Abstract
Background
Nucleophosmin/nucleoplasmin 3 (NPM3), a member of the NPM protein family, is widely expressed in various human tissues. Although previous studies identified elevated NPM3 expression in several cancers, a systematic pan-cancer analysis remains lacking. In this study, we conducted a comprehensive analysis of NPM3 to determine its role in tumorigenesis and tumor development.
Methods
Using data from The Cancer Genome Atlas (TCGA) and various bioinformatics analysis tools, we conducted a pan-cancer analysis of NPM3. Additionally, we collected gene expression and clinical data from 890 patients with lung adenocarcinoma (LUAD) from TCGA and the Gene Expression Omnibus database. We performed Cox regression analyses to explore the independent prognostic value of NPM3 expression in LUAD and plotted a nomogram to predict patient survival. We also used real-time quantitative polymerase chain reaction (RT-qPCR) to examine the expression levels of NPM3 in seven pairs of LUAD and paraneoplastic tissue samples.
Results
NPM3 expression was significantly increased in 20 types of cancer and was associated with poor prognosis in five types (P < 0.05). NPM3 expression was negatively correlated with DNA methylation and positively correlated with copy number variation. NPM3 was also significantly associated with immune cell infiltration in various cancers. Cox regression analyses revealed that NPM3 expression could serve as an independent prognostic marker of LUAD. Moreover, our nomogram demonstrated good predictive ability for the prognosis of patients with LUAD. Finally, the high expression of NPM3 in LUAD was verified using RT-qPCR.
Conclusion
NPM3 is a promising biomarker for predicting pan-cancer prognosis and immunotherapeutic efficacy.
Keywords: NPM3, Pan-cancer, Lung adenocarcinoma, Prognosis, Immune infiltration, Biomarker
Graphical abstract
Highlights
-
•
A systematic pan-cancer analysis revealed increased nucleophosmin/nucleoplasmin 3 (NPM3) expression in 20 types of cancer.
-
•
Overall survival analysis indicates that NPM3 expression was correlated with poor prognosis in five types of cancer.
-
•
NPM3 expression was significantly associated with the infiltration of various immune and stromal cells in various cancers.
-
•
NPM3 expression is an independent prognostic marker for lung adenocarcinoma.
-
•
NPM3 could serve as a potential biomarker for predicting prognosis and immunotherapy efficacy in various cancers.
Introduction
In the 21st century, cancer is a prominent cause of death in several countries.1 The morbidity and mortality burdens of cancer are rapidly growing,2 with lung cancer serving as the second most common cancer and leading cause of death worldwide.2 However, cancer treatment has made great progress, and the 5-year survival rate has consistently improved over the last decade.3 Particularly, immunotherapy has revolutionized cancer treatments4 and achieved promising clinical results in several cancer types.5, 6, 7, 8 Unfortunately, its complex, heterogeneous, and uncertain nature poses significant challenges to effective cancer treatment.9 Therefore, a thorough understanding of its underlying mechanisms is key to achieving breakthroughs in tumor immunotherapy.
The tumor immune microenvironment (TME) plays a critical role in anticancer immune mechanisms.10 The TME consists of tumor cells, innate immune cells, adaptive immune cells, endothelial cells, stromal cells, and extracellular components that interact with and regulate responses to promote tumor growth.11 The TME is divided into different subclasses that are crucial for tumor development, metastasis, and immune escape.12 Consequently, a comprehensive analysis of the TME is necessary to elucidate its regulatory mechanisms and to identify new targets for immunotherapy to achieve accurate and effective cancer treatment.
Nucleophosmin/nucleoplasmin 3 (NPM3) is primarily a nuclear protein, widely expressed in various somatic tissues.13 NPM3 belongs to the NPM protein family, which comprises NPM1 and NPM3, commonly expressed in somatic tissues, and NPM2, specifically expressed in oocytes.14 The NPM family plays vital roles in DNA replication, histone chaperone activity, ribosome biogenesis, and chromatin remodeling,15 which may influence tumorigenesis and progression.16 Numerous studies have suggested that NPM3 is strongly associated with cancer, as evidenced by its upregulation in various types of cancers, such as myxoinflammatory fibroblastic sarcoma (MIFS),17 gastric cancer (GC),18 and diffuse large B-cell lymphoma (DLBCL).19 NPM3 is a potential biomarker for various malignant tumors; however, its specific association and underlying mechanisms remain unclear. While previous studies each focus on only one specific form of cancer, a systematic analysis of the function of NPM3 in pan-cancer is currently lacking.
Our study comprehensively analyzed for the first time the pan-cancer role of NPM3 using multiple databases to analyze correlations between the differential expression of NPM3 and survival, DNA methylation, copy number variation (CNV), immune infiltration, related cellular pathways, and drug sensitivity rarely. Our findings suggest that NPM3 expression is an independent risk factor for lung adenocarcinoma (LUAD) prognosis and that NPM3 could serve as a promising target for tumor treatment as it is connected with poor tumor prognosis and efficacy of tumor immunotherapy.
Methods
Data collection and processing
We obtained ribonucleic acid (RNA) sequencing data and clinical information, including survival status, age, sex, tumor stage, tumor size, presence or absence of lymph node metastasis, and presence of distant metastasis from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The dataset GSE68465 was downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov). The two datasets were merged, and the “sva” package in R software version 4.1.3 was employed for batch correction. Samples with incomplete clinical information were excluded from the analysis, resulting in the inclusion of 890 LUAD and 59 normal samples in this study.
Gene and protein expression analysis
Tumor immune estimation resource version 2 (Timer 2.0; http://timer.cistrome.org/) is a detailed platform for systematically analyzing immune infiltrates in various cancers. It allows for the investigation of various genetic, immunological, and clinical aspects of tumors.20, 21, 22 In our study, we used the “Gene Differential Expression (DE)” module of TIMER 2.0 to analyze NPM3 differential expression in tumor and paraneoplastic tissues of 33 tumors from TCGA.
Gene Expression Profiling Interactive Analysis 2 (GEPIA 2.0; http://gepia2.cancer-pku.cn/) is a web-based tool that conducts extensive analysis using RNA sequencing data from TCGA and the Genotype-Tissue Expression (GTEx) database.23 In this study, we used GEPIA 2.0, matching TCGA normal data and GTEx data, to perform an NPM3 differential expression analysis for cancers lacking normal tissue samples in TCGA. In addition, we assessed the association between NPM3 expression and the pathological stage of cancer using GEPIA 2.0. The results are presented as violin plots or box plots using log 2[transcripts per million (TPM) + 1] transformed expression data. Statistical significance was set at P < 0.05.
The University of Alabama at Birmingham Cancer (UALCAN) data analysis portal (http://ualcan.path.uab.edu/analysis-prot.html) is a web-based tool that performs genetic and proteomic analyses using various databases, including the Clinical Proteomic Tumor Analysis Consortium (CPTAC).24,25 We used this tool to examine the differential expression of NPM3 in cancer and paraneoplastic tissues.
Immunohistochemical staining analysis
The Human Protein Atlas (HPA; http://www.proteinatlas.org/) is an interactive platform that maps numerous proteins across cells, tissues, and organs by leveraging multiomics data and various omics technologies.26,27 We obtained and analyzed immunohistochemistry (IHC) images of NPM3 protein expression in normal tissues and five types of carcinoma tissues: breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), liver hepatocellular carcinoma (LIHC), LUAD, and prostate adenocarcinoma (PRAD).
Survival prognosis analysis
We investigated the association between NPM3 expression and cancer prognosis using the “Survival” module of GEPIA 2.0. We used Kaplan–Meier (KM) plots, made with the KM plotter tool (kmplot.com), and survival maps to demonstrate this association. The statistical significance of our hypothesis was tested using the log-rank test, with statistical significance set at P < 0.05.
Analysis of the relationship between nucleophosmin/nucleoplasmin 3 expression and methylation
Gene Set Cancer Analysis (GSCA; http://bioinfo.life.hust.edu.cn/GSCA/) is a public platform for genomic, pharmacogenomic, and immunogenomic cancer analysis.28 Using this platform, we examined the relationship between NPM3 methylation and messenger RNA (mRNA) expression levels in TCGA tumors.
Analysis of the relationship between nucleophosmin/nucleoplasmin 3 expression and copy number variations
We assessed the correlation between the mRNA expression levels and CNVs of NPM3 in genomic data using the GSCA platform.
Immune infiltration analysis
We used the TIMER 2.0 tool to comprehensively evaluate the association between NPM3 and immune cell infiltration. Specifically, we input “NPM3” into the “gene” module of the “immune” section, then applied different algorithms to derive the Spearman correlation. The algorithms used were CIBERSORT, CIBERSORT-ABS, EPIC, MCPCOUNTER, QUANTISEQ, TIDE, TIMER, and XCELL. We analyzed B cells, cluster of differentiation (CD)4+ and CD8+ T cells, regulatory T Cells (Treg) cells, mast cells, cancer-associated fibroblasts, myeloid-derived suppressor cells (MDSCs), monocytes, macrophages, and dendritic cells (DCs).
Analysis of the relationship between nucleophosmin/nucleoplasmin 3 expression and both tumor mutation load and microsatellite instability
Tumor mutation load (TMB) refers to the overall number of somatic gene coding errors, base substitutions, insertions, and deletions discovered per million bases.29 Microsatellite instability (MSI) is a heritable mutational state resulting from defects in DNA mismatch repair function.30 We downloaded TMB and MSI data from TCGA. Next, we explored the relationship among NPM3 mRNA expression, TMB expression, and MSI using Spearman's correlation analysis.
Nucleophosmin/nucleoplasmin 3-related gene enrichment analysis
The STRING website (https://string-db.org/) was used to identify proteins that bind to NPM3 and conduct a follow-up protein–protein interaction (PPI) network analysis. We inserted NPM3 into the “protein by name” section under the “search” module and selected “homo intelligence.” Subsequently, we established several primary conditions: the meaning of network edges (“evidence”), active interaction sources (“experiments”), minimum required interaction score (“low confidence [0.150]”), and max number of interactors to show (“no more than 20 interactors” in the first shell).
Based on the datasets from TCGA and GTEx, we utilized GEPIA 2.0, to retrieve the top 100 NPM3-related genes. Subsequently, we plotted a heatmap of the Spearman correlations between NPM3 and its top-5 correlated genes using TIMER 2.0.
We utilized “cluster Profiler” R package to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of the 100 NPM3-related genes.
Drug sensitivity analysis
CellMiner (http://discover.nci.nih.gov/cellminer/) is an online cancer analysis tool that integrates and studies the molecular and pharmacological data of the United States National Cancer Institute (NCI)-60 cancer cell line. The NCI-60 cell line database contains comprehensive analytical data for 60 distinct human cancer cell lines.31 Using this resource, we extracted data on the correlation between NPM3 expression and drug sensitivity. The statistical analysis was performed via R4.1.3, and “ggplot 2” R package was applied for visualization.
Prognostic value and association with clinical characteristics of nucleophosmin/nucleoplasmin 3 in lung adenocarcinoma
Univariate and multivariate Cox regression analyses were conducted on NPM3 expression and associated clinical characteristics to evaluate the independent prognostic significance of NPM3 expression and associated clinical characteristics. To predict the survival outcomes at 1, 3, and 5 years for patients with LUAD, the independent prognostic factors were employed as variables to develop a nomogram utilizing the “rms” R package. Additionally, calibration curves of the column line graphs were generated using R software (version 4.1.3) to assess the reliability of the nomogram.
Tissue sample collection and real-time quantitative polymerase chain reaction analysis
We collected seven pairs of LUAD tumor tissue and corresponding paraneoplastic tissue samples from patients who underwent lung cancer resection at the Tianjin Medical University Cancer Hospital.
Total RNA was extracted from fresh frozen tissues using TRIzol reagent (#9109, TaKaRa, Tokyo, Japan). Total RNA was reverse transcribed to complementary DNA by the StarScript III All-in-one RT Mix with genomic DNA remover kit (#A230-10, GenStar, Beijing, China) with a 20 μL system (37 °C, 2 min; 50 °C, 15 min; 85 °C, 2 min). Real-time quantitative polymerase chain reaction (RT-qPCR) analysis was performed on an ABI QuantStudio 5 (Q5) system (Applied Biosystems, United States). The internal control was glyceraldehyde 3-phosphate dehydrogenase (GADPH). The relative expression was determined using the 2−ΔΔCt method. The primer sequences were 5′-GAGGGAGCCAAAGACGAGTG-3′ (forward) and 5′-CATCCAGACTGAGCATGGG-3′ (reverse). A paired Student's t-test was used to compare the relative expression of NPM3 in LUAD tumor and paraneoplastic samples.
Results
Expression levels of nucleophosmin/nucleoplasmin 3 in pan-cancer
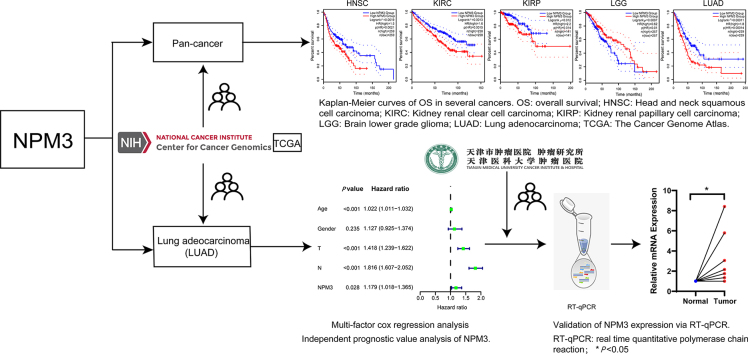
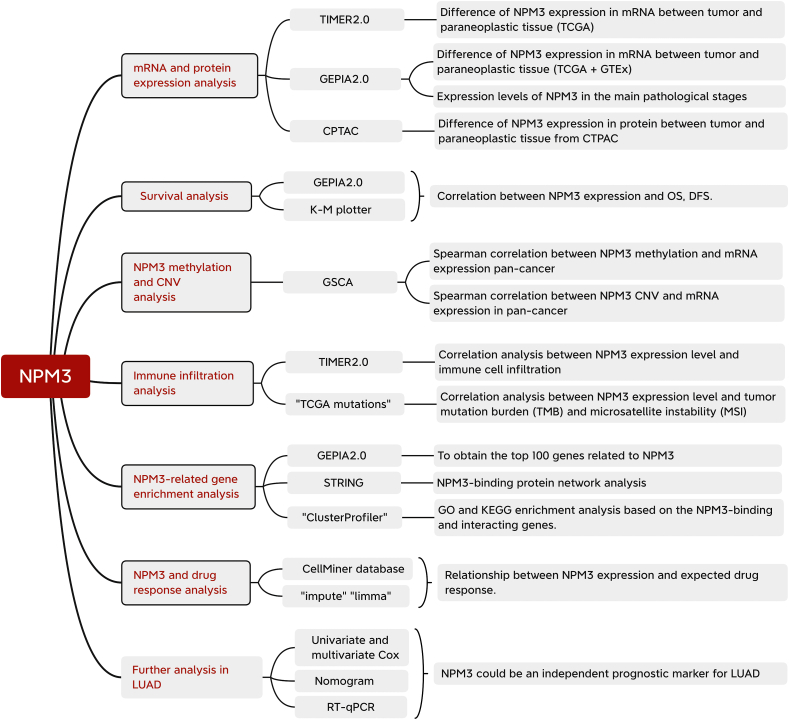
We utilized multiple databases, including TCGA, GEO, Clinical Proteomic Tumor Analysis Consortium (CTPAC), and CellMiner, to conduct a thorough gene- and protein-level pan-cancer analysis of NPM3. We performed analyses of the correlations between NPM3 expression and survival, methylation, CNV, immune cell infiltration, gene enrichment, and drug sensitivity in pan-cancer. Our goal was to investigate the possible pathogenic mechanisms underlying the role of NPM3 in tumorigenesis and to identify novel targets and potential drugs to improve tumor therapy. The methodology and process framework employed in this study are shown in Figure 1.
Figure 1.
Layout of this study, including the parameters investigated and databases or tools used for the analyses. CNV: Copy number variation; CPTAC: Clinical Proteomic Tumor Analysis Consortium; DFS: Disease-free survival; GEPIA 2.0: Gene Expression Profiling Interactive Analysis 2; GO: Gene Ontology; GSCA: Gene Set Cancer Analysis; GTEx: Genotype-tissue expression; KEGG: Kyoto Encyclopedia of Genes and Genomes; LUAD: Lung adenocarcinoma; NPM3: Nucleophosmin/nucleoplasmin 3; OS: Overall survival; RT-qPCR: Real-time quantitative polymerase chain reaction; TCGA: The Cancer Genome Atlas; TIMER 2.0: Tumor immune estimation resource, version 2.
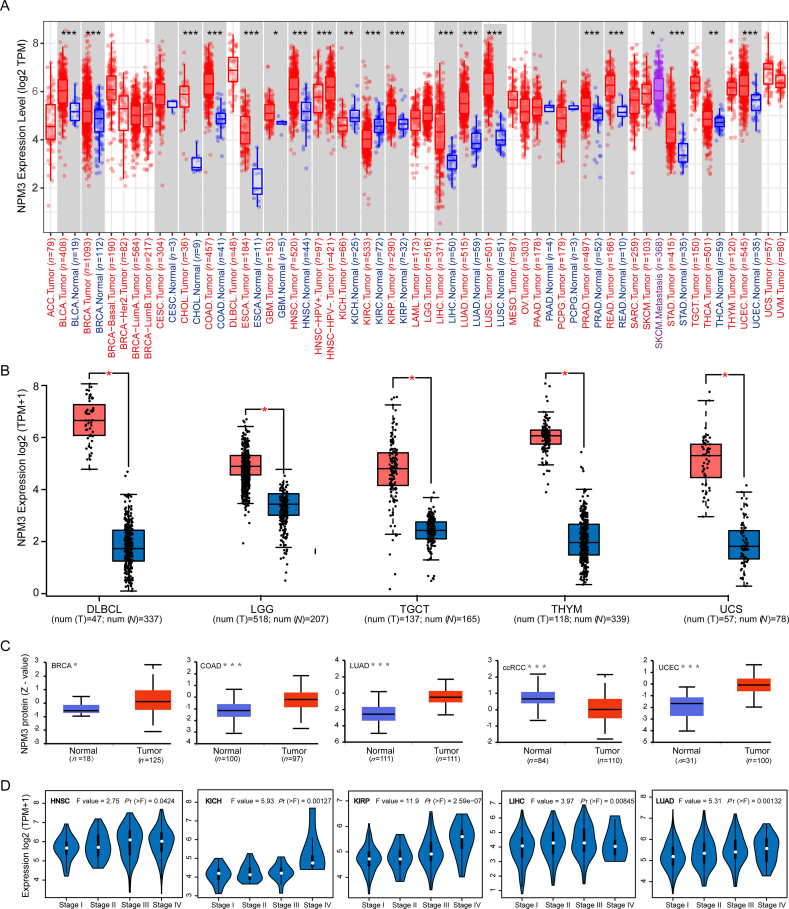
We conducted a pan-cancer analysis of NPM3 expression using the TIMER 2.0 database. As shown in Figure 2A, NPM3 has significantly expressed in tumor tissues of bladder urothelial carcinoma (BLCA), BRCA, cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), kidney renal papillary cell carcinoma (KIRP), LIHC, LUAD, lung squamous cell carcinoma (LUSC), PRAD, rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC) (P < 0.05). Notably, NPM3 was weakly expressed in the tumor tissues of kidney chromophobes (KICH) and kidney renal clear cell carcinoma (KIRC), whereas there was no differential expression in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), mesothelioma (MESO), pancreatic adenocarcinoma (PAAD), and pheochromocytoma and paraganglioma (PCPG).
Figure 2.
Messenger ribonucleic acid expression and protein levels of nucleophosmin/nucleoplasmin 3 in pan-cancer. (A) Comparison of NPM3 expression levels in tumor and paraneoplastic tissues (where available) from TCGA in TIMER 2.0. ∗P < 0.05; ∗∗P < 0.01; ∗ ∗ ∗P < 0.001. (B) Box plot presentation of NPM3 mRNA expression levels in DLBC, LGG, TGCT, THYM, and UCS from TCGA and the GTEx database in GEPIA 2.0. ∗P < 0.05. (C) Differences in protein expression levels between normal tissue and tumor samples of BRCA, COAD, LUAD, ccRCC, and UCEC in CPTAC. The results are presented in box plots; the Z-value represents the standard deviation of the sample from the median for the type of cancer, and “n” represents the number of samples. ∗P < 0.05; ∗ ∗ ∗P < 0.001. (D) Expression levels of NPM3 in the main pathological stages (stage I, II, III, and IV) of HNSC, KICH, KIRP, LIHC, and LUAD. Log 2(TPM+1) was used for logarithmic scaling. ACC: Adrenocortical carcinoma; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; ccRCC: Clear cell renal cell carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; CPTAC: Clinical Proteomic Tumor Analysis Consortium; DLBCL: Lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; GEPIA 2.0: Gene Expression Profiling Interactive Analysis 2; GTEx: Genotype-tissue expression; HNSC: Head and neck squamous cell carcinoma; KICH: Kidney chromophobe; KIRP: Kidney renal papillary cell carcinoma; KIRC: Kidney renal clear cell carcinoma; LAML: Acute myeloid leukemia; LGG: Lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MECO: Mesothelioma; NPM3: Nucleophosmin/nucleoplasmin 3; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; SKCM: Skin cutaneous melanoma; STAD: Stomach adenocarcinoma; TCGA: The Cancer Genome Atlas; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; TIMER 2.0: Tumor immune estimation resource, version 2; TPM: Transcripts per million; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma; UVM: Uveal melanoma.
To evaluate the differential expression of NPM3 in cancers without paraneoplastic tissues in TCGA, we used normal tissues from the GTEx database for comparison. As shown in Figure 2B, NPM3 was highly expressed in lymphoid neoplasm DLBCL, brain lower grade glioma (LGG), testicular germ cell tumors (TGCT), thymoma (THYM), uterine carcinosarcoma (UCS), and skin cutaneous melanoma (SKCM) (P < 0.05), whereas NPM3 expression was reduced for acute myeloid leukemia (LAML; P < 0.05). Additionally, no significant differences were observed for adrenocortical carcinoma (ACC), ovarian serous cystadenocarcinoma (OV), or sarcoma (SARC) [Supplementary Figure 1].
At the protein level, we used large-scale proteomic data made available by CPTAC to compare NPM3 expression levels across several cancers. Our analysis revealed that NPM3 was significantly overexpressed in BRCA, COAD, LUAD, and UCEC and significantly reduced in clear cell renal cell carcinoma (ccRCC; P < 0.05; Figure 2C).
The association between NPM3 expression and tumor pathological stage was then evaluated using GEPIA 2.0, which revealed a positive correlation between NPM3 expression and pathological stage progression in head and neck squamous cell carcinoma (HNSC), KICH, KIRP, LIHC, and LUAD, with significantly higher NPM3 expression levels in late-stage tumors than in early-stage tumors (P < 0.05; Figure 2D). These findings suggest a potential role for NPM3 in cancer development. No significant correlation between NPM3 expression and the pathological stage was observed in other cancer types [Supplementary Figure 2].
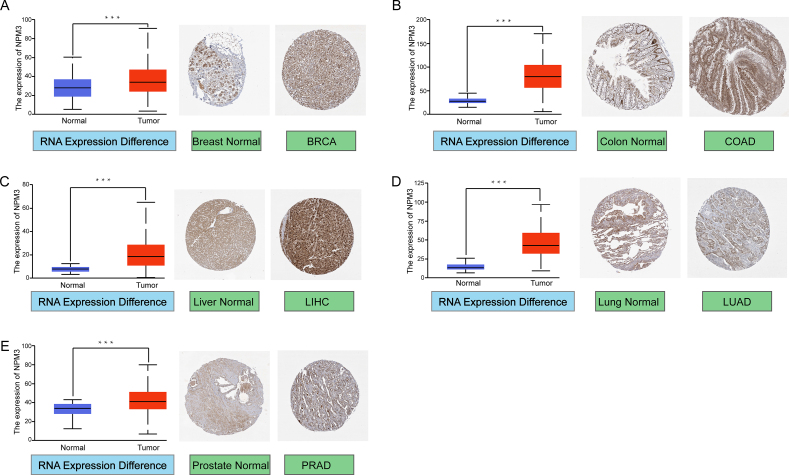
In addition, we analyzed the protein expression of NPM3 using immunohistochemistry (IHC) data from the HPA database and compared it with NPM3 RNA differential expression data from TCGA. Notably, for BRCA, COAD, LIHC, LUAD, and PRAD, IHC analysis revelated that NPM3 was low or moderately positive in normal tissues but moderately or strongly positive in tumor tissues [Figures 3A–E].
Figure 3.
Comparison of Nucleophosmin/nucleoplasmin 3 messenger ribonucleic acid expression between normal and tumor tissues in TCGA (left), and immunohistochemical images of NPM3 expression in normal (middle) and tumor (right) tissues from the HPA database. Representative immunohistochemical images of NPM3 were taken at 100X magnification. (A) Breast (B) Colon (C) Liver (D) Lung (E) Prostate. ∗∗∗P < 0.001. BRCA: Breast invasive carcinoma; COAD: Colon adenocarcinoma; HPA: Human Protein Atlas; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; NPM3: Nucleophosmin/nucleoplasmin 3; PRAD: Prostate adenocarcinoma; TCGA: The Cancer Genome Atlas.
Survival analysis of nucleophosmin/nucleoplasmin 3 in pan-cancer
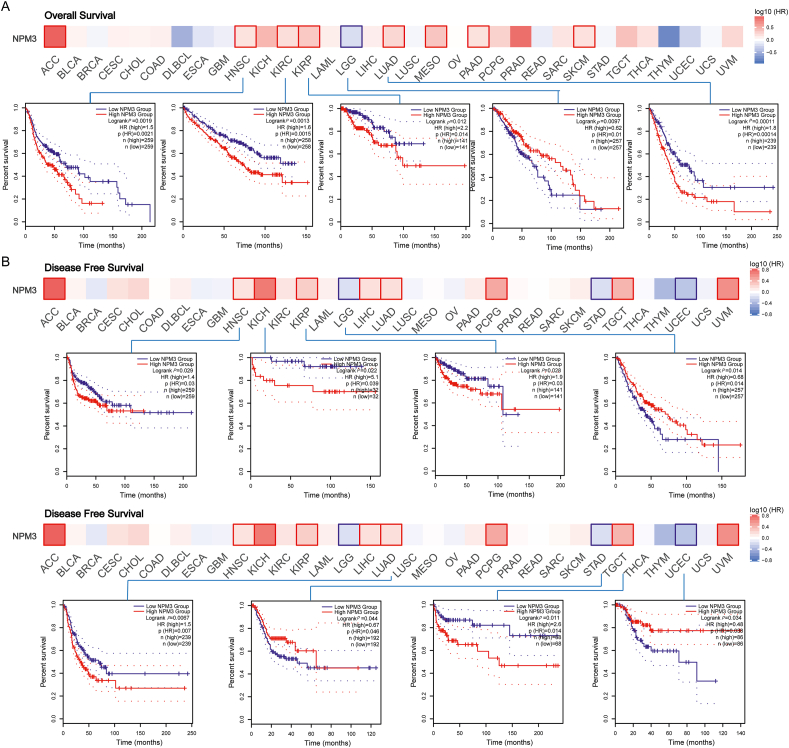
We analyzed the relationship between NPM3 expression and prognosis by conducting overall survival (OS) and disease-free survival (DFS) analyses using GEPIA 2.0. Patients were categorized into high and low NPM3 mRNA expression groups. Furthermore, we investigated the relationship between NPM3 expression and patient prognosis by integrating data from the GEO and TCGA databases. The results of the OS analysis [Figure 4A] revealed that high NPM3 expression levels were associated with poor prognosis in HNSC (P = 0.0019), KIRC (P = 0.0013), KIRP (P = 0.012), and LUAD (P = 0.00011), and low NPM3 expression was observed in LGG (P = 0.0097). Additionally, our DFS analysis [Figure 4B] revealed that high NPM3 expression was associated with worse prognosis in HNSC (P = 0.029), KICH (P = 0.022), KIRP (P = 0.028), LUAD (P = 0.007), and TGCT (P = 0.011). Conversely, low NPM3 expression was associated with a poor prognosis in LGG (P = 0.014), STAD (P = 0.044), and UCEC (P = 0.034).
Figure 4.
The prognostic significance of nucleophosmin/nucleoplasmin 3 in multiple human cancers. GEPIA 2.0 was utilized to evaluate the connection between NPM3 and the overall survival (A) and disease-free survival (B) of various tumors in TCGA. The results were presented with Survival Map and Kaplan–Meier plots. ACC: Adrenocortical carcinoma; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; ccRCC: Clear cell renal cell carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; DLBCL: Lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: Esophageal carcinoma; GEPIA 2.0: Gene Expression Profiling Interactive Analysis; GBM: Glioblastoma multiforme; HNSC: Head and neck squamous cell carcinoma; KICH: Kidney chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute myeloid leukemia; LGG: Lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MECO: Mesothelioma; NPM3: Nucleophosmin/nucleoplasmin 3; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PRAD: Prostate adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; READ: Rectum adenocarcinoma; SARC: Sarcoma; SKCM: Skin cutaneous melanoma; STAD: Stomach adenocarcinoma; TCGA: The Cancer Genome Atlas; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma; UVM: Uveal melanoma.
Subsequently, we performed a survival analysis of the selected cancer types using the KM plotter tool. According to these findings, higher NPM3 expression was associated with worse OS (P = 0.045), disease-specific survival (DSS; P = 0.034), progression-free survival (PFS; P = 0.0014), and recurrence-free survival (RFS; P = 0.01) in LIHC, as well as worse OS (P = 1.6e-08) and first progression (FP; P = 0.00028) in LUAD. In contrast, lower NPM3 expression was associated with poor post-progression survival (PPS; P = 0.033) in OV and poor OS (P = 0.031), FP (P = 0.019), and PPS (P = 1.6e-08) in STAD [Supplementary Figure 3].
Correlation between nucleophosmin/nucleoplasmin 3 expression and methylation
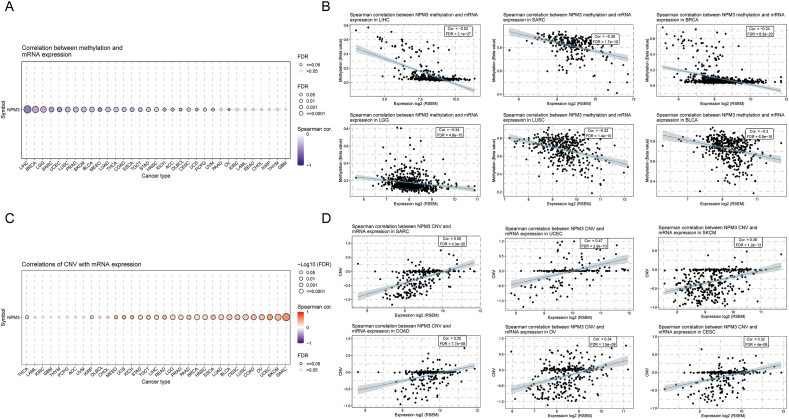
DNA methylation is common in numerous cancer types and is closely associated with tumorigenesis.32 Therefore, we used GSCA to investigate the association between NPM3 methylation and mRNA expression. As shown in Figure 5A, NPM3 methylation was negatively correlated with mRNA expression in most cancer types, and its relevance in LIHC, SARC, BRCA, LGG, LUSC, and BLCA was particularly significant (P < 0.01; Figure 5B). However, in only a few cancers, including PAAD, KIRC, LAML, READ, CHOL, KIRP, THYM, and GBM, no significant relationship was observed [Figure 5A].
Figure 5.
Association between Nucleophosmin/nucleoplasmin 3 expression and Nucleophosmin/nucleoplasmin 3 methylation and copy number variation. (A) Spearman correlation between NPM3 methylation and mRNA expression in pan-cancer. (B) The top six cancers with the highest correlation score between NPM3 methylation and mRNA expression. (C) Spearman correlation between NPM3 CNV and mRNA expression in pan-cancer. (D) The top six cancers with the highest correlation score between NPM3 CNV and mRNA expression. ACC: Adrenocortical carcinoma; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; ccRCC: Clear cell renal cell carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; CNV: Copy number variation; DLBCL: Lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; HNSC: Head and neck squamous cell carcinoma; KICH: Kidney chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute myeloid leukemia; LGG: Lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MECO: Mesothelioma; NPM3: Nucleophosmin/nucleoplasmin 3; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; SKCM: Skin cutaneous melanoma; STAD: Stomach adenocarcinoma; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma; UVM: Uveal melanoma.
Correlation between nucleophosmin/nucleoplasmin 3 expression and copy number variation
We also used GSCA to examine the Spearman's correlation between NPM3 CNV and mRNA expression. NPM3 CNV showed a positive association with mRNA expression, except in LAML, KIRC, GBM, THYM, PCPG, ACC, uveal melanoma (UVM), DLBCL, and CHOL (P < 0.05, Figure 5C). This correlation was significantly higher in SARC, UCEC, SKCM, COAD, OV, and CESC (P < 0.05; Figure 5D).
Analysis of nucleophosmin/nucleoplasmin 3 expression and immune cell infiltration
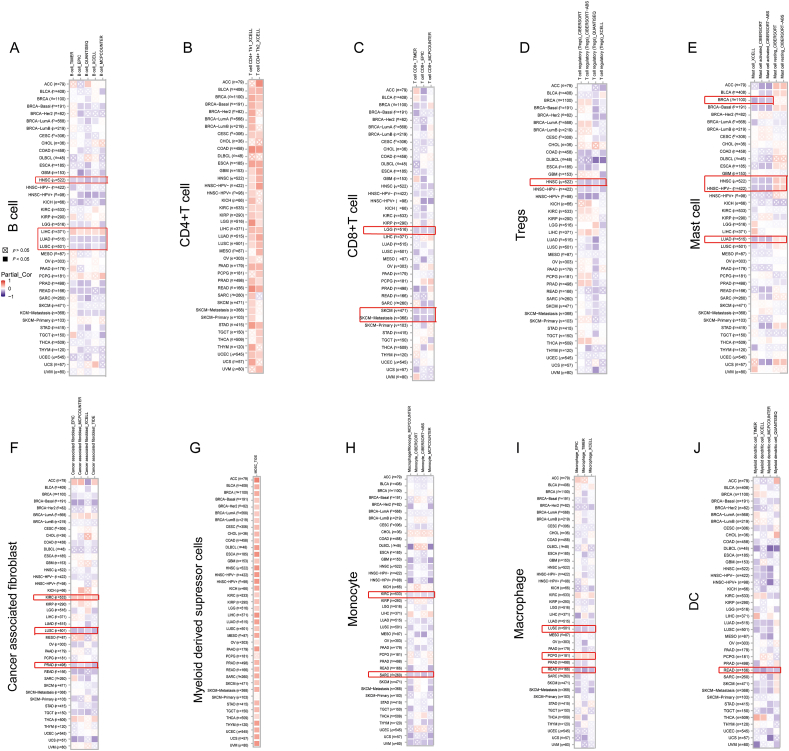
The level of immune cell infiltration plays a significant role in predicting the prognosis of patients with tumors and determining the efficacy of immunotherapy.33 We used several different algorithms to investigate the connection between NPM3 expression and the infiltration of various immune and stromal cells in pan-cancer. As shown in Figure 6A, NPM3 expression was negatively associated with B-cell infiltration in HNSC, LUAD, and LUSC, whereas it was positively correlated with LIHC. For CD4+T cells [Figure 6B], the correlation between NPM3 expression and cell infiltration was mostly positive in various types of cancer. However, for CD8+T cells, the connection was negative in LGG and SKCM [Figure 6C], and for Treg cells, it was negative in HNSC [Figure 6D]. For mast cells, NPM3 expression was negatively correlated with activated mast cells but positively correlated with resting mast cells in HNSC and LUAD [Figure 6E]. For cancer-associated fibroblasts, a negative relationship between NPM3 expression and cell infiltration in LUSC and PRAD was observed, but a positive relationship was observed in KIRC [Figure 6F]. The association between NPM3 expression and MDSC cell infiltration was generally positive in various cancer types [Figure 6G]. Furthermore, NPM3 expression negatively correlated with monocyte infiltration in both KIRC and SARC [Figure 6H]. In macrophages, the relationship was negative for LUSC and READ, whereas PCPG showed a positive relationship [Figure 6I]. The expression of NPM3 in READ was negatively correlated with DCs [Figure 6J].
Figure 6.
Correlation analysis between nucleophosmin/nucleoplasmin 3 expression level and immune cell infiltration. (A–J) Correlative analysis between NPM3 expression level and the infiltration of B cells, CD4+T cells, CD8+ T cells, Tregs, mast cells, cancer-associated fibroblasts, MDSCs, monocytes, macrophages, and DCs using TIMER, CIBERSORT, CIBERSORT-ABS, EPIC, MCPCOUNTER, QUANTISEQ, and XCELL algorithms. Correlations with P values < 0.05 are considered statistically significant and are indicated by black rectangles, and statistically insignificant correlation values are marked by crosses. Red indicates a positive correlation (0–1), while blue indicates a negative correlation (-1-0). ACC: Adrenocortical carcinoma; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; ccRCC: Clear cell renal cell carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; DC: Dendritic cell; DLBCL: Lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; HNSC: Head and neck squamous cell carcinoma; KICH: Kidney chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute myeloid leukemia; LGG: Lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MECO: Mesothelioma; MDSC: Myeloid-derived suppressor cell; NPM3: Nucleophosmin/nucleoplasmin 3; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; SKCM: Skin cutaneous melanoma; STAD: Stomach adenocarcinoma; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; TIMER: Tumor immune estimation resource; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine Carcinosarcoma; UVM: Uveal melanoma.
Association of nucleophosmin/nucleoplasmin 3 expression and tumor mutation load and microsatellite instability in various cancers
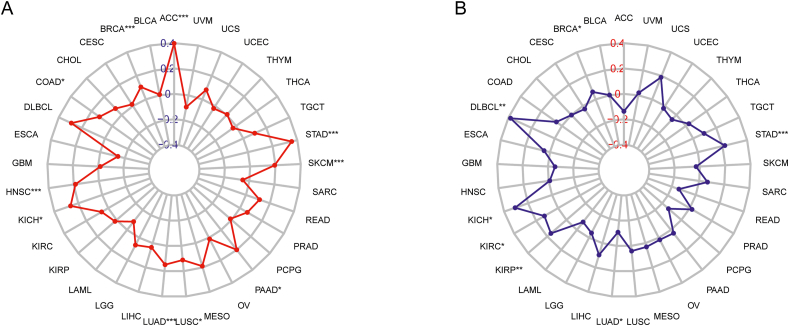
TMB and MSI have recently identified biomarkers for tumor diagnosis that have shown a strong correlation with the effectiveness of immunotherapy in patients with cancer.34 Therefore, we explored the correlation between NPM3 expression, TMB, and MSI in pancreatic cancers. Figure 7A illustrates the positive correlation between NPM3 expression and TMB in 10 cancer types: ACC, BRCA, COAD, HNSC, KICH, LUAD, LUSC, SKCM, PAAD, and STAD. Figure 7B shows a positive connection between NPM3 expression and MSI in BRCA, DLBCL, KICH, KIRC, KIRP, and STAD and a negative connection with LUAD.
Figure 7.
Correlation of nucleophosmin/nucleoplasmin 3 expression with tumor mutation burden and microsatellite instability in pan-cancer. The results are presented with radar diagrams. (A) Correlation between TMB and NPM3 expression. (B) Association between MSI and NPM3 expression. Spearman's correlation coefficients are shown above (P < 0.05 was considered statistically significant, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). ACC: Adrenocortical carcinoma; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; ccRCC: Clear cell renal cell carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; DLBCL: Lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; HNSC: Head and neck squamous cell carcinoma; KICH: Kidney chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute myeloid leukemia; LGG: Lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MECO: Mesothelioma; MSI: Microsatellite instability; NPM3: Nucleophosmin/nucleoplasmin 3; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; SKCM: Skin cutaneous melanoma; STAD: Stomach adenocarcinoma; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; TMB: Tumor mutation burden; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma; UVM: Uveal melanoma.
Protein–protein interaction network of nucleophosmin/nucleoplasmin 3-binding proteins and enrichment analysis
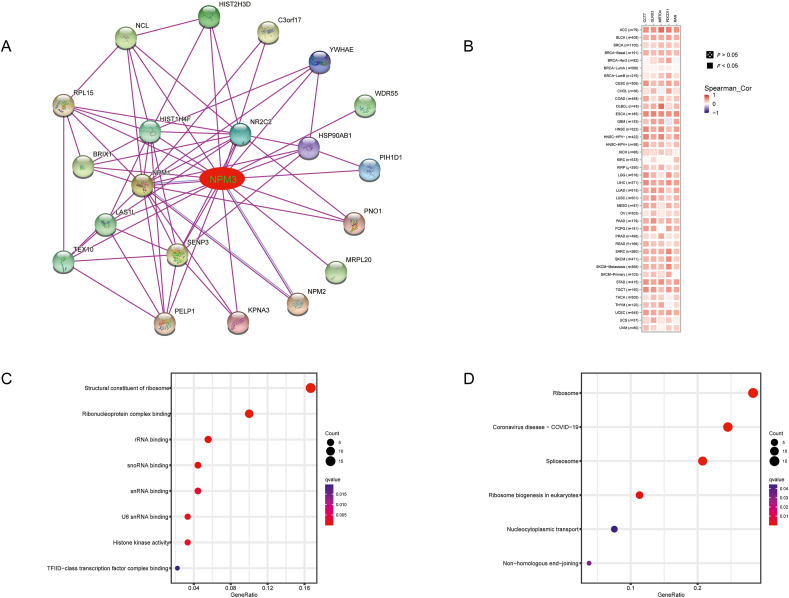
To investigate the possible involvement of NPM3 expression in carcinogenesis and development, we utilized STRING to obtain 20 NPM3-binding proteins supported by experimental data and established a PPI network [Figure 8A]. Subsequently, we identified the top five genes with the highest relevance to NPM3 and generated a heatmap of the association between NPM3 and these five genes across all cancers. The results revealed that NPM3 expression positively correlated with chaperonin containing TCP1 subunit 7 (CCT7), glutaredoxin 3 (GLRX3), MRT4 homolog ribosome maturation factor (MRTO4), programmed cell death 11 (PDCD11), and Ras-related nuclear (RAN) expression in various cancers [Figure 8B].
Figure 8.
Nucleophosmin/nucleoplasmin 3-related gene enrichment analysis. (A) A STRING PPI network for NPM3. Colored nodes represent experimentally confirmed proteins that interact with NPM3. (B) Correlations of NPM3 with the top-5 related genes in a heatmap. (C) KEGG enrichment analysis based on NPM3-binding and interacting genes. (D) GO analysis based on NPM3-interacting and correlated genes. ACC: Adrenocortical carcinoma; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; ccRCC: Clear cell renal cell carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; DLBCL: Lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; GO: Gene Ontology; HNSC: Head and neck squamous cell carcinoma; KEGG: Kyoto Encyclopedia of Genes and Genomes; KICH: Kidney chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute myeloid leukemia; LGG: Lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MECO: Mesothelioma; NPM3: Nucleophosmin/nucleoplasmin 3; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; PPI: Protein–protein interaction; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; SKCM: Skin cutaneous melanoma; STAD: Stomach adenocarcinoma; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine Carcinosarcoma; UVM: Uveal melanoma.
We utilized the “cluster Profiler” R package to perform GO and KEGG enrichment analyses to explore the possible molecular functions, cellular environment, biological processes, and potential carcinogenic pathways associated with NPM3. GO analysis indicated that NPM3 was mainly related to biological processes such as “structural constituent of ribosome,” “ribonucleoprotein complex binding,” “ribosomal RNA binding,” “small nucleolar RNA binding,” “small nuclear RNA binding,” “U6 small nuclear RNA binding,” and “histone kinase activity” [Figure 8C]. KEGG analysis revealed that NPM3 may be involved in “ribosome,” “Coronavirus disease-COVID-19,” “spliceosome,” “ribosome biogenesis in eukaryotes,” “nucleocytoplasmic transport,” and “non-homologous end-joining” pathways [Figure 8D].
Nucleophosmin/nucleoplasmin 3 expression and drug response
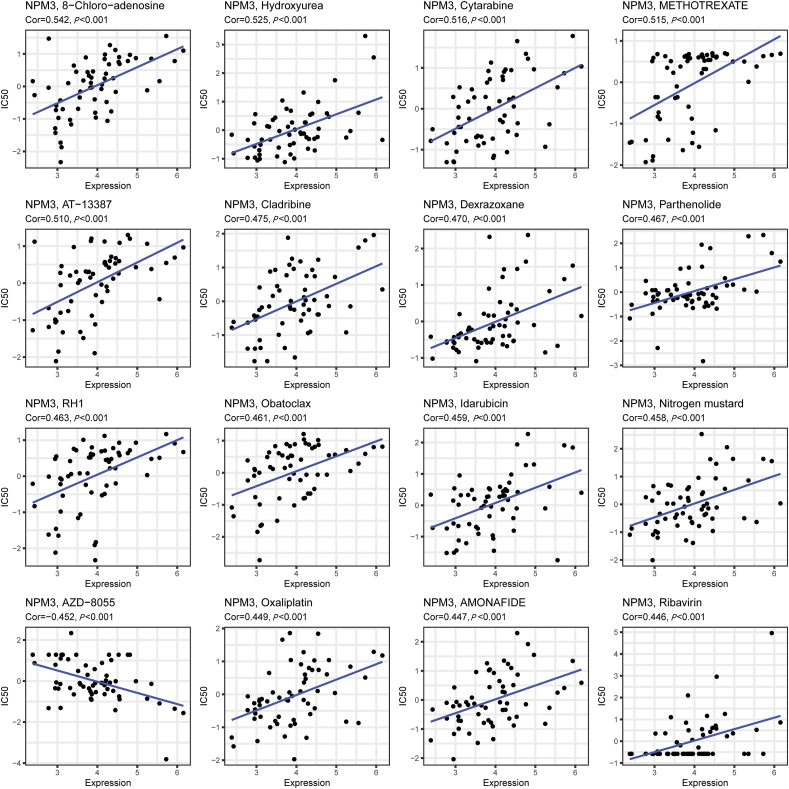
We utilized the CellMiner database and R4.1.3 to investigate the connection between NPM3 expression and sensitivity to antineoplastic agents. This analysis revealed a significant positive correlation between NPM3 expression and sensitivity to 8-Chloro-adenosine, cytarabine, methotrexate, AT-13387, cladribine, dexrazoxane, parthenolide, RH1, obatoclax, idarubicin, nitrogen mustard, oxaliplatin, amonafide, and ribavirin. Conversely, a negative correlation was observed between NPM3 expression and the response to AZD-8055 [Figure 9].
Figure 9.
Scatter plots of the connection between nucleophosmin/nucleoplasmin 3 expression and expected drug response.
Prognostic value and association with clinical characteristics of nucleophosmin/nucleoplasmin 3 expression in lung adenocarcinoma
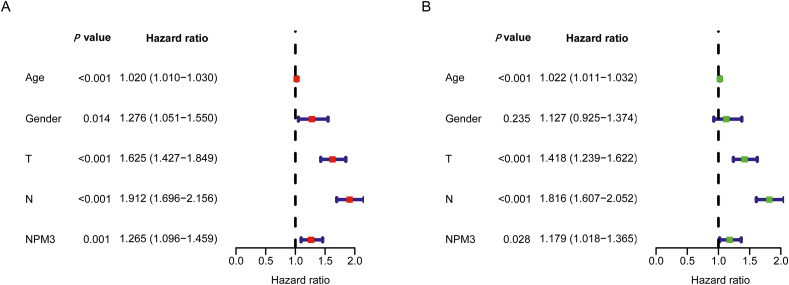
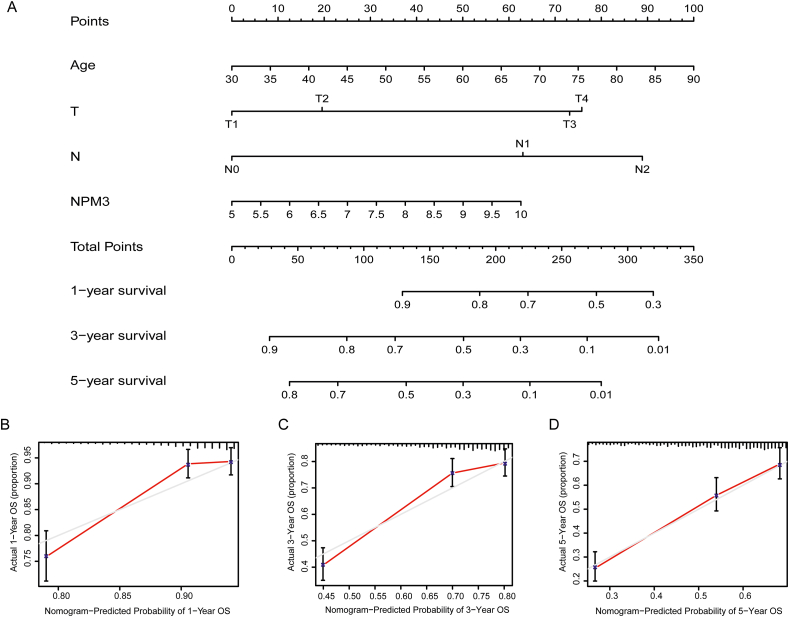
NPM3 expression, age, sex, tumor size, and the presence of lymph node metastasis were included as variables in the univariate and multivariate Cox regression analyses. The results showed that age, tumor size, presence of lymph node metastasis, and NPM3 expression were independent risk factors affecting the prognosis of patients with LUAD (P < 0.05; Figures 10A and B). These risk factors were included in the construction of a nomogram to predict the survival of patients with LUAD at 1, 3, and 5 years [Figure 11A]. The calibration curves of the nomogram demonstrated reliable predictive performance for predicting the actual likelihood of 1-year, 3-year, and 5-year OS in patients with LUAD [Figures 11B–D].
Figure 10.
Independent prognostic value analysis of nucleophosmin/nucleoplasmin 3. (A) Single-factor Cox regression analysis. (B) Multi-factor Cox regression analysis.
Figure 11.
Nomogram and calibration curves for patients with lung adenocarcinoma. (A) Nomogram. (B) Calibration curve for 1-year OS predicted by nomogram. (C) calibration curve for 3-year OS predicted by nomogram. (D) calibration curve for 5-year OS predicted by nomogram (the gray line represents the ideal prediction, and the red line denotes the actual fit). LUAD: Lung adenocarcinoma; N: Lymph node; OS: Overall survival; T: Tumor.
Validation of the relative expression of nucleophosmin/nucleoplasmin 3 in lung adenocarcinoma by real-time quantitative polymerase chain reaction
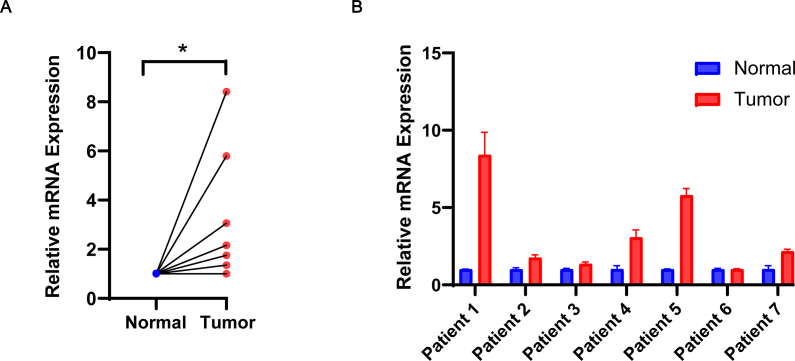
To confirm the differential expression of NPM3 in LUAD, we quantified the mRNA levels of NPM3 in seven tumor and matched normal tissue samples using RT-qPCR. The results demonstrated a significant upregulation of NPM3 expression in tumor tissues compared to the corresponding normal tissues (P < 0.05; Figures 12A and B).
Figure 12.
Validation of nucleophosmin/nucleoplasmin 3 expression via real-time quantitative polymerase chain reaction. (A) The relative mRNA expression of NPM3 in tumor and paraneoplastic tissues is presented in the form of a paired scatter graph. (B) Comparison of the relative mRNA expression of NPM3 in tumor and paraneoplastic tissues of seven patients, respectively. P < 0.05 was considered statistically significant; ∗P < 0.05. NPM3: Nucleophosmin/nucleoplasmin 3; RT-qPCR: Real-time quantitative polymerase chain reaction.
Discussion
NPM3 was initially identified among the genes activated by pro-viral insertions of the mouse tumor virus and is closely linked to the proto-oncogene fibroblast growth factor 8 (FGF8), which is often co-activated.35 NPM3 belongs to the NPM family and is involved in many important biological processes such as DNA replication and transcriptional regulation, histone storage, histone chaperone activity, ribosome biogenesis, and chromatin remodeling.15 Several studies have investigated the association between NPM3 expression and cancer. For example, deep sequencing of MIFS has shown significant upregulation of NPM3.17 Additionally, a study conducted on a transgenic mouse model suggested that c-Myc could enhance gene expression through chromatin remodeling and post-transcriptional RNA editing and identified NPM3 as a new c-Myc-responsive gene. Presently, changes in NPM3 levels have been linked to human LUAD.36 Similarly, a high-resolution gene microarray analysis of 27 patients with GC revealed deletions in NPM3.18 Moreover, a study analyzing gene expression in follicular and diffuse large B-cell lymphomas using RT-qPCR showed a significant upregulation of NPM3 in patients with DLBCL.19
In line with previous studies, our findings indicate that NPM3 expression is upregulated in many malignancies, including DLBCL and LUAD.18,19 NPM3 may be a valuable prognostic marker due to the significant correlation between high NPM3 expression and poor prognosis in several malignancies, as shown by KM analysis. Conversely, we observed the downregulation of NPM3 expression in KICH, KIRC, and LAML cells, suggesting that NPM3 may have different functions in different malignancies. Furthermore, we found significant changes in NPM3 expression levels during HNSC, KIRP, and LUAD progression, implying a role for NPM3 in malignancy progression. Notably, NPM3 expression levels were higher in the late stages of these tumors than in the early stages.
Tumor development is largely affected by genetic variations.37 Therefore, we investigated the relationship between NPM3 methylation, CNV, and NPM3 expression. In most cancer types, our findings depicted a positive association between NPM3 expression and methylation but showed a negative relationship with CNV.
A deeper understanding of immune cell infiltration is crucial for improving the outcomes of patients with tumors and increasing their response rates to immunotherapy.4 Immunological cell-infiltrating tumors are generally comprised of T and B lymphocytes, natural killer cells, DCs, macrophages, neutrophils, eosinophils, MDSCs, and mast cells.33 T cells have a strong tumor-killing ability and serve as the cellular basis of cancer immunotherapy.38 According to our research, NPM3 expression was positively correlated with the proportion of infiltrating Th1 and Th2 cells in most cancers but negatively correlated with the infiltration of CD8+T cells in LGG and SKCM. Th cells can remove tumor cells by direct cytolysis or indirectly regulating the tumor microenvironment.39 CD8+ T cells can be primed and activated into effector cytotoxic T cells, generating a potent and durable antitumor immune response.40 Additionally, our investigation revealed a significant correlation between NPM3 expression and MDSC infiltration in most malignancies. MDSCs are strongly linked to negative clinical outcomes of cancer and have been biologically established as potent immunosuppressive cells.41 Furthermore, we observed a correlation between NPM3 expression and levels of immune and stromal cell infiltration in certain types of cancer. Our findings suggest that NPM3 may play a role in modulating the TME, but the specific regulation mechanism seems to be intricate and unexplainable with the present data. Recently, single-cell technologies, including single-cell RNA sequencing and mass spectrometry, have advanced dramatically4 and would enable obtaining a more systematic and thorough understanding of the interactions between different TME components in future research.
TMB and MSI are biomarkers that provide insights into potential responses to immunotherapy.42,43 In various types of cancers, a combination of TMB and programmed cell death ligand 1 (PD-L1) is frequently employed to determine the need for immune checkpoint blockade selection, whereas MSI can serve as a biosignature for programmed cell death protein 1 (PD-1) blockade.44 Our study revealed a significant correlation between NPM3 and TMB/MSI in various cancers, including LUAD, suggesting a potential link between NPM3 and tumor immunity, which motivated our in-depth study of NPM3.
To understand the potential role of NPM3 in cancer development, we established a PPI network for NPM3 and conducted GO and KEGG enrichment analyses. PPI network analysis revealed a strong association between NPM3 and NPM1. During the cell cycle, nucleolar phosphoprotein NPM1 shuttles continuously between the cytoplasm and nucleus.45 NPM1 has dual functions as a proto-oncogene and a tumor suppressor.46 NPM1 regulates the ARF-p53 pathway.45 Numerous studies have shown a strong correlation between NPM1 expression and several types of cancer. NPM1 has been identified as a prognostic biomarker for breast cancer,47 colon cancer,48, 49, 50 GC,51,52 hepatocellular carcinoma,53 ovarian cancer54 and prostate cancer55,56 and is one of the most commonly affected genes in hematological cancers.57 Notably, some studies have shown that NPM3 promotes the nucleocytoplasmic shuttling of NPM1 in the somatic nucleus by inhibiting the RNA-binding activity of NPM1. Furthermore, the NPM1-NPM3 complex may regulate the chromatin structure at specific sites that are RNA polymerase II promoter-driven.58 Other studies have shown that NPM3 inhibits ribosome biogenesis and prevents NPM1 from acting as a histone chaperone.59 In summary, the optimal balance between NPM1 and NPM3 expression levels plays a critical role in maintaining the dynamic equilibrium of cells and preventing malignant transformation, which is closely linked to oncogenesis.59 The results of our GO and KEGG analyses confirmed previous findings by demonstrating that high levels of NPM3 expression in eukaryotes are primarily linked to rRNA binding and ribosome biogenesis.59 Tumorigenesis is characterized by the uncontrolled proliferation of cancer cells60 and is intimately linked to ribosome biogenesis,61 and ribosome biogenesis is thought to play an important role in tumor metastasis and drug resistance.62 Several traditional, highly efficient chemotherapeutic drugs have been shown to target ribosome biogenesis.63 However, the underlying mechanism remains unclear.64 Therefore, our analysis of NPM3 is highly significant because it provides opportunities to understand the complex mechanisms of ribosome biogenesis and opens the door for new targeted cancer therapies.
Lung cancer is the leading cause of cancer-related deaths worldwide,2 with LUAD as the most prevalent pathological type.65 Therefore, it is imperative to investigate the fundamental mechanisms that drive the progression of LUAD and identify novel biomarkers. Our Cox regression analyses identified NPM3 expression as an independent risk factor for poor LUAD prognosis. Therefore, we constructed a nomogram based on independent prognostic factors for LUAD, such as NPM3 expression, which can effectively predict the survival of patients with LUAD. Furthermore, the high expression of NPM3 was validated using RT-qPCR in seven pairs of LUAD and paraneoplastic tissues. Our study indicates that NPM3 expression is a potential biomarker for the prognosis of LUAD.
Despite conducting an initial exploration of the carcinogenic role of NPM3 via several bioinformatics approaches, our research has certain limitations. Our study was based on several public databases with varying data collection and processing methods, which may have influenced our findings. Although we used biomolecular methods to validate the high expression of NPM3 in LUAD, further in vivo and in vitro experiments are necessary to confirm and investigate our findings.
In conclusion, we performed a novel comprehensive pan-cancer analysis of NPM3 that revealed significant associations between NPM3 expression and multiple cancer prognoses, DNA methylation, CNV, immune cell infiltration, TMB, and MSI. Our investigation also included a preliminary exploration of the underlying mechanisms involved. Notably, we identified NPM3 expression as an independent prognostic marker for LUAD and developed a nomogram that could effectively predict the prognosis and clinical outcomes of patients with LUAD. Our study indicates that NPM3 could be a promising biomarker for predicting prognosis and immunotherapy efficacy in pan-cancer, thus potentially providing an antitumor strategy that targets NPM3.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82172620) and the Nature Science Foundation of Tianjin City (No. 18JCZDJC98800).
Authors contribution
Qianhui Wei: Conceptualization, Writing- Original draft preparation. Jing Zhou: Data curation. Xinyue Wang: Methodology, Software. Zhaona Li: Visualization. Xiuqiong Chen: Software, Validation. Kaidi Chen: Data curation. Richeng Jiang: Supervision.
Ethics statement
This project was reviewed and approved by the Institutional Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (No. bc2022263). All participants provided written informed consent to participate in this study. All the procedures were in accordance with the ethical standards of the responsible committee on human experimentation (institutional or regional) and with the Helsinki Declaration of 1975, as revised in 2000.
Data availability statement
The proposed dataset can be found in an online repository. Please refer to the article/supplementary material for the name and accession number of the repository. For further inquiries, the appropriate author can be contacted directly.
Conflicts of interest
None.
Acknowledgments
The authors thank all the patients from the Tianjin Medical University Cancer Institute & Hospital. The authors also thank TCGA, GTEx, GEO, TIMER 2.0, and other databases for online data sharing.
Managing Editor: Peng Lyu
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cpt.2023.06.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Bray F., Laversanne M., Weiderpass E., Soerjomataram I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer. 2021;127:3029–3030. doi: 10.1002/cncr.33587. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., et al. Global Cancer Statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. Ca - Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y., Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17:807–821. doi: 10.1038/s41423-020-0488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mamdani H., Matosevic S., Khalid A.B., Durm G., Jalal S.I. Immunotherapy in lung cancer: current landscape and future directions. Front Immunol. 2022;13:823618. doi: 10.3389/fimmu.2022.823618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emens L.A. Breast cancer immunotherapy: facts and hopes. Clin Cancer Res. 2018;24:511–520. doi: 10.1158/1078-0432.Ccr-16-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johdi N.A., Sukor N.F. Colorectal cancer immunotherapy: options and strategies. Front Immunol. 2020;11:1624. doi: 10.3389/fimmu.2020.01624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C., Xia B.R., Zhang Z.C., Zhang Y.J., Lou G., Jin W.L. Immunotherapy for ovarian cancer: adjuvant, combination, and neoadjuvant. Front Immunol. 2020;11:577869. doi: 10.3389/fimmu.2020.577869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hegde P.S., Chen D.S. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52:17–35. doi: 10.1016/j.immuni.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Tang T., Huang X., Zhang G., Hong Z., Bai X., Liang T. Advantages of targeting the tumor immune microenvironment over blocking immune checkpoint in cancer immunotherapy. Signal Transduct Targeted Ther. 2021;6:72. doi: 10.1038/s41392-020-00449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oura K., Morishita A., Tani J., Masaki T. Tumor immune microenvironment and immunosuppressive therapy in hepatocellular carcinoma: a review. Int J Mol Sci. 2021;22:5801. doi: 10.3390/ijms22115801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binnewies M., Roberts E.W., Kersten K., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shackleford G.M., Ganguly A., MacArthur C.A. Cloning, expression, and nuclear localization of human NPM3, a member of the nucleophosmin/nucleoplasmin family of nuclear chaperones. BMC Genom. 2001;2:8. doi: 10.1186/1471-2164-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eirín-López J.M., Frehlick L.J., Ausió J. Long-term evolution and functional diversification in the members of the nucleophosmin/nucleoplasmin family of nuclear chaperones. Genetics. 2006;173:1835–1850. doi: 10.1534/genetics.106.058990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frehlick L.J., Eirín-López J.M., Ausió J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29:49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- 16.Finn R.M., Ellard K., Eirín-López J.M., Ausió J. Vertebrate nucleoplasmin and NASP: egg histone storage proteins with multiple chaperone activities. Faseb J. 2012;26:4788–4804. doi: 10.1096/fj.12-216663. [DOI] [PubMed] [Google Scholar]
- 17.Arbajian E., Hofvander J., Magnusson L., Mertens F. Deep sequencing of myxoinflammatory fibroblastic sarcoma. Genes Chromosomes Cancer. 2020;59:309–317. doi: 10.1002/gcc.22832. [DOI] [PubMed] [Google Scholar]
- 18.Kang J.U., Koo S.H., Kwon K.C., Park J.W. AMY2a: a possible tumor-suppressor gene of 1p21.1 loss in gastric carcinoma. Int J Oncol. 2010;36:1429–1435. doi: 10.3892/ijo_00000628. [DOI] [PubMed] [Google Scholar]
- 19.Sakhinia E., Glennie C., Hoyland J.A., et al. Clinical quantitation of diagnostic and predictive gene expression levels in follicular and diffuse large B-cell lymphoma by RT-PCR gene expression profiling. Blood. 2007;109:3922–3928. doi: 10.1182/blood-2006-09-046391. [DOI] [PubMed] [Google Scholar]
- 20.Li T., Fu J., Zeng Z., et al. Timer2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T., Fan J., Wang B., et al. Timer: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.Can-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B., Severson E., Pignon J.C., et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Z., Kang B., Li C., Chen T., Zhang Z. Gepia2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Chen F., Chandrashekar D.S., Varambally S., Creighton C.J. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat Commun. 2022;13:2669. doi: 10.1038/s41467-022-30342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen F., Chandrashekar D.S., Varambally S., Creighton C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun. 2019;10:5679. doi: 10.1038/s41467-019-13528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhlén M., Fagerberg L., Hallström B.M., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 27.Uhlen M., Zhang C., Lee S., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 28.Liu C.J., Hu F.F., Xia M.X., Han L., Zhang Q., Guo A.Y. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34:3771–3772. doi: 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]
- 29.Sha D., Jin Z., Budczies J., Kluck K., Stenzinger A., Sinicrope F.A. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov. 2020;10:1808–1825. doi: 10.1158/2159-8290.Cd-20-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dal Buono A., Gaiani F., Poliani L., Correale C., Laghi L. Defects in MMR genes as a seminal example of personalized medicine: from diagnosis to therapy. J Personalized Med. 2021;11:1333. doi: 10.3390/jpm11121333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinhold W.C., Sunshine M., Liu H., et al. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012;72:3499–3511. doi: 10.1158/0008-5472.Can-12-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klutstein M., Nejman D., Greenfield R., Cedar H. DNA methylation in cancer and aging. Cancer Res. 2016;76:3446–3450. doi: 10.1158/0008-5472.Can-15-3278. [DOI] [PubMed] [Google Scholar]
- 33.Pagès F., Galon J., Dieu-Nosjean M.C., Tartour E., Sautès-Fridman C., Fridman W.H. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29:1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 34.Shum B., Larkin J., Turajlic S. Predictive biomarkers for response to immune checkpoint inhibition. Semin Cancer Biol. 2022;79:4–17. doi: 10.1016/j.semcancer.2021.03.036. [DOI] [PubMed] [Google Scholar]
- 35.Kuriki K., Kamiakito T., Yoshida H., Saito K., Fukayama M., Tanaka A. Integration of proviral sequences, but not at the common integration sites of the FGF8 locus, in an androgen-dependent mouse mammary Shionogi carcinoma. Cell Mol Biol. 2000;46:1147–1156. [PubMed] [Google Scholar]
- 36.Ciribilli Y., Singh P., Inga A., Borlak J. C-MYC targeted regulators of cell metabolism in a transgenic mouse model of papillary lung adenocarcinoma. Oncotarget. 2016;7:65514–65539. doi: 10.18632/oncotarget.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia S. Genetic variation as a modifier of association between therapeutic exposure and subsequent malignant neoplasms in cancer survivors. Cancer. 2015;121:648–663. doi: 10.1002/cncr.29096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest. 2015;125:3335–3337. doi: 10.1172/jci83871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy R., Celis E. Multiple roles for CD4+ T cells in anti-tumor immune responses. Immunol Rev. 2008;222:129–144. doi: 10.1111/j.1600-065X.2008.00616.x. [DOI] [PubMed] [Google Scholar]
- 40.Farhood B., Najafi M., Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: a review. J Cell Physiol. 2019;234:8509–8521. doi: 10.1002/jcp.27782. [DOI] [PubMed] [Google Scholar]
- 41.Grover A., Sanseviero E., Timosenko E., Gabrilovich D.I. Myeloid-derived suppressor cells: a propitious road to clinic. Cancer Discov. 2021;11:2693–2706. doi: 10.1158/2159-8290.Cd-21-0764. [DOI] [PubMed] [Google Scholar]
- 42.Kim S.I., Cassella C.R., Byrne K.T. Tumor burden and immunotherapy: impact on immune infiltration and therapeutic outcomes. Front Immunol. 2020;11:629722. doi: 10.3389/fimmu.2020.629722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang L., Chang M., Chang H.M., Chang F. Microsatellite instability: a predictive biomarker for cancer immunotherapy. Appl Immunohistochem Mol Morphol. 2018;26:e15–e21. doi: 10.1097/pai.0000000000000575. [DOI] [PubMed] [Google Scholar]
- 44.Dudley J.C., Lin M.T., Le D.T., Eshleman J.R. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res. 2016;22:813–820. doi: 10.1158/1078-0432.Ccr-15-1678. [DOI] [PubMed] [Google Scholar]
- 45.Lim M.J., Wang X.W. Nucleophosmin and human cancer. Cancer Detect Prev. 2006;30:481–490. doi: 10.1016/j.cdp.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grisendi S., Mecucci C., Falini B., Pandolfi P.P. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 47.Zeng D., Xiao Y., Zhu J., Peng C., Liang W., Lin H. Knockdown of nucleophosmin 1 suppresses proliferation of triple-negative breast cancer cells through activating CDH1/Skp2/P27kip1 pathway. Cancer Manag Res. 2019;11:143–156. doi: 10.2147/cmar.S191176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y., Zhang F., Zhang X.F., et al. Expression of nucleophosmin/NPM1 correlates with migration and invasiveness of colon cancer cells. J Biomed Sci. 2012;19:53. doi: 10.1186/1423-0127-19-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grbčić P., Fučkar Čupić D., Gamberi T., Kraljević Pavelić S., Sedić M. Proteomic profiling of BRAFV600E mutant colon cancer cells reveals the involvement of nucleophosmin/C-Myc axis in modulating the response and resistance to BRAF inhibition by vemurafenib. Int J Mol Sci. 2021;22:6174. doi: 10.3390/ijms22126174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong J.C., Hasan M.R., Rahman M., et al. Nucleophosmin 1, upregulated in adenomas and cancers of the colon, inhibits P53-mediated cellular senescence. Int J Cancer. 2013;133:1567–1577. doi: 10.1002/ijc.28180. [DOI] [PubMed] [Google Scholar]
- 51.Guo C.A., Su X.L., Wang W.J., et al. NPM1 is a diagnostic and prognostic biomarker associated with the clinicopathological characteristics of gastric cancer. Neoplasma. 2022;69:965–975. doi: 10.4149/neo_2022_220303N237. [DOI] [PubMed] [Google Scholar]
- 52.You B.J., Huang I.J., Liu W.H., Hung Y.B., Chang J.H., Yung B.Y. Decrease in nucleophosmin/B23 mRNA and telomerase activity during indomethacin-induced apoptosis of gastric KATO-III cancer cells. Naunyn-Schmiedeberg’s Arch Pharmacol. 1999;360:683–690. doi: 10.1007/s002109900145. [DOI] [PubMed] [Google Scholar]
- 53.Yun J.P., Miao J., Chen G.G., et al. Increased expression of nucleophosmin/B23 in hepatocellular carcinoma and correlation with clinicopathological parameters. Br J Cancer. 2007;96:477–484. doi: 10.1038/sj.bjc.6603574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shields L.B., Gerçel-Taylor C., Yashar C.M., et al. Induction of immune responses to ovarian tumor antigens by multiparity. J Soc Gynecol Invest. 1997;4:298–304. doi: 10.1177/107155769700400606. [DOI] [PubMed] [Google Scholar]
- 55.Subong E.N., Shue M.J., Epstein J.I., Briggman J.V., Chan P.K., Partin A.W. Monoclonal antibody to prostate cancer nuclear matrix protein (PRO:4-216) recognizes nucleophosmin/B23. Prostate. 1999;39:298–304. doi: 10.1002/(sici)1097-0045(19990601)39:4<298::aid-pros11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 56.Ruan Y., Xu H., Ji X. High expression of NPM1 via the Wnt/Β-catenin signalling pathway might predict poor prognosis for patients with prostate adenocarcinoma. Clin Exp Pharmacol Physiol. 2022;49:525–535. doi: 10.1111/1440-1681.13628. [DOI] [PubMed] [Google Scholar]
- 57.Karimi Dermani F., Gholamzadeh Khoei S., Afshar S., Amini R. The potential role of nucleophosmin (NPM1) in the development of cancer. J Cell Physiol. 2021;236:7832–7852. doi: 10.1002/jcp.30406. [DOI] [PubMed] [Google Scholar]
- 58.Okuwaki M., Sumi A., Hisaoka M., et al. Function of homo- and hetero-oligomers of human nucleoplasmin/nucleophosmin family proteins NPM1, NPM2 and NPM3 during sperm chromatin remodeling. Nucleic Acids Res. 2012;40:4861–4878. doi: 10.1093/nar/gks162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gadad S.S., Shandilya J., Kishore A.H., Kundu T.K. NPM3, a member of the nucleophosmin/nucleoplasmin family, enhances activator-dependent transcription. Biochemistry. 2010;49:1355–1357. doi: 10.1021/bi9021632. [DOI] [PubMed] [Google Scholar]
- 60.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 61.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elhamamsy A.R., Metge B.J., Alsheikh H.A., Shevde L.A., Samant R.S. Ribosome biogenesis: a central player in cancer metastasis and therapeutic resistance. Cancer Res. 2022;82:2344–2353. doi: 10.1158/0008-5472.Can-21-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burger K., Mühl B., Harasim T., et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010;285:12416–12425. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zisi A., Bartek J., Lindström M.S. Targeting ribosome biogenesis in cancer: lessons learned and way forward. Cancers. 2022;14:2126. doi: 10.3390/cancers14092126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Molina J.R., Yang P., Cassivi S.D., Schild S.E., Adjei A.A. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proposed dataset can be found in an online repository. Please refer to the article/supplementary material for the name and accession number of the repository. For further inquiries, the appropriate author can be contacted directly.