Abstract
Giant Axonal Neuropathy (GAN) is a pediatric-neurodegenerative disease caused by KLHL16 mutations. KLHL16 encodes gigaxonin, which regulates intermediate filament turnover. Previous neuropathological studies and examination of postmortem brain tissue in the current study revealed involvement of astrocytes in GAN. To develop a clinically relevant model, we reprogrammed skin fibroblasts from seven GAN patients to pluripotent stem cells (iPSCs), which were used to generate neural progenitor cells (NPCs), astrocytes, and brain organoids. Multiple isogenic control clones were derived via CRISPR/Cas9 gene editing of one patient line carrying the G332R gigaxonin mutation. All GAN iPSCs were deficient for gigaxonin and displayed patient-specific increased vimentin expression. GAN NPCs had lower nestin expression and fewer nestin-positive cells compared with isogenic controls, but nestin morphology was unaffected. GAN brain organoids were marked by the presence of neurofilament and GFAP aggregates. GAN iPSC-astrocytes displayed striking dense perinuclear vimentin and GFAP accumulations and abnormal nuclear morphology. In overexpression systems, GFAP oligomerization and perinuclear aggregation were augmented in the presence of vimentin. GAN patient cells with large perinuclear vimentin aggregates accumulated significantly more nuclear KLHL16 mRNA compared with cells without vimentin aggregates. As an early effector of KLHL16 mutations, vimentin may be a potential target in GAN.
INTRODUCTION
Giant Axonal Neuropathy (GAN) is a rare, progressive, pediatric neurodegenerative disease that affects both the peripheral nervous system (PNS) and the central nervous system (CNS; Johnson-Kerner et al., 2014; Armao et al., 2021; Bharucha-Goebel et al., 2021). In children with GAN, early milestone development is normal (Hentati et al., 2013). Disease onset is usually 3–4 years of age and is often heralded by clumsiness of gait. By the end of the second decade of life, most patients are nonambulatory and have limited use of their arms and little to no use of their legs (Armao et al., 2021). Death nearly always occurs by the third decade. The clinical findings and pathologic distribution of lesions in GAN are consistent with a central-peripheral distal axonopathy (Dubeau et al., 1985; Thomas et al., 1987).
GAN is caused by recessive loss-of-function mutations in the KLHL16 gene (also known as GAN) that encodes gigaxonin, an E3 ubiquitin ligase adaptor protein (Bomont et al., 2000). While the exact function of gigaxonin remains to be defined, current understanding is that gigaxonin facilitates degradation of many members of the intermediate filament (IF) gene family, which are cell-type specific cytoskeleton components (Mahammad et al., 2013; Lin et al., 2016; Büchau et al., 2018). In the absence of functional gigaxonin, IF proteins accumulate in many cell types, including melanocytes, endothelial cells, lens epithelial cells, Schwann cells, astrocytes, and neurons (Peiffer et al., 1977; Kretzschmar et al., 1987; Mohri et al., 1998; Armao et al., 2019). In 1972, the first reported case of GAN was described in a six-year-old child with an unusual, slowly progressive, sensorimotor neuropathy, and the details of the nerve biopsy–fiber loss and “masses of neurofilaments distending many of the axons to giant proportions” –christened the disease (Asbury et al., 1972). Since that time, validation of therapeutic efficacy and viral vector delivery systems in GAN knock-out (KO) rodent models (Ding et al., 2006; Dequen et al., 2008; Ganay et al., 2011) have provided the springboard for a first-in-human phase I clinical trial of intrathecal gene transfer of KLHL16. To date, most attention has been directed at IF aggregates within GAN neurons. Far less attention has been directed at IF aggregates within GAN astrocytes, despite the fact that conspicuous and abundant IF aggregates (Rosenthal fibers [RFs]) are described within astrocytes in every reported autopsy case of GAN (Peiffer et al., 1977; Dubeau et al., 1985; Kretzschmar et al., 1987; Thomas et al., 1987; Kumar et al., 1990). The pathogenesis of IF proteostasis failure in GAN astrocytes, and its relationship to IF proteostasis failure in GAN neurons, remains underinvestigated.
In this work, we use patient-derived induced pluripotent stem cells (iPSCs) and directed differentiation to develop human astrocyte and brain organoid models of GAN. These models reproduce astrocyte IF protein aggregation comparable to astrocytes of GAN patients in vivo. The GFAP IF protein inclusions observed in GAN iPSC-astrocytes are accompanied by nuclear abnormalities, similar to those recently reported in Alexander Disease (AxD) iPSC-astrocytes (Battaglia et al., 2019), suggesting possible shared mechanisms. Additionally, we reveal new mechanisms regarding the role of vimentin in GFAP aggregation and KLHL16 mRNA localization in cells.
RESULTS
Astrocytopathy in human GAN
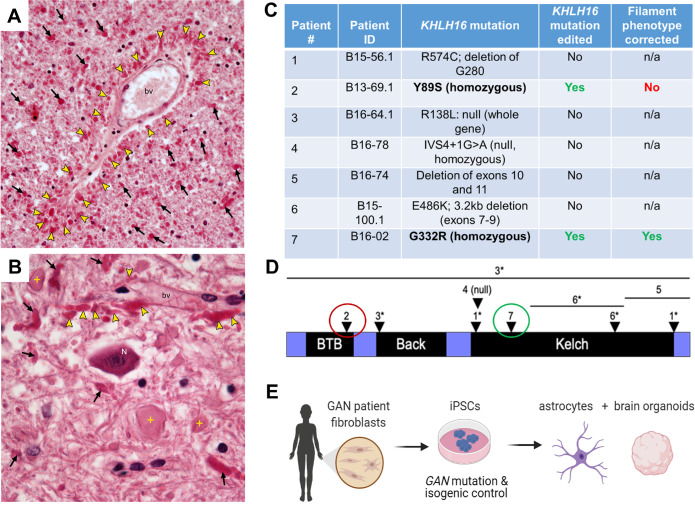
Postmortem histologic examination of cerebral white matter in a GAN patient revealed extensive astrocyte involvement with the presence of innumerable RFs and reactive astrocytosis (Figure 1, A and B). RFs were concentrated in astrocyte end feet around blood vessels (bv; Figure 1A) and dispersed in cerebellar white matter in proximity to giant axonal swellings (Figure 1B). These data clearly demonstrate that KLHL16 loss-of-function mutation leads to astrocytes acquiring marked histopathologic features, in agreement with early neuropathologic observations in GAN (Peiffer et al., 1977; Dubeau et al., 1985; Kretzschmar et al., 1987; Thomas et al., 1987; Kumar et al., 1990). To examine the link between KLHL16/gigaxonin dysfunction and astrocytopathy, we reprogrammed seven skin fibroblast lines from GAN patients with different KLHL16 mutations (Figure 1C; Supplemental Figure S1) to iPSCs. The GAN cell lines contained unique KLHL16 mutations that span all functional domains of gigaxonin (Lescouzères and Bomont, 2020), including null mutations (patients 3, 4), mutations in the BTB (patient 2), Back (patient 3), and Kelch (patients 1, 5, 6, and 7) domains (Figure 1D). Isogenic controls of the two lines carrying homozygous missense mutations: patient 2 (Y89S) and patient 7 (G332R) were generated by CRISPR/Cas9 gene editing (Supplemental Figure S2, A–F). The isogenic control cells from patient 7 showed the expected phenotypic correction of IF morphology (Supplemental Figure S2G). Despite the correction of the known Y89S mutation in patient 2, gigaxonin was not restored and IF protein aggregation persisted in the corresponding isogenic control iPSC-astrocytes (Supplemental Figure S2, H and I). According to the ClinVar archive (Landrum et al., 2016), nearly 40% of KLHL16/GAN gene mutations are in the untranslated region (UTR) and most of these mutations are of unknown significance. At this time we cannot rule out the possibility of noncoding KLHL16 mutations being present in GAN patient 2. For these reasons, the majority of this work focused on patient 7 carrying the G332R Kelch-domain mutation and corresponding isogenic clones that had a revertant phenotype. Because the Kelch domain mediates the associations between gigaxonin and IF proteins (Mahammad et al., 2013; Johnson-Kerner et al., 2015b), we specifically focused on IF phenotypes in iPSC, NPCs, astrocytes, and brain organoids (Figure 1E) generated from patient 7 cells and isogenic controls.
FIGURE 1:
Astrocytopathy in human GAN, and modeling GAN-related astrocytopathy using patient-derived iPSCs. (A) Abundant, bright red, oval and club-shaped astrocyte inclusions, termed Rosenthal fibers (RFs; arrowheads) are clustered around a blood vessel (bv) in GAN. Cerebral white matter shows large numbers of RFs (arrows), which are indicative of IF proteostasis failure and an astrocytopathy (H&E;40X). (B) RFs are concentrated in astrocyte endfeet (arrowheads) around a blood vessel (bv). RFs and giant axonal swellings (crosses) are dispersed in cerebellar white matter surrounding the dentate nucleus. A neuronal cell body (N) is morphologically unremarkable (H&E; 60X). Decedent was a young child with phenotypically classic GAN. (C) Summary of GAN patient cell lines with their known mutations that were used in the study. The two lines carrying homozygous missense mutations (Y89S and G332R) were edited to generate isogenic clones. Only one (patient 7) of the two isogenic lines displayed normal filaments upon differentiation and was used in subsequent experiments. (D) Schematic of human KLHL16 mutations mapped to the protein, gigaxonin. Numbers 1–7 refer to the patient donor cells that were used in this study; listed in Panel C. Asterisks indicate compound heterozygous mutations. Red circle indicates isogenic line was generated but retained abnormal IF phenotype. Green circle indicated isogenic line was generated and resulted in restored IF phenotype. Please also see Supplemental Figure 2. (E) Schematic diagram of methods used to generate isogenic control and GAN iPSC-astrocytes and brain organoids from Patient 7 (G332R).
Patient-specific upregulation and perinuclear bundling of vimentin in GAN iPSCs
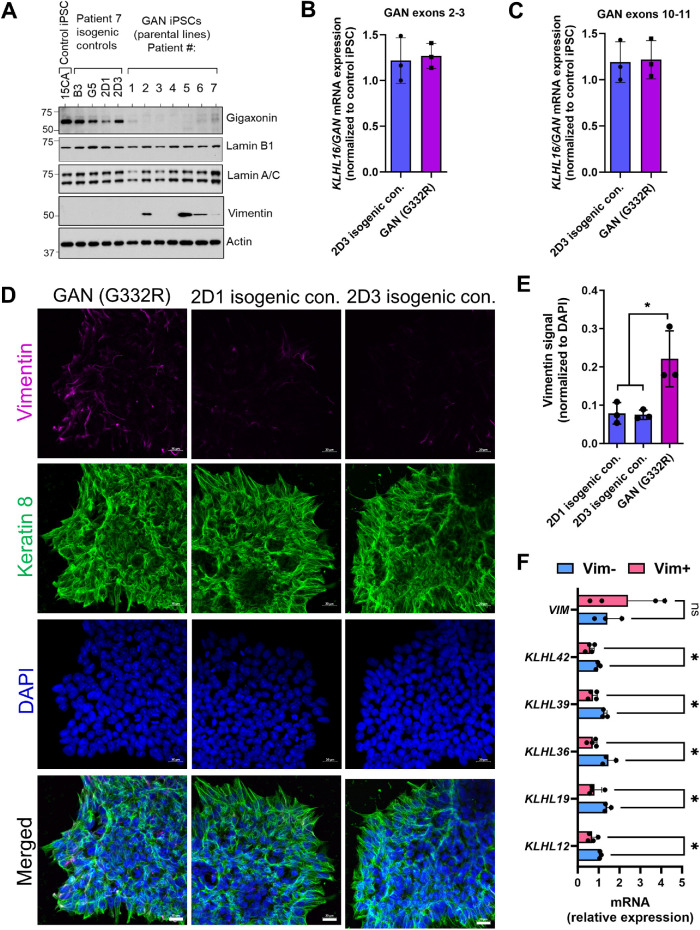
We observed by immunoblot analysis that gigaxonin was largely undetectable or extremely low in all seven GAN iPSC lines when compared with a non-GAN control iPSC line (15CA) as a reference (Molina et al., 2020). Gigaxonin expression in the four independent isogenic clones of patient 7 (B3, G5, 2D1, and 2D3) was restored to levels similar of those in the 15CA non-GAN control (Figure 2A). Notably, there was no difference in the total KLHL16 mRNA expression between GAN and isogenic control cells (Figure 2, B and C), ruling out upregulation in transcription as the mechanism behind restored gigaxonin expression. We did not observe major differences in lamin-A/C and B1 protein expression (Figure 2A) or Lamin B1 localization (Supplemental Figure S3) between the parental GAN G332R cells versus the isogenic 2D3 control cells. However, by immunoblot we observed variable vimentin expression, such that four patient lines (#2, 5, 6, and 7) expressed detectable levels of vimentin (Vim+), while the remaining patient lines (#1,3, and 4) did not express vimentin (Vim–), similar to the 15CA non-GAN control and the patient 7 isogenic clones (Figure 2A). Immunofluorescence analysis of the GAN patient G332R iPSCs and two isogenic clones (2D1 and 2D3) also showed that vimentin expression was significantly higher in iPSCs colonies of the unedited parental line compared with the isogenic controls (p < 0.05; one-way ANOVA) (Figure 2, D and E). Keratin 8 filament staining was used as another cytoplasmic IF marker of the iPSC colonies (Figure 2D), appearing filamentous across lines, although detailed ultrastructural analysis and quantitative examination of the keratin network dynamics were not performed as part of this study. Differences in vimentin mRNA (VIM) expression did not account for vimentin protein variation among patient lines (Figure 2F). We also tested by qPCR the expression of all KLHL genes using sequence-specific primers (Supplemental Table S3) and observed that the expression of five KLHL genes (KLHL-12, 19, 36, 39, and 42) was significantly decreased in the Vim+ compared with the Vim– iPSCs (Figure 2F). These results suggest that the phenotypic variability in vimentin expression in the setting of KLHL16 mutations may potentially be influenced by the expression of related KLHL genes.
FIGURE 2:
Human KLHL16 loss-of-function mutations selectively alter vimentin expression in iPSCs. (A) Immunoblotting for gigaxonin and IF proteins lamin B1, lamin-A/C and vimentin in total lysates from non-GAN control (15CA), four isogenic iPSC controls of GAN Patient #7 (B3,G5,2D1,2D3) and seven unedited GAN iPSCs parental lines (patients 1–7). Actin blot serves as protein loading control. (B) KLHL16/GAN gene expression in GAN (G332R) and 2D3 isogenic control iPSCs by qRT-PCR using primers targeting exons 2–3. (C) KLHL16/GAN gene expression in GAN (G332R) and 2D3 isogenic control iPSCs by qRT-PCR using primers targeting exons 10-11. (D) Immunofluorescence analysis of vimentin (magenta), Keratin 8 (green) and DAPI (blue) in patient 7 (G332R) and isogenic control iPSC clones 2D1 and 2D3 from the same patient. Scale bars = 20 μm. (E) Vimentin immunofluorescence intensity quantified by Image J. n = 3 fields of view per condition (equal 1228800 pixel areas measured and normalized to DAPI signal) *p < 0.05; One-way ANOVA. (F) Quantitative real-time PCR analysis of VIM and KLHL gene expression in Vim– (n = 3; patients 1,3,4) vs Vim+ (n = 4; patients 2,5,6,7) GAN iPSCs. n.s., not significant; *p < 0.05; multiple t-tests.
Reduced nestin expression in GAN neural progenitor cells (NPCs) compared with isogenic control
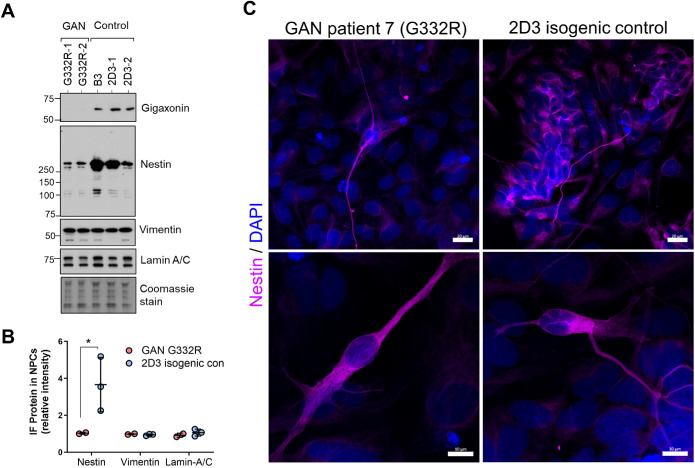
Next, we differentiated the iPSCs to NPCs, which express the type IV IF protein nestin. Immunoblot analysis of IF proteins revealed similar expression of lamin-A/C and vimentin, but three-fold higher (p < 0.05; two-way ANOVA) nestin expression in the 2D3 isogenic controls compared with the parental line G332R (Figure 3, A and B), which was corroborated by immunofluorescence analysis (Figure 3C). These data suggest GAN iPSCs have reduced potential to differentiate to nestin-positive NPCs in vitro, which may be linked to the previously reported abnormality in the Sonic hedgehog (Shh) pathway in GAN (Arribat et al., 2019b).
FIGURE 3:
Reduced nestin expression in GAN mutant neural progenitor cells (NPCs) compared to isogenic controls. (A) Immunoblotting for gigaxonin, nestin, vimentin, and lamin A/C in GAN (n = 2) and isogenic (n = 3) NPCs derived from Patient #7 (G332R). Coomassie stain serves as total protein loading control. (B) Quantification of band intensities for G332R (n = 2) and control (n = 3) from panel A. *p < 0.05 two-way ANOVA. (C) Confocal images of immunofluorescence of NPCs stained for nestin (magenta) and DAPI (blue). Scale bars = 20 μm (top) and 10 μm (bottom).
Perinuclear accumulations of vimentin and GFAP in GAN iPSC-astrocytes
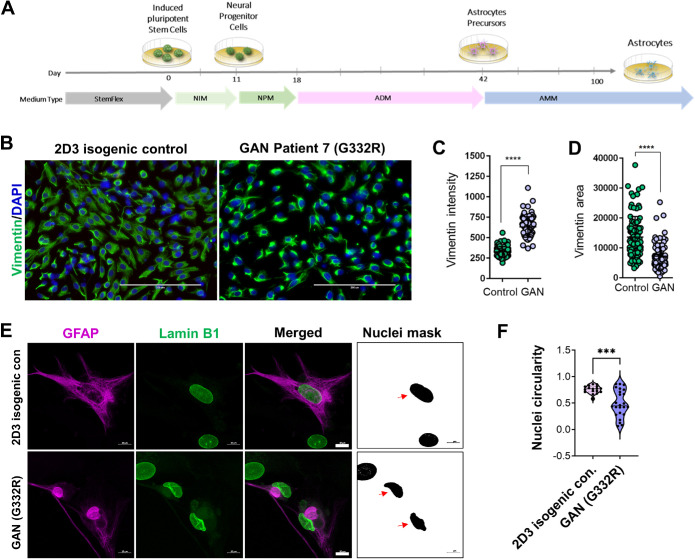
Given the observed early progenitor phenotypes, we next asked whether GAN NPCs could be differentiated to astrocytes in culture. Using defined differentiation media (Battaglia et al., 2019), we generated GAN (G332R) and corresponding 2D3 isogenic control vimentin-positive astrocytes, which were analyzed at day 60 of the differentiation procedure (Figure 4A). One major difference between control and GAN iPSC-astrocytes at this stage was the presence of large, intensely stained, perinuclear vimentin accumulations in the GAN astrocytes (Figure 4, B and C) that was not due to cell death or overall change in cell size (Supplemental Figure S4). Consistent with this perinuclear concentration, the vimentin-occupied areas were significantly smaller in the GAN mutant cells (Figure 4D). At this time point of differentiation (60 d), the marker of mature astrocytes aldehyde dehydrogenase 1 family, member L1 (ALDHL1) was present in 12 and 10% in the isogenic and mutant cells, respectively (Supplemental Figure S5, A and B). ALDH1L1 is normally expressed throughout the cytoplasm, but appeared to coaggregate with vimentin in some GAN cells (Supplemental Figure S5C). This may be due to the severely altered cytoplasmic structure of GAN cells, but will need to be addressed in detail in future studies. GFAP-positive cells comprised ∼10 and 7% in the isogenic and mutant cells, respectively, compared with the total number of cells (Supplemental Figure S6, A and B). The GFAP-positive GAN iPSC-astrocytes contained large perinuclear GFAP aggregates and displayed misshapen nuclei along with weak and diffuse abnormal cytoplasmic lamin B1 staining (Figure 4E). This phenotype was in stark contrast to the isogenic control astrocytes, which displayed normal GFAP IF organization, normal nuclear morphology, and normal lamin B1 staining. Quantitative analysis of the lamin B1-stained nuclei in the 2D3 isogenic control and G332R GAN GFAP+ astrocytes revealed significant decrease in the circularity of GAN nuclei (Figure 4F; Supplemental Figure S6C).
FIGURE 4:
GAN iPSC-astrocytes display perinuclear vimentin and GFAP aggregates and abnormal nuclei. (A) Schematic illustrating the workflow for iPSC differentiation to astrocytes. (B) Vimentin bundling in immature GAN iPSC-astrocytes analyzed at day 60 of the differentiation timeline. Scale bars = 200 μm. (C) Quantification of the vimentin signal intensity in control and GAN iPSC-astrocytes. ****p < 0.0001; unpaired t-test. (D) Quantification of vimentin area in control and GAN astrocytes. p < 0.0001; unpaired t-test. (E) GAN and isogenic control iPSC-astrocytes were stained for GFAP (magenta) and lamin B1 (green). Scale bars = 10 μm. Nuclei mask reveals changes in nuclear shape that were quantified by ImageJ. (F) Circularity of nuclei in GAN compared to 2D3 isogenic control cells. ***p < 0.001; unpaired t-test.
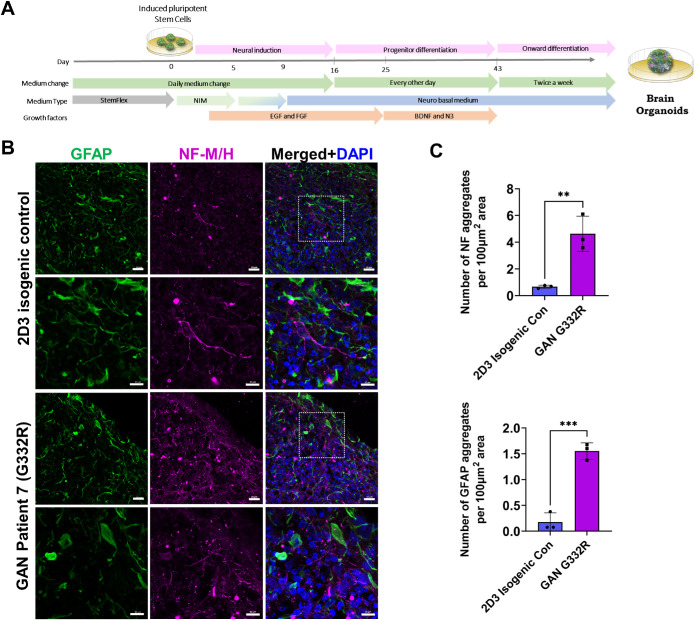
GAN brain organoids express GFAP and neurofilaments (NF) and exhibit cytoskeletal and nuclear abnormalities
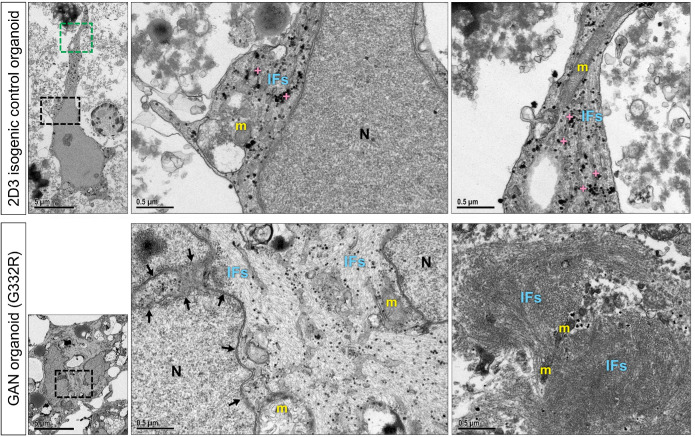
Because few iPSC-astrocytes expressed GFAP in the monoculture system, we sought a new model to generate greater numbers of GFAP-positive astrocytes. To that end, we differentiated iPSCs to brain organoids using an established protocol (Figure 5A; Pasca et al., 2015). After 7 mo of in vitro culture, the isogenic (2D3 clone) and the corresponding mutant (G332R mutation) GAN brain organoids were enriched in GFAP-positive astrocytes that were integrated with other cell types, including NF-expressing neurons (Figure 5B). The GAN organoids had increased numbers of NF and GFAP aggregates compared with the 2D3 isogenic control organoids (Figure 5C; Supplemental Figure S7). At the ultrastructural level, cellular morphology appeared normal in the isogenic organoids, reflected in evenly spaced IFs running in parallel to one another and unremarkable organelles, like mitochondria (Figure 6; top panels). In contrast, GAN organoids displayed tightly bundled, swirling, abnormal IF accumulations that trapped smaller organelles, like mitochondria (Figure 6; bottom panels). Furthermore, and in line with the phenotype of the iPSC-astrocyte monoculture system, the organoids contained abnormal IF accumulations encroaching upon irregularly contoured nuclei displaying compromised integrity of the nuclear envelope. Although the ultrastructural examination of the organoids reveals GAN phenotypes that are observed in vivo, our present analysis did not distinguish between neurons and astrocytes, as both are affected in GAN cells.
FIGURE 5:
Generation of GAN and isogenic control iPSC-derived brain organoids. (A) Schematic diagram of methods for generating brain organoids from iPSCs. (B) Confocal images of immunofluorescence staining for GFAP (green), neurofilaments-M/H (magenta) and DAPI (blue) in GAN patient 7 (G332R) and corresponding isogenic control brain organoid clone 2D3. Scale bars = 20 μm (top and third row) and 10 μm (second and fourth row). (C) Quantification of NF and GFAP aggregates in GAN and isogenic control brain organoids based on average measurements across 3 areas (shown in Supplemental Fig.7). **p < 0.01; ***p < 0.001 unpaired t-test.
FIGURE 6:
GAN brain organoids recapitulate IF dysregulation and nuclear abnormalities. Top left panel shows EM image of an isogenic control brain organoid revealing a star-shaped cell with a regularly contoured nucleus and multiple, slender cytoplasmic processes. [5kx; scale bar = 5 μm]. Top middle panel shows enlargement of black boxed area. Note abundant glycogen granules (crosses), ultrastructurally unremarkable mitochondria (m), smoothly contoured nucleus (N), and evenly dispersed, parallel arrays of intermediate filaments (IFs) [50 kx; scale bar = 0.5 μm]. Top right panel shows enlargement of green boxed area. Note unremarkable mitochondria (m), glycogen granules (crosses) and normally distributed and aligned intermediate filaments (IFs) [50 kx; scale bar = 0.5 μm] Bottom left panel shows EM image of a GAN brain organoid revealing a cell with an irregularly contoured nucleus. [5kx; scale bar = 5 μm]. Bottom middle panel shows enlargement of boxed area. Note abnormal intermediate filament accumulations (IFs) encroaching upon an irregularly contoured nuclear envelope (arrows) and enmeshed mitochondria (m). [50kx; scale bar = 0.5 μm]. Bottom right panel shows a GAN brain organoid revealing tightly bundled, swirling abnormal IF accumulations and IF-entrapped organelles, including mitochondria (m). [50kx; scale bar = 0.5 μm]
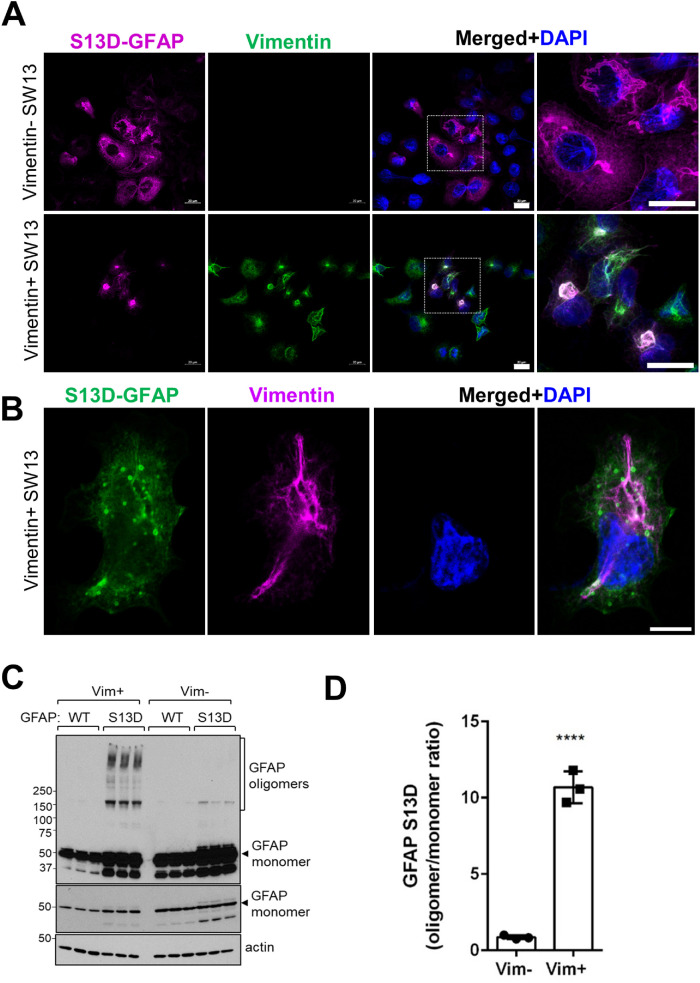
Increased perinuclear GFAP accumulation in the presence of vimentin IFs
To probe the role of vimentin in GAN astrocytes, we utilized an assembly compromised and aggregation-prone S13D-GFAP phosphomimic mutant, which we characterized previously (Battaglia et al., 2019). As detailed previously (Battaglia et al., 2019), we used SW13 cells specifically because the Vim+ SW13 subclone of this cell line expresses only vimentin IFs (and no other cytoplasmic IFs), while the Vim– SW13 subclone lacks all cytoplasmic IFs (Hedberg and Chen, 1986; Sarria et al., 1990), making these cells a suitable transfection host to ask how vimentin affects GFAP aggregation. We observed that the perinuclear aggregation of S13D-GFAP was augmented in the presence of vimentin (Figure 7A). Notably, smaller spherical S13D-GFAP aggregates appeared to associate with the vimentin network in and around the perinuclear region (Figure 7B). In addition, we observed 10-fold elevated levels of high molecular mass S13D-GFAP oligomers in vimentin-positive compared with vimentin-negative SW13 cells (Figure 7, C and D). These results reveal an aggregation-promoting role of vimentin on GFAP in an over-expression system.
FIGURE 7:
Increased GFAP aggregation in the presence of vimentin IFs and perinuclear aggregates. (A) Confocal images of immunofluorescence staining for GFAP (magenta), vimentin (green), and DAPI (blue) in 24h-transfected Vim– (top) and Vim+ (bottom) SW13 cells. Scale bars = 20 μm. Images on the far right show the selected enlarged areas. (B) Immunofluorescence analysis of S13D-GFAP (green), vimentin (magenta) and DAPI (DNA; blue) after 12h of transfection. Note the co-localization of the smaller spherical GFAP aggregates along vimentin filaments. (C) Immunoblot analysis of GFAP and actin (loading control) of lysates from SW13 vimentin-positive and vimentin-negative SW13 cells. Note the abundance of S13D-GFAP oligomers in the presence, but not in the absence, of vimentin. Middle panel is a lighter exposure of the top membrane to show the GFAP monomer. (D) Densitometry quantification of the GFAP oligomer/monomer ratios from the panel B. *p < 0.0001; unpaired t-test.
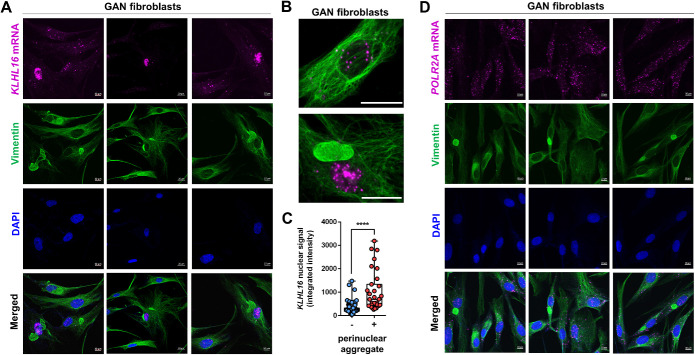
Nuclear enrichment of KLHL16 mRNA in the presence of perinuclear vimentin aggregates
We observed that human KLHL16 mRNA (NM_022041.4) contains an exceptionally long (∼13kb) 3′UTR. This feature makes human KLHL16 mRNA distinct from Klhl16/Gan in other species used to model GAN, including mouse (NM_001081151.2) and zebrafish (XM_003200434.5). The function of the long KLHL16 3′UTR is not known. In general, 3′UTRs regulate mRNA stability, translation, localization, and protein–protein interactions (Mayr, 2019). Given the deficiency of gigaxonin protein in the GAN patient cells, irrespective of the mutation and independent of the total mRNA level, we asked whether the KLHL16 mRNA is sequestered inside IF aggregates. To address this question, we analyzed GAN patient fibroblasts. GAN fibroblasts are a relevant cellular model system reflecting vimentin accumulation (Mahammad et al., 2013), which we observed in astrocytes. Importantly, GAN fibroblasts in the same culture can express normal vimentin IFs alone, or together with large perinuclear aggregates, making direct, side-by-side comparisons of cells with and without vimentin aggregates possible. Although we did not observe colocalization between KLHL16 and vimentin aggregates, there was a strong nuclear presence of KLHL16 in the cells with large perinuclear vimentin aggregates compared with cells without such inclusions (Figure 8, A–C). Other mRNAs, including UBC9, PPIB, and POLR2A did not show altered distribution in GAN fibroblasts (Figure 8D and Supplemental Figure S8). Our results here suggest that the morphologic perturbations induced by IF protein accumulation may have functional consequences on the aberrant processing of KLHL16 mRNA in GAN patient cells. This remains to be rigorously tested. Collectively, our observations highlight the importance of human-specific disease models of GAN, such as the one described herein, to uncover disease mechanisms and identify molecular targets for therapy.
FIGURE 8:
Increased nuclear KLHL16 mRNA accumulation in the presence of vimentin perinuclear aggregates. (A) Co-staining of vimentin (green), KLHL16 mRNA encoding gigaxonin (magenta) and DAPI (blue) in GAN patient fibroblasts. Shown are three representative images. Note the significant nuclear accumulation of KLHL16 mRNA in the vimentin aggregate-containing cells compared to the surrounding cells without aggregates. Scale bars = 20 μm. (B) Magnified representative images of KLHL16 mRNA (magenta) in a cell without (top) and with (bottom) vimentin (green) aggregate. (C) Quantification of KLHL16 mRNA nuclear signal intensity in GAN fibroblasts with (n = 31) and without (n = 143) perinuclear vimentin aggregates. p < 0.0001; unpaired t-test. (D) Co-staining of vimentin (green), POLR2A mRNA as a control mRNA (magenta) and DAPI (blue) in GAN patient fibroblasts. Shown are three representative images. Note there is no difference in distribution of the POLR2A mRNA in the vimentin aggregate-containing cells compared to the surrounding cells without aggregates.
DISCUSSION
In this study, we developed a model to investigate IF proteostasis failure in human astrocytes from GAN patients. Utilizing GAN patient-derived iPSCs and genetically engineered isogenic controls of a single patient line (G332R), we showed that KLHL16 mutations depleted gigaxonin protein and affected vimentin and nestin expression in iPSCs and NPCs, respectively. We also detected large-perinuclear cytoplasmic aggregates of vimentin and GFAP, and the latter were associated with the presence of nuclear abnormalities. We extended this work to 3D iPSC-brain organoids, which reflected IF pathology that is observed in vivo in GAN patients. Finally, we showed increased oligomerization and aggregation of assembly compromised GFAP and selective accumulation of nuclear KLHL16 in the presence of perinuclear vimentin aggregates. Thus, our findings confirm that GAN iPSC-astrocytes and brain organoids are a valuable experimental tool to examine uniquely human cellular disease mechanisms in GAN–mechanisms that implicate vimentin as an underappreciated contributor to the IF proteostasis failure observed in astrocytes in human GAN.
Our model expands upon a previous human iPSC-neuron model of GAN by directing attention to the involvement of astrocytes (Johnson-Kerner et al., 2015a). The human iPSC-astrocyte and organoid models used in this work offer several advantages in the study of GAN and other neurologic diseases associated with pathologic alterations of astrocytes. For example, iPSC-derived patient astrocytes show potential for expressing disease-relevant phenotypes, accelerating characterization of pathologic pathways and providing a tractable platform for development of astrocyte-specific drug therapy. Human astrocytes differ from their rodent counterparts in size, shape, and function (Oberheim et al., 2009). To examine astrocyte-related disease mechanisms, it is necessary to complement animal models with a cellular model faithful to mirroring distinctly human properties. Advances in directed differentiation methods in combination with iPSC technology provide a minimally invasive option to generate human astrocytes (Pasca et al., 2015; Chandrasekaran et al., 2016). Further, organoid-derived astrocytes have demonstrated several astrocyte functions, and thus, this model can be utilized in the future to determine astrocyte-functional deficits in GAN (Sloan et al., 2017). The use of patient-derived cells also allows for tailoring of background genetics. This is especially helpful in studies of the role of IF proteostasis failure in astrocytes in human GAN, where there is phenotypic variability in disease severity likely due to genetic modifiers that may not be reflected in animal models.
Species differences in the functional and pathologic phenotype of GAN are apparent when comparing humans to canines, rodents, and zebrafish. From both a clinical and pathologic standpoint, canine GAN most closely mirrors human GAN (Duncan and Griffiths, 1981). Canine GAN disease onset is early in life (∼15 mo), heralded by hindlimb signs including dragging of toes, diminished proprioception, progressive ataxia, and paresis (Duncan and Griffiths, 1977; Griffith et al., 1980; King et al., 1993). In canine GAN, death is usually by 2 y of age, and the clinical and pathologic phenotype is compatible with a central-peripheral distal axonopathy (Duncan and Griffiths, 1977, 1981). In contrast, while rodent models demonstrate similar human GAN pathology, they have normal lifespans and only mild motor and sensory deficits that are late onset (Ding et al., 2006; Dequen et al., 2008; Ganay et al., 2011). The zebrafish model of GAN has locomotion defects but also displays developmental defects in motor neurons that are not seen in other models (Arribat et al., 2019a). The availability of human cellular disease models of GAN can help bridge the gap in our understanding of disease mechanisms based on other species. However, we acknowledge that disparities in GAN severity across species are multifactorial, including lifespan, length differences in axons, and differing complexity of cells (e.g., astrocyte complexity), such that the various aspects of cell biology, including astrocyte homeostasis and astrocyte-neuron interactions, must be tested experimentally.
The recessive nature of GAN and the absence of detectable gigaxonin protein has led to the understanding that the disorder is caused primarily by loss of function of gigaxonin. In the case of null mutations, the cause of gigaxonin depletion is clear, but for missense mutations, it is not fully understood why gigaxonin protein is lost. It has been proposed that the defect occurs posttranslationally because missense mutations could destabilize the gigaxonin protein (Boizot et al., 2014). However, this hypothesis is yet to be tested experimentally, and we suggest that a key intermediate step–RNA processing–deserves to be closely examined. It is plausible that KLHL16 is transcribed and stalled at the mRNA transcript stage, an idea that is supported by the finding that KLHL16 mRNA can be stored in stress granules in human cells (Khong et al., 2017). The possibility that posttranscriptional mechanisms may contribute to GAN raises the question of whether GAN can be caused by mutations outside the KLHL16 coding region. Although some mutations have been identified in splicing sites, sequencing is not routinely performed on noncoding regions of KLHL16 in suspected GAN patients, which is understandable considering the remarkable length of the KLHL16 gene (Lescouzères and Bomont, 2020). This patient-derived iPSC model will provide a useful tool to address the mechanism of loss of gigaxonin in GAN.
Finally, our results indicate that vimentin and GFAP IFs are involved in cytoskeletal proteostasis failure in GAN astrocytes. The presence of deep nuclear indentations juxtaposed to cytoplasmic GFAP+ aggregates suggests that GFAP could promote enhanced and possibly toxic interactions between IF aggregates and the nucleus. One possible cause for this could be related to plectin, which is an IF-associated protein that crosslinks IFs to other structures, including microtubules, actin, and the nuclear envelope (Castañón et al., 2013; Ketema et al., 2013). While both vimentin and GFAP are known to bind to plectin, it is possible that each IF develops distinct posttranslational modifications, which can alter plectin binding (Foisner et al., 1988; Favre et al., 2011; Castañón et al., 2013; Snider and Omary, 2014). Another potential mechanism could be disrupted nuclear transport. GFAP and vimentin both contain a predicted nuclear localization signal in the C-terminal tail domain, but GFAP harbors an additional predicted domain in the N-terminal head domain (Hobbs et al., 2016). However, it has yet to be shown that GFAP is transported into the nucleus, as has been demonstrated with keratin-17, an epithelial IF (Hobbs et al., 2016). Using our SW13 overexpression system, we also show evidence of enhanced oligomerization and aggregation of assembly-compromised GFAP in the presence of vimentin. We suspect, as others have posited, that vimentin promotes aggregation by providing a scaffold on which aggregates are seeded (Sosunov et al., 2017). Astrocytes are not the only cell type that express more than one IF in development and disease. In fact, neurons can express at least four IFs: NFs light, medium, and heavy, as well as peripherin or α-internexin, which are assembled into filaments at very specific ratios (Yuan et al., 2006, 2012). It is likely that different IFs cooperate to promote aggregation in other IF-associated disorders where IF proteostasis and assembly are compromised. The abundance of IF aggregates in a variety of cell types in GAN makes our iPSC model a clinically relevant and versatile tool to investigate the effects of IF proteostasis failure in diverse human cellular contexts.
A limitation of this study is that we compared a single GAN patient line to its corresponding isogenic control lines. In the future, it will be important to generate isogenic control lines from multiple GAN patients to rigorously examine the role of astrocytes in GAN. Collectively, our results provide a new model to study the impact of KLHL16 gene mutations on astrocyte cell biology that may contribute to disease pathogenesis in GAN. Future studies will implement this model to characterize the role of vimentin and posttranscriptional mechanisms in GAN-associated IF proteostasis failure in astrocytes.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Antibodies
The following primary antibodies and concentrations were used in this study: rabbit anti-GFAP (DAKO, Agilent, clone Z0334; IF 1:500, WB 1:10,000), mouse anti-GFAP (Sigma, clone GA5; IF 1:300), mouse anti-Gigaxonin (Santa Cruz Biotechnology, F3, WB 1:200), rabbit anti-Vimentin (Cell-Signaling Technology, D21H3, IF 1:100), mouse anti-Vimentin (Thermo Fisher Scientific, V9, WB 1:1000), mouse anti-Keratin 8 (Thermo Fisher Scientific, TS1, IF 1:100), rat anti-K8 (Developmental Studies Hybridoma Bank, Troma I, WB 1:5000), rabbit anti-Lamin A/C (Santa Cruz Biotechnology), rabbit anti-Lamin B1 (Abcam ab16048, IF 1:10,000, WB 1:10,000), mouse anti-Nestin (Thermo Fisher Scientific, 10C2, IF 1:200), mouse anti-NF-M/H (clone RMdO-20; IF 1:100), mouse anti-Tra-1-60 (Thermo Fisher Scientific, 41-1000, IF 1:300), mouse anti-Tra-1-81 (Thermo Fisher Scientific, 41-1100, IF 1:300), rabbit anti-Oct4 (Abcam, ab19857, IF 1:40), and rabbit anti-Sox2 (Thermo Fisher Scientific, 48-1400, IF 1:125). The following secondary antibodies and concentrations were used: Alexa 488- and Alexa 594-conjugated goat-, anti-mouse-, and rabbit-antibodies (Thermo Fisher Scientific, IF 1:500), and peroxidase-conjugated goat-, anti-mouse-, and rabbit-antibodies (Sigma, WB 1:5000).
Cell lines
SW13vim+ and vim– cells (Hedberg and Chen, 1986; Sarria et al., 1990) were provided by Bishr Omary and cultured in DMEM with 10% fetal bovine serum and 1% penicillin-streptomycin. Authentication was done by short tandem repeat (STR) profiling by ATCC. Transfection were performed as described previously (Battaglia et al., 2019) and cells analyzed after 24 h, unless noted otherwise in the (Figure 7B) Legend.
Human tissue and cells
Human postmortem specimens (Figure 1, A and B) were obtained through a validated autopsy consent from the legal next of kin, which explicitly stated that tissues could be used for research purposes. The consent and diagnostic autopsy report were filed in the deceased patient’s medical record. Skin fibroblasts were obtained from seven GAN patients (numbered 1–7 in the current study; Figure 1C) that were enrolled in a natural history study under an approved protocol (Bharucha-Goebel et al., 2021). These seven patients were part of a larger clinical cohort that was previously described by Bharucha-Goebel et al., (2021). Please see supplement in Bharucha-Goebel et al., (2021) for genotype, phenotype, and functional data. The IDs of patients #1–7 in the current study: B15-56.1; B13-69.1; B16-64.1; B16-78; B16-74; B15-100.1; B16-02 correspond, respectively, to the following patient IDs from the larger GAN cohort that are listed in the supplement of the Bharucha-Goebel (Bharucha-Goebel et al., 2021) study: Patient 2; Patient 17; Patient 21; Patient 23; Patient 4; Patient 9; Patient 10. Four different isogenic-control cell lines (named B3, G5, 2D1, and 2D3 in this study) were derived from patient ID B16-02 carrying the G332R mutation (patient #7 in this study; please see Figure 1C).
Cellular reprogramming, characterization, and gene editing of iPSCs
iPSCs were generated from GAN (OMIM #256850) patient fibroblasts and characterized using the methods previously described (Beltran et al., 2020). Fibroblast reprogramming to iPSCs, karyotyping, CRISPR/Cas9 gene editing, screening of edited clones, and off-target analyses were performed as we described in detail previously in studies on AxD iPSC-astrocytes (Battaglia et al., 2019) with the addition of FGF-coated beads from StemCultures. Pluripotency was evaluated by the Thermo Fisher Scientific Scorecard (Bock et al., 2011; Tsankov et al., 2015; Fergus et al., 2016), which compares gene expression of various self-renewal/pluripotency genes to that of genes distinct to the three germ layers (Supplemental Figure S1A, Supplemental Figure S2, E and F). We followed the manufacturer protocol for the Scorecard data collection and analysis. The pluripotency was also assessed in each cell line via immunofluorescence staining for several markers of pluripotency (Supplementary Figures S1B; S2D). Finally, karyotyping was performed to ensure that no genomic abnormalities had arisen during reprogramming (Supplemental Figure S1C). After gene editing, we used an allele-specific PCR screen to select for edited clones, which were verified by Sanger sequencing (Supplemental Figure S2A). Chromatograms from the target region are shown for the parental line as well as the corrected clones (Supplemental Figure S2B). We confirmed that there were no off-target mutations via Sanger sequencing of the top 20 off-target regions within exons (Supplemental Table S1). KLHL16 gene expression (Supplemental Figure S2C) was measured via sequence-specific primers (Supplemental Table S3) and pluripotency and trilineage differentiation were assessed in the edited clones (Supplemental Figure S2, D–F).
iPSC culture and differentiation
iPSCs were maintained on Matrigel (Corning, Catalogue No. 354480) in StemFlex Medium (Thermo Fisher Scientific, Catalogue No. A3349401) and passaged every 3–4 day with 0.5 mM EDTA dissociation solution. Astrocytes were differentiated as described (Battaglia et al., 2019). To generate NPCs, embryoid bodies (EBs) were plated on polyornithine and laminin-coated plates in Neural Induction Medium (StemCell Technologies) and rosettes were selected after 12 day and expanded in Neural Progenitor Medium (StemCell Technologies). NPCs were differentiated to astrocytes using the Stem Cell Technologies STEMdiff astrocyte differentiation and maturation kits during which they were split weekly with Accutase (Millipore).
Organoids were generated using an established protocol (Paşca et al., 2015) with a few following modifications for feeder-free conditions. The EBs were generated as described above on day 0–6. On day 7, the EBs were moved to neural medium (NM, Invitrogen) containing serum-free Neurobasal medium with B-27 without vitamin A (Invitrogen), GlutaMax (Life Technologies). NM was supplemented with FGF2 (20 ng/ml, R&D Systems), and EGF (20 ng/ml, R&D Systems). Media was changed daily for day 7–16 and every other day for day 17–25. After neural induction, the media was replaced with NM supplemented with BDNF (20 ng/ml, Peprotech), and NT3 (20 ng/ml, Peprotech) every other day to promote differentiation. On day 43, NM without growth factors was used for medium changes every 4 d.
Preparation of protein lysates and immunoblotting
Total lysates were prepared from 60–80% confluent iPSCs plated on six-well plates by rinsing cells quickly with 1 ml of 1× phosphate-buffered saline (PBS) and adding 2× Novex Tris-Glycine SDS Sample Buffer (Thermo Fisher Scientific, Catalogue #LC2676) directly to the plate, collected into Eppendorf tubes, and heated at 95°C for 5 min. Protein lysates were normalized by running Coomassie staining before running western blots. For immunoblotting, samples were separated on a 4–20% gradient SDS–PAGE gel and transferred either for 1 h at 110V or overnight at 40V onto an activated polyvinylidene-difluoride membrane. Membranes were blocked in 5% nonfat milk dissolved into 0.1% Tween 20/PBS (PBST) for 1 h at room temperature and then incubated in primary and secondary antibodies diluted into blocking solution at the concentrations listed above. Antibodies were detected via ECL reagents (PerkinElmer).
RNA isolation and quantitative real-time PCR (qRT-PCR)
RNA was isolated from iPSCs using the Thermo Fisher Scientific PureLink RNA mini kit (Thermo Fisher Scientific, Catalogue #12183025) following all manufacturer guidelines and immediately converted to cDNA. The High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Catalogue#4368814) was used to convert 2 μg of total RNA to cDNA. To measure gene expression, qRT-PCR was performed using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, Catalogue#A25778) and the Applied Biosystems QuantStudio 6 Flex Real-Time PCR System. The primers used to detect gene expression in this study are listed in Supplemental Table S2.
Fluorescent RNA in situ hybridization (ISH) to detect KLHL16 mRNA
Custom RNAScope target-specific oligonucleotide (ZZ) probe (19ZZ length) was designed to target the region spanning nucleotides 2–997 of human KLHL16 mRNA (NM_022041.4). The cells were treated with hydrogen peroxidase to minimize background (10 min, RT), followed by three PBST washes and permeabilization using Protease III digestion (1:15 dilution; 322,000, ACD), at 40°C for 10 min. Subsequently, the expression of target probe was assessed using RNAscope Multiplex Fluorescent v2 Assay protocol (323,100, ACD). Positive (POLR2A, PPIB, and UBC) and negative (dapB) control probes were included to ensure specificity of signal detection. Following ISH signal development, the cells were washed three times in PBS for 5 min each and stained for vimentin as described below.
Immunofluorescence, imaging, and analysis
Cells were fixed in methanol at –20°C for 10 min, washed three times in PBS, and blocked in Buffer B (2.5% Bovine Serum Albumin [Sigma], 2% normal goat serum [Life Technologies]/PBS) for 1 h at room temperature. Next, cells were incubated in primary antibodies overnight at 4°C after which they were washed three times in PBS and incubated with Alexa Fluor-conjugated secondary antibodies 1 h at room temperature. Cells were washed three times in PBS, incubated in DAPI (Invitrogen), washed twice in PBS, and mounted in Fluoromount-G (SouthernBiotech) overnight. Cells were imaged on Zeiss 880 confocal laser scanning microscope using a 40× oil immersion objective or for widefield on the EVOS-FL auto cell-imaging system (Thermo Fisher Scientific) using a 20× (0.75 NA) objective. NIH ImageJ software was used to measure vimentin signal, size, and intensity of the vimentin cytoskeleton as shown in Figure 4 by using the polygon tool to circle the vimentin signal and the measure tool, which provides a measure of area and average signal intensity. The circularity of lamin B1-stained nuclei was measured using the analyze particles function of ImageJ. Data shown in Figure 4F represent 10 images from each condition; representative images that were quantified are shown in Supplemental Figure S6C. For analysis of GAN mRNA nuclear content in Figure 7, 3D Images were acquired using Zeiss confocal LSM880 microscope (Carl Zeiss) using a 63× oil immersion objective with NA 0.95 providing a 257 nm resolution and using z-stack function at multiple areas. CellProfiler (v. 4.2.1) object identification pipelines were used to determine nuclear and perinuclear area, and to determine cells with and without vimentin aggregates (clumps) within a predetermined perinuclear area. This was followed by identifying and counting GAN mRNA objects in nuclear area of cells with and without perinuclear clumps.
Statistics
The graph data were presented using GraphPad Prism software and statistics were performed using the tests described in each figure legend. Adobe Photoshop was used to perform densitometry on immunoblots and Image J was used for signal quantification of immunofluorescence data.
Supplementary Material
Acknowledgments
This study was funded by Hannah’s Hope Fund and National Institute of Health grants F99AG068523 (RAB), R21NS121578 (NTS, DA), R01NS127204 (PO), and R01NS082351 (PO). We thank Anne Messer (Neural Stem Cell Institute) for feedback and advice on the studies and manuscript.
Abbreviations used:
- ALDHL1
aldehyde dehydrogenase 1 family, member L1
- AxD
Alexander disease
- BDNF
brain-derived neurotrophic factor
- EB
embryoid body
- FGF2
fibroblast growth factor 2
- GAN
giant axonal neuropathy
- GFAP
glial fibrillary acidic protein
- IF
intermediate filament
- iPSC
induced pluripotent stem cell
- KLHL
Kelch-like gene family
- NF
neurofilament
- NIM
neural induction medium
- NM
neural medium
- NPC
neural progenitor cell
- NT3
Neurotrophin-3
- RF
Rosenthal fiber
- UTR
untranslated region
- VIM
vimentin
- WB
western blot
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-03-0094) on September 6, 2023.
REFERENCES
- Armao D, Bouldin TW, Bailey RM, Hooper JE, Bharucha DX, Gray SJ (2019). Advancing the pathologic phenotype of giant axonal neuropathy: early involvement of the ocular lens. Orphanet J Rare Dis 14, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armao D, Bouldin TW, Bailey RM, Gray SJ (2021). Extensive rod and cone photoreceptor-cell degeneration in rat models of giant axonal neuropathy: implications for gene therapy of human disease. Ophthalmic Genet 42, 600–603. [DOI] [PubMed] [Google Scholar]
- Arribat Y, Mysiak KS, Lescouzéres L, Boizot A, Ruiz M, Rossel M, Bomont P (2019a). Sonic Hedgehog repression underlies gigaxonin mutation-induced motor defecits in giant axonal neuropathy. J Clin Invest 129, 5312–5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribat Y, Mysiak KS, Lescouzères L, Boizot A, Ruiz M, Rossel M, Bomont P (2019b). Sonic Hedgehog repression underlies gigaxonin mutation–induced motor deficits in giant axonal neuropathy. J Clin Invest 129, 5312–5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury A, Gale M, Cox S, Baringer J, Berg B (1972). Giant axonal neuropathy—a unique case with segmental neurofilamentous masses. Acta Neuropathol 20, 237–247. [DOI] [PubMed] [Google Scholar]
- Battaglia RA, Beltran AS, Delic S, Dumitru R, Robinson JA, Kabiraj P, Herring LE, Madden VJ, Ravinder N, Willems E (2019). Site-specific phosphorylation and caspase cleavage of GFAP are new markers of Alexander disease severity. eLife 8, e47789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran AA, Molina SG, Beltran AS (2020). Derivation of induced pluripotent stem cells from human fibroblasts using a non-integrative system in feeder-free conditions. Bio-Protocol 10, e3788–e3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha-Goebel DX, Norato G, Saade D, Paredes E, Biancavilla V, Donkervoort S, Kaur R, Lehky T, Fink M, Armao D (2021). Giant axonal neuropathy: cross-sectional analysis of a large natural history cohort. Brain 144, 3239–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. (2011). Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent stem cell lines. Cell 144, 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boizot A, Talmat-Amar Y, Morrogh D, Kuntz NL, Halbert C, Chabrol B, Houlden H, Stojkovic T, Schulman BA, Rautenstrauss B, Bomont P (2014). The instability of the BTB-KELCH protein Gigaxonin causes Giant Axonal Neuropathy and constitutes a new penetrant and specific diagnostic test. Acta Neuropathol Commun 2, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomont P, Cavalier L, Blondeau F, Hamida CB, Belal S, Tazir M, Demir E, Topaloglu H, Korinthenberg R, Tüysüz B, et al. (2000). The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet 26, 370–374. [DOI] [PubMed] [Google Scholar]
- Büchau F, Munz C, Has C, Lehmann R, Magin TM (2018). KLHL16 degrades epidermal keratins. J Invest Dermatol 138, 1871–1873. [DOI] [PubMed] [Google Scholar]
- Castañón MJ, Walko G, Winter L, Wiche G (2013). Plectin-intermediate filament partnership in skin, skeletal muscle, and peripheral nerve. Histochem Cell Biol 140, 33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran A, Avci HX, Leist M, Kobolak J, Dinnyes A (2016). Astrocyte differentiation of human pluripotent stem cells: New tools for neurological disorder research. Front Cell Neurosci 10, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequen F, Bomont P, Gowing G, Cleveland DW, Julien J-P (2008). Modest loss of peripheral axons, muscle atrophy and formation of brain inclusions in mice with targeted deletion of gigaxonin exon 1. J Neurochem 107, 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Allen E, Wang W, Valle A, Wu C, Nardine T, Cui B, Yi J, Taylor A, Jeon NL, et al. (2006). Gene targeting of GAN in mouse causes toxic accumulation of microtubule-associated protein 8 and impairment of retrograde axonal transport. Hum Mol Genet 15, 1451–1463. [DOI] [PubMed] [Google Scholar]
- Dubeau F, Michaud J, Lamarre L, Larbrisseau A (1985). Giant axonal neuropathy-a complete autopsy study. J Neuropathol Exp Neurol 44, 355–355. [Google Scholar]
- Duncan ID, Griffiths IR (1977). Canine giant axonal neuropathy. Vet Rec 101, 438–441. [DOI] [PubMed] [Google Scholar]
- Duncan I, Griffiths I (1981). Canine giant axonal neuropathy; some aspects of its clinical, pathological and comparative features. J Small Anim Pract 22, 491–501. [DOI] [PubMed] [Google Scholar]
- Favre B, Schneider Y, Lingasamy P, Bouameur J-E, Begré N, Gontier Y, Champliaud M-FS, Fria MA, Borradori L, Fontao L (2011). Plectin interacts with the rod domain of type III intermediate filament proteins desmin and vimentin. Eur J Cell Biol 90, 390–400. [DOI] [PubMed] [Google Scholar]
- Fergus J, Quintanilla R, Lakshmipathy U (2016). Characterizing Pluripotent Stem Cells Using the TaqMan(R) hPSC Scorecard(TM) Panel. Methods Mol Biol 1307, 25–37. [DOI] [PubMed] [Google Scholar]
- Foisner R, Leichtfried FE, Herrmann H, Small JV, Lawson D, WIche G (1988). Cytoskeleton-associated Plectin: In situ localization, in vitro reconstitution, and binding to immonibilzed intermediate filament proteins. J Cell Biol 106, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganay T, Boizot A, Burrer R, Chauvin JP, Bomont P (2011). Sensory-motor deficits and neurofilament disorganization in gigaxonin-null mice. Mol Neurodegener 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith IR, Duncan ID, McCulloch M, Carmichael S (1980). Further studies of the central nervous system in canine giant axonal neuropathy. Neuropathol App Neurobiol 6, 421–432. [DOI] [PubMed] [Google Scholar]
- Hedberg KK, Chen LB (1986). Absence of intermediate filaments in a human adrenal cortex carcinoma-derived cell line. Exp Cell Res 163, 509–517. [DOI] [PubMed] [Google Scholar]
- Hentati F, Hentati E, Amouri R (2013). Giant axonal neuropathy. Handb Clin Neurol 115, 933–938. [DOI] [PubMed] [Google Scholar]
- Hobbs RP, Jacob JT, Coulombe PA (2016). Keratins are going nuclear. Dev Cell 38, 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Kerner BL, Roth L, Greene JP, Wichterle H, Sproule DM (2014). Giant axonal neuropathy: An updated perspective on its pathology and pathogenesis. Muscle Nerve 50, 467–476. [DOI] [PubMed] [Google Scholar]
- Johnson-Kerner BL, Ahmad FS, Diaz AG, Greene JP, Gray SJ, Samulski RJ, Chung WK, Coster RV, Maertens P, Noggle SA, et al. (2015a). Intermediate filament protein accumulation in motor neurons derived from giant axonal neuropathy iPSCs rescued by restoration of gigaxonin. Hum Mol Genet 24, 1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Kerner BL, Garcia Diaz A, Ekins S, Wichterle H (2015b). Kelch domain of gigaxonin interacts with intermediate filament proteins affected in giant axonal neuropathy. PLoS One 10, e0140157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketema M, Kreft M, Secades P, Janssen H, Sonnenberg A (2013). Nesprin-3 connects plectin and vimentin to the nuclear envelope of Sertoli cells but is not required for Sertoli cell function in spermatogenesis. Mol Biol Cell 24, 2454–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 68, 808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RHM, Sarsilmaz M, Thomas PK, Muddle JM, Duncan ID (1993). Axonal neurofilamentous accumulations: a comparison between human and canine giant axonal neuropathy and 2, 5-HD neuropathy. Neuropathol App Neurobiol 19, 224–232. [DOI] [PubMed] [Google Scholar]
- Kretzschmar HA, Berg BO, Davis RL (1987). Giant axonal neuropathy. Acta Neuropathol 73, 138–144. [DOI] [PubMed] [Google Scholar]
- Kumar K, Barre P, Nigro M, Jones MZ (1990). Giant axonal neuropathy: clinical, electrophysiologic, and neuropathologic features in two siblings. J Child Neurol 5, 229–234. [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J (2016). ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44, D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescouzères L, Bomont P (2020). E3 ubiquitin ligases in neurological diseases: focus on gigaxonin and autophagy. Front Physiol 11, 1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N-H, Huang Y-S, Opal P, Goldman RD, Messing A, Perng M-D (2016). The role of gigaxonin in the degradation of the glial-specific intermediate filament protein GFAP. Mol Biol Cell 27, 3980–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahammad S, Murthy SNP, Didonna A, Grin B, Israeli E, Perrot R, Bomont P, Julien J-P, Kuczmarski E, Opal P, Goldman RD (2013). Giant axonal neuropathy-associated gigaxonin mutations impair intermediate filament protein degradation. J Clin Invest 123, 1964–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C (2019). What are 3′ UTRs doing? Cold Spring Harb Perspect Biol 11, a034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri I, Taniike M, Yoshikawa H, Higashiyama M, Itami S, Okada S (1998). A case of giant axonal neuropathy showing focal aggregation and hypophosphorylation of intermediate filaments. Brain Dev 20, 594–597. [DOI] [PubMed] [Google Scholar]
- Molina SG, Beltran AA, Beltran AS (2020). Generation of an integration-free induced pluripotent stem cell line (UNC001-A) from blood of a healthy individual. Stem Cell Res 49, 102015. [DOI] [PubMed] [Google Scholar]
- Oberheim NA, Takano T, Han X, Lin JHC, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, Ransom BR, et al. (2009). Uniquely hominid features of adult human astrocytes. J Neurosci 29, 3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park J-Y, O’Rourke NA, Nguyen KD, et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 12, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer J, Schlote W, Bischoff A, Boltschauser E, Müller G (1977). Generalized giant axonal neuropathy: A filament-forming disease of neuronal, endothelial, glial, and schwann cells in a patient without kinky hair. Acta Neuropathol 40, 213–218. [DOI] [PubMed] [Google Scholar]
- Sarria AJ, Nordeen SK, Evans RM (1990). Regulated expression of vimentin cDNA in cells in the presence and absence of a preexisting vimentin filament network. J Cell Biol 111, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA, Pasca SP (2017). Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 95, 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Omary MB (2014). Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat Rev Mol Cell Biol 15, 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosunov AA, McKhann GM, 2nd, Goldman JE (2017). The origin of Rosenthal fibers and their contributions to astrocyte pathology in Alexander disease. Acta Neuropathol Commun 5, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Love S, Powell H, Schultz P, Lampert P (1987). Giant axonal neuropathy: correlation of clinical findings with postmortem neuropathology. Ann Neurol 22, 79–84. [DOI] [PubMed] [Google Scholar]
- Tsankov AM, Akopian V, Pop R, Chetty S, Gifford CA, Daheron L, Tsankova NM, Meissner A (2015). A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat Biotechnol 33, 1182–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Rao MV, Sasaki T, Chen Y, Kumar A, Veeranna, Liem RKH, Eyer J, Peterson AC, Julien J-P, Nixon RA (2006). alpha-internexin is structurally and functionally associated with the neurofilament triplet proteins in the mature CNS. J Neurosci 26, 10006–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Sasaki T, Kumar A, Peterhoff CM, Rao MV, Liem RK, Julien J-P, Nixon RA (2012). Peripherin is a subunit of peripheral nerve neurofilaments: Implications for differential vulnerability of CNS and peripheral system axons. J Neurosci 32, 8501–8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.