Abstract
To develop a simple, speedy, economical and widely applicable method for multiple-site mutagenesis, we have substantially modified the Quik-Change™ Site-Directed Mutagenesis Kit protocol (Stratagene, La Jolla, CA). Our new protocol consists of (i) a PCR reaction using an in vitro technique, LDA (ligation-during-amplification), (ii) a DpnI treatment to digest parental DNA and to make megaprimers and (iii) a synthesis of double-stranded plasmid DNA for bacterial transformation. While the Quik Change™ Kit protocol introduces mutations at a single site, requiring two complementary mutagenic oligonucleotides, our new protocol requires only one mutagenic oligonucleotide for a mutation site, and can introduce mutations in a plasmid at multiple sites simultaneously. A targeting efficiency >70% was consistently achieved for multiple-site mutagenesis. Furthermore, the new protocol allows random mutagenesis with degenerative primers, because it does not use two complementary primers. Our mutagenesis strategy was successfully used to alter the fluorescence properties of green fluorescent protein (GFP), creating a new-color GFP mutant, cyan-green fluorescent protein (CGFP). An eminent feature of CGFP is its remarkable stability in a wide pH range (pH 4–12). The use of CGFP would allow us to monitor protein localization quantitatively in acidic organelles in secretory pathways.
INTRODUCTION
Site-directed mutagenesis is a powerful tool for producing mutants to assess the importance of specific amino acid residues in a protein’s structure and function (1,2). A variety of protocols have been established to achieve efficient mutagenesis, including several that use the polymerase chain reaction (PCR) (3). Among the PCR-based protocols, the Quik-ChangeTM Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) is widely used. The kit protocol utilizes a supercoiled, double-stranded (ds) DNA plasmid and two complementary synthetic oligonucleotide primers containing a desired mutation. The oligonucleotide primers extend during temperature cycling by means of the high-fidelity Pfu TurboTM DNA polymerase. The product is then treated with DpnI endonuclease, which cuts fully- or hemi-methylated 5′-GATC-3′ sequences in duplex DNA, resulting in the selective digestion of the template DNA. The in vitro synthesized and nicked plasmid DNA including the desired mutation is then transformed into Escherichia coli. Although this protocol is simple, rapid, and efficient, it suffers from the following disadvantages.
(i) Mutations are introduced at only one site at a time (4,5); introduction of more than one mutation at different sites takes longer, since it requires a transformation and a DNA preparation step between consecutive rounds of mutagenesis.
(ii) Two complementary mutagenic oligonucleotide primers are needed for a mutation site. Also it does not allow random mutagenesis using degenerative primers.
The protocol presented here bypasses the above limitations, while keeping the simplicity and efficiency of the original protocol. The versatility of our new protocol was tested using Aequorea green fluorescent protein (GFP) (6) as a template for site-directed and semi-random mutagenesis.
The Aequorea GFP is a 238 amino acid, spontaneously fluorescent protein that has become popular in molecular and cell biology. Mutants of GFP with a variety of fluorescence properties have been produced using mutational strategies: random mutagenesis by error-prone PCR (7,8), DNA shuffling (9) and deliberate site-directed mutagenesis (10). The use of GFP for many applications strongly depends on its spectral properties. As a reporter of gene expression, a fusion tag and a partner for fluorescence resonance energy transfer (FRET) (11,12), GFP should be indifferent to environmental changes. Although GFP is stable at high temperature (Tm = 78°C), in chaotropic reagents (8 M urea) and against proteolysis, its spectrum is more or less affected by some factors, such as pH (13), ionic strength and protein concentration (14). Particularly the sensitivity to quenching by acidification prevents quantitative imaging; careful checking or clamping of the ambient pH has to be done to avoid pH-related artifacts (12). In this paper, using the new mutagenesis protocol we have evolved CFP (cyan fluorescent protein) into CGFP (cyan-green fluorescent protein), which shows slightly longer wavelengths of excitation and emission and is more resistant to acidification than the parental CFP.
MATERIALS AND METHODS
GFP variants
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with the accession number AB041904. Relative to wild-type GFP, the mutations in the GFP variants that were used as templates are: enhanced GFP (EGFP), F64L/S65T; ECFP, F64L/S65T/Y66W/N146I/M153T/V163A/N164H.
A fluorescence image analyzing system
We developed a fluorescence image analyzing system for bacterial colonies based on that reported by Baird et al. (15) with some modifications. An LB plate (9 cm in diameter) with fluorescent colonies was excited with light from a 150 W xenon lamp. The excitation wavelength was selected by a grating monochromator (CT-10, JASCO Corporation, Tokyo, Japan). To illuminate the plate evenly, a pair of fiber optic light guides (1 m) was used. The light emitted from the colonies was passed through an interference band-pass filter (Omega, Brattleboro, VT, 1 inch in diameter), collected with a lens (Nikon, Tokyo, Japan, AF NIKKOR). The image was captured by the SenSys cooled CCD camera (Photometrics, Tucson, AZ) and was analyzed using MetaMorph 3.0 software (Universal Imaging, West Chester, PA).
Site-directed mutagenesis for substitutions Y66W and T203Y on EGFP
The anti-sense primers for substitutions Y66W and T203Y were 5′-GCACTGCACGCCCCAGGTCAGGGTGGT-3′ and 5′-GGGCGGACTGGTAGCTCAGGTAGTGG-3′, respectively. They were phosphorylated at their 5′ end with T4 polynucleotide kinase. PCR was carried out in a 50 µl mixture using 50 ng template plasmid DNA [pRSETB (Invitrogen, Carlsbad, CA)/EGFP], 14 pmol of each primer, 10 nmol of dNTPs, 2.5 U of cloned Pfu DNA polymerase (Stratagene) in 0.5× Pfu polymerase reaction buffer, and 20 U of Taq DNA ligase (New England Biolabs, Beverly, MA) in 0.5× Taq DNA ligase buffer containing 50 nmol of NAD. The thermal cycler was programmed as follows: pre-incubation at 65°C for 5 min allowing the ligase to repair any nicks in the template; initial denaturation at 95°C for 2 min; 18 cycles at 95°C for 30 s, 55°C for 30 s and 65°C for 7 min; post-incubation at 75°C for 7 min. The time at 65°C was relatively long, so that the extended primers could be fully ligated. One microliter (20 U) of DpnI (New England Biolabs) was added to the sample (50 µl), and incubated at 37°C for 1 h. Then the sample (51 µl) was subjected to denaturation at 95°C for 30 s, followed by 2 cycles at 95°C for 30 s, 55°C for 1 min and 70°C for 7 min. Two microliters of the final sample was used to transform competent E.coli cells [JM109 (DE3)] (16) by the Ca2+-co-precipitation technique.
Random substitution at residue of 203 in ECFP
The degenerative primer was 5′-GCGGACTGNNNGCTCAGGTAG-3′. The three mutagenic positions (N) of the oligonucleotide were synthesized using equimolar concentrations of the four nucleoside phosphoramidites.
Protein expression and purification
Recombinant fluorescent proteins were expressed using the T7 expression system [pRSETB/JM109(DE3)]. Escherichia coli cells were transformed with the plasmids and selected at 37°C on LB plates containing 100 µg/ml ampicillin. A single colony was picked into 2 ml of LB medium containing 100 µg/ml ampicillin, and grown overnight at 37°C. A 100-fold dilution was made, and the culture was grown at room temperature until it reached a density of approximately OD600 = 0.5, and then protein expression was induced by isopropyl β-d-thiogalactoside (IPTG) at a final concentration of 0.2 mM. Growth was induced for a further 18 h. The cells were lysed by a French press, and the polyhistidine-tagged proteins were purified from the cleared lysates on nickel-chelate columns (Qiagen, Hilden, Germany). The protein samples in the eluates (100 mM imidazole, 50 mM Tris–Cl pH 7.5, 300 mM NaCl) were concentrated by Centricon 10 (Amicon, Bedford, MA), and were further purified by gel filtration.
pH titrations
A series of pH buffers were prepared with pHs ranging from 3.8 to 11 in either 50 mM acetate (pH 3.8–5.6), Na2HPO4-NaH2PO4 (pH 5.5–7.0), 2-(N-morpholino)ethanesulfonate (pH 5.5–6.0), 3-(N-morpholino)propanesulfonate (pH 6.5–7.0), HEPES (pH 7.0–7.9), glycine (pH 9.0–10.0), and phosphate (pH 11). The protein was concentrated in a weakly buffered solution, and mixed with an equal volume of the corresponding buffer solution.
Quantum yield (Φ) and molar extinction coefficient (ɛ)
These values were measured in 50 mM HEPES, pH 7.5. A quantum yield of enhanced cyan-green fluorescent protein (ECGFP) was determined by comparison to the standards: fluorescein (Φ = 0.91) and ECFP (Φ = 0.40). For the calculation of molar extinction coefficients, protein concentrations were measured using the Bradford assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard.
RESULTS AND DISCUSSION
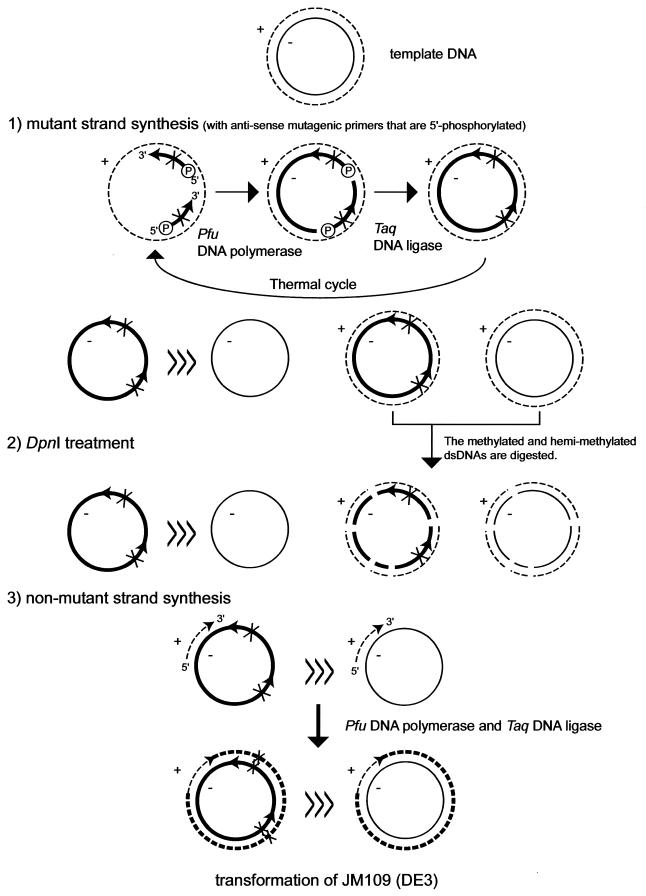
Figure 1 shows an overview of the protocol which is composed of the following three steps: (i) a PCR reaction, (ii) a DpnI digestion and (iii) a synthesis of ds plasmid DNA.
Figure 1.
Schematic diagram of the multiple-site mutagenesis protocol. The two mutagenic primers are designed on a minus strand. The methylated template DNA strands are shown by thin lines; the in vitro-synthesized DNA strands by thick lines. The plus and minus strands are drawn by broken and solid lines, respectively. Xs indicate introduced mutations. ‘>>>’ means that the species on the left is dominant in the reaction mixture.
The PCR reaction includes a thermostable DNA ligase (17,18) to ligate the multiple extended primers on a circular template in every cycle, generating circular single-stranded (ss) DNA molecules that carry mutations. The mutant strands dominate as ssDNA molecules in the reaction mixture. A small fraction of the species anneals to the sense strand of parental DNA, making a hemi-methylated dsDNA. Although Figure 1 shows two phosphorylated mutagenic primers on an anti-sense strand, more than two primers on either strand can be designed.
Wild-type template DNA is eliminated by DpnI digestion. The methylated (wild-type plasmid DNA) and hemi-methylated (a hybrid of the mutant strand and the anti-sense strand from wild-type DNA) dsDNAs are digested.
The non-mutant strands of some DNA fragments produced by DpnI digestion anneal to the ssDNAs synthesized in the PCR reaction, and serve as megaprimers to complete the synthesis of the remainder of the plasmid DNAs that are replicatively competent in bacteria. Also this reaction utilizes the Pfu polymerase, Taq DNA ligase, dNTPs, and NAD carried over from the PCR reaction.
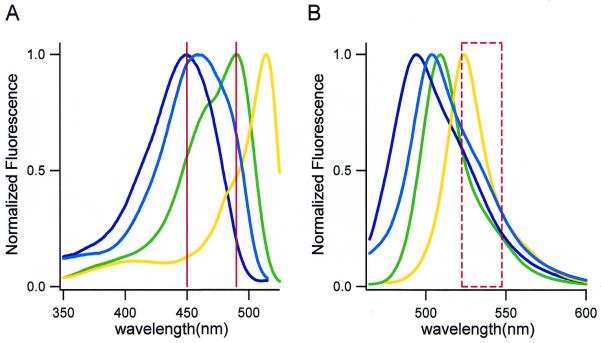
This protocol was used to carry out site-directed mutagenesis of GFP cloned in pRSETB (Invitrogen). An advantage of using GFP is that the efficiency of mutagenesis can be scored by the fluorescence or color of bacterial colonies producing the proteins. EGFP (Clontech) has a phenolate anion in the chromophore, showing excitation and emission peaks at 488 and 509 nm (Fig. 2, green lines). Of the 238 amino acids of EGFP, residues 66 (tyrosine) and 203 (threonine) were chosen for the mutagenesis, since the identity of the residue at these positions is known to be critical for the spectra. Substitution of Trp for Tyr66 (Y66W) produces a new chromophore with an indole. Because the major excitation and emission wavelengths are 450 and 494 nm (blue-green or cyan emission), the protein belongs to the CFP family (Fig. 2, blue lines). On the other hand, substitution of Tyr for Thr203 (T203Y) results in a π–π interaction (stacking) between the Tyr203 and the phenolate anion (Tyr66), shifting excitation and emission wavelengths to 514 and 524 nm, respectively. The protein is called yellow fluorescent protein (YFP) (Fig. 2, yellow lines). In a random mutagenesis experiment described later in this paper, we have shown that both the substitutions created a novel GFP mutant, CGFP, with excitation and emission wavelengths at 458 and 504 nm (Fig. 2, cyan lines).
Figure 2.
Fluorescence excitation (A) and emission (B) spectra of EGFP and its mutants: EGFP-Y66W, EGFP-T203Y and EGFP-Y66W/T203Y in cuvettes. Fluorescence was measured in 50 mM HEPES/KOH pH 7.5 at a protein concentration of 1 µM. They are depicted by green, blue, yellow and cyan lines, respectively. All spectra were normalized to a maximal value of 1.0. Differential excitation of fluorescent colonies was performed using two wavelengths, 450 and 490 nm, which are indicated by vertical red lines in (A). A band-pass filter (535DF25) is shown by a box drawn by a red dashed line.
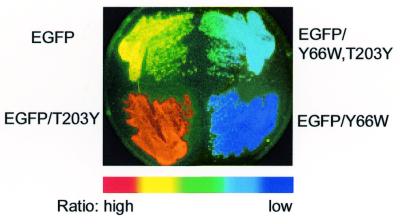
We have created a fluorescence image analyzing system to characterize the spectral qualities of mutant GFPs expressed in individual bacterial colonies growing on an agar plate. The colonies expressing EGFP, EGFP-Y66W, EGFP-T203Y and EGFP-Y66W/T203Y were illuminated at the excitation wavelengths at 450 and 490 nm (indicated by vertical lines in Fig. 2). The fluorescence images were taken using a band-pass filter (535DF25). The ratio image was obtained by dividing the 490 by the 450 nm image. As expected from their excitation spectra (Fig. 2), the ratio values: EGFP-T203Y > EGFP > EGFP-Y66W/T203Y > EGFP-Y66W (Fig. 3). Compared with an interference filter (15), a grating monochrometer permitted us to change and choose excitation wavelengths with very narrow width. Such differential excitation using a monochrometer in this system distinguished effectively among the jammed spectra of the four-colored GFP variants.
Figure 3.
An excitation ratio image (490/450) of fluorescent streaked bacterial cells producing EGFP (top left), EGFP-Y66W (bottom right), EGFP-T203Y (bottom left) and EGFP-Y66W/T203Y (top right).
We tried two simultaneous mutations (Y66W and T203Y) on EGFP using our new mutagenesis protocol. Figure 4 represents one of the typical images of colonies. Of 42 colonies that appeared on a LB plate containing ampicillin, 32 showed cyan-green fluorescence (EGFP-Y66W/T203Y); the efficiency of the double mutagenesis was calculated to be 76%. The rest of the colonies were as follows: four with EGFP-T203Y; four with EGFP-Y66W; and two with EGFP, the parental protein. There were no non-fluorescent colonies. Also, DNA sequencing concluded that no mutations had occurred outside of the mutagenic primers. The protocol (for Y66W and T203Y) was repeated four times; the number of the double mutants/total number of clones analyzed was 21/30, 42/50, 50/72, 32/40. The overall efficiency of the double mutagenesis was 75.8 ± 4.2% (n = 5).
Figure 4.
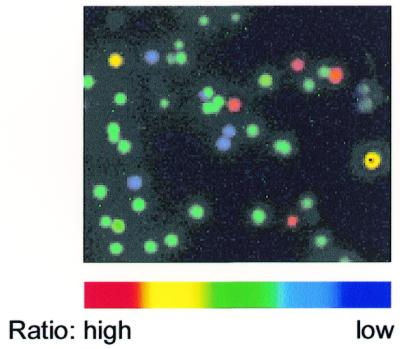
An excitation ratio image (490/450) of fluorescent colonies expressing EGFP with the mutagenized substitutions Y66W and T203Y.
The target sequence of the DpnI endonuclease (5′-Gm6ATC-3′) is predicted to occur, on average, every 256 bp. Complete cleavage of a 3672 bp plasmid pRSETB/EGFP gives 23 DNA fragments of 6, 8, 11, 12, 17, 18, 24, 31, 36, 38, 46, 47, 75, 78, 105, 133, 148, 160, 258, 341, 506, 740 and 789 bp. Following denaturation at 95°C, sense strands of the digested DNAs annealed to the circular ssDNAs (anti-sense strands), then worked as primers. Because all the sense strands are derived from parental DNA, the primers should anneal in the plasmid outside the mutated sites. In our experiment, the sense strands of 506 and 148 bp fragments are required not to work as the primers, since their coding regions contain the residues 66 and 203, respectively. Addition of a common sense-primer that anneals outside the cloning site (14 pmol of T7 primer) after DpnI digestion increased the efficiency of the mutagenesis to >90%. In an experiment, out of 200 colonies, 196 showed the fluorescence of EGFP-Y66W/T203Y.
Site-directed mutagenesis by whole-plasmid amplification (the Quik-ChangeTM Site-Directed Mutagenesis Kit) relies on the principle that an entirely complementary primer pair containing a desired mutation generates a PCR product with 5′ overhangs at both ends, which can anneal, and subsequently be circularized and propagated in E.coli. For introduction of more than one point mutation at different sites, however, this method requires time-consuming intermittent transformation and screening steps (4,5). Two separate protocols to circumvent these steps have been described previously.
The use of non-overlapping oligonucleotides for making megaprimers was shown to mutagenize two separate sites simultaneously (4). Although the method saves money (two primers for two mutation sites), simultaneous mutagenesis at more than two sites cannot be performed.
In vitro methylation/ligation at 37°C using DpnI and T4 DNA ligase between successive PCR amplifications with different primer pairs can increase the number of independent mutations with a single transformation and DNA preparation step (5). However, isolation of the DpnI-resistant DNA from agarose gel slices in each round is cumbersome and time consuming. Also this protocol requires as many oligonucleotides as the basic Quik ChangeTM protocol.
Our new protocol, in contrast, requires one mutagenic primer at one site. The primers are designed to be complimentary to either strand of a template DNA. To make circular ssDNAs containing mutations at different sites in a PCR reaction, we utilized an in vitro technique, LDA (ligation during amplification) (18). After the mutagenic primer(s) are fully extended on a template, the gaps must be closed, otherwise the mutant strands are scattered in the following denaturation step at 95°C. In fact, we obtained no bacterial colonies when we omitted the thermo-stable ligase in our protocol. Mutation of EGFP at up to four different sites was achieved efficiently (>70%), when the second strand synthesis was primed by the standard sense primer (T7 primer) (data not shown).
Directed evolution is effective for exploring and optimizing protein functions (19). Random mutagenesis is a powerful means of creating gene libraries for accumulation of beneficial mutations. The use of degenerative primers in the Quik ChangeTM Kit protocol produces circular dsDNA molecules with ‘bubbles’; the randomized regions cannot anneal completely (20). The formation of bubble blocks replication of the PCR products in bacterial cells. By contrast, since our method utilizes a single mutagenic primer for a mutation site, there is no concern about the bubble formation. It is well suited for random mutagenesis using degenerative primers. When more than one degenerative primer is used, it is possible to introduce random mutagenesis at multiple specific sites simultaneously. Our protocol can expand the diversity of products. Our combination and/or modification of existing protocols allows efficient, semi-random mutagenesis to be completed within several hours, making this a method amenable to automation.
Random-mutant libraries were constructed using ECFP as a template. The CFP has a chromophore with an indole, and shows double-humped excitation and emission spectra (excitation peaks, 434 and 452 nm; emission peaks, 476 and 505 nm), suggesting there are two equilibrated quantum states. It is interesting to introduce the amino acid substitutions that affect the equilibrium, thereby shifting the spectra of ECFP. We hypothesized that the replacement of Thr203 with other amino acids, particularly with aromatic ones, could achieve that. A degenerative primer for the random mutagenesis (see Materials and Methods) was used. Sixty-two clones were randomly picked up and sequenced. Among them 28 clones produced non-fluorescent proteins with substitution of K, N, H, R, L for Thr203. Thirty-four clones produced fluorescent proteins, of which 23 contained mutations that substituted P, V, I, W, F, S, C, Y for the Thr203. A remarkable spectral shift was observed in the mutant of T203Y. This substitution in EGFP caused π–π stacking between Tyr66 and Try203, and increased both the excitation and emission wavelengths of EGFP (10). Likewise, the same substitution in ECFP shifted the wavelengths longer, presumably due to π–π stacking between Trp66 and Try203. The mutant was referred to as ECGFP (enhanced cyan-green fluorescent protein), because its overall excitation and emission spectra were intermediate between those of ECFP and EGFP.
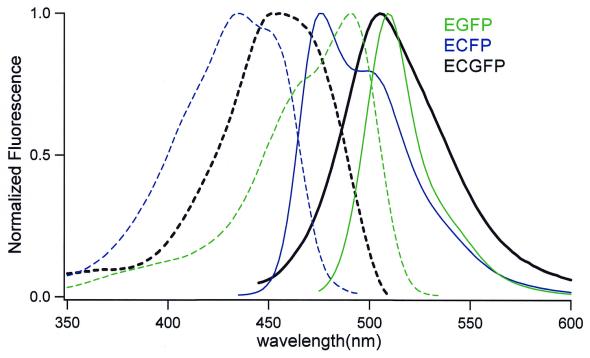
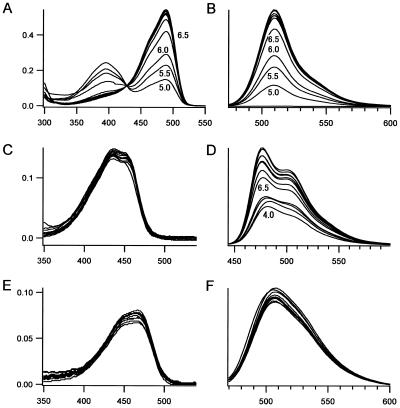
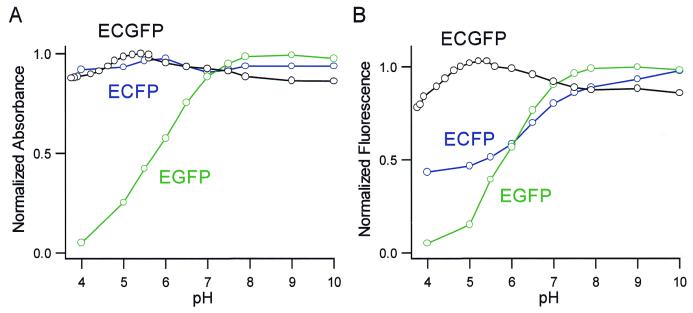
The fluorescence excitation and emission spectra of ECGFP are compared with those of ECFP and EGFP in Figure 5. Interestingly, the excitation and emission peaks of ECGFP (455 and 506 nm) were fairly close to those of the long wavelength component of ECFP (452 and 505 nm). It appeared that the π–π stacking favored the quantum state in which the longer wavelength predominated. Figure 5 also indicates that ECGFP and EGFP have similar emission peaks (ECGFP, 506 nm; EGFP, 511 nm), but that ECGFP exhibits a larger Stokes’ shift (the gap in wavelength between excitation and emission peaks). Compared with EGFP-Y66W/T203Y (Fig. 2), ECGFP has additional substitutions (N146I, M153T, V163A) that help folding and maturation of the protein. The absorbance extinction coefficient (ɛ) and quantum yield (Φ) of ECGFP were measured at pH 7.5, and compared with those of EGFP and ECFP (Table 1). Since the overall brightness is the product of ɛ and Φ, ECGFP is dimmer than EGFP or ECFP at neutral pH. It is agreed that almost all GFP mutants are quenched to some extent by acidic pH. We examined pH-dependency of the three GFP mutants in vitro. EGFP showed an acidification-dependent decrease in the absorbance peak at 488 nm and a concomitant increase in the absorbance at 398 nm (Fig. 6A). The emission (511 nm peak) and excitation (488 nm peak) spectra decreased with decreasing pH, but the fluorescence excitation spectrum showed no compensating increase at 398 nm. Thus the species absorbing at 398 nm was non-fluorescent. EGFP showed a pKa of 6.0 for both its absorbance and emission spectra (Fig. 6A and B), indicating that its absorbance not its quantum yield is pH-sensitive (Fig. 7) (21). ECFP was relatively less pH-sensitive. Decreasing pH quenched the emission (Fig. 6D) with little effect on its absorbance spectra (Fig. 6C), indicating that its quantum yield is depressed by acid, but to a lesser extent compared with the pH-sensitivity of EGFP (Fig. 7) (21). By contrast, ECGFP was outstandingly stable in a wide pH range (pH 4–12) (Fig. 6E and F). Even when pH was decreased to 4, there was only 10% reduction in its fluorescence (Fig. 7). For the analysis of intracellular membrane dynamics of acidic organelles, including secretory and endocytic organelles, the fluorescence of ECGFP would faithfully report the number of the molecules being imaged.
Figure 5.
Excitation (dotted lines) and emission (solid lines) spectra of ECFP (cyan), ECGFP (black) and EGFP (green). All amplitudes have been normalized to a maximum value of 1.0.
Table 1. Spectra characterizations of EGFP, ECFP and ECGFP.
| Name | Mutation | λabs(ɛ)a | λem(Φ)b | pKac | References |
|---|---|---|---|---|---|
| EGFP | F64L, S65T | 488 (55–57) | 507–509 (0.60) | 6 | 22 |
| ECFP | F64L, S65T, Y66W, N146I, M153T, V163A, N164H | 434 (33.9) 452 | 476 (0.40) 505 | 4–6 | 22 |
| ECGFP | F64L, S65T, Y66W, N146I, M153T, V163A, N164H, T203Y | 463 (23.9) | 506 (0.14) | <4 |
aλabs is the peak of the absorbance spectrum in units of nanometers. ɛ in parentheses is the absorbance extinction coefficient in units of 103 M–1 cm–1.
bλem is the peak of emission spectrum in units of nanometers. Φ in parentheses is the fluorescence quantum yield.
cKa is the acid dissociation constant, and pKa = –log10Ka. pKas were estimated from Figure 7B.
Figure 6.
pH-dependency of absorbance (A, C and E) and fluorescence emission (B, D and F) spectra of EGFP (A and B), ECFP (C and D) and ECGFP (E and F). Excitation was at 450 nm for EGFP, 430 nm for ECFP and 450 nm for ECGFP.
Figure 7.
(A) pH-dependency of normalized peak amplitudes of absorbance of EGFP (488 nm), ECFP (435 nm) and ECGFP (463 nm). (B) pH-dependency of fluorescence emission of EGFP, ECFP and ECGFP. Because the emission spectrum of ECFP changes its shape with pH, the peak areas were normalized and plotted.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs David Zacharias and Kevin Truong for critical comments on the manuscript. We thank Drs H. Mizuno and T. Nagai for fruitful discussions. This work was partly supported by grants from CREST of JST (Japan Science and Technology) and the Japanese Ministry of Education, Science and Culture.
DDBJ/EMBL/GenBank accession no. AB041904
REFERENCES
- 1.Ishii T.M., Zerr,P., Xia,X., Bond,C.T., Maylie,J. and Adelman,J.P. (1998) Methods Enzymol., 293, 53–71. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel T.A., Bebenek,K. and McClary,J. (1991) Methods Enzymol., 204, 125–139. [DOI] [PubMed] [Google Scholar]
- 3.Saiki R.K., Scharf,S., Faloona,F., Mullis,K.B., Horn,G.T., Erlich,H.A. and Arnheim,N. (1985) Science, 230, 1350–1354. [DOI] [PubMed] [Google Scholar]
- 4.Kirsch R.D. and Joly,E. (1998) Nucleic Acids Res., 26, 1848–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y.-G. and Maas,S. (2000) Biotechniques, 28, 196–198. [DOI] [PubMed] [Google Scholar]
- 6.Tsien R.Y. (1998) Annu. Rev. Biochem., 67, 509–544. [DOI] [PubMed] [Google Scholar]
- 7.Heim R., Prasher,D.C. and Tsien,R.Y. (1994) Proc. Natl Acad. Sci. USA, 91, 12501–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heim R. and Tsien,R.Y. (1996) Curr. Biol., 6, 178–182. [DOI] [PubMed] [Google Scholar]
- 9.Crameri A., Whitehorn,E.A., Tate,E. and Stemmer,W.P.C. (1996) Nature Biotechnol., 14, 315–319. [DOI] [PubMed] [Google Scholar]
- 10.Ormö M., Cubitt,A.B., Kallio,K., Gross,L.A., Tsien,R.Y. and Remington,S.J. (1996) Science, 273, 1392–1395. [DOI] [PubMed] [Google Scholar]
- 11.Miyawaki A., Llopis,J., Heim,R., McCaffery,J.M., Adams,J.A., Ikura,M. and Tsien,R.Y. (1997) Nature, 388, 882–887. [DOI] [PubMed] [Google Scholar]
- 12.Miyawaki A., Griesbeck,O., Heim,R. and Tsien,R.Y. (1999) Proc. Natl Acad. Sci. USA, 96, 2135–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llopis J., McCaffery,J.M., Miyawaki,A., Farquhar,M.G. and Tsien,R.Y. (1998) Proc. Natl Acad. Sci. USA, 95, 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward W.W., Prentice,H.J., Roth,A.F., Cody,C.W. and Reeves,S.C. (1982) Photochem. Photobiol., 35, 803–808. [Google Scholar]
- 15.Baird G.S., Zacharias,D.A. and Tsien,R.Y. (1999) Proc. Natl Acad. Sci. USA, 96, 11241–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Studier F.W., Rosenberg,A.H., Dunn,J.J. and Dubendorff,J.W. (1990) In Goeddel,D.V. (ed.), Methods in Enzymology, Vol. 185. Academic Press, San Diego, CA, pp. 60–89. [DOI] [PubMed]
- 17.Michael F.S. (1994) Biotechniques, 16, 411–412. [Google Scholar]
- 18.Chen Z. and Ruffner,D.E. (1998) Nucleic Acids Res., 26, 1126–1127. [PMC free article] [PubMed] [Google Scholar]
- 19.Shao Z. and Arnold,F.H. (1996) Curr. Opin. Struct. Biol., 6, 513–518. [DOI] [PubMed] [Google Scholar]
- 20.Parikh A. and Guengerich,F.P. (1998) Biotechniques, 24, 428–431. [DOI] [PubMed] [Google Scholar]
- 21.Miyawaki A. and Tsien,R.Y. (2000) In Thorner,J., Emr,S.D. and Abelson,J.N. (eds), Methods in Enzymology, Vol. 327. Academic Press, San Diego, CA, pp. 472–500. [DOI] [PubMed]
- 22.Cubitt A.B., Heim,R. and Woollenweber,L.A. (1999) In Sullivan,K.F. and Kay,S.A. (eds), Methods in Cell Biology, Vol. 58. Academic Press, pp. 19–30. [DOI] [PubMed]