ABSTRACT
Carbapenemase-producing Enterobacterales (CPE) are a growing threat to global health and the economy. Understanding the interactions between resistance and virulence mechanisms of CPE is crucial for managing difficult-to-treat infections and informing outbreak prevention and control programs. Here, we report the characterization of 21 consecutive, unique clinical isolates of CPE collected in 2018 at a tertiary hospital in Lima, Peru. Isolates were characterized by phenotypic antimicrobial susceptibility testing and whole-genome sequencing to identify resistance determinants and virulence factors. Seven Klebsiella pneumoniae isolates were classified as extensively drug-resistant. The remaining Klebsiella, Enterobacter hormaechei, and Escherichia coli isolates were multidrug-resistant. Eighteen strains carried the metallo-β-lactamase NDM-1, two the serine-carbapenemase KPC-2, and one isolate had both carbapenemases. The blaNDM-1 gene was located in the truncated ΔISAba125 element, and the blaKPC-2 gene was in the Tn4401a transposon. ST147 was the most frequent sequence type among K. pneumoniae isolates. Our findings highlight the urgent need to address the emergence of CPE and strengthen control measures and antibiotic stewardship programs in low- and middle-income settings.
IMPORTANCE
Genomic surveillance of antimicrobial resistance contributes to monitoring the spread of resistance and informs treatment and prevention strategies. We characterized 21 carbapenemase-producing Enterobacterales collected at a Peruvian tertiary hospital in 2018, which exhibited very high levels of resistance and carried numerous resistance genes. We detected the coexistence of carbapenemase-encoding genes (blaNDM-1 and blaKPC-2) in a Klebsiella pneumoniae isolate that also had the PmrB(R256G) mutation associated with colistin resistance. The blaKPC-2 genes were located in Tn4401a transposons, while the blaNDM-1 genes were in the genetic structure Tn125 (ΔISAba125). The presence of high-risk clones among Klebsiella pneumoniae (ST11 and ST147) and Escherichia coli (ST410) isolates is also reported. The study reveals the emergence of highly resistant bacteria in a Peruvian hospital, which could compromise the effectiveness of current treatments and control.
KEYWORDS: carbapenems, carbapenem-producing Enterobacterales, drug resistance, Whole-genome sequencing, Peru
INTRODUCTION
Antimicrobial resistance (AMR) poses a significant threat to human health and the global economy. The rise of multidrug-resistant (MDR) and pan-resistant (PDR) bacteria jeopardizes the advancements in modern medicine, potentially leading us toward a post-antibiotic era (1). Carbapenems, vital antibiotics often reserved for severe infections caused by MDR Gram-negative bacilli, are now threatened by the emergence of carbapenemase-producing Enterobacterales (CPE) (2). These pathogens are notably difficult to treat due to the limited effectiveness of available drugs, forcing clinicians to revert to older medications such as colistin (2, 3).
Carbapenemases, as categorized by Ambler’s classification, include Class A Klebsiella pneumoniae carbapenemase (KPC), Class B metallo-β-lactamases (MBLs) like New Delhi MBL (NDM), and Class D oxacillinases (OXA) such as OXA-48-like enzymes found in Enterobacterales (2). Klebsiella pneumoniae and Escherichia coli, among the CPE, have emerged as significant global health threats due to their high mortality rates (4). The widespread dissemination of CPE is primarily attributed to the horizontal transfer of antibiotic-resistance genes via mobile genetic elements, including plasmids and transposons (5). Various virulence factors enhance the pathogenicity of CPE. While there is extensive knowledge about the interplay between these factors and AMR in many bacterial pathogens (6), the specific genetic determinants that drive virulence and their interaction with resistance mechanisms are not fully understood (7).
In Peru, the first CPE infections were identified in late 2013 as a carbapenem-resistant K. pneumoniae, sequence type (ST) 340 of the high-risk clonal complex (CC) 258 (8, 9). Subsequent reports indicate the spread of various carbapenemases in clinical settings and non-traditional hosts such as Providencia stuartii (10, 11). By 2020, the spread of the metallo-β-lactamase NDM-1 in K. pneumoniae was evident, involving newer lineages like ST348 and ST147 (12). Despite numerous CPE reports in Peru and Latin America, comprehensive genomic data are needed to conduct in-depth evolutionary, molecular, and epidemiological studies. Understanding the relationships between different CPE strains, both locally and globally, is essential. To address this gap, we sequenced the genomes and analyzed the resistance and virulence determinants of 21 CPE samples from a tertiary hospital in Lima collected in 2018.
RESULTS
Antimicrobial susceptibility profiles
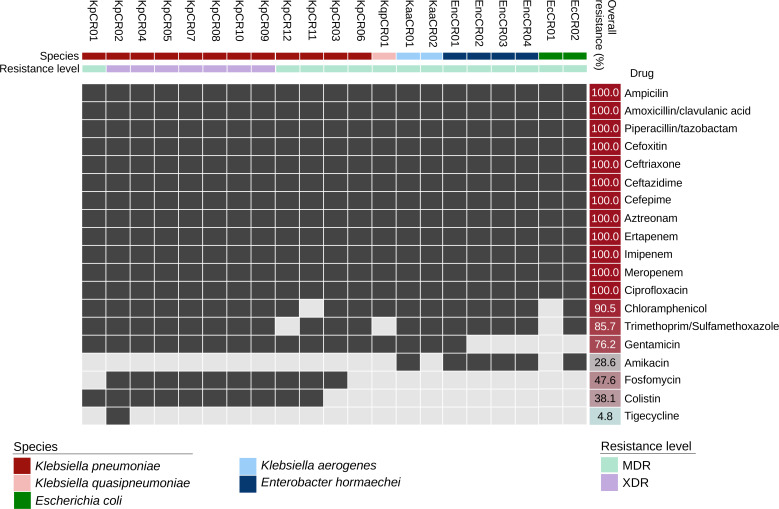
The results of antibiotic susceptibility testing of 12 K. pneumoniae, 1 Klebsiella quasipneumoniae, 2 Klebsiella aerogenes, 2 E. coli, and 4 Enterobacter hormaechei are shown in Fig. 1. Overall, all CPE isolates were resistant to penicillins plus β-lactamase inhibitors, cephalosporins, monobactams, and fluoroquinolones. Furthermore, nearly all CPE isolates were resistant to phenicols (19/21, 90.5%) and folate pathway inhibitors (18/21, 85.7%). All Klebsiella species were resistant to gentamicin, and almost all were susceptible to amikacin, whereas all E. hormaechei were resistant to amikacin but mainly susceptible to gentamicin. E. coli isolates were predominantly susceptible to both aminoglycosides. Resistance to fosfomycin (10/12, 83.3%) and colistin (10/12, 83.3%) was observed only among K. pneumoniae isolates, whereas the other CPE were susceptible to both drugs. Phenotypic screening of carbapenemase production resulted in the identification of 19 metallo-β-lactamase-producers and 3 serine-carbapenemase producers. Notably, strain KpCR02 tested positive for serine and metallo-carbapenemase enzymes using phenotypic methods. Seven K. pneumoniae strains were classified as extensively drug-resistant (XDR), and the remaining CPE as MDR (Fig. 1; Table S1).
Fig 1.
Antimicrobial susceptibility profiles of Enterobacterales by antimicrobial category. Black and gray boxes indicate resistance and susceptibility, respectively.
Genomic diversity
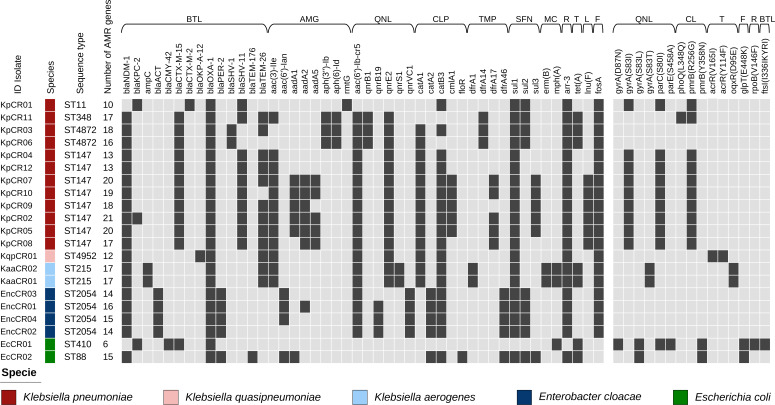
Four STs were identified among K. pneumoniae isolates: ST11 (n = 1), ST147 (n = 8), ST348 (n = 1), and ST4872 (n = 2) (Fig. 2). Among the K. pneumoniae ST147 genomes, they diverged by 21–115 single nucleotide polymorphisms (SNP). The K. quasipneumoniae isolate was classified as ST4952, and both K. aerogenes isolates were ST215. ST88 and ST471 were identified among E. coli isolates. The four E. hormaechei isolates were classified as ST2054.
Fig 2.
Resistance genes of carbapenem-resistant Enterobacterales. Total AMR genes are summarized by bacterial isolate, and individual annotated genes conferring resistance are also summarized by antimicrobial category. BTL, β-lactams; AMG, aminoglycoside; QNL, quinolone; CLP, chloramphenicol; TMP, trimethoprim; SFN, sulfonamide; MC, macrolide; R, rifamycin; T, tetracycline; L, lincosamide; CL, colistin; and F, florfenicol.
Antimicrobial resistance genes
K. pneumoniae isolates harbored more AMR genes (average = 16.8, SD = 3.3) than other species in this study (average = 14.0, SD = 3.4). The blaOXA-1 gene was detected in all CPE isolates, while sul1 and arr-3 (20/21, 95.2%), catB3 (19/21, 90.5%), and aac(6′)-lb-cr5 (18/21, 85.7%) genes were detected in nearly all isolates. Several AMR genes responsible for resistance to non-carbapenem β-lactams, including AmpC β-lactamases, extended-spectrum β-lactamases (ESBLs), aminoglycosides, fluoroquinolones, and other antimicrobial classes, were also detected among CPE isolates. Regarding carbapenemase-encoding genes, blaNDM-1 (19/21, 90.5%) was detected in almost all isolates, whereas blaKPC-2 (3/21, 14.3%) was detected in two K. pneumoniae and one E. coli. The K. pneumoniae strain KpCR02 tested positive for both blaNDM-1 and blaKPC-2 genes.
Genetic context of carbapenemase genes
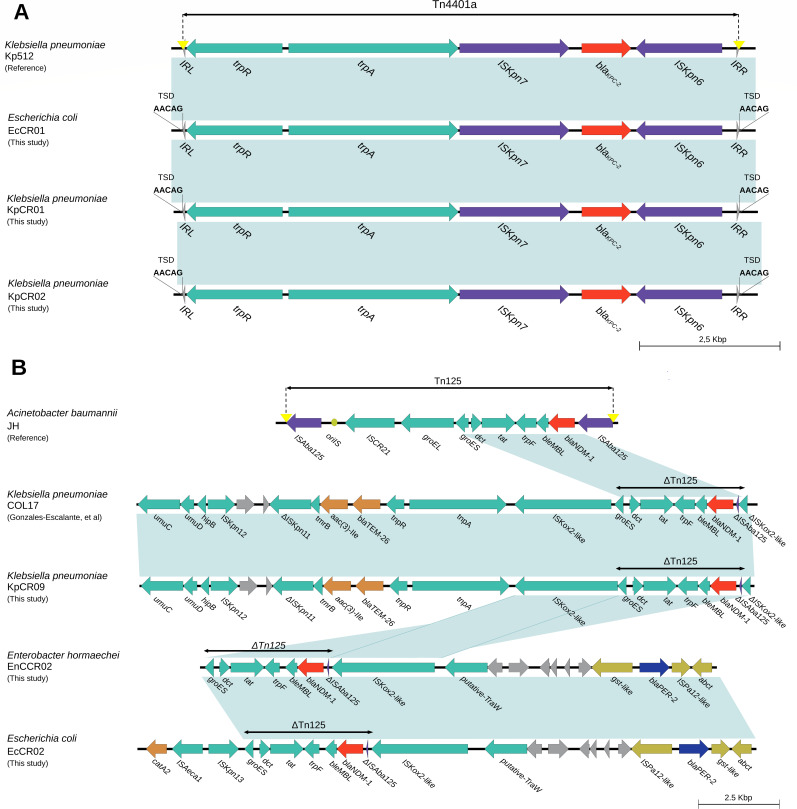
The blaKPC-2 gene was found in K. pneumoniae KpCR01 and KpCR02, and E. coli EcCR01, located in a Tn4401a transposon of the Tn3 family. The gene was flanked by ISKpn6 and ISkpn7 elements near the tnpA and tnpR transposase genes (Fig. 3A). The blaNDM-1 gene in K. pneumoniae strain KpCR09, E. hormaechei strain EnCCR02, and E. coli strain EcCR02 was found within the ΔTn125 element (Fig. 3B). The blaNDM-1 gene was flanked by ISA-ba125 and the blaMBL gene (Fig. 3B) in both reference strains and the three isolates described here. In K. pneumoniae strain KpCR09, the blaTEM-26 and aac (3)-lle genes, which encode resistance to third- and fourth-generation cephalosporin and aminoglycoside-modifying enzyme (AME), respectively, were identified upstream of ΔTn125.
Fig 3.
(A) Genetic context of the structures of the Tn4401a transposon carrying blaKPC-2 (GenBank accession number KT378596) and (B) genetic context of Tn125 (GenBank reference number JN872328) in our Enterobacterales isolates. Shading indicates 100% sequence similarity.
Virulence factors
K. pneumoniae isolate KpCR01 presented the yersiniabactin ybt9 gene located on the integrative conjugative element (ICE) Kp3, along with the genes encoding for the wzi50 capsular type and antigenic determinants K15 and O4. In contrast, isolate KpCR11 exhibited the ybt0 gene within the ICEKp12 element, accompanied by the wzi94 capsular type gene and antigenic determinants K62 and O1. Notably, all K. pneumoniae ST147 isolates in our study had the aerobactin (iuc5) gene, genes encoding for the wzi64 capsular type, and antigenic determinants KL64 and O2a. Additionally, in our two E. coli isolates, we identified several virulence genes, including fimH, associated with type 1 fimbriae; iucC, related to aerobactin synthetase; iutA, encoding the ferric aerobactin receptor; lpfA, indicative of long polar fimbriae; and sitA, associated with iron acquisition (Table S2).
DISCUSSION
The rapid spread of CPE poses a significant threat to global public health. Our study of CPE from one Peruvian hospital in 2018 confirms the increasing prevalence of blaNDM-1 over blaKPC-2 in Peru since 2013, alongside the emergence of OXA-48-like carbapenemases, particularly OXA-181 (3–5, 13).
Our findings confirm high resistance levels in CPE, particularly in Klebsiella pneumoniae. Notably, the KpCR02 isolate exhibited up to 21 AMR genes, including NDM-1 and KPC-2. All NDM-1-producing CPE showed resistance to β-lactams, including aztreonam, often due to AmpC and ESBL genes, with the blaCTX-M-15 gene being particularly prevalent in our data set (14, 15). The co-production of carbapenemases NDM-1 and KPC-2 in K. pneumoniae has been previously described in South America, including Brazil, Argentina, Uruguay, Ecuador, and Paraguay (16). Our report provides further evidence of this occurrence and regional dispersion of CPE co-producing NDM-1 and KPC-2 carbapenemases.
The aminoglycoside resistance observed in our isolates could be attributed to genes that encode AMEs. The rmtG gene involved in 16S rRNA methylation confers high-level resistance to aminoglycosides (minimum inhibitory concentration, MIC ≥ 256 µg/mL) and is widely spread in K. pneumoniae found in South America (17). Among the AMEs, the acetyltransferases aac(3)-lle and aac(6')Ib-cr5 were the most frequently detected. The aac (3)-IIa has been associated with resistance to gentamicin, tobramycin, and netilmicin, as described elsewhere (18). The aac(6')Ib-cr is characterized by W102R and D179Y/G substitutions [both substitutions being present in aac(6')Ib-cr5], which, in addition to its capacity to inactivate several aminoglycosides, also result in the ability to inactivate several fluoroquinolones such as ciprofloxacin and norfloxacin (19). Nine K. pneumoniae and all E. coli isolates had amino acid substitutions at GyrA and ParC, the most common quinolone resistance mechanism (20). The remaining CPE did not present mutations in the established quinolone targets. However, all presented at least two transferable mechanisms of quinolone resistance and alterations in efflux pump regulators in one case. While highly unusual several years ago, this scenario is increasingly being described (19, 20).
Resistance to colistin observed in K. pneumoniae isolates could be explained by the amino acid substitution detected in the PmrB (R256G) of 10 isolates, one of which also had the PhoQ(L348Q) mutation. Remarkably, both mutations are associated with lipid A modification (21). PmrB amino acid substitution (Y358N) was detected in two E. coli isolates; however, both were susceptible to colistin. Although the polymorphism may not be ruled out, this amino acid substitution has been previously described in E. coli isolates with high levels of colistin resistance (MIC >32 µg/mL) in the absence of mcr genes or other alteration in PmrAB, PhoPQ, or MgrB. Therefore, this single substitution may play a role in the development of colistin resistance (22).
Our genomic analysis found ST147 to be the dominant clone in K. pneumoniae, known for its high drug resistance and global dissemination. This includes the KpCR02 isolate, exhibiting both KPC-2 and NDM-1 and colistin resistance through pmrB mutations. The global emergence of KPC has been influenced by the wide dispersion of K. pneumoniae CC258, including ST11 and ST340 (23). ST11 is endemic in Brazil and several European countries (24, 25), and ST340 has been previously reported in Peru (7). Several K. pneumoniae ST11, ST147, and ST348 outbreaks have also been reported in Peru (10, 26). Our report of K. pneumoniae KPC-producing ST11 adds evidence of a high-risk international clone in Peru. In contrast to other reports, K. pneumoniae ST348 isolate KpCR11 described in this study significantly differs from other ST348 strains characterized by CTX-M-15 and KPC-3 production (27). The study reports K. pneumoniae ST4872, K. quasipneumoniae ST4952, and K. aerogenes ST215 for the first time in Peru. Among E. coli strains, ST471 and ST88 were identified, known for their resistance and virulence, alongside the first reported cases of E. hormaechei ST2054 in the region.
Analysis of the genetic context of the blaKPC-2 gene revealed that it was located in the Tn4401a transposon, a member of the Tn3 transposon family (28, 29). To our knowledge, this is one of the few reports on Tn4401a in E. coli (30). Of note, current data differ from the previous genetic environment of blaKPC-2 detected in Peru, highlighting the presence of different genetic structures carrying this gene in the country (7). Furthermore, the analysis of the genetic context of the blaNDM-1 gene revealed that it was located in a truncated structure of Tn125. The architecture of the characterized ∆Tn was identical to those previously reported in Peru and other Latin American countries (31, 32), and no differences were observed among the other CPE described here.
Hypervirulence is associated with the presence of additional siderophores, yersiniabactin (ybt), aerobactin (iuc), salmochelin (iro), and specific capsular serotypes (K1, K2, and K5) (33). Interestingly, the aerobactin iuc5 gene was detected in all K. pneumoniae ST147 samples. Besides potentially playing a critical role in vitro and in vivo virulence, it is a marker to identify highly virulent strains (33). This finding is compatible with that described in other CC258 isolates (34); however, in our isolate, neither characteristic virulence determinants such as salmochelin or colibactin nor genetic determinants associated with hypermucoviscosity (rmpA) were detected. To date, 12 distinct O loci have been identified, with O1 and O2 as the most common antigens (35), while K-type variation has been associated with six conserved genes (galF, orf2, wzi, wza, wzb, and wzc). The wzi gene best predicts K antigen-related virulence (36).
The study’s limitations include a need for gene expression data and a limited sampling period from one healthcare center in 2018, which restricts the broader application of our findings. Instead, the results described here should be considered reference data and a starting point for further studies using more complex designs and sampling methods.
Overall, our comprehensive analysis of 21 CPE isolates underscores the significant challenge posed by drug-resistant CPE strains, emphasizing the need for expanded genomic surveillance, effective infection control, and antimicrobial stewardship programs to curb the spread of CPE and maintain the efficacy of antimicrobial agents, particularly in the understudied Latin American region.
MATERIALS AND METHODS
Clinical isolates
From January to December 2018, 21 consecutive, non-replicated, and unique suspected CPE isolates were chosen based on their phenotypic resistance to meropenem at an MIC of ≥1 µg/mL using disk-diffusion assays. Clinical isolates were obtained from blood, urine, and other biological samples from inpatients and outpatients admitted to the tertiary public hospital, Hospital Nacional Arzobispo Loayza in Lima, Peru. The hospital’s laboratory characterized all isolates at the species level using a standard biochemical test panel (37) and sent them to Universidad Peruana Cayetano Heredia (UPCH) for phenotypic and genomic characterization.
Carbapenemase screening and identification
The phenotypic screening of carbapenemase production was done with the Triton Hodge Test and the carbapenem inactivation method as previously described (38). Inhibition tests using EDTA and phenylboronic acid were used to distinguish classes of carbapenemases (39).
Antimicrobial susceptibility testing
Resistance phenotypes were determined using the VITEK2 System with AST-N249 cards (bioMérieux, France) following the manufacturer’s instructions. MIC values were interpreted using clinical breakpoints established by the Clinical and Laboratory Standards Institute (CLSI) M100 guidelines (40). The European Committee for Antimicrobial Susceptibility Testing breakpoints were used for tigecycline interpretation since CLSI breakpoints were not available at the time of the study (41). Colistin susceptibility was assessed by the colistin broth disk elution test as previously described (42). Intermediate resistance results were analyzed and reported as resistant, and the criteria suggested by Magiorakos et al. were used to classify bacteria as MDR, XDR, or PDR (43).
DNA extraction and whole-genome sequencing
Genomic DNA was extracted using the GeneJET DNA purification kit (Thermo Fisher Scientific, USA) from single colonies that were incubated in 1 mL of tryptic soy broth (Becton & Dickinson, USA) at 37°C and shaking at 300 rpm for 8 hours. The extracted gDNA was quantified with a Qubit 4.0 fluorometer, and Illumina libraries were constructed using 1 ng of gDNA with the Nextera XT kit (Illumina, USA). Paired-end libraries were sequenced on an Illumina MiSeq instrument generating 250-nt reads resulting in a mean per-base coverage of 75× (minimum 51× and maximum 108×).
Bioinformatics analysis
Raw reads were assessed using FastQC v0.11.9 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), trimmed with Trimmomatic v0.36.6 (LEADING:10, TRAILING:10, SLIDINGWINDOW:4:20, and MINLEN:40) (44), assembled de novo with SPAdes v3.10.0 (45), and evaluated for assembly quality using Quast v5.0.2 (46). Bacterial identification was performed using PathogenWatch (https://pathogen.watch/), and ST assignment was determined using the PubMLST (https://pubmlst.org/) scheme. Bacterial genomes with novel alleles were submitted to BIGSdb (https://pubmlst.org/software/bigsdb) for analysis and ST assignment. Virulence factors of E. coli were screened using VirulenceFinder v2.0 (https://cge.food.dtu.dk/services/VirulenceFinder/), and virulence factors and predicted capsule (K) and lipopolysaccharide (O) profiles of Klebsiella species were determined using Kleborate v2.3.2. AMR genes were identified using AMRFinderPlus (coverage length ≥ 90%, nucleotide identity ≥ 90%, without gaps) (47). E. coli phylogroups were determined in silico using the ClermonTyping algorithm (http://clermontyping.iame-research.center/), and O and flagellar (H) serotypes were determined using SeroTypeFinder v2 (https://cge.food.dtu.dk/services/SerotypeFinder/).
Genome comparisons
K. pneumoniae genomes belonging to ST147 were aligned to the reference genome NTUH-K2044, and recombinant regions were filtered out using Gubbins v3.3.0. Pairwise SNP differences between genomes were calculated and estimated using snp-dists (https://github.com/tseemann/snp-dists) (48). Contigs bearing the KPC-2 and NDM-1 genes were extracted from de novo assemblies and annotated with Prokka v1.5 (49). Since both genes were detected in transposable genetic elements, they were examined and annotated using MobileElementFinder v1.0.3 (https://pypi.org/project/MobileElementFinder/) and ISfinder (https://isfinder.biotoul.fr/) and then characterized using TETyper v1.1 (https://github.com/aesheppard/TETyper). Transposons were characterized using the Transposon Registry (http://transposon.lstmed.ac.uk/tn-registry) (50).
Supplementary Material
ACKNOWLEDGMENTS
The study was funded by Prociencia grant number 088-2018. D.C. and P.T. are supported by a D43 TW007393 training grant awarded to UPCH by the Fogarty International Center of the U.S. National Institutes of Health. S.L. is supported by a competitive award received from Universidad de Cartagena–UNIMOL, as part of a Ph.D. scholarship.
Contributor Information
Pablo Tsukayama, Email: pablo.tsukayama@upch.pe.
Cheryl P. Andam, University at Albany, Albany, New York, USA
Dennis Nurjadi, Universitat zu Lubeck, Lübeck, Germany.
ETHICS APPROVAL
The study protocol was approved by the Institutional Review Board of the Universidad Peruana Cayetano Heredia (SIDISI 100535) and approved by the Hospital Arzobispo Loayza research board. Patient information was anonymized and deidentified before analysis.
DATA AVAILABILITY
The raw read files and assemblies for the five isolates are available at NCBI under BioProject accession number PRJNA865026.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.02503-23.
MIC values for the studied isolates of carbapenemase-producing Enterobacterales (CPE).
Virulence and antimicrobial resistance genes, along with assembly statistics of studied isolates of CPE.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Tilahun M, Kassa Y, Gedefie A, Ashagire M. 2021. Emerging carbapenem-resistant Enterobacteriaceae infection, its epidemiology and novel treatment options: a review. Infect Drug Resist 14:4363–4374. doi: 10.2147/IDR.S337611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sheu C-C, Chang Y-T, Lin S-Y, Chen Y-H, Hsueh P-R. 2019. Infections caused by carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Front Microbiol 10:80. doi: 10.3389/fmicb.2019.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khuntayaporn P, Thirapanmethee K, Chomnawang MT. 2022. An update of mobile colistin resistance in non-fermentative gram-negative bacilli. Front Cell Infect Microbiol 12:882236. doi: 10.3389/fcimb.2022.882236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. GBD 2019 Antimicrobial Resistance Collaborators . 2022. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the global burden of disease study 2019. Lancet 400:2221–2248. doi: 10.1016/S0140-6736(22)02185-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beceiro A, Tomás M, Bou G. 2013. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world Clin Microbiol Rev 26:185–230. doi: 10.1128/CMR.00059-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amaretti A, Righini L, Candeliere F, Musmeci E, Bonvicini F, Gentilomi GA, Rossi M, Raimondi S. 2020. Antibiotic resistance, virulence factors, phenotyping, and genotyping of non-Escherichia coli Enterobacterales from the gut microbiota of healthy subjects. Int J Mol Sci 21:1847. doi: 10.3390/ijms21051847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Velásquez J, Hernández R, Pamo O, Candiotti M, Pinedo Y, Sacsaquispe R, Cáceres L, Fernández N. 2013. Klebsiella pneumoniae resistente a los carbapenemes. primer caso de carbapenemasa tipo KPC en Perú. Rev Soc Peru Med Interna 26:192–196. [Google Scholar]
- 9. Horna G, Velasquez J, Fernández N, Tamariz J, Ruiz J. 2017. Characterisation of the first KPC-2-producing Klebsiella pneumoniae ST340 from Peru. J Glob Antimicrob Resist 9:36–40. doi: 10.1016/j.jgar.2016.12.011 [DOI] [PubMed] [Google Scholar]
- 10. Mayta-Barrios MM, Ramirez-Illescas JJ, Pampa-Espinoza L, Yagui-Moscoso MJA. 2021. Molecular characterization of carbapenemases in Peru during 2019. Rev Peru Med Exp Salud Publica 38:113–118. doi: 10.17843/rpmesp.2021.381.5882 [DOI] [PubMed] [Google Scholar]
- 11. Lezameta L, Cuicapuza D, Dávila-Barclay A, Torres S, Salvatierra G, Tsukayama P, Tamariz J. 2020. Draft genome sequence of a new Delhi metallo-β-lactamase (NDM-1)-producing Providencia stuartii strain isolated in Lima, Peru. Microbiol Resour Announc 9:e00788-20. doi: 10.1128/MRA.00788-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roach D, Waalkes A, Abanto J, Zunt J, Cucho C, Soria J, Salipante SJ. 2020. Whole genome sequencing of Peruvian Klebsiella pneumoniae identifies novel plasmid vectors bearing carbapenem resistance gene NDM-1. Open Forum Infect Dis 7:faa266. doi: 10.1093/ofid/ofaa266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cuicapuza D, Alvarado L, Tocasca N, Aguilar D, Gómez-de-la-Torre JC, Salvatierra G, Tsukayama P, Tamariz J. 2023. First report of OXA-181-producing Enterobacterales isolates in Latin America. Microbiol Spectr 11:e0458422. doi: 10.1128/spectrum.04584-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. doi: 10.1093/jac/dkx146 [DOI] [PubMed] [Google Scholar]
- 15. Gonzales E, Patiño L, Ore E, Martínez V, Moreno S, Cruzado NB, Rojas R, Quispe M del C, Carbonell I, Villarreal F, Maza G, Olivo J, Vicuña R, Bustamante D. 2020. β-lactamasas de espectro extendido tipo CTX-M en aislamientos clínicos de Escherichia coli y Klebsiella pneumoniae en el Instituto Nacional de Salud del Niño-Breña, Lima Perú. Rev Med Hered 30:242–248. doi: 10.20453/rmh.v30i4.3659 [DOI] [Google Scholar]
- 16. Vásquez-Ponce F, Dantas K, Becerra J, Melocco G, Esposito F, Cardoso B, Rodrigues L, Lima K, de Lima AV, Sellera FP, Mattos R, Trevisoli L, Vianello MA, Sincero T, Di Conza J, Vespero E, Gutkind G, Sampaio J, Lincopan N. 2022. Detecting KPC-2 and NDM-1 coexpression in Klebsiella pneumoniae complex from human and animal hosts in South America. Microbiol Spectr 10:e0115922. doi: 10.1128/spectrum.01159-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang W, Hu F. 2022. Research updates of plasmid-mediated aminoglycoside resistance 16S rRNA methyltransferase. Antibiotics (Basel) 11:906. doi: 10.3390/antibiotics11070906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krause KM, Serio AW, Kane TR, Connolly LE. 2016. Aminoglycosides: an overview. Cold Spring Harb Perspect Med 6:a027029. doi: 10.1101/cshperspect.a027029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruiz J. 2019. Transferable mechanisms of quinolone resistance from 1998 onward. Clin Microbiol Rev 32:e00007-19. doi: 10.1128/CMR.00007-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruiz J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 51:1109–1117. doi: 10.1093/jac/dkg222 [DOI] [PubMed] [Google Scholar]
- 21. Huang J, Li C, Song J, Velkov T, Wang L, Zhu Y, Li J. 2020. Regulating polymyxin resistance in gram-negative bacteria: roles of two-component systems PhoPQ and PmrAB. Future Microbiol 15:445–459. doi: 10.2217/fmb-2019-0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo Q, Yu W, Zhou K, Guo L, Shen P, Lu H, Huang C, Xu H, Xu S, Xiao Y, Li L. 2017. Molecular epidemiology and colistin resistant mechanism of mcr-positive and mcr-negative clinical isolated Escherichia coli. Front. Microbiol 8:2262. doi: 10.3389/fmicb.2017.02262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Castanheira M, Costello AJ, Deshpande LM, Jones RN. 2012. Expansion of Clonal complex 258 KPC-2-producing Klebsiella pneumoniae in Latin American hospitals: Report of the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother 56:1668–1669. doi: 10.1128/AAC.05942-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nascimento T, Cantamessa R, Melo L, Fernandes MR, Fraga E, Dropa M, Sato MIZ, Cerdeira L, Lincopan N. 2017. International high-risk clones of Klebsiella pneumoniae KPC-2/Cc258 and Escherichia coli CTX-M-15/CC10 in urban lake waters. Sci Total Environ 598:910–915. doi: 10.1016/j.scitotenv.2017.03.207 [DOI] [PubMed] [Google Scholar]
- 25. David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, et al. 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. doi: 10.1038/s41564-019-0492-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pons MJ, Marí-Almirall M, Ymaña B, Moya-Salazar J, Muñoz L, Sauñe S, Salazar-Hernández R, Vila J, Roca I. 2020. Spread of ST348 Klebsiella pneumoniae producing NDM-1 in a Peruvian hospital. Microorganisms 8:1392. doi: 10.3390/microorganisms8091392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trigo da Roza F, Couto N, Carneiro C, Cunha E, Rosa T, Magalhães M, Tavares L, Novais Â, Peixe L, Rossen JW, Lamas LP, Oliveira M. 2019. Commonality of multidrug-resistant Klebsiella pneumoniae ST348 isolates in horses and humans in Portugal. Front Microbiol 10:1657. doi: 10.3389/fmicb.2019.01657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen L, Chavda KD, Mediavilla JR, Jacobs MR, Levi MH, Bonomo RA, Kreiswirth BN. 2012. Partial excision of blaKPC from Tn4401 in carbapenem-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother 56:1635–1638. doi: 10.1128/AAC.06182-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 55:5370–5373. doi: 10.1128/AAC.05202-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Piekar M, Álvarez VE, Knecht C, Leguina C, García Allende N, Carrera Páez L, Gambino AS, González Machuca A, Campos J, Fox B, Carpio E, Aguilar A, Alonso FM, Fernández Canigia L, Quiroga MP, Centrón D. 2023. Genomic data reveals the emergence of the co-occurrence of blaKPC-2 and blaCTX-M-15 in an Escherichia coli ST648 strain isolated from rectal SWAB within the framework of hospital surveillance. J Glob Antimicrob Resist 32:108–112. doi: 10.1016/j.jgar.2022.12.012 [DOI] [PubMed] [Google Scholar]
- 31. Gonzales-Escalante E, Ruggiero M, Cerdeira L, Esposito F, Fontana H, Lincopan N, Gutkind G, Conza J. 2022. Whole-genome analysis of a high-risk clone of Klebsiella pneumoniae ST147 carrying both mcr-1 and blaNDM-1 genes in Peru. Microb Drug Resist 28:171–179. doi: 10.1089/mdr.2021.0128 [DOI] [PubMed] [Google Scholar]
- 32. Marquez-Ortiz RA, Haggerty L, Olarte N, Duarte C, Garza-Ramos U, Silva-Sanchez J, Castro BE, Sim EM, Beltran M, Moncada MV, Valderrama A, Castellanos JE, Charles IG, Vanegas N, Escobar-Perez J, Petty NK. 2017. Genomic epidemiology of NDM-1-encoding plasmids in Latin American clinical isolates reveals insights into the evolution of multidrug resistance. Genome Biol Evol 9:1725–1741. doi: 10.1093/gbe/evx115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lam MMC, Wyres KL, Judd LM, Wick RR, Jenney A, Brisse S, Holt KE. 2018. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med 10:77. doi: 10.1186/s13073-018-0587-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wyres KL, Lam MMC, Holt KE. 2020. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 18:344–359. doi: 10.1038/s41579-019-0315-1 [DOI] [PubMed] [Google Scholar]
- 35. Wick RR, Heinz E, Holt KE, Wyres KL. 2018. Kaptive web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J Clin Microbiol 56:e00197-18. doi: 10.1128/JCM.00197-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decré D. 2013. Wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol 51:4073–4078. doi: 10.1128/JCM.01924-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Garrity G, Brenner DJ, Krieg NR, Staley JR. 2005. The proteobacteria, part B: the gamma proteobacteria, p 1106. In Bergey’s manual of systematic bacteriology. Springer, New York, NY. [Google Scholar]
- 38. Pasteran F, Gonzalez LJ, Albornoz E, Bahr G, Vila AJ, Corso A. 2016. Triton hodge test: improved protocol for modified hodge test for enhanced detection of NDM and other carbapenemase producers. J Clin Microbiol 54:640–649. doi: 10.1128/JCM.01298-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsakris A, Poulou A, Pournaras S, Voulgari E, Vrioni G, Themeli-Digalaki K, Petropoulou D, Sofianou D. 2010. A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J Antimicrob Chemother 65:1664–1671. doi: 10.1093/jac/dkq210 [DOI] [PubMed] [Google Scholar]
- 40. Clinical and Laboratory Standards Institute . 2018. Performance standards for antimicrobial susceptibility testing. In CLSI supplement M100, 30th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 41. European Committee on Antimicrobial Susceptibility Testing . 2023. Clinical breakpoints and dosing of antibiotics. Available from: https://www.eucast.org/clinical_breakpoints. Retrieved 18 Apr 2023.
- 42. Simner PJ, Bergman Y, Trejo M, Roberts AA, Marayan R, Tekle T, Campeau S, Kazmi AQ, Bell DT, Lewis S, Tamma PD, Humphries R, Hindler JA. 2019. Two-site evaluation of the colistin broth disk elution test to determine colistin in vitro activity against gram-negative bacilli. J Clin Microbiol 57:e01163-18. doi: 10.1128/JCM.01163-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 44. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB, Prasad AB, Tillman GE, Tyson GH, Klimke W. 2021. Amrfinderplus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 11:12728. doi: 10.1038/s41598-021-91456-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 49. Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roberts AP, Chandler M, Courvalin P, Guédon G, Mullany P, Pembroke T, Rood JI, Smith CJ, Summers AO, Tsuda M, Berg DE. 2008. Revised nomenclature for transposable genetic elements. Plasmid 60:167–173. doi: 10.1016/j.plasmid.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MIC values for the studied isolates of carbapenemase-producing Enterobacterales (CPE).
Virulence and antimicrobial resistance genes, along with assembly statistics of studied isolates of CPE.
An accounting of the reviewer comments and feedback.
Data Availability Statement
The raw read files and assemblies for the five isolates are available at NCBI under BioProject accession number PRJNA865026.