TO THE EDITOR:
Allogeneic hematopoietic cell transplantation (alloHCT) is an important part of the therapeutic algorithm of myeloid neoplasms. Risk of acute myeloid leukemia (AML)/myelodysplastic syndrome (MDS) relapse after alloHCT continues to be significant, and recent efforts have been devoted to dissecting the molecular pathways leading to post-transplant relapse (1). The dynamic process of clonal evolution is shaped by the nature of ancestral hits and emergence of subclones to create a highly diverse clonal architecture (2, 3). During the course of disease, both evolution drives and types of therapies mold the trajectory of this architectural change with extinction of less fit (more sensitive) subclones and appearance of new subclonal mutations as a result of a selection process (1). However, if not eradicated by the therapy, ancestral mutations may remain present throughout the subsequent clinical course, alone or with other subclones, may initiate disease recurrence or be a source of persistent disease (3).
Previously, we investigated the impact of mutational landscapes in AML and MDS undergoing alloHCT (4). In addition to molecular drivers, a limited normal hematopoietic stem cell compartment and allogenic immunogenicity play a fundamental role in determining the fate of the malignant process in the post-transplant setting. So far, studies comparing mutational landscape changes at post-alloHCT relapse to those at post-chemotherapy relapse are lacking. We theorized that the clonal selection process following alloHCT is distinct from that of post-chemotherapy relapse, as it is shaped not only by the exposure to the conditioning regimen, but also by graft-vs-leukemia (GVL) responses. These factors may have implications for our understanding of the biology and timing of relapse, help to assign prognosis, and inform appropriate clinical management.
Hence, using a next-generation sequencing (NGS)-based method targeting the most frequently mutated myeloid genes, we comprehensively analyzed clinical and molecular characteristics (i.e., types of mutation, variant allelic frequency [VAF]) associated with post-transplant relapse and compared the observed somatic changes to those occurring at relapse post-chemotherapy. Among patients transplanted at our program between 2005 and 2018, we identified 76 AML /MDS cases with available sampling at diagnosis and full clinical and molecular annotation (cohort 1). Among them, a subset of 49 cases had available samples for molecular analysis at post-transplant relapse (Supp. Fig.1). Additionally, we reviewed a well annotated cohort of 259 patients with AML or MDS who relapsed after chemotherapy from 2006 to 2017 and identified additional 35 cases with serial samples for comparison with post-transplant cohort (cohort 2). The study was approved by the Cleveland Clinic Foundation’s Institutional Review Board.
Multiamplicon deep sequencing was used to identify 30 genes most frequently mutated in myeloid disorders in the selected DNA samples (Supp. Table 1). The sequencing libraries were generated as previously described (3). The sequencing libraries were generated according to manufacturer protocol and instructions. Analyses were performed to investigate molecular, patient, disease, and transplant characteristics associated with long-term survival without relapse after first alloHCT, defined as 5 years in this study (relapse-free survival [RFS]) and mutational changes associated with post-transplant relapse were analyzed by comparing relapse after transplant and relapse after chemotherapy. The transplant cohort was further categorized by early relapse (relapse <6 months from alloHCT) and late relapse (relapse ≥6 months) based on findings from our prior analysis (5). Changes in molecular features at relapse were categorized as [1] complete recapitulation of pre-treatment landscape (recapitulation), [2] additional subclonal loss without subclonal gain (loss only), [3] additional subclonal gain without subclonal loss (gain only) or [4] additional subclonal loss with subclonal gain (loss & gain). Descriptive statistics, logistic regression, Bootstrap analysis, and two-sample t-test were performed using SAS® software, version 9.4 (SAS Institute, Inc., Cary, NC, USA).
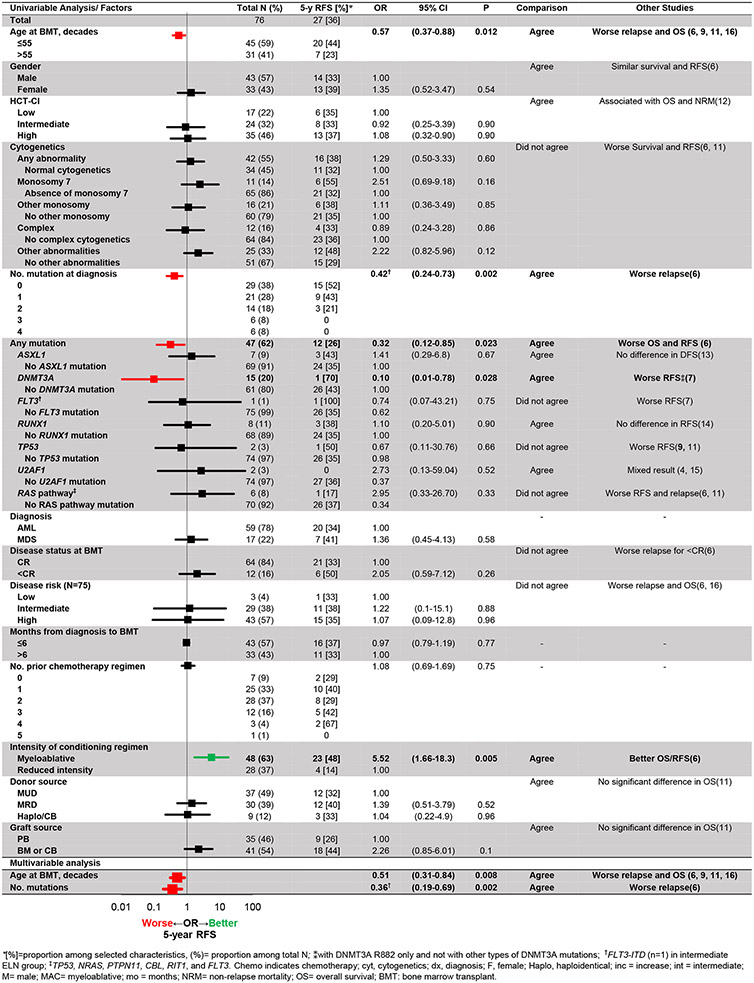
Most patients (64/76, 84%) were in a complete remission at the time of pre-alloHCT evaluation. With a median follow up of 67 months post-alloHCT, 64% (49/76) experienced post-transplant relapse while the remainder of patients were alive 5 years post-alloHCT without relapse (4). Disease- and transplant-related characteristics were similar among cohort 1 as well as cohort 2 (Supp. Tables 2A-B). Myeloablative conditioning and presence of extensive stage chronic graft-versus-host disease were associated with higher RFS in univariate analysis but was not confirmed in multivariate analyses (Fig. 1). As demonstrated previously (6), older age (OR 0.51 per 10-year increase, CI 0.31-0.84, P=0.008) and higher number of mutations at baseline (OR 0.36 per 1 mutation increase, CI 0.19-0.69, P=0.002) were independent negative predictors of 5-year RFS (Fig. 2A).
Figure 1. Pre-transplant factors associated with relapse-free survival at 5 years after transplant.
Mutations that were unevaluable due to 0% or 100% presence were not listed. From multivariable analysis, age at transplant and number of mutation present before transplant were associated with 5-year RFS.
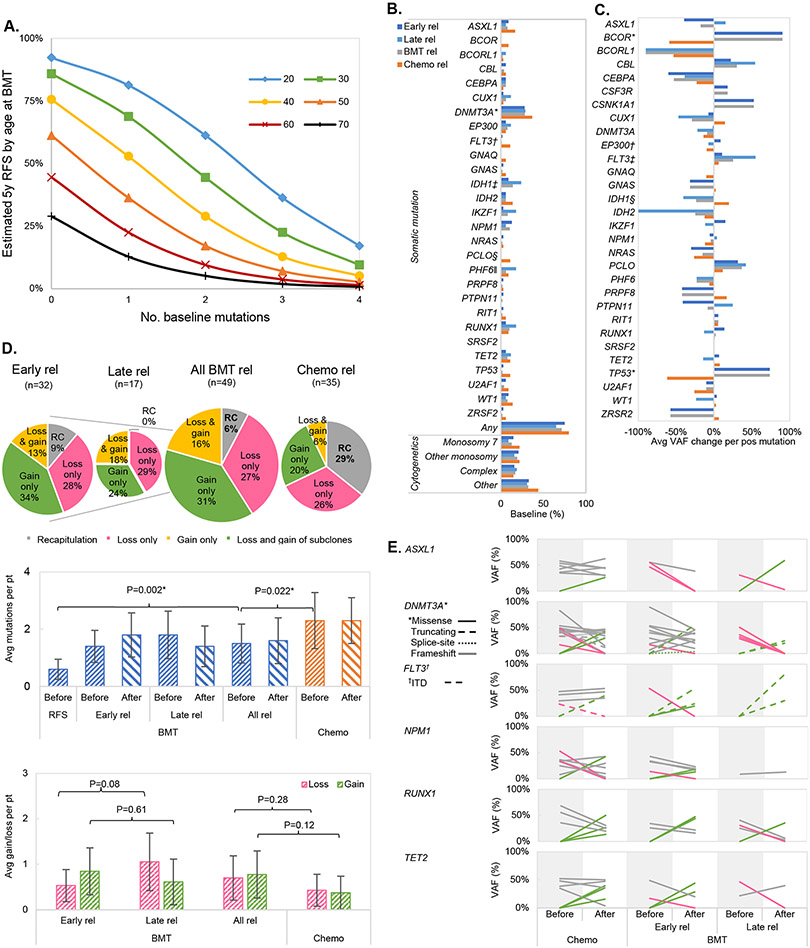
Figure 2. Relapse after allogeneic transplant.
(A) Estimated probability of achieving RFS5 based on number of mutations and age at BMT.
(B) Frequency of selected somatic mutations and cytogenetic abnormalities at diagnosis. *DNMT3A was the most commonly mutated genes in both post-transplant relapses (29%) and post-chemotherapy relapses (37%). DNMT3A mutations were less frequently seen in cases with 5-year RFS post BMT (4% vs. 29% in BMT rel, P=0.028). †25% (n=1 in early BMT relapse) and 50% (n=1) of baseline FLT3 mutations were internal tandem duplication (ITD) at BMT and chemotherapy relapses, respectively. ‡IDH1 mutations were enriched in relapses post BMT than those post-chemotherapy (14% vs. 0%, P=0.038) while §PCLO mutations were seen more often at chemotherapy relapses (11% vs. 0%, P=0.027). ∥Presence of PHF6 mutations was registered more often at late (18%) than early relapse (0%, P=0.037).
(C) Changes in average variant allelic frequency (VAF) for each positive mutation. *Gain of BCOR and TP53 mutations were seen at BMT relapse than chemotherapy relapse (BCOR +91% at BMT relapse vs. −59% at chemotherapy relapse, P=0.011; TP53 +74% at BMT relapse vs. −62% at chemotherapy relapse, P=0.004). †There were more VAF increase of EP300 at early relapse (+9%) than at late relapse (−7%, P=0.003). §The greater decrease in VAF of IDH1 was seen with late relapses (−1%) than in early relapse (−41%, P=0.033). Overall, ǂ3% of chemotherapy (n=1), 6% of early transplant (n=2), and 12% of late transplant relapses (n=2) were seen with FLT3-ITD subclonal gain (P=0.48). Morover, 3%, 3%, and 6% of chemotherapy (n=1), early (n=1), and late transplant relapses (n=1) were seen with FLT3-ITD subclonal loss, respectively (P=0.84).
(D) Overall molecular changes at relapse. Top = pattern of somatic mutation changes at relapses after BMT and after chemo. Middle= Somatic mutations before and after relapses (rel). A lower number of mutations was found in transplanted patients surviving without relapse. Also, patients who relapsed from chemotherapy had higher number of mutations before treatment than those who relapsed after transplant. Bottom = Subclonal gains and losses of individual mutations at relapse. A two-sample t-test was used for statistical comparisons.
(E) VAF changes in selected 6 most positive gene mutations: ASXL1, DNMT3A, FLT3, NPM1, RUNX1, and TET2 (alphabetical order). Grey = reappearance, green =subclonal gain, pink =subclonal loss.
Focusing on pre-alloHCT leukemic mutations, presence of any mutation (OR 0.32, CI 0.12-0.85, P=0.023) and DNMT3A lesions before transplant (OR 0.10, CI 0.01-0.78, P=0.028), regardless of R882 status, were also associated with worse RFS. This finding aligned with the results described by Ahn et al. (7) that DNMT3A R882 mutation but no other lesions in this gene is associated with worse event-free survival. However, among cases with DNMT3A mutations at baseline (of which 1 splice site [7%], 2 truncating [13%], and the remainder [80%] missense), only 1 had a long-term RFS, while 15 patients experienced post-transplant relapse, thereby precluding any further analysis on the prognostic impact of the type of DNMT3A mutations at baseline. Older age is likely to be a confounding factor considering the variables of DNMT3A mutation rate and reduced-intensity conditioning use. Advanced age at transplant has been associated with DNMT3A mutation in myeloid neoplasms (8), and older age is an indication to consider reduced-intensity conditioning. No other molecular signature/pathways or cytogenetic abnormalities were otherwise associated with RFS. Specific comparisons with previous studies are listed in Fig. 1.
The types and VAFs of pre-treatment leukemic mutations varied widely (Figs. 2B, 2C). Mean number of mutations at baseline was higher in the post-chemotherapy relapse group (2.3 ± 1.6 vs. 1.6±1.5, P=0.022 compared to the post-alloHCT relapse group; Fig. 2D). Recapitulation of original clonal architecture was seen less frequently at post-transplant relapse (6%) than after chemotherapy relapse (29%, P=0.011). Of note is that new IDH1 mutations were registered only in post-transplant relapse (41% vs. 0% in post-chemotherapy group, P=0.038) while new PCLO mutations were characteristics of relapse following chemotherapy (11% vs. 0% in post-transplant group, P=0.027; Fig. 2B). Moreover, VAFs of BCOR and TP53 mutations increased in post-transplant relapses (by on average +91% and +74%, respectively), while they decreased in post-chemotherapy relapses (for BCOR by −59%, P=0.011, TP53 by −74%, P=0.004; Fig. 2C). Overall, no specific patterns of molecular changes were identified at post-transplant relapse, as illustrated by an analysis of clonal dynamics (Fig. 2D) and exemplificative cases (Supp. Fig. 2). No specific pattern of mutational changes based on conditioning regimen or graft-versus-host disease prophylaxis was found.
When analyzing the possible effects of time from alloHCT on the molecular makeup of relapse, no recapitulation of the baseline mutational landscape was found in late relapses, possibly suggesting a specific imprint dictated by the immunological forces of the post-transplant period. There was also a trend towards more subclonal losses was noticed in the late relapse group (mean 1.1 vs. 0.5 in early relapses, P=0.08) in particular with disappearance of all PHF6 subclones (18%, P=0.037; Fig. 2C). The burden of subclonal gains after early and late relapse were instead not significantly different (Fig. 2E). Focusing on individual gene mutations, the clonal burden of EP300 increased in early relapses (mean +9% in VAF) and decreased in the late relapse group (mean −7%, P=0.003; Fig. 2C), whereas VAFs of IDH1 mutations decreased more heavily at late relapse (−41%) than at early relapse (−1%, P=0.033).
Our results confirm that a greater number of baseline mutations are associated with an increased risk of relapse after alloHCT, as an increasing number of subclones can enhance the leukemic cell’s ability to resist treatment. Conversely, no specific cytogenetic or molecular abnormality was associated with RFS, unlike what was previously reported (4, 6, 9, 11). Evolution of the mutational landscape, represented by addition or loss of subclonal mutation(s), was seen more frequently at alloHCT relapse than at chemotherapy relapse. Specifically, there was no recapitulation of baseline mutations and a trend towards more subclonal losses at late transplant relapse. This may implicate the emergence of a dynamic evolution after alloHCT that contributes to overcome the GvL effects to possibly acquire a fitness advantage. It has been hypothesized that mutations with tumor suppressive or proliferative function can drive mutations arising post-alloHCT, but our study did not find a specific type or pathway of gene mutations associated with such a scenario (6, 9, 10). Limitations of our study include the retrospective nature prone to selection bias, the small sample size, and a possibility of missing some somatic mutations or epigenetic drivers by not including whole genome or exome sequencing. Our study included a serial paired samples and, in this setting, lower numbers of patients were available to achieve appropriate power. Particularly, due to a small number of cases, a specific analysis on the impact of FLT3-ITD, TP53, or mutations in the RAS pathway, or of allelic burden changes on RFS was not feasible. Nonetheless, the overall clonal architecture after alloHCT was reconfirmed to be distinctive from the relapse after chemotherapy, possibly due to the intervention of selective immune pressure.
In conclusion, we demonstrate that AML/MDS relapses after alloHCT represent a more complex and dynamic molecular evolution than chemotherapy relapse, possibly as a result of the intervention of GVL forces. The multifaceted nature of this immune-pressure is reflected in the absence of specific patterns of somatic mutation changes observed at the time of transplant relapse. Our results add to the current literature concerning the genomic underpinnings of transplant relapse and highlight the importance of re-testing for mutations at disease recurrence to update the genomic landscape that may guide targeted therapy choices. Larger scale analyses that incorporate an appreciation of the complex evolutionary patterns of myeloid neoplasm are warranted to pinpoint more specific genomic data and better exploit the vulnerabilities of such disorders post-alloHCT, ultimately improving treatment outcomes.
Supplementary Material
Footnotes
Competing Interests Statement: The authors declare no competing interests.
Data Availability Statement:
Data available on request from the authors.
REFERENCES
- 1.Hirsch P, Zhang Y, Tang R, Joulin V, Boutroux H, Pronier E, et al. Genetic hierarchy and temporal variegation in the clonal history of acute myeloid leukaemia. Nat Commun. 2016. Aug 18;7:12475. Epub 2016/08/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagata Y, Zhao R, Awada H, Kerr CM, Mirzaev I, Kongkiatkamon S, et al. Machine learning demonstrates that somatic mutations imprint invariant morphologic features in myelodysplastic syndromes. Blood. 2020. Nov 12;136(20):2249–62. scientific advisor for Presagia. The remaining authors declare no competing financial interests. Epub 2020/09/23. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017. Feb;49(2):204–12. Epub 2016/12/19. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton BK, Rybicki L, Hirsch C, Przychodzen B, Nazha A, Gerds AT, et al. Mutation clonal burden and allogeneic hematopoietic cell transplantation outcomes in acute myeloid leukemia and myelodysplastic syndromes. Bone Marrow Transplant. 2019. August;54(8):1281–6. Epub 2019/01/17. eng. [DOI] [PubMed] [Google Scholar]
- 5.Hong S, Rybicki L, Corrigan D, Hamilton BK, Sobecks R, Kalaycio M, et al. Survival following relapse after allogeneic hematopoietic cell transplantation for acute leukemia and myelodysplastic syndromes in the contemporary era. Hematology/Oncology and Stem Cell Therapy. 2020. 2020/December/05/. [DOI] [PubMed] [Google Scholar]
- 6.Nazha A, Hu ZH, Wang T, Lindsley RC, Abdel-Azim H, Aljurf M, et al. A Personalized Prediction Model for Outcomes after Allogeneic Hematopoietic Cell Transplant in Patients with Myelodysplastic Syndromes. Biol Blood Marrow Transplant. 2020. Nov;26(11):2139–46. Epub 2020/08/12. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn JS, Kim HJ, Kim YK, Lee SS, Jung SH, Yang DH, et al. DNMT3A R882 Mutation with FLT3-ITD Positivity Is an Extremely Poor Prognostic Factor in Patients with Normal-Karyotype Acute Myeloid Leukemia after Allogeneic Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant. 2016. Jan;22(1):61–70. Epub 20150731. eng. [DOI] [PubMed] [Google Scholar]
- 8.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014. Dec;20(12):1472–8. Epub 2014/10/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshizato T, Nannya Y, Atsuta Y, Shiozawa Y, Iijima-Yamashita Y, Yoshida K, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017. Apr 27;129(17):2347–58. Epub 20170221. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernasconi P, Borsani O. Immune Escape after Hematopoietic Stem Cell Transplantation (HSCT): From Mechanisms to Novel Therapies. Cancers (Basel). 2019. Dec;12(1). Epub 2019/12/25. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindsley RC, Saber W, Mar BG, Redd R, Wang T, Haagenson MD, et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N Engl J Med. 2017. February;376(6):536–47. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorror ML, Logan BR, Zhu X, Rizzo JD, Cooke KR, McCarthy PL, et al. Prospective Validation of the Predictive Power of the Hematopoietic Cell Transplantation Comorbidity Index: A Center for International Blood and Marrow Transplant Research Study. Biol Blood Marrow Transplant. 2015. Aug;21(8):1479–87. Epub 2015/04/12. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou L, An J, Hou C, Ding Z, Qiu H, Tang X, et al. Allogeneic hematopoietic stem cell transplantation could improve the survival of acute myeloid leukemia patients with ASXL1 mutations. Hematology. 2021. Dec;26(1):340–7. eng. [DOI] [PubMed] [Google Scholar]
- 14.Waidhauser J, Labopin M, Esteve J, Kröger N, Cornelissen J, Gedde-Dahl T, et al. Allogeneic stem cell transplantation for AML patients with RUNX1 mutation in first complete remission: a study on behalf of the acute leukemia working party of the EBMT. Bone Marrow Transplant. 2021. Oct;56(10):2445–53. Epub 20210531. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton BK, Visconte V, Jia X, Tabarroki A, Makishima H, Hasrouni E, et al. Impact of allogeneic hematopoietic cell transplant in patients with myeloid neoplasms carrying spliceosomal mutations. Am J Hematol. 2016. Jun;91(4):406–9. Epub 2016/01/23. eng. [DOI] [PubMed] [Google Scholar]
- 16.Jimenez Jimenez AM, De Lima M, Komanduri KV, Wang TP, Zhang MJ, Chen K, et al. An adapted European LeukemiaNet genetic risk stratification for acute myeloid leukemia patients undergoing allogeneic hematopoietic cell transplant. A CIBMTR analysis. Bone Marrow Transplant. 2021. Dec;56(12):3068–77. Epub 20210928. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available on request from the authors.