Y, Yu S, Yang K, Zhang F, Yang Y, Wang T, He S, Zhu Y, Fan Z, Du J. 2023. A polypeptide coating for preventing biofilm on implants by inhibiting antibiotic resistance genes. Biomaterials 293:121957. doi: 10.1016/j.biomaterials.2022.121957 [DOI] [PubMed] [Google Scholar]
- 31. Palau M, Muñoz E, Larrosa N, Gomis X, Márquez E, Len O, Almirante B, Gavaldà J. 2023. Hyperthermia prevents in vitro and in vivo biofilm formation on endotracheal tubes. Microbiol Spectr 11:e0280722. doi: 10.1128/spectrum.02807-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clinton A, Carter T. 2015. Chronic wound biofilms: pathogenesis and potential therapies. Lab Med 46:277–284. doi: 10.1309/LMBNSWKUI4JPN7SO [DOI] [PubMed] [Google Scholar]
- 33. Tran NN, Morrisette T, Jorgensen SCJ, Orench-Benvenutti JM, Kebriaei R. 2023. Current therapies and challenges for the treatment of Staphylococcus aureus biofilm-related infections. Pharmacotherapy 43:816–832. doi: 10.1002/phar.2806 [DOI] [PubMed] [Google Scholar]
- 34. Huan Y, Kong Q, Mou H, Yi H. 2020. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front Microbiol 11:582779. doi: 10.3389/fmicb.2020.582779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Q-Y, Yan Z-B, Meng Y-M, Hong X-Y, Shao G, Ma J-J, Cheng X-R, Liu J, Kang J, Fu C-Y. 2021. Antimicrobial peptides: mechanism of action, activity and clinical potential. Mil Med Res 8:48. doi: 10.1186/s40779-021-00343-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kmeck A, Tancer RJ, Ventura CR, Wiedman GR. 2020. Synergies with and resistance to membrane-active peptides. Antibiotics (Basel) 9:620. doi: 10.3390/antibiotics9090620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wong P, Houry WA. 2004. Chaperone networks in bacteria: analysis of protein homeostasis in minimal cells. J Struct Biol 146:79–89. doi: 10.1016/j.jsb.2003.11.006 [DOI] [PubMed] [Google Scholar]
- 38. Arita-Morioka K, Yamanaka K, Mizunoe Y, Ogura T, Sugimoto S. 2015. Novel strategy for biofilm inhibition by using small molecules targeting molecular chaperone Dnak. Antimicrob Agents Chemother 59:633–641. doi: 10.1128/AAC.04465-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Gerven N, Van der Verren SE, Reiter DM, Remaut H. 2018. The role of functional amyloids in bacterial virulence. J Mol Biol 430:3657–3684. doi: 10.1016/j.jmb.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang M-B, Gonzalez RR, Lillard J, Bond VC. 2017. Secretion modification region-derived peptide blocks exosome release and mediates cell cycle arrest in breast cancer cells. Oncotarget 8:11302–11315. doi: 10.18632/oncotarget.14513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang M-B, Brena D, Wu JY, Roth WW, Owusu S, Bond VC. 2022. Novel secretion modification region (SMR) peptide exhibits anti-metastatic properties in human breast cancer cells. Sci Rep 12:13204. doi: 10.1038/s41598-022-17534-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Henriques ST, Melo MN, Castanho M. 2006. Cell-penetrating peptides and antimicrobial peptides: how different are they? Biochem J 399:1–7. doi: 10.1042/BJ20061100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guo Z, Peng H, Kang J, Sun D. 2016. Cell-penetrating peptides: possible transduction mechanisms and therapeutic applications. Biomed Rep 4:528–534. doi: 10.3892/br.2016.639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao K, Su B, Dai J, Li P, Wang R, Yang X. 2022. Anti-biofilm and anti-hemolysis activities of 10-hydroxy-2-decenoic acid against Staphylococcus aureus. Molecules 27:1485. doi: 10.3390/molecules27051485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rather MA, Gupta K, Mandal M. 2021. Microbial biofilm: formation, architecture, antibiotic resistance, and control strategies. Braz J Microbiol 52:1701–1718. doi: 10.1007/s42770-021-00624-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lister JL, Horswill AR. 2014. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol 4:178. doi: 10.3389/fcimb.2014.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Idrees M, Sawant S, Karodia N, Rahman A. 2021. Staphylococcus aureus biofilm: morphology, genetics, pathogenesis and treatment strategies. Int J Environ Res Public Health 18:7602. doi: 10.3390/ijerph18147602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zheng J, Shang Y, Wu Y, Zhao Y, Chen Z, Lin Z, Li P, Sun X, Xu G, Wen Z, Chen J, Wang Y, Wang Z, Xiong Y, Deng Q, Qu D, Yu Z. 2022. Loratadine inhibits Staphylococcus aureus virulence and biofilm formation. iScience 25:103731. doi: 10.1016/j.isci.2022.103731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gangwar B, Kumar S, Darokar MP. 2020. Glabridin averts biofilms formation in methicillin-resistant Staphylococcus aureus by modulation of the surfaceome. Front Microbiol 11:1779. doi: 10.3389/fmicb.2020.01779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Singh VK, Syring M, Singh A, Singhal K, Dalecki A, Johansson T. 2012. An insight into the significance of the Dnak heat shock system in Staphylococcus aureus. Int J Med Microbiol 302:242–252. doi: 10.1016/j.ijmm.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 51. Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR. 2007. Role for DnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology (Reading) 153:3162–3173. doi: 10.1099/mic.0.2007/009506-0 [DOI] [PubMed] [Google Scholar]
- 52. Rosa da Luz BS, Azevedo V, Le-loir Y, Guedon E. 2021. Extracellular Vesicles and their role in Staphylococcus aureus resistance and virulence, in insights into drug resistance in Staphylococcus aureus. In Amjad A (ed), Insights into drug resistance in Staphylococcus aureus. IntechOpen: Rijeka. [Google Scholar]
- 53. Bose S, Aggarwal S, Singh DV, Acharya N. 2020. Extracellular vesicles: an emerging platform in gram-positive bacteria. Microb Cell 7:312–322. doi: 10.15698/mic2020.12.737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vahdani F, Ghafouri H, Sarikhan S, Khodarahmi R. 2019. Molecular cloning, expression, and functional characterization of 70-kDa heat shock protein, DnaK, from bacillus halodurans. Int J Biol Macromol 137:151–159. doi: 10.1016/j.ijbiomac.2019.06.217 [DOI] [PubMed] [Google Scholar]
- 55. Mohammadi-Ostad-Kalayeh S, Hrupins V, Helmsen S, Ahlbrecht C, Stahl F, Scheper T, Preller M, Surup F, Stadler M, Kirschning A, Zeilinger C. 2017. Development of a microarray-based assay for efficient testing of new Hsp70/DnaK inhibitors. Bioorg Med Chem 25:6345–6352. doi: 10.1016/j.bmc.2017.10.003 [DOI] [PubMed] [Google Scholar]
- 56. Stetz G, Verkhivker GM. 2016. Probing allosteric inhibition mechanisms of the Hsp70 chaperone proteins using molecular dynamics simulations and analysis of the residue interaction networks. J Chem Inf Model 56:1490–1517. doi: 10.1021/acs.jcim.5b00755 [DOI] [PubMed] [Google Scholar]
- 57. Zhang H, Yang J, Wu S, Gong W, Chen C, Perrett S. 2016. Glutathionylation of the bacterial Hsp70 chaperone DnaK provides a link between oxidative stress and the heat shock response. J Biol Chem 291:6967–6981. doi: 10.1074/jbc.M115.673608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Knappe D, Ruden S, Langanke S, Tikkoo T, Ritzer J, Mikut R, Martin LL, Hoffmann R, Hilpert K. 2016. Optimization of oncocin for antibacterial activity using a SPOT synthesis approach: extending the pathogen spectrum to Staphylococcus aureus. Amino Acids 48:269–280. doi: 10.1007/s00726-015-2082-2 [DOI] [PubMed] [Google Scholar]
- 59. Chiappori F, Fumian M, Milanesi L, Merelli I. 2015. Dnak as antibiotic target: Hot spot residues analysis for differential inhibition of the bacterial protein in comparison with the human Hsp70. PLoS One 10:e0124563. doi: 10.1371/journal.pone.0124563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. El-Kasaby A, Koban F, Sitte HH, Freissmuth M, Sucic S. 2014. A cytosolic relay of heat shock proteins Hsp70-1A and Hsp90Beta monitors the folding trajectory of the serotonin transporter. J Biol Chem 289:28987–29000. doi: 10.1074/jbc.M114.595090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Leu J-J, Zhang P, Murphy ME, Marmorstein R, George DL. 2014. Structural basis for the inhibition of Hsp70 and Dnak chaperones by small-molecule targeting of a C-terminal allosteric pocket. ACS Chem Biol 9:2508–2516. doi: 10.1021/cb500236y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Borges JC, Seraphim TV, Mokry DZ, Almeida FCL, Cyr DM, Ramos CHI. 2012. Identification of regions involved in substrate binding and Dimer stabilization within the central domains of yeast Hsp40 Sis1. PLoS One 7:e50927. doi: 10.1371/journal.pone.0050927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Al Refaii A, Alix JH. 2008. Inhibition of chaperone-dependent bacterial ribosome biogenesis. Methods Mol Med 142:75–85. doi: 10.1007/978-1-59745-246-5_7 [DOI] [PubMed] [Google Scholar]
- 64. Karzai AW, McMacken R. 1996. A bipartite signaling mechanism involved in Dnaj-mediated activation of the Escherichia coli DnaK protein. J Biol Chem 271:11236–11246. doi: 10.1074/jbc.271.19.11236 [DOI] [PubMed] [Google Scholar]
- 65. Wickner S, Nguyen T-L, Genest O. 2021. The bacterial Hsp90 chaperone: cellular functions and mechanism of action. Annu Rev Microbiol 75:719–739. doi: 10.1146/annurev-micro-032421-035644 [DOI] [PubMed] [Google Scholar]
- 66. Zahn M, Kieslich B, Berthold N, Knappe D, Hoffmann R, Strater N. 2014. Structural identification of DnaK binding sites within bovine and sheep bactenecin Bac7. Protein Pept Lett 21:407–412. doi: 10.2174/09298665113206660111 [DOI] [PubMed] [Google Scholar]
- 67. Zeymer C, Werbeck ND, Schlichting I, Reinstein J. 2013. The molecular mechanism of Hsp100 chaperone inhibition by the prion curing agent guanidinium chloride. J Biol Chem 288:7065–7076. doi: 10.1074/jbc.M112.432583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chang L, Miyata Y, Ung PMU, Bertelsen EB, McQuade TJ, Carlson HA, Zuiderweg ERP, Gestwicki JE. 2011. Chemical screens against a reconstituted multiprotein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem Biol 18:210–221. doi: 10.1016/j.chembiol.2010.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cellitti J, Zhang Z, Wang S, Wu B, Yuan H, Hasegawa P, Guiney DG, Pellecchia M. 2009. Small molecule DnaK modulators targeting the beta-domain. Chem Biol Drug Des 74:349–357. doi: 10.1111/j.1747-0285.2009.00869.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ben-Zvi A, De Los Rios P, Dietler G, Goloubinoff P. 2004. Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual Hsp70 chaperones. J Biol Chem 279:37298–37303. doi: 10.1074/jbc.M405627200 [DOI] [PubMed] [Google Scholar]
- 71. Verderosa AD, Totsika M, Fairfull-Smith KE. 2019. Bacterial biofilm eradication agents: a current review. Front Chem 7:824. doi: 10.3389/fchem.2019.00824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stepanović S, Cirković I, Ranin L, Svabić-Vlahović M. 2004. Biofilm formation by salmonella spp. and listeria monocytogenes on plastic surface. Lett Appl Microbiol 38:428–432. doi: 10.1111/j.1472-765X.2004.01513.x [DOI] [PubMed] [Google Scholar]
- 73. Davey ME, O’toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867. doi: 10.1128/MMBR.64.4.847-867.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bénard L, Litzler P-Y, Cosette P, Lemeland J-F, Jouenne T, Junter G-A. 2009. Proteomic analysis of Staphylococcus aureus biofilms grown in vitro on mechanical heart valve leaflets. J Biomed Mater Res A 88:1069–1078. doi: 10.1002/jbm.a.31941 [DOI] [PubMed] [Google Scholar]
- 75. Cunningham SA, Jeraldo PR, Schuetz AN, Heitman AA, Patel R. 2020. Staphylococcus aureus whole genome sequence-based susceptibility and resistance prediction using a clinically amenable workflow. Diagn Microbiol Infect Dis 97:115060. doi: 10.1016/j.diagmicrobio.2020.115060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. O’Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x [DOI] [PubMed] [Google Scholar]
- 77. Vasconcelos MA, Arruda FVS, Carneiro VA, Silva HC, Nascimento KS, Sampaio AH, Cavada B, Teixeira EH, Henriques M, Pereira MO. 2014. Effect of algae and plant lectins on planktonic growth and biofilm formation in clinically relevant bacteria and yeasts. Biomed Res Int 2014:365272. doi: 10.1155/2014/365272 [DOI] [PMC free article] [PubMed] [Google Scholar]