Abstract
The green fluorescent protein (GFP) gene was expressed on a plasmid in B. henselae, and GFP-expressing bacteria were visualized by fluorescence microscopy. HEp-2 cells infected with GFP-expressing bacteria were separated from uninfected cells with a fluorescence activated cell sorter. Promoter fusions of B. henselae chromosomal DNA to gfp were examined by flow cytometry, and a B. henselae groEL promoter fusion which induced expression at 37°C was isolated.
Bartonella henselae, a gram-negative, fastidious, rod-shaped bacterium, is the infectious agent responsible for several human diseases including cat scratch disease in immunocompetent hosts and bacillary angiomatosis and bacillary peliosis hepatis in immunocompromised hosts (13, 15). The domestic cat is the primary reservoir of B. henselae (9, 11, 14), and the cat flea, Ctenocephalides felis, has been shown experimentally to be the vector in cat-to-cat transmission (2, 10). Transmission to humans is associated with traumatic contact with an infected cat (15, 19).
B. henselae interaction with human cells has been investigated in vitro with HEp-2 and HUVEC cell lines (1, 3, 6). Specific bacterial virulence genes have yet to be identified for this organism due to a lack of genetic tools. Introduction into and maintenance in B. henselae of a broad-host-range plasmid was the first important step toward developing genetic tools (5). Here we describe a green fluorescent protein (GFP) reporter system that has been instrumental in the study of several bacterial pathogens (20, 22).
A promoterless gfp gene, gfpmut3 (4), was cloned into the BamHI and SphI sites downstream of the ptac promoter in pCom100, making pANT4 (see Table 1 for descriptions of all strains and plasmids). GFP was expressed constitutively because a kanamycin cassette disrupted the lacIq repressor gene. pANT4 in SM10λpir was transferred to 882str via bacterial conjugation. Our conjugal transfer technique was similar to that described previously (5) with these exceptions: cultures were pelleted and washed twice in Luria-Bertani broth for Escherichia coli and RPMI for B. henselae; bacteria were spotted together onto a rabbit blood (RB) agar plate and incubated at 26°C for 2 to 6 h; and exconjugates were selected by streaking them onto RB agar (200 μg of streptomycin and 50 μg of kanamycin per ml) and incubating them at 37°C in 5% CO2. Exconjugate frequencies were approximately 3 × 10−4. Southern blotting confirmed that exconjugates (882-ANT4) maintained pANT4 as episomal DNA with antibiotic selection.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsc | Source or reference |
|---|---|---|
| E. colia | ||
| DH5α | end-1 hsdR17 (rK−/mK+)supE44 thi-1 λ− recA1 gyrA96 relA1 Δ(lacIZYA-argF)U169 deoR [F80 Δlacd(lacZ)M15] | 8 |
| SM10λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr λpir | 16 |
| B. henselaeb | ||
| 882str | Strr derivative of ATCC 49882 selected with 200 μg of streptomycin per ml | J. Peek |
| 882-ANT4 | First 882str strain carrying pANT4 plasmid; neither stably expresses GFP nor maintains plasmid | This study |
| 882-ANT5 | Selected 882-GFP that stably expresses GFP and stably maintains plasmid | This study |
| JK13 | Human isolate from cutaneous BA lesion | J. Koehler |
| JK13str | Strr strain of JK13 selected with 200 μg of streptomycin per ml | G. Murakawa |
| JK13-ANT5 | JK13str carrying pANT5; stably expresses GFP and maintains plasmid | This study |
| Bhcon-1, Bhcon-2, Bhcon-3 | JK13str carrying pANT-3 with B. henselae DNA fragment containing a constitutive promoter cloned upstream of gfpmut3 | This study |
| BhGroEL | JK13str carrying pANT-3 with B. henselae groEL promoter cloned upstream of gfpmut3 | This study |
| Plasmids | ||
| pCom100 | oriRSF1010 bla+mob+lacIq::kan+ | R. Valdivia |
| pANT4 | pCom100 with gfpmut3 under ptac control | This study |
| pANT5 | pANT4 selected to be stable in B. henselae | This study |
| pRS201 | oriRSF1010 mob+kan+strep+ | 18 |
| pRK600 | oriR6K cam+tra+ | 12 |
| pANT3 | oriRSF1010 mob+kan+strep; promoterless gfpmut3 | This study |
All E. coli strains were grown in Luria-Bertani media supplemented with appropriate antibiotics at 37°C.
B. henselae strains were grown on heart infusion agar supplemented with 5% RB and appropriate antibiotics at 37 or 26°C and 5% CO2.
Strr, streptomycin resistant; BA, bacillary angiomatosis.
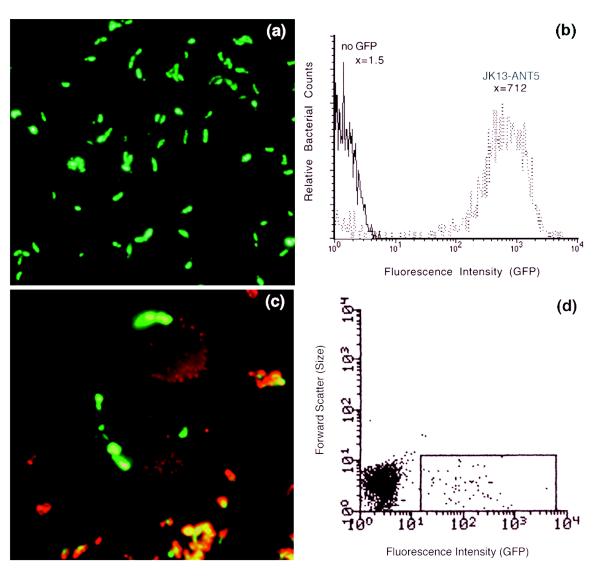
882-ANT4 exconjugates were highly fluorescent when viewed by fluorescent microscopy (Fig. 1a). An analysis using a FACScalibur cell scanner (Becton Dickinson) showed that 882-ANT4 had a mean fluorescence of 1,000 versus a mean fluorescence 1.5 for 882str (data not shown). However, the 882-ANT4 population lost most of its fluorescence with increasing passage without antibiotic selection, suggesting that GFP overexpression was toxic to the bacteria. We then selected for an 882-ANT4 clone that fulfilled the criteria of maintaining pANT4 stably and keeping constitutive GFP expression in the absence of antibiotic selection. After three consecutive passages on nonselective media, only 5% of the 882-ANT4 population had a mean fluorescence above 700. From this minority population, a single, highly fluorescent clone, 882-ANT5, was selected. After five serial passages on nonselective media, the 882-ANT5 population maintained consistently a mean fluorescence of about 700. Sequence analysis comparing the ptac promoter region in pCom100 to the ptac promoter region in pANT5 (the plasmid isolated from 882-ANT5) revealed a single-base-pair transition in the −35 consensus sequence from TTGACA to TTGGCA. The gfpmut3 gene sequence in pANT5 was unchanged compared to the original gfpmut3 sequence (4). pANT5 was conjugated into strain JK13str, a more invasive strain for HEp-2 cells (17), creating JK13-ANT5, which constitutively and stably expresses GFP after two passages on nonselective media. By flow cytometry, we could distinguish and separate JK13-ANT5 from nonfluorescent B. henselae (Fig. 1b).
FIG. 1.
(a) B. henselae strain 882-ANT4 expresses GFP. Bacteria were visualized with an epifluorescence microscope with a fluorescein isothiocyanate (FITC) filter. (b) JK13-ANT5 has about a 475-fold-higher mean fluorescence intensity (x) than non-GFP-expressing bacteria. Fluorescence levels were examined with a FACScalibur scanner (Becton Dickinson). Cell Quest software (Becton Dickinson) was used for the analysis and quantitation of fluorescence. (c) Intracellular bacteria can be distinguished from extracellular bacteria by antibody labelling. All bacteria express GFP. However, only extracellular bacteria are labelled red. Colocalization of GFP and TRITC markers renders extracellular bacteria yellow, while intracellular bacteria remain green. Cells were visualized with an epifluorescence microscope with FITC and TRITC filters to visualize green and red fluorescence, respectively. Images were superimposed with Adobe Photoshop 3.0.5 software. (d) HEp-2 cells infected with JK13-ANT5 can be distinguished from uninfected cells with the FACStar cytometer (Becton Dickinson). Cells within the gated region have cell-associated, GFP-expressing B. henselae and were separated from uninfected cells.
B. henselae was reported to enter HEp-2 cells (1). GFP-expressing B. henselae was used to visualize intracellular bacteria. HEp-2 cells seeded onto glass coverslips were infected with JK13-ANT5 for 2 h at a multiplicity of infection (MOI) of 10, and then the cells were fixed with 3.7% formaldehyde. Polyclonal rabbit anti-B. henselae antibody was added for 30 min, and the cells were washed with phosphate-buffered saline. Then, tetramethyl rhodamine B isothiocyanate (TRITC)-conjugated anti-rabbit antibody was added for 30 min, and the cells were again washed. Only extracellular bacteria were labeled with antibody because the cells were not permeabilized. JK13-ANT5 bacteria inside cells were visualized easily by fluorescence microscopy (Fig. 1c).
GFP-expressing B. henselae could be used as a fluorescent marker for separating infected cells from noninfected cells with a fluorescence-activated cell sorter (FACS). HEp-2 cells were infected with JK13-ANT5 at a MOI of 10 for 2 h. Whole cells were resuspended in RPMI (5% fetal calf serum), and infected cells were separated from uninfected cells with the FACStar cytometer (Becton Dickinson) (Fig. 1d).
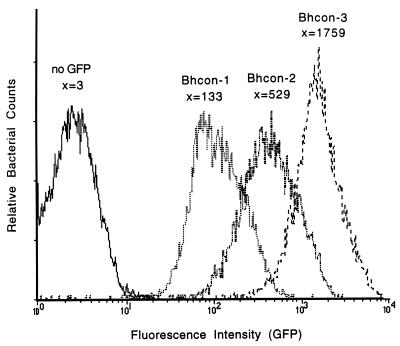
Little is known about B. henselae gene regulation and expression. To identify constitutive and inducible B. henselae promoters, a promoterless GFP vector, pANT3, was constructed from pRS201. A promoterless gfpmut3 gene was cloned into the PstI and EcoRI sites of pRS201, and the streptomycin resistance gene was partially removed by exonuclease deletion. A B. henselae promoter library was generated by cloning Sau3AI fragments (0.4 to 1.4 kb) of JK13 chromosomal DNA into the BamHI site immediately upstream of the promoterless gfpmut3. This library, in DH5α, was conjugated into JK13str as described above, with the addition of the helper plasmid pRK600 in DH5α. By FACS analysis, 1.33% of the library was found to be fluorescent, indicating that some DNA fragments contained constitutive promoters. Three clones, Bhcon-1, Bhcon-2, and Bhcon-3, expressed GFP at varying levels of fluorescence, as measured by flow cytometry (Fig. 2). Bhcon-1, Bhcon-2, and Bhcon-3 maintained their fluorescence when passaged on nonselective media. A sequence analysis of these three fragments showed no homology to sequences in the GenBank database.
FIG. 2.
Native B. henselae promoters drove GFP expression at varying levels of fluorescence intensity. x, mean fluorescence intensity for the given clonal population of bacteria, Bhcon-1, Bhcon-2, or Bhcon-3.
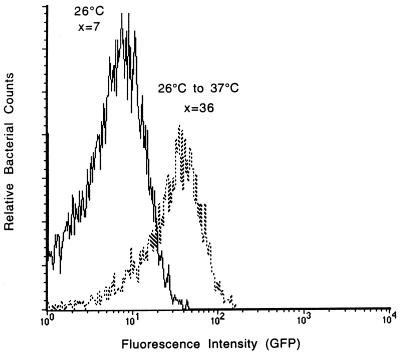
We then used the B. henselae promoter library to identify inducible promoters by differential fluorescence induction (21). In accordance with this technique, promoters that were activated at high temperature were selected as follows. The library population was grown at 26°C for 4 weeks and shifted to 37°C for 24 h, and bacteria expressing GFP were sorted by flow cytometry and plated on RB agar. This sequence was repeated to amplify clones carrying promoters that were activated only at high temperature. Most clones expressed GFP constitutively. However, one clone, BhGroEL, had a low level of GFP expression at 26°C but when shifted to 37°C experienced a fivefold increase in mean fluorescence (Fig. 3). A sequence analysis of the 500-bp insertion fragment showed that a 117-bp partial open reading frame adjacent to the GFP junction was 99% identical to the 5′ region of the gene encoding Bartonella heat shock protein GroEL (7).
FIG. 3.
GFP could be expressed differentially when the B. henselae groEL promoter was fused to gfpmut3. At 26°C, little GFP was expressed, but when shifted to 37°C, the groEL gene was turned on, and GFP expression was induced fivefold. x, mean fluorescence intensity.
The stable expression of GFP in B. henselae provides a useful method for studying the interactions of B. henselae with different cell types in vitro and in vivo. Animal studies, especially, will benefit from the stable GFP-expressing B. henselae. In addition, we have been able to identify a differentially expressed B. henselae gene by the differential fluorescence induction technique (21). We hope that these new genetic tools will help identify B. henselae virulence genes and that the pathogenesis of the organism can be studied more easily.
Acknowledgments
We are grateful to Lusijah Rott and Tim Knaak for their technical expertise with the FACS machines. We would like to extend special thanks to Joan Mecsas and Lalita Ramakrishnan for critical readings of the manuscript and to Denise Monack for graphics assistance. JK13 was a generous gift from Jane E. Koehler, and Gerhard Miksch provided pRS201.
Anthea Lee was funded by NIH training grants 5T32 GM07276-22 and 5T32 GM07276-23.
REFERENCES
- 1.Batterman H J, Peek J A, Loutit J S, Falkow S, Tompkins L S. Bartonella henselae and Bartonella quintana adherence to and entry into cultured human epithelial cells. Infect Immun. 1995;63:4553–4556. doi: 10.1128/iai.63.11.4553-4556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chomel B, Kasten R, Floyd-Hawkins K, Chi B, Yamamoto K, Roberts-Wilson J, Gurfield A, Abbott R, Pedersen N, Koehler J. Experimental transmission of Bartonella henselae by the cat flea. J Clin Microbiol. 1996;34:1952–1956. doi: 10.1128/jcm.34.8.1952-1956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conley T, Slater L, Hamilton K. Rochalimaea species stimulate human endothelial cell proliferation and migration in vitro. J Lab Clin Med. 1994;124:521–528. [PubMed] [Google Scholar]
- 4.Cormack B P, Valdivia R, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 5.Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol. 1997;179:538–540. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dehio C, Meyer M, Berger J, Schwarz H, Lanz C. Interaction of Bartonella henselae with endothelial cells results in bacterial aggregation on the cell surface and the subsequent engulfment and internalisation of the bacterial aggregate by a unique structure, the invasome. J Cell Sci. 1997;110:2141–2154. doi: 10.1242/jcs.110.18.2141. [DOI] [PubMed] [Google Scholar]
- 7.Haake D, Summers T, McCoy A, Schwartzman W. Heat shock response and groEL sequence of Bartonella henselae and Bartonella quintana. Microbiology. 1997;143:2807–2815. doi: 10.1099/00221287-143-8-2807. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 9.Heller R, Artois M, Xemar V, De Briel D, Gehin H, Jaulhac B, Monteil H, Piemont Y. Prevalence of Bartonella henselae and Bartonella clarridgeiae in stray cats. J Clin Microbiol. 1997;35:1327–1331. doi: 10.1128/jcm.35.6.1327-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins J A, Radulovic S, Jaworski D C, Azad A F. Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera: Pulicidae) J Med Entomol. 1996;33:490–495. doi: 10.1093/jmedent/33.3.490. [DOI] [PubMed] [Google Scholar]
- 11.Jameson P, Greene C, Regnery R, Dryden M, Marks A, Brown J, Cooper J, Glaus B, Greene R. Prevalence of Bartonella henselae antibodies in pet cats throughout regions of North America. J Infect Dis. 1995;172:1145–1149. doi: 10.1093/infdis/172.4.1145. [DOI] [PubMed] [Google Scholar]
- 12.Kessler B, de Lorenzo V, Timmis K. A general system to integrate lacZ fusions into the chromosomes of gram-negative Eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol Gen Genet. 1992;233:293–301. doi: 10.1007/BF00587591. [DOI] [PubMed] [Google Scholar]
- 13.Koehler J E. Bartonella infections. Adv Pediatr Infect Dis. 1996;11:1–27. [PubMed] [Google Scholar]
- 14.Koehler J E, Glaser C A. Rochalimaea henselae infections. JAMA. 1994;271:531–535. doi: 10.1001/jama.271.7.531. [DOI] [PubMed] [Google Scholar]
- 15.Midani S, Ayoub E M, Anderson B. Cat scratch disease. Adv Pediatr. 1996;43:397–422. [PubMed] [Google Scholar]
- 16.Miller V, Mekalanos J. A novel suicide vector and its use in constructions of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakawa, G. J., A. K. Lee, and S. Falkow. Unpublished data.
- 18.Schroder R, Maassen A, Lippoldt A, Borner T, von Baehr R, Dobrowolski P. Expression of the core antigen gene of hepatitis B virus (HBV) in Acetobacter methanolicus using broad-host-range vectors. Appl Microbiol Biotechnol. 1991;35:631–637. doi: 10.1007/BF00169628. [DOI] [PubMed] [Google Scholar]
- 19.Shinall E. Cat-scratch disease: a review of the literature. Pediatr Dermatol. 1990;7:11–18. doi: 10.1111/j.1525-1470.1990.tb01066.x. [DOI] [PubMed] [Google Scholar]
- 20.Suarez A, Guttler A, Stratz M, Staendne R L, Timmis K, Guzman C. Green fluorescent protein-based reporter systems for genetic analysis of bacteria including monocopy applications. Gene. 1997;196:69–74. doi: 10.1016/s0378-1119(97)00197-2. [DOI] [PubMed] [Google Scholar]
- 21.Valdivia R, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 22.Valdivia R H, Hromockyj A E, Monack D, Ramakrishnan L, Falkow S. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene. 1996;173:47–52. doi: 10.1016/0378-1119(95)00706-7. [DOI] [PubMed] [Google Scholar]