ABSTRACT
In 2015, Staphylococcus argenteus and Staphylococcus schweitzeri were proposed as new species, distinct from Staphylococcus aureus and collectively referred to as the S. aureus complex. However, no clinical reports of these new species exist in Korea. Upon the application of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) for all bloodstream isolates since September 2022, S. argenteus was identified in one patient. Therefore, we aimed to search for new species among the archives of the S. aureus bacteremia cohort and describe their clinical and microbiological characteristics. Among the 691 archived S. aureus isolates between 2012 and 2018, one was identified as S. argenteus via MALDI-TOF MS. Both S. argenteus isolates (one in 2022) were obtained from patients with extensive pneumonia accompanied by bacteremia and both cases had fatal outcomes. They harbored multiple virulence genes (clfA, clfB, fnbpA, sdrC, sdrD, sdrE, bbp, cna, see, seg, sei, blaZ, fnbpB, and map) but did not harbor mecA and pvl. No matched sequence type (ST) was found in either isolate, and both S. argenteus isolates were closely related to ST1594, ST1593, ST1793, and ST1303, which belonged to S. argenteus. S. argenteus accounted for <1% of the S. aureus complex but had clinical characteristics similar to S. aureus. Therefore, clinicians should be aware of these factors to avoid misidentifying these strains as coagulase-negative staphylococci, and appropriate reporting is required to minimize confusion.
IMPORTANCE
Staphylococcus argenteus, a member of Staphylococcus aureus complex, has been reported as an important pathogen that causes clinically invasive infections in humans similar to S. aureus. Clinical isolates of S. argenteus have been reported across the world, showing a large geographical difference in prevalence and genomic profile. However, there have been no clinical reports regarding this new species in Korea. This is the first report to investigate the clinical and genetic characteristics of S. argenteus identified in patients with bacteremia, and the proportion of S. argenteus bacteremia among S. aureus bacteremia cohort in Korea.
KEYWORDS: Staphylococcus aureus, Staphylococcus argenteus, bacteremia, MALDI-TOF MS, Republic of Korea
OBSERVATION
Staphylococcus aureus is a clinically important pathogen that causes various infections ranging from skin and soft tissue infections to infective endocarditis (1). In 2015, Staphylococcus argenteus and Staphylococcus schweitzeri were identified as new species, distinct from S. aureus (2). These new species are indistinguishable from S. aureus using conventional routine diagnostics, such as microscopy, colony morphology, and coagulase assays and are collectively referred to as the S. aureus complex. However, they can be distinguished via peptidoglycan composition analysis, molecular typing, such as matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), genotyping, including multi-locus sequence typing (MLST), and whole-genome sequencing (3). Since new diagnostic methods based on molecular typing or genotyping have been introduced and are widely used, clinicians may encounter S. argenteus more frequently based on clinical microbiology laboratory results. However, no clinical reports on these new species exist in Korea; thus, they remain largely unrecognized. Therefore, we aimed to investigate the presence of these species in an archived S. aureus bacteremia cohort and describe their clinical and microbiological characteristics.
Since September 2022, MALDI-TOF MS has been applied to all positive blood cultures of patients with bacteremia at the study hospital. S. argenteus was identified in the blood culture of one patient (strain 1) on 4 December 2022. Stored isolates from the S. aureus bacteremia cohort were re-evaluated using MALDI-TOF MS to identify possible misidentifications. This study involved the S. aureus bacteremia cohort treated between May 2012 and December 2018. Four cohorts from which data were prospectively collected over the study period were included, and their detailed information is described in the Acknowledgments section (4–7).
Isolates were evaluated using a MALDI Biotyper Sirius (Bruker Daltonics GmbH & Co. KG, Bremen, Germany) with the MBT compass reference library (version 11.0) containing S. argenteus and S. schweitzeri in the strain list, which was highly sensitive and specific for distinguishing S. argenteus from S. aureus (8). MLST was performed and the sequence type (ST) was analyzed based on the MLST scheme for S. aureus using the S. aureus pubMLST database (https://pubmlst.org/organisms/staphylococcus-aureus) (9). Phylogenetic analysis was performed by aligning concatenated MLST data using CLUSTALW and constructing the phylogenetic tree using the neighbor-joining method in MEGA version 11. S. argenteus isolates used in this study were distinguished from the reference collection based on phylogenetic clustering. Furthermore, 73 STs were downloaded from the MLST database and selected as reference sequences based on the initial analysis of each S. aureus MLST locus using the same database (not shown).
Genomic DNA was extracted using an E.Z.N.A. Stool DNA Kit (Omega Bio-tek, USA) according to the manufacturer’s protocol. Library preparation from isolated DNA and de novo whole-genome sequencing (WGS) were performed by Macrogen (Seoul, Republic of Korea). High-quality DNA was used for constructing the library with the help of the TruSeq Nano DNA kit. The WGS was done on the Illumina sequencing by synthesis platform (Illumina Inc., San Diego, CA, USA).
The antimicrobial susceptibility of S. argenteus isolates was determined using the Sensititre Gram-positive GPALL1F AST Plate (Thermo Fisher Scientific, MA, USA), a commercially available broth microdilution method-based kit. The test was performed manually according to the manufacturer’s instructions, and the results were interpreted based on the Clinical and Laboratory Standards Institute breakpoints for S. aureus (10). The isolates were investigated for the presence of genes encoding mecA, blaZ, Panton-Valentine leukocidin, adhesins, enterotoxins, toxic shock syndrome toxin, and exfoliative toxins using primers as described in previous reports (11–13). The 95% confidence interval of the proportion of S. argenteus among S. aureus complexes was estimated using the binomial exact method.
Among the 691 isolates of S. aureus bacteremia cohort from the study hospital, one was identified as S. argenteus (strain 2). The estimated proportion of S. argenteus among S. aureus complex was 0.14% (95% confidence interval 0%–0.8%). The clinical and microbiological characteristics of both S. argenteus isolates (strains 1 and 2) are presented in Table 1. Both isolates were identified from the blood cultures of patients with extensive pneumonia accompanied by bacteremia. Both patients died from multi-organ failure despite intensive care.
TABLE 1.
Clinical and microbiological characteristics of two cases of Staphylococcus argenteus bacteremiab
| Clinical characteristics | Case 1 (2022) | Case 2 (2017) |
|---|---|---|
| Age | 70 | 81 |
| Sex | Male | Male |
| Relevant comorbidities | Video-assisted pulmonary lobectomy, chemotherapy, neutropenia (ANC 980 cells/mm3) | Chronic obstructive pulmonary disease |
| Symptom | ||
| Onset | 5 days before admission | 1 day before admission |
| Fever | + | + |
| Dyspnea | + | + |
| Cough | + | − |
| Infection type | Healthcare-associated pneumonia | Community-acquired pneumonia |
| Clinical sample | Blood | Blood |
| Date of sample collection | 04.12.2022 | 20.02.2017 |
| Treatment | ||
| Empirical antibiotics | Ceftizoxime, amikacin | Piperacillin/tazobactam |
| Definitive antibiotics | Cefazolin | Nafcillin |
| Mechanical ventilation | + | + |
| Continuous renal replacement therapy | − | + |
| Outcome | Deceased | Deceased |
| Microbiological characteristics | ||
| Multi-locus sequence typing (allelic profile) | ||
| arcC | 151 | 151 |
| aroE | 755 | 47 |
| glpF | 20 | 8 |
| gmk | 101 | 34 |
| Pta | 145 | 175 |
| Tpi | 150 | 180 |
| yqiL | 131 | 169 |
| Sequence type | 8342 | 8343 |
| Antimicrobial susceptibility test (MIC, mg/L)a | ||
| Oxacillin | 0.5 (S) | ≤0.25 (S) |
| Rifampin | ≤0.5 (S) | ≤0.5 (S) |
| Clindamycin | ≤0.5 (S) | ≤0.5 (S) |
| Vancomycin | 1 (S) | 2 (S) |
| Linezolid | 4 (S) | 4 (S) |
| Levofloxacin | >4 (R) | ≤0.25 (S) |
| Trimethoprim-sulfamethoxazole | ≤0.5/9.5 (S) | ≤0.5/9.5 (S) |
Antimicrobial susceptibility test was performed using the Sensititre Gram-positive GPALL1F AST Plate (Thermo Fisher Scientific, MA, USA), a commercially available broth microdilution method-based kit, and interpreted according to Clinical and Laboratory Standards Institute breakpoints for Staphylococcus aureus.
ANC, absolute neutrophil count and MIC, minimal inhibitory concentration.
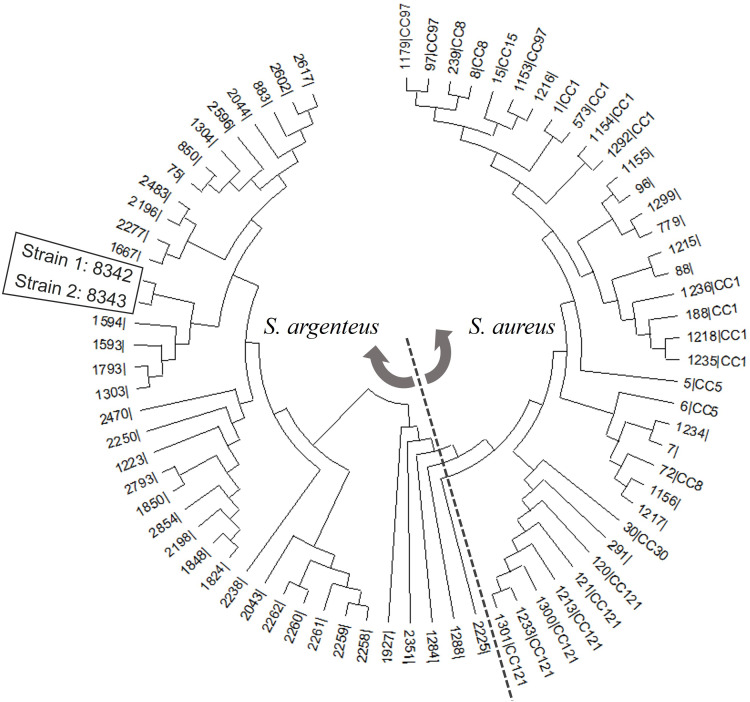
No ST was consistent with both isolates using the S. aureus pubMLST database on 17 April 2023. Therefore, the allelic profiles of both isolates were deposited in the MLST database, and the new STs were classified as 8342 and 8343. Phylogenetic analysis revealed that strains 1 and 2 were closely related to ST1594, ST1593, ST1793, and ST1303, which belonged to S. argenteus (Fig. 1). Both isolates were susceptible to oxacillin, rifampin, and vancomycin (Table 1). Strain 1 was resistant to levofloxacin. Both isolates harbored genes associated with virulence (clfA, clfB, fnbpA, sdrC, sdrD, sdrE, bbp, cna, see, seg, and sei). Additionally, blaZ, fnbpB, and map existed only in strain 1. Neither isolate had mecA and pvl.
Fig 1.
Phylogenetic tree of two S. argenteus isolates and 73 STs downloaded from the MLST website (http://www.mlst.net). The tree was constructed based on concatenated sequences of seven MLST loci using the neighbor-joining method.
This is the first report to investigate the clinical and microbiological characteristics of S. argenteus identified in patients with bacteremia and the proportion of S. argenteus bacteremia among S. aureus bacteremia in Korea. S. argenteus has been reported worldwide, particularly high in Australia, Thailand, and Taiwan accounting for 11.9%–25% of S. aureus-associated infections (14–16). However, <1% have been reported in Europe and East Asia, including the Netherlands, Belgium, Sweden, Denmark, China, and Japan, revealing a large geographical difference (17–22). The proportion of S. argenteus among S. aureus complex in this study was consistent with that of reports from neighboring countries, China and Japan (20, 21). The estimated proportion of S. argenteus in this study was low; nonetheless, the incidence of S. argenteus infection may change as global interaction/immigration increases and requires further monitoring.
Both S. argenteus isolates were associated with community- and healthcare-acquired pneumonia resulting in fatal outcomes, consistent with previously reported high virulence (15, 16). Strains 1 and 2 were classified as new ST8342 and ST8343, respectively. The most common ST in S. argenteus was ST2250, followed by ST1123 (23). As ST composition indicates geographical diversity, further studies are needed to determine whether the S. argenteus ST in Korea differs from those of other regions (23). Various virulence genes encoding adhesins, enterotoxins, toxic shock syndrome toxin, and exfoliative toxin were identified in this study, consistent with previous reports that S. argenteus and S. aureus share a significant proportion of genes encoding virulence factors (3, 24). Both isolates were methicillin-susceptible, as S. argenteus is associated with lower antimicrobial resistance than S. aureus (16, 17). However, penicillin-resistant S. argenteus is relatively common and carries blaZ, similar to strain 1 (23).
Although, S. argenteus accounted for <1% of the S. aureus complex in Korea, it was highly virulent and associated with severe infections as S. aureus. However, S. argenteus may be misrecognized as coagulase-negative staphylococci in clinical settings because it is an unfamiliar species, especially in regions with a low incidence, such as Korea. Therefore, we believe reporting S. argenteus as a member of the S. aureus complex is necessary to avoid confusion with coagulase-negative staphylococci, as suggested by the ESCMID Study Group for Staphylococci and Staphylococcal Diseases in 2019 (3).
ACKNOWLEDGMENTS
This study included three multicenter SAB cohorts. The cohorts comprise SAB cases occurred between May and December 2012 (grant 2012-E44003-00 from the Korea Centers for Disease Control and Prevention) (4), between September 2013 and March 2015 [grant HI10C2020 from the National Strategic Coordinating Centre for Clinical Research for the cohort study Korea Infectious Diseases Study Group/Staphylococcus aureus Bacteremia 2013 (KIND-SAB 2013)] (5, 6), and between September 2017 and February 2018 (grant 2017-E280301 from the Korea Disease Control and Prevention Agency) (7). This study was supported by the SNUBH Research Fund (grant 02-2023-0010).
The authors thank the Division of Statistics at the Medical Research Collaborating Center at Seoul National University Bundang Hospital for statistical analyses.
AFTER EPUB
[This article was published on 10 January 2024 with an error in Acknowledgments. The error was corrected in the current version, posted on 16 January 2024.]
Contributor Information
Jeong Su Park, Email: mdmicrobe@gmail.com.
Kyoung-Ho Song, Email: khsongmd@gmail.com.
Deena R. Altman, Icahn School of Medicine at Mount Sinai, New York, New York, USA
DATA AVAILABILITY
The complete genome sequences of the two S. argenteus strains have been deposited in GenBank under accession numbers SAMN38109731 (SNUBHSAr1) and SAMN38109732 (SNUBHSAr2).
ETHICS APPROVAL
The study protocol (no. B-2302-810-101) was approved by the institutional review board of the Seoul National University Bundang Hospital. The study mainly focused on archived clinical information and isolates of S. aureus bacteremia. The need for informed consent was waived owing to the study’s retrospective nature and minimal risk.
REFERENCES
- 1. Song KH, Kim ES, Sin HY, Park KH, Jung SI, Yoon N, Kim DM, Lee CS, Jang HC, Park Y, Lee KS, Kwak YG, Lee JH, Park SY, Song M, Park SK, Lee YS, Kim HB, Korea IDsg . 2013. Characteristics of invasive Staphylococcus aureus infections in three regions of Korea, 2009-2011: a multi-center cohort study. BMC Infect Dis 13:581. doi: 10.1186/1471-2334-13-581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tong SYC, Schaumburg F, Ellington MJ, Corander J, Pichon B, Leendertz F, Bentley SD, Parkhill J, Holt DC, Peters G, Giffard PM. 2015. Novel Staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int J Syst Evol Microbiol 65:15–22. doi: 10.1099/ijs.0.062752-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Becker K, Schaumburg F, Kearns A, Larsen AR, Lindsay JA, Skov RL, Westh H. 2019. Implications of identifying the recently defined members of the Staphylococcus aureus complex S. argenteus and S. schweitzeri: a position paper of members of the ESCMID study group for staphylococci and staphylococcal diseases (ESGS). Clin Microbiol Infect 25:1064–1070. doi: 10.1016/j.cmi.2019.02.028 [DOI] [PubMed] [Google Scholar]
- 4. Kim ES, Kim HB, Kim G, Kim KH, Park KH, Lee S, Choi YH, Yi J, Kim CJ, Song KH, Choe PG, Kim NJ, Lee YS, Oh MD, Korea IDsg . 2014. Clinical and epidemiological factors associated with methicillin resistance in community-onset invasive Staphylococcus aureus infections: prospective multicenter cross-sectional study in Korea. PLoS One 9:e114127. doi: 10.1371/journal.pone.0114127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee S, Song KH, Jung SI, Park WB, Lee SH, Kim YS, Kwak YG, Kim YK, Kiem SM, Kim HI, et al. 2013. Comparative outcomes of cefazolin versus nafcillin for methicillin-susceptible Staphylococcus aureus bacteraemia: a prospective multicentre cohort study in Korea. Clin Microbiol Infect 13:581. doi: 10.1186/1471-2334-13-581 [DOI] [PubMed] [Google Scholar]
- 6. Song KH, Jung SI, Lee S, Park S, Kiem SM, Lee SH, Kwak YG, Kim YK, Jang HC, Kim YS, Kim HI, Kim CJ, Park KH, Kim NJ, Oh MD, Kim HB, Korea IDSG . 2017. Characteristics of cefazolin Inoculum effect-positive methicillin-susceptible Staphylococcus aureus infection in a multicentre bacteraemia cohort. Eur J Clin Microbiol Infect Dis 36:285–294. doi: 10.1007/s10096-016-2799-1 [DOI] [PubMed] [Google Scholar]
- 7. Song KH, Kim CJ, Choi NK, Ahn J, Choe PG, Park WB, Kim NJ, Choi HJ, Bae JY, Kim ES, Lee H, Park JS, Jung Y, Lee SS, Park KH, Jung SI, Kim YS, Bang JH, Lee S, Kang YM, Kwak YG, Kim HB, Korea IDSG . 2022. Clinical and economic burden of bacteremia due to multidrug-resistant organisms in Korea: a prospective case control study. J Glob Antimicrob Resist 31:379–385. doi: 10.1016/j.jgar.2022.11.005 [DOI] [PubMed] [Google Scholar]
- 8. Chen SY, Lee H, Teng SH, Wang XM, Lee TF, Huang YC, Liao CH, Teng LJ, Hsueh PR. 2018. Accurate differentiation of novel Staphylococcus argenteus from Staphylococcus aureus using MALDI-TOF MS. Future Microbiol 13:997–1006. doi: 10.2217/fmb-2018-0015 [DOI] [PubMed] [Google Scholar]
- 9. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. CLSI . 2023. Performance standards for antimicrobial susceptibility testing. In CLSI supplement M100. Clinical and laboratory standards Institute, 33rd ed [Google Scholar]
- 11. Song KH, Jung SI, Lee S, Park S, Kim ES, Park KH, Park WB, Choe PG, Kim YK, Kwak YG, Kim YS, Jang HC, Kiem S, Kim HI, Kim HB, IDsg K. 2019. Inoculum effect of methicillin-susceptible Staphylococcus aureus against broad-spectrum beta-lactam antibiotics. Eur J Clin Microbiol Infect Dis 38:67–74. doi: 10.1007/s10096-018-3392-6 [DOI] [PubMed] [Google Scholar]
- 12. Campbell SJ, Deshmukh HS, Nelson CL, Bae IG, Stryjewski ME, Federspiel JJ, Tonthat GT, Rude TH, Barriere SL, Corey R, Fowler VG. 2008. Genotypic characteristics of Staphylococcus aureus isolates from a multinational trial of complicated skin and skin structure infections. J Clin Microbiol 46:678–684. doi: 10.1128/JCM.01822-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. 1999. Involvement of panton-valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29:1128–1132. doi: 10.1086/313461 [DOI] [PubMed] [Google Scholar]
- 14. McDonald M, Dougall A, Holt D, Huygens F, Oppedisano F, Giffard PM, Inman-Bamber J, Stephens AJ, Towers R, Carapetis JR, Currie BJ. 2006. Use of a single-nucleotide polymorphism genotyping system to demonstrate the unique epidemiology of methicillin-resistant Staphylococcus aureus in remote aboriginal communities. J Clin Microbiol 44:3720–3727. doi: 10.1128/JCM.00836-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chantratita N, Wikraiphat C, Tandhavanant S, Wongsuvan G, Ariyaprasert P, Suntornsut P, Thaipadungpanit J, Teerawattanasook N, Jutrakul Y, Srisurat N, Chaimanee P, Anukunananchai J, Phiphitaporn S, Srisamang P, Chetchotisakd P, West TE, Peacock SJ. 2016. Comparison of community-onset Staphylococcus argenteus and Staphylococcus aureus sepsis in Thailand: a prospective multicentre observational study. Clin Microbiol Infect 22:458. doi: 10.1016/j.cmi.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen SY, Lee H, Wang XM, Lee TF, Liao CH, Teng LJ, Hsueh PR. 2018. High mortality impact of Staphylococcus argenteus on patients with community-onset Staphylococcal bacteraemia. Int J Antimicrob Agents 52:747–753. doi: 10.1016/j.ijantimicag.2018.08.017 [DOI] [PubMed] [Google Scholar]
- 17. Witteveen S, Hendrickx APA, de Haan A, Notermans DW, Landman F, van Santen-Verheuvel MG, de Greeff SC, Kuijper EJ, van Maarseveen NM, Vainio S, Schouls LM. 2022. Genetic characteristics of methicillin-resistant Staphylococcus argenteus isolates collected in the dutch national MRSA surveillance from 2008 to 2021. Microbiol Spectr 10:e0103522. doi: 10.1128/spectrum.01035-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Argudín MA, Dodémont M, Vandendriessche S, Rottiers S, Tribes C, Roisin S, de Mendonça R, Nonhoff C, Deplano A, Denis O. 2016. Low occurrence of the new species Staphylococcus argenteus in a Staphylococcus aureus collection of human isolates from Belgium. Eur J Clin Microbiol Infect Dis 35:1017–1022. doi: 10.1007/s10096-016-2632-x [DOI] [PubMed] [Google Scholar]
- 19. Tång Hallbäck E, Karami N, Adlerberth I, Cardew S, Ohlén M, Engström Jakobsson H, Svensson Stadler L. 2018. Methicillin-resistant Staphylococcus argenteus misidentified as methicillin-resistant Staphylococcus aureus emerging in Western Sweden. J Med Microbiol 67:968–971. doi: 10.1099/jmm.0.000760 [DOI] [PubMed] [Google Scholar]
- 20. Zhang DF, Xu X, Song Q, Bai Y, Zhang Y, Song M, Shi C, Shi X. 2016. Identification of Staphylococcus argenteus in Eastern China based on a nonribosomal peptide synthetase (NRPS) gene. Future Microbiol 11:1113–1121. doi: 10.2217/fmb-2016-0017 [DOI] [PubMed] [Google Scholar]
- 21. Aung MS, Urushibara N, Kawaguchiya M, Sumi A, Takahashi S, Ike M, Ito M, Habadera S, Kobayashi N. 2019. Molecular epidemiological characterization of Staphylococcus argenteus clinical isolates in Japan: identification of three clones (ST1223, ST2198, and ST2550) and a novel Staphylocoagulase genotype XV.. Microorg 7:389. doi: 10.3390/microorganisms7100389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hansen TA, Bartels MD, Høgh SV, Dons LE, Pedersen M, Jensen TG, Kemp M, Skov MN, Gumpert H, Worning P, Westh H. 2017. Whole genome sequencing of danish Staphylococcus argenteus reveals a genetically diverse collection with clear separation from Staphylococcus aureus. Front Microbiol 8:1512. doi: 10.3389/fmicb.2017.01512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goswami C, Fox S, Holden M, Leanord A, Evans TJ. 2021. Genomic analysis of global Staphylococcus argenteus strains reveals distinct lineages with differing virulence and antibiotic resistance gene content. Front Microbiol 12:795173. doi: 10.3389/fmicb.2021.795173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang DF, Zhi XY, Zhang J, Paoli GC, Cui Y, Shi C, Shi X. 2017. Preliminary comparative genomics revealed pathogenic potential and international spread of Staphylococcus argenteus. BMC Genomics 18:808. doi: 10.1186/s12864-017-4149-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequences of the two S. argenteus strains have been deposited in GenBank under accession numbers SAMN38109731 (SNUBHSAr1) and SAMN38109732 (SNUBHSAr2).