Abstract
Variations in the dynein axonemal heavy chain gene, dynein axonemal heavy chain 6 (DNAH6), lead to multiple morphological abnormalities of the flagella. Recent studies have reported that these deficiencies may result in sperm head deformation. However, whether DNAH6 is also involved in human acrosome biogenesis remains unknown. The purpose of this study was to investigate DNAH6 gene variants and their potential functions in the formation of defective sperm heads and flagella. Whole-exome sequencing was performed on a cohort of 375 patients with asthenoteratozoospermia from the First Affiliated Hospital of Anhui Medical University (Hefei, China). Hematoxylin and eosin staining, scanning electron microscopy, and transmission electron microscopy were performed to analyze the sperm morphology and ultrastructure. Immunofluorescence staining and Western blot analysis were conducted to examine the effects of genetic variants. We identified three novel deleterious variants in DNAH6 among three unrelated families. The absence of inner dynein arms and radial spokes was observed in the sperm of patients with DNAH6 variants. Additionally, deficiencies in the acrosome, abnormal chromatin compaction, and vacuole-containing sperm heads were observed in these patients with DNAH6 variants. The decreased levels of the component proteins in these defective structures were further confirmed in sperm from patients with DNAH6 variants using Western blot. After intracytoplasmic sperm injection (ICSI) treatment, the partner of one patient with a DNAH6 variant achieved successful pregnancy. Overall, novel variants in DNAH6 genes that contribute to defects in the sperm head and flagella were identified, and the findings indicated ICSI as an effective clinical treatment for such patients.
Keywords: asthenoteratospermia, DNAH6, intracytoplasmic sperm injection, male infertility
INTRODUCTION
Infertility has become a major health issue, affecting approximately 70 million couples worldwide.1 Numerous causes of infertility exist; however, approximately 50% of the confirmed cases are due to male infertility owing to defective sperm quality and function.2 Male infertility is a multifactorial disease, with genetic causes accounting for approximately 30% of confirmed cases, and approximately 40% are due to an unexplained etiology classified as idiopathic.3,4 Asthenoteratospermia, a common cause of male infertility, is characterized by morphological abnormalities in the sperm head, neck, or flagella and decreased sperm motility.5,6 Multiple morphological abnormalities of the sperm flagella (MMAF) is the predominant type of asthenoteratospermia, and it is characterized by absent, coiled, bent, short, or irregular flagella.7
The basic structure of mature spermatozoa is composed of the sperm head, neck, and flagellum.8 The sperm flagellum is divided into three parts, including the mid-, principal, and terminal pieces. The mitochondrial sheath (MS) is formed through the accumulation of mitochondria around the outer dense fibers and the axoneme, composed of the middle segment.9 The MS was replaced by a fibrous sheath in the principal piece, and the axoneme was only surrounded by a cytoplasmic membrane in the terminal piece. The axoneme runs throughout the sperm flagellum, which comprises a typical “9 + 2” microtubule structure (i.e., two central singlet microtubules are encircled by nine outer doublet microtubules),10 and the disruption of any of its structures may cause MMAF, which is genetically heterogeneous. There are two types of microtubules, A and B. A-type microtubules contain inner dynein arms (IDA) and outer dynein arms (ODA), which are collectively known as multiprotein ATPase complexes and constitute the motor protein complexes that are essential for flagellar motility.9 As a result, many pathogenic mutations of genes producing proteins involved in sperm centrosome assembly, flagella structure components, or intraflagellar transport led to flagellar abnormalities. In recent years, numerous genes associated with MMAF have been reported. Ben Khelifa et al.7 reported the dynein axonemal heavy chain 1 (DNAH1) gene, which expresses the axoneme kinetic protein of the IDA, as the initial genetic cause of MMAF. Subsequently, about 43 genes related to MMAF, including centrosomal protein genes (DAZ interacting zinc finger protein 1 [DZIP1],11 and centrosomal protein 70 [CEP70]12), flagella structure proteins genes (DNAH6,13 dynein axonemal heavy chain 10 [DNAH10],14 dynein heavy chain domain 1 [DNHD1],15 and dynein axonemal light intermediate chain 1 [DNALI1]16), and intraflagellar transport or related genes (tetratricopeptide repeat domain 21A [TTC21A],17 coiled-coil domain containing 34 [CCDC34],18 sperm flagellar 2 [SPEF2],19 cilia and flagella associated protein 69 [CFAP69],20 cilia and flagella associated protein 43 [CFAP43],21 and cilia and flagella associated protein 44 [CFAP44]21), were identified successively. All of these are involved in the structure and assembly of the sperm flagellum, which account for approximately 30%–60% of MMAF patients;22 however, numerous genes remain unexplored.
The DNAH6 proteins are components of the IDA; they are predominantly expressed in the testis and lung tissues and mainly control the size and shape of the flagellar bend. Variations in the DNAH6 gene are associated with primary ciliary dyskinesia (PCD), as reported previously.23 Li et al.24 found that DNAH6 is concentrated at the head-neck junction, and its deficiency resulted in sperm head anomalies. Additionally, Tu et al.13 observed that the DNAH6 protein signal is distributed along the flagella, and its absence was responsible for MMAF. Furthermore, almost of patients with DNAH6 variants failed pregnancy after intracytoplasmic sperm injection (ICSI) treatment.13,24 Gershoni et al.25 identified DNAH6 variants in men with azoospermia. These findings suggest that variations in DNAH6 may cause cilia and sperm flagella dysfunctions and even affect ICSI outcomes.
As a result, in the present study, we aimed to identify novel variants in the DNAH6 gene in patients with asthenoteratospermia using whole-exome sequencing.
PARTICIPANTS AND METHODS
Study participants
This study was approved by the Ethics Committee of the First Affiliated Hospital of Anhui Medical University (Hefei, China; Approval No. 20200048), and informed consent was obtained from all the study participants. Three hundred and seventy-five men who received a diagnosis of asthenoteratozoospermia (sperm progressive motilities estimated in replicate counts of at least 200 spermatozoa were <32%, and the percentage of normal sperm morphology, including sperm head, excess residual cytoplasm, and sperm tail, was <4%) and infertility for more than 1 year at the Reproductive Center of the First Affiliated Hospital of Anhui Medical University were recruited. No abnormalities in external genitalia, bilateral testicular size, hormone levels, or Y chromosome microdeletions after basic clinical examination were observed in all participants in this study. Furthermore, their chromosomal karyotype was normal (46,XY), and none of the affected patients had sinusitis, bronchiectasis, or pulmonary disease associated with PCD. Normal spermatozoa were obtained from fertile, healthy volunteers from the Human Sperm Bank of the First Affiliated Hospital of Anhui Medical University.
Whole-exome sequencing, Sanger sequencing, and in silico analyses
We collected 5 ml peripheral blood from each of the participants with asthenoteratozoospermia (n = 375). After that, 1 ml peripheral blood was used for extracting genomic DNA, which was subjected to whole-exome sequencing using the MGISEQ-2000 platform (MGI Tech Co., Ltd., Shenzhen, China) in different companies. The original FASTQ data were mapped to the human genome using Burrows-Wheeler Aligner (BWA) software, and Sequence Alignment/Mapping (SAM) tools and Genome Analysis Toolkit (GATK) were used to call genetic variants.26 The sequencing coverage and the total number of reads for patients in our study were 99.9% and 40 207 727 in AY0334, 99.1% and 207 015 777 in AY0159, and 97.3% and 214 225 218 in AY0283, respectively. Functional annotation was performed according to a previously described method by Li et al.27 We specifically focused on splicing within two bases, deleterious missense variants, frameshift variants, and stop gain or loss variants. We used the human genome database to predict the allele frequency in the population, and these variants were predicted for deleteriousness using Sorting Intolerant From Tolerant (SIFT), Polymorphism Phenotyping v2 (PolyPhen-2), and MutationTaster. Three patients with DNAH6 variants were further verified using Sanger sequencing. The primers used for Sanger sequencing are listed in Supplementary Table 1. We analyzed the impact of DNAH6 variants on protein structure using the SWISS-MODEL website (https://swissmodel.expasy.org/; last accessed on June 01, 2023).
Supplementary Table 1.
Primers used for amplification and verification of dynein axonemal heavy chain 6 variants
| Primer names | Primer sequences (5’–3’) | Temperature (°C) |
|---|---|---|
| M1-F | GGAGTCCAAATTCCAGAACATGA | 60 |
| M1-R | AGCATCTACTGGCCCATCACT | |
| M2-F | CATAAAGCCTCAGATATTGGCAAAG | 60 |
| M2-R | CAATGACAGGGTTAAGCTGGTTCTA | |
| M3-F | TTCAACCAGAGTAATGGAGTATC | 55 |
| M3-R | GACATTCCTCCCCATTCTTCG | |
| M4-F | AGACAGAAAATCTACAACAGCG | 58 |
| M4-R | TGGTCCAGGTAGGATGTCGTG | |
| M5-F | CAGCTTATAGAGTGTTGGATCC | 56 |
| M5-R | CCTTTCTTACTTTCTCATCCAC |
Semen routine and sperm morphology analysis
According to the World Health Organization (WHO) laboratory manual (6th edition),6 semen samples were collected by masturbation from three patients carrying the DNAH6 mutations after 2–7 days of sexual abstinence and liquefied at 37°C for 30 min. Semen volumes were calculated by weighing. Then, 10 μl semen sample was added to the sperm counter plate, and the sperm motility and concentration were measured by the Spanish Sperm Class Analyzer (SCA) computer-aided sperm analysis (CASA) system (MICROPTIC, Barcelona, Spain). At the same time, semen samples were washed with saline, followed by smearing and fixing with 4% paraformaldehyde (PFA) and staining of spermatozoa with Papanicolaou or hematoxylin and eosin (H&E; Beyotime, Shanghai, China). Sperm morphology was observed under a light microscope at 100× (Axio Scope A1, Carl Zeiss AG, Oberkochen, Germany). More than 200 spermatozoa were evaluated for each patient, and abnormalities in the sperm head and flagellum were analyzed.
Sperm morphology was further examined by scanning electron microscopy (SEM; GeminiSEM 300, Carl Zeiss AG). Fresh sperm samples from patients AY0334 and AY0283 with the DNAH6 mutations and two normal fertile men were liquefied at 37°C for 30 min, washed thrice using phosphate buffer saline (PBS), and fixed with 2.5% glutaraldehyde (G7651, Sigma-Aldrich, St. Louis, MO, USA) for 4–12 h. After washing thrice with PBS, the sperm was dehydrated with gradients of 30%, 50%, 70%, 90%, and 100% ethanol and finally dried using hexamethyldisilazane (H810965, MACKLIN, Shanghai, China) for 8–12 h. The dried samples were coated under vacuum using a 108 Auto Sputter Coater (Cressington, Watford, UK). Spermatozoa were observed through SEM at a magnification of 2500× and an accelerating voltage of 3 kV.
Ultrastructural analysis of spermatozoa
For transmission electron microscopy (TEM; Talos L120C G2, Thermo Scientific, Waltham, MA, USA), sperm samples from patients AY0334 and AY0283 and two normal fertile men were pre-fixed in 2.5% glutaraldehyde at 4°C for 24–48 h. The samples were then washed four times with PBS and post-fixed with 1% osmium acid (18456, Ted Pella, Inc., Altadena, CA, USA) at 4°C for 2 h. Following that, they were washed twice with ultrapure water and block-stained in a 2% aqueous uranyl acetate solution (22400, Electron Microscopy Sciences, Hatfield, PA, USA) for 2 h. The samples were dehydrated in a gradient and permeabilized with a mixture of 100% acetone and EPON 812 resin (18010, Ted Pella, Inc.) in equal volumes for 2 h. After embedding in EPON 812 resin, the samples were placed in a thermostat at 37°C and 45°C for 12 h each and then at 60°C for 36–48 h. The samples were then sectioned using an ultra-thin sectioning machine (EM UC7, Leica, Wetzlar, Germany) at a thickness of 70 nm to 100 nm, then placed on a copper mesh and stained in lead citrate solution, dried, and observed under a 120 kV transmission electron microscope at 18 500×.
Western blot (WB)
Spermatozoa samples from two normal fertile men and patient AY0334 were washed thrice with PBS; the samples were then added to 1× loading buffer (P0015L, Beyotime) diluted with radio immunoprecipitation assay (RIPA) buffer (50 mmol l−1 Tris-HCl, pH 7.4, 150 mmol l−1 NaCl, 1% Triton X-100, 1% sodium dodecyl sulfate, 1% sodium deoxycholate, and 1 mmol l−1 ethylenediaminetetraacetic acid [EDTA; P0013B, Beyotime]) together with a complete EDTA-free protease inhibitor cocktail (P1005, Beyotime) and 1 mmol l−1 phenylmethylsulfonyl fluoride (ST506, Beyotime) and 5 mmol l−1 phosphatase inhibitors (P1081, Beyotime) followed by cooking for 10 min at 100°C in a metal pan. The proteins were separated on a 10% polyacrylamide gel at a constant voltage of 120 V for 90 min; the gel was transferred to a polyvinylidene fluoride membrane at a constant current of 230 mA for 100 min and blocked with 5% milk at 25°C for 1 h. Next, the samples were incubated with outer dynein arm-associated protein (dynein axonemal intermediate chain 2 [DNAI2]) antibody (1:1000, 17533-1-AP, Proteintech, Wuhan, China), radial spoke-associated protein (radial spoke head component 1 [RSPH1]) antibody (1:1000, HPA017382, Sigma-Aldrich), center pair-associated protein (sperm associated antigen 6 [SPAG6]) antibody (1:1000, HPA038440, Sigma-Aldrich), fibrous sheath-associated protein (A-kinase anchoring protein 4 [AKAP4]) antibody (1:1000, HPA020046, Sigma-Aldrich), acrosome-associated protein (actin like 7A [ACTL7A]) antibody (1:1000, HPA021624, Sigma-Aldrich), and acrosin antibody (1:1000, NBP2-14260, Novus, Littleton, CO, USA) separately at 4°C for 12 h. On the second day, they were washed three times with tris buffered saline with Tween-20 (TBST; BS100, Biosharp, Hefei, China) and incubated with the secondary antibodies at 25°C for 1 h. Images were taken with a Tanon 5200 fully automated chemiluminescence image analysis system (Tanon, Shanghai, China). β-actin (TA-09, ZSGB-BIO, Beijing, China) was used as the internal control.
Immunofluorescence (IF) analysis
A sperm sample from patient AY0283 and two normal sperm samples fixed with 4% PFA were applied to slides, permeabilized with 0.1% Triton X-100 (BS084, Biosharp) for 10 min, and then blocked with 10% donkey serum for 2 h at 37°C. To analyze the location and expression of DNAH6 in spermatozoa and the changes in flagellar-associated and acrosome-associated proteins, anti-DNAH6 (rabbit, 1:100, ab122333, Abcam, Cambridge, UK), anti-DNAH1 (rabbit, 1:100, ab122367, Abcam), anti-DNAI2 (rabbit, 1:200, 17533-1-AP, Proteintech), anti-SPAG6 (rabbit, 1:200, HPA038440, Sigma-Aldrich), anti-RSPH1 (rabbit, 1:100, HPA017382, Sigma-Aldrich), and anti-AKAP4 (rabbit, 1:200, HPA020046, Sigma-Aldrich) antibody were co-incubated with monoclonal anti-acetylated-tubulin antibody (mouse, 1:500, T6793, Sigma-Aldrich) at 4°C for 16 h. Anti-ACTL7A (rabbit, 1:100, HPA021624, Sigma-Aldrich) and anti-acrosin antibodies (rabbit, 1:200, NBP2-14260, Novus) were also incubated separately at 4°C for 16 h. We used Alexa Fluor 488 anti-mouse antibody (1:800, Jackson, Lancaster, PA, USA), Alexa Fluor 594 anti-rabbit antibody (1:800, Jackson), and Hoechst 33 342 (1:500, Thermo Scientific) as secondary antibodies. The stained samples were observed with a laser scanning confocal microscope (LSM800, Carl Zeiss AG) in selected channels (Alexa Fluor 594, Alexa Fluor 488, and Hoechst 33 342).
Assisted reproductive technology
The female partner of patient AY0159 underwent standard controlled ovarian hyperstimulation with the gonadotropin-releasing hormone (GnRH) agonist long protocol, and a total of 8 oocytes were obtained. Semen samples were prepared through discontinuous density gradient centrifugation as previously described,28 and seven mature oocytes and motile spermatozoa were selected for ICSI using a micromanipulator system (Olympus, Tokyo, Japan). According the embryo assessment system reported previously, we obtained three day-5 blastocysts (4BB, 4BC, and 4BC) and one day-6 (4BC) blastocyst for the patient.29,30 One day-5 (4BB) blastocyst was transferred fresh to the patient’s partner, and the rest were preserved by vitrification for freezing. Biochemical pregnancy was determined by measuring serum β-human chorionic gonadotropin (hCG) levels on day 14 after embryo transfer. Clinical pregnancy was confirmed via ultrasound 28 days after embryo transfer.
RESULTS
Identification of DNAH6 variants in patients with MMAF and infertility
Following whole-exome sequencing, we were able to identify three individuals with DNAH6 variants (Figure 1). Two novel compound heterozygous variants of DNAH6 from patients AY0334 (c.5264C>T, p.T1755I; and c.8726A>G, p.Q2909R) and AY0159 (c.8852-1G>A; and c.10127T>A, p.V3376D) were isolated from their parents, which were confirmed by using Sanger sequencing. AY0283, from a consanguineous family, was found to carry a homozygous variant (c.9250C>G, p.Q3084E). These variations were rare or absent in the public human genome database and deleterious as predicted by at least two of the SIFT, MutationTaster, and PolyPhen-2 software (Table 1). The affected residues of the DNAH6 protein located between the dynein heavy chain and N-terminal region 2 (DHC-N2) and dynein-heavy chain domains were found to be highly conserved among species and may disturb the tertiary or quaternary protein structure (Figure 1). As a result, we assumed that variants of DNAH6 might be responsible for the infertility phenotypes of these individuals.
Figure 1.
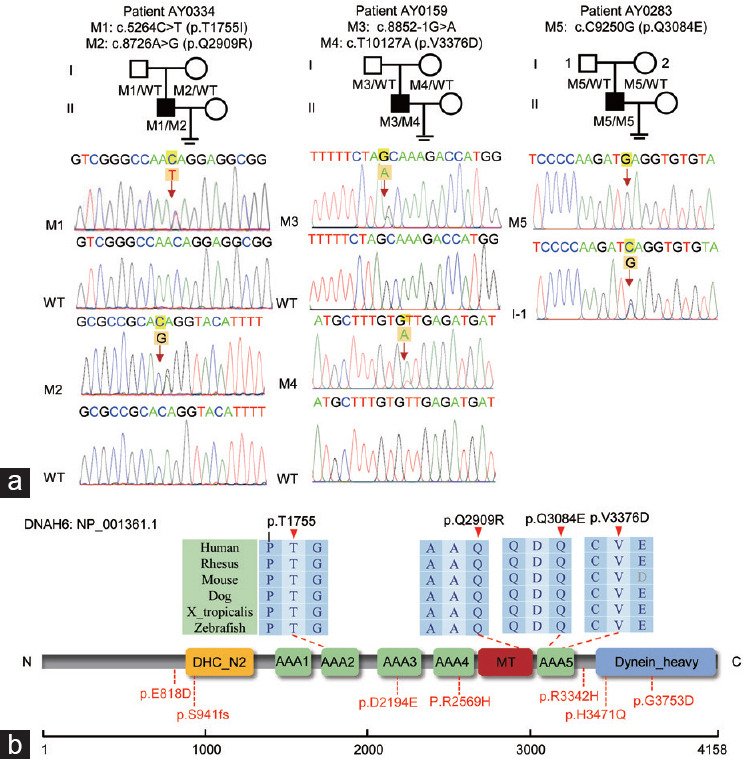
Identification of DNAH6 variants in male infertility. (a) Lineage maps and Sanger sequencing results of patients carrying the DNAH6 variants are shown under their respective families. Black squares indicate mutant patients, namely AY0334, AY0159, and AY0283. Red arrows indicate the positions of point variants or the splicing mutation sites. (b) Protein structure diagram of DNAH6. Positions of the affected amino acid residue sites were illustrated on DNAH6 protein structure, and red arrows indicated the affected amino acid residue sites, which were highly conserved across species. The positions of the affected amino acid residues reported previously are marked in red. Colored squares indicated the different structural domains. M: mutation; WT: wild type; DHC_N2: dynein heavy chain, and N-terminal region 2; dynein heavy: dynein heavy chain and region D6 of dynein motor; MT: microtubule-binding stalk of dynein motor; AAA_1-6: hydrolytic ATP-binding dynein motor region; DNAH6: dynein axonemal heavy chain 6.
Table 1.
Bi-allelic variants of dynein axonemal heavy chain 6 identified in patients with multiple morphological abnormalities of the sperm flagella and the clinical outcomes following intracytoplasmic sperm injection
| Characteristic | Patient AY0334 | Patient AY0159 | Patient AY0283 | Patient in Li et al.24 | Patient in Tu et al.13 | Patient in Gershoni et al.25 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cDNA mutation | c. 5264C>T | c. 8726A>G | c. 8852-1G>A | c. 10127T>A | c. 9250C>G | c. 2454A>T | c. 7706G>A | c. 6582C>A | c. 11258G>A | c. 2823dupT | c. 10025G>A | c.C10413A |
| Protein alteration | p.T1755I | p.Q2909R | / | p.V3376D | p.Q3084E | p.E818D | p.R2569H | p.D2194E | p.G3753D | p.S941fs | p.R3342H | p.H3471Q |
| Mutation type | Missense | Missense | Splicing | Missense | Missense | Missense | Missense | Missense | Missense | Frameshift | Missense | Missense |
| Affected allele | Heterozygous | Heterozygous | Heterozygous | Heterozygous | Homozygous | Heterozygous | Heterozygous | Heterozygous | Heterozygous | Heterozygous | Heterozygous | Homozygous |
| Allele frequency in population | ||||||||||||
| 1KGP | 0.001682 | 0 | 0 | 0 | 0 | 0 | 0.0003994 | / | / | / | 0.001 | / |
| gnomAD | 0 | 0.00001396 | 0 | 0 | 0 | 0.00003227 | 0.001647 | / | / | / | / | / |
| gnomAD-EAS | 0 | 0.0006 | 0 | 0 | 0 | / | / | / | / | / | / | / |
| Function prediction | ||||||||||||
| SIFT | Damaging | Damaging | NA | NA | Damaging | Tolerable | Damaging | Damaging | Damaging | / | Damaging | / |
| PolyPhen-2 | Damaging | Benign | NA | NA | Damaging | Benign | Probably damaging | Probably damaging | Probably damaging | / | Probably damaging | / |
| Mutation taster | Damaging | Damaging | NA | NA | Damaging | / | / | Damaging | Damaging | / | Damaging | / |
| ICSI outcomes | ||||||||||||
| Male age (year) | 30 | 38 | 28 | / | / | / | / | |||||
| Female age (year) | / | 33 | / | / | / | / | / | |||||
| ICSI cycles (n) | / | 1 | / | 2 | 1 | / | / | |||||
| Oocytes retrieved (n) | / | 8 | / | / | / | / | / | |||||
| Oocytes injected (n) | / | 7 | / | / | / | / | / | |||||
| Fertilization rate, % (n/total) | / | 100.0 (7/7) | / | / | / | / | / | |||||
| Cleavage rate, % (n/total) | / | 100.0 (7/7) | / | / | / | / | / | |||||
| 8-cell formation rate, % (n/total) | / | 71.4 (5/7) | / | / | / | / | / | |||||
| Blastocyst formation rate, % (n/total) | / | 57.1 (4/7) | / | / | / | / | / | |||||
| High quality blastocyst rate, % (n/total) | / | 14.3 (1/7) | / | / | / | / | / | |||||
| Transfer cycles (n) | / | 1 | / | / | / | / | / | |||||
| Embryos transferred per cycle (n) | / | 1 | / | / | / | / | / | |||||
| Implantation rate (%) | / | 100.0 | / | / | / | / | / | |||||
| Clinical pregnancy rate per transfer cycle (%) | / | 100.0 | / | 0 | 100.0 | / | / | |||||
| Miscarriage rate (%) | / | 0 | / | / | 100.0 | / | / | |||||
Reference sequence accession number of DNAH6 is NM_001370.2. 1KGP: 1000 Genomes Project; gnomAD: the Genome Aggregation Database; NA: not available; /: not applicable; ICSI: intracytoplasmic sperm injection; DNAH6: dynein axonemal heavy chain 6; SIFT: Sorting Intolerant From Tolerant; EAS: East Asian
Defects in sperm morphology and ultrastructure in patients with DNAH6 variants
The semen samples obtained from study participants were analyzed according to the criteria of the WHO. The semen volume and sperm concentration were normal. However, sperm from individuals carrying DNAH6 variants had very low motility and almost no progressive motility (Table 2). Sperm morphology was observed using Papanicolaou or H&E staining under a light microscope (Figure 2). Compared with the regular oval sperm head and long smooth flagellum in normal sperm, approximately 50% of sperm in patients with DNAH6 variants presented with amorphous heads, and nearly 30% of sperm had tapered heads (Table 2). The morphological abnormalities of the sperm flagella of the mutants were mainly absent or coiled.
Table 2.
Semen routine parameters and sperm morphology of men harboring bi-allelic dynein axonemal heavy chain 6 variants
| Characteristic | Patient AY0334 | Patient AY0159 | Patient AY0283 | Reference values |
|---|---|---|---|---|
| Semen parameters | ||||
| Semen volume (ml) | 3.2 | 2.5 | 5.4 | >1.5a |
| Sperm concentration (106 ml−1) | 12.8 | 20.2 | 120.5 | >15.0a |
| Motility (%) | 15.3 | 8.9 | 5.1 | >40.0a |
| Progressive motility (%) | 2.1 | 2.3 | 1.4 | >32.0a |
| Sperm morphology | ||||
| Normal head (%)c | 19.1 | 16.7 | 13.1 | NA |
| Amorphous head (%)c | 42.6 | 46.6 | 72.5 | NA |
| Pyramid head (%)c | 27.3 | 26.2 | 0.5 | <3.0b |
| Vacuolar head (%)c | 2.5 | 1.4 | 0 | NA |
| Small acrosome (%)c | 8.1 | 9.0 | 13.9 | NA |
| Normal flagella (%)d | 33.3 | 27.9 | 25.4 | >23.0b |
| Absent flagella (%)d | 35.8 | 46.6 | 2.0 | <5.0b |
| Short flagella (%)d | 16.9 | 9.8 | 13.9 | <1.0b |
| Coiled flagella (%)d | 12.9 | 13.7 | 49.3 | <17.0b |
| Angulation (%)d | 1.0 | 2.0 | 7.9 | <13.0b |
| Irregular caliber (%)d | 0.1 | 0 | 1.5 | <2.0 |
aThe reference limit set according to WHO 6th edition standards.6 bReference values shown by observation of morphologically abnormal sperm in fertile individuals.37 cSperm head morphology in patients. dSperm flagellar morphology in patients. NA: not available; WHO: World Health Organization
Figure 2.
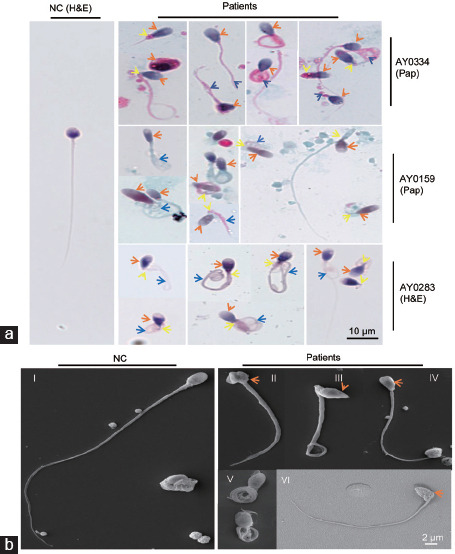
The morphology of spermatozoa from men with DNAH6 variants. (a) Normal spermatozoa and those of patient AY0223 were stained with H&E, and sperm of patients AY0334 and AY0159 were stained with Pap, and it was observed that the sperm morphology of the mutant patients was significantly abnormal compared to normal spermatozoa. The head of the spermatozoa in the mutant patients appeared malformed (orange arrows), such as amorphous head, tapered head or small acrosome; the flagella were mostly coiled (blue arrows). Yellow arrows indicate the abnormal sperm necks. Scale bar=10 µm. (b) Scanning electron microscopy was applied to analyze the morphology of spermatozoa from normal (I) and mutant patients. Sperm from patients show malformations of the sperm head (II, III, IV and VI) with short flagella and normal head with coiled flagella (V). The orange arrows indicate the abnormal sperm heads. Scale bar=2 µm. NC: normal control. Pap: papanicolaou; H&E: hematoxylin and eosin; DNAH6: dynein axonemal heavy chain 6.
To investigate the sperm head and flagellar ultrastructure of men with bi-allelic DNAH6 variants, we performed TEM analyses of samples obtained from three men. The normal sperm head comprises a tightly packed nucleus surrounded by three layers: the outer acrosomal membrane (OAM), acrosome (AC), and inner acrosomal membrane (IAM). The absence of the OAM and AC, loose and vacuolar nuclei, and irregular sperm heads were mainly observed, and a proportion of the disordered mitochondrial sheath was also observed in sperm from patients with DNAH6 variants (Figure 3a). Additionally, an analysis of the ultrastructure of the sperm flagella using TEM revealed the absence of a central pair in sperm from patients AY0334 and AY0283 carrying DNAH6 variants, and the radial spokes (RS) were also affected. The abundance of the outer dense fibers (ODF) and peripheral microtubule doublets (MTs) was not significantly altered (Figure 3b). Overall, these findings indicate that DNAH6 deficiencies cause sperm morphology defects associated with a disruption in the sperm head and flagellar ultrastructure and affect different components of the flagellar structure.
Figure 3.
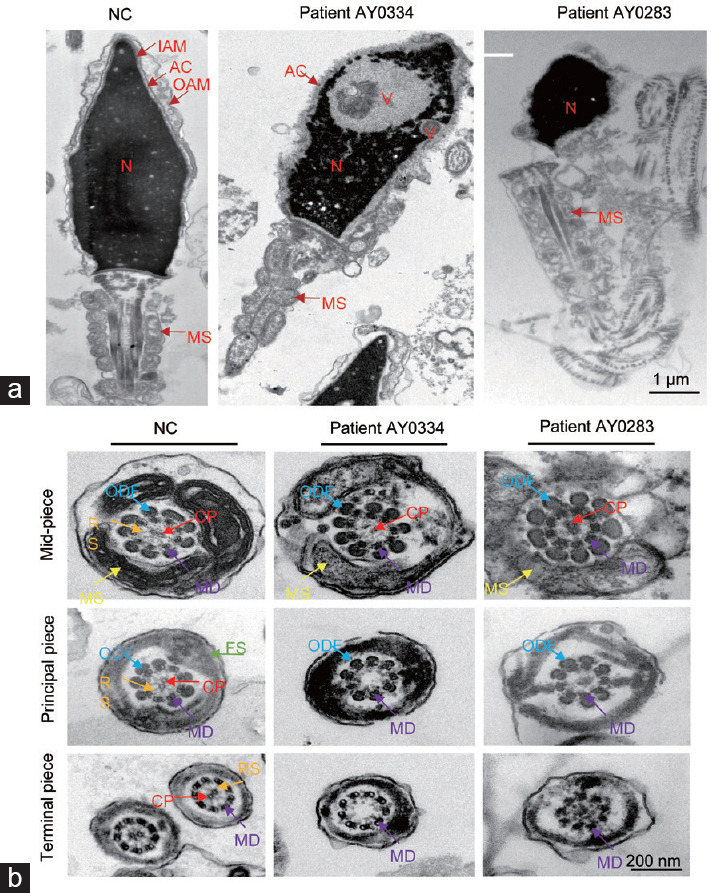
The ultrastructure analysis of sperm defects in patients with DNAH6 variants. (a) Transmission electron microscopy longitudinal sections of sperm from fertile man and subjects with DNAH6 variants revealed the absence or disruption of the acrosome and outer acrosome membrane of spermatozoa heads, vacuoles in the head, head malformations, and disorganized mitochondrial sheaths in mutant patients. Scale bar=1 µm. (b) Transmission electron microscopy of a cross-section of sperm from variants and normal control sperm. The normal sperm have a typical “9+2” microtubule structure in the mid-piece, principal piece and terminal piece; whereas in DNAH6 patients AY0334 and AY0283, there was no central pair. Scale bar=200 nm MD: doublet microtubules; ODF: outer dense fibers; CP: central pair of microtubules; RS: radial spoke; FS: fibrous sheath; MS: mitochondrial sheath; IAM: inner acrosome membrane; OAM: outer acrosome membrane; AC: acrosome; N: nuclei; V: vacuoles; DNAH6: dynein axonemal heavy chain 6.
Molecular alterations in the flagella associated with DNAH6 deficiencies
To further assess the potential contribution of DNAH6 variants to infertility and sperm defects, we conducted an IF staining analysis of the localization and expression of DNAH6 and additional acrosomal- and flagella-associated proteins. DNAH6 was localized throughout the flagella of normal sperm, and the signals were reduced in patient AY0283 carrying a DNAH6 variant (Figure 4a). Furthermore, to investigate the impact of DNAH6 variants on sperm flagella, we analyzed the internal dynein arm-associated protein DNAH1. In normal sperm, the DNAH1 signals were distributed along the sperm flagella and concentrated on the mid-piece. The signals on the sperm mid-piece decreased remarkably in sperm from patient AY0283, and the signals of tubulin did not change (Figure 4b). DNAI2, a component of ODAs, was also examined using IF staining in patient AY0283 and WB analyses in patient AY0334. Notably, we found that the expression of DNAI2 was reduced or almost absent in sperm from AY0283 (Figure 4c), which was confirmed in patient AY0334 using a WB assay (Figure 4d). These findings implied that variants in DNAH6 reduced the expression levels of DNAH1 without affecting each other’s protein level or location, as well as influenced the DNAI2 expression.
Figure 4.
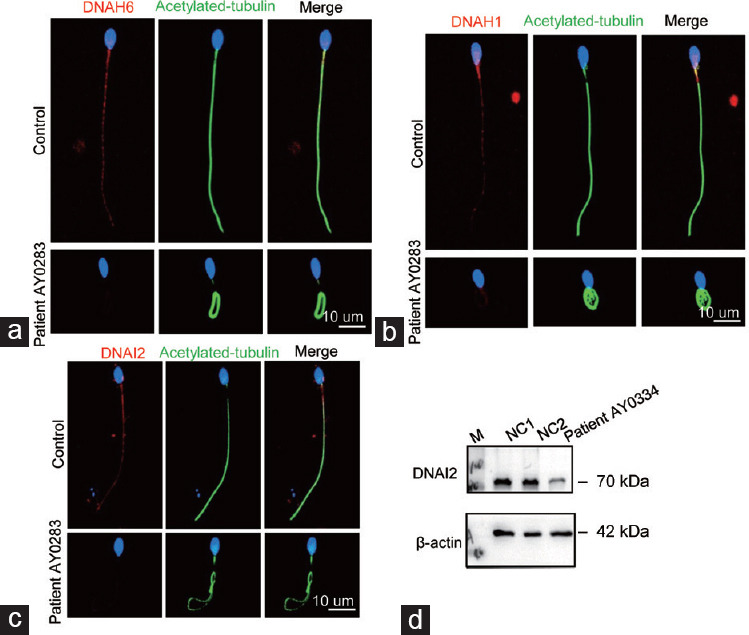
Illustration of the immunofluorescence staining localization of DNAH6 and the results of DNAH1 and DNAI2 staining. (a) Spermatozoa from normal and mutant patients were stained for DNAH6 (red). DNAH6 was localized throughout the flagellum of spermatozoa, whereas patient AY0283 expression was reduced in DNAH6 mutant patients. (b) Normal and mutant patients’ spermatozoa were stained using the internal dynamin arm -associated protein DNAH1 (red). In contrast, DNAH1 expression was reduced in mutant patients. (c) The analysis of spermatozoa from normal and mutant patients using the outer dynamin arm-associated protein DNAI2 (red) showed that DNAI2 expression was reduced in patients with DNAH6 mutation. Scale bar=10 µm. (d) DNAI2 protein level in patient AY0334 was detected by Western blot and found to be reduced. β-actin was used as internal reference. DNAH6: dynein axonemal heavy chain 6; DNAH1: dynein axonemal heavy chain 1; DNAI2: dynein axonemal intermediate chain 2: M: maker; NC: normal control.
To investigate the ultrastructural defects of sperm flagella observed using TEM, we examined the presence and levels of several proteins associated with flagellar substructures using IF staining and WB, including the component of the center pair (CP; SPAG6), radial spoke-associated protein (RSPH1), and fibrous sheath (FS) component (AKAP4). We found that the SPAG6 and RSPH1 signals were almost absent in most of the sperm from patient AY0283 with a DNAH6 variant compared with those in the control by IF staining (Supplementary Figure 1a (192.7KB, tif) and 1b (192.7KB, tif) ). Moreover, the AKAP4 signal was distributed along the principal piece of the sperm in the control, which was slightly reduced in sperm from patient AY0283 (Supplementary Figure 1c (192.7KB, tif) ). These results were also confirmed by WB in sperm from patient AY0334 (Supplementary Figure 1d (192.7KB, tif) ). Together, our data suggest that CP is directly affected by the DNAH6 variant.
Defective acrosome in sperm from patients with DNAH6 variants
Most sperm acrosomes were defective in patients with DNAH6 variants, as observed in TEM. To further explore the possible role of DNAH6 in sperm head development, we performed IF and WB assays of acrosin and ACTL7A acrosome-associated proteins. We observed that acrosin and ACTL7A were localized on the acrosome of normal sperm heads but were nearly absent in a large proportion of sperm from patient AY0283 with DNAH6 variant (Supplementary Figure 2a (100.5KB, tif) and 2b (100.5KB, tif) ). The protein levels of acrosin and ACTL7A were significantly decreased in the sperm of patient AY0334 (Supplementary Figure 2c (100.5KB, tif) ). These observations suggest that sperm heads were severely affected by DNAH6 variants.
Outcomes of ICSI in patients with DNAH6 variants
In the present study, only one patient with a DNAH6 variant (AY0159) received ICSI treatment, while his partner was treated with a GnRH agonist to induce ovulation. Eight oocytes were retrieved, of which seven were successfully injected and cleaved. Only one high-quality blastocyst was formed and transferred. A healthy girl was born after 38 weeks (Table 1). These results suggest that patients with MMAF resulting from DNAH6 variants can be effectively treated with ICSI, a discovery that represents a major step forward in the clinical treatment of MMAF.
DISCUSSION
DNAH6 is a member of the kinesin family, which is involved in cilia and flagellar beating by hydrolyzing ATPase. The expression levels of DNAH6 are significantly upregulated in human testicular tissues. Genetic alterations in DNAH6 are pathogenic for MMAF;13,14 in this study, we identified novel DNAH6 variants in patients with MMAF who were excluded from the PCD phenotype. Sperm from these patients presented with the MMAF phenotype and displayed head malformations. Additionally, the findings suggest that the inheritance of DNAH6 is consistent with an autosomal recessive pattern. The mutant sites identified in this study were highly conserved among species. We identified variants in DNAH6 that are rare or absent in public human databases, and they were predicted to be pathogenic or absent at these mutated sites.
The critical component of the sperm flagellum is the axoneme. Currently, the genes identified to be associated with the IDA include DNAH1,7 dynein axonemal heavy chain 2 (DNAH2),31 DNAH6,13 dynein axonemal heavy chain 7 (DNAH7),32 and DNAH10.14 Furthermore, peripheral duplexes are attached to the central pair by a T-shaped multiprotein structure called radial spoke (RS).33 However, the integrity and stability of the axoneme structure serve as the basis of flagellar motility.
Variations in DNAH6 are reportedly associated with PCD; these might act both recessively and possibly through trans-heterozygous interactions with dynein axonemal heavy chain 5 (DNAH5), DNAH1, or dynein axonemal intermediate chain 1 (DNAI1).23 Subsequently, Tu et al.13 found that DNAH6 signals are localized in the sperm flagellum, and compound heterozygous variants of DNAH6 cause protein structure misfolding, which reduces ATPase activity and microtubule-binding ability, leading to the MMAF phenotype. Moreover, Li et al.24 discovered that DNAH6 is localized in the sperm neck, and compound heterozygous missense variants of DNAH6 cause acephalic spermatozoa associated with globozoospermia. In this study, we identified novel DNAH6 variants that contribute to a decrease in protein levels, which we speculate have significant implications for the sperm head and flagella. According to the ultrastructure observed, the central pairs of sperm flagella, IDA, and RS are all absent in patients with DNAH6 variants. Furthermore, various defects exist in the sperm head morphology of these patients. We observed a significant decrease in the expression of DNAH1 of the IDA in patients with DNAH6 variants. We also found a remarkable reduction in SPAG6 and RSPH1, which are markers of central pairs and RS, respectively, and the FS marker, AKAP4, which was unevenly distributed in sperm from patients with DNAH6 variants. Thus, it was confirmed that IDA deletion directly affects RS, which consequently causes CP deficiency and disorganization of FSs, indicating that DNAH6 variants lead to MMAF. Furthermore, the presence and localization of acrosome-associated proteins acrosin and ACTL7A were severely disturbed in patients with DNAH6 variants, which was consistent with the results observed in a previous study by Li et al.24 These findings imply that DNAH6 plays a key role in sperm head deformation and flagellar development during spermatogenesis.
In recent years, with an increasing number of couples being subjected to infertility, assisted reproduction techniques have become an attractive option for couples with infertility. The International Committee for Monitoring Assisted Reproductive Technology (ICMAR) retrospective report for 2013 showed that the percentage of ICSI in the total number of in vitro treatments (ICSI vs in vitro fertilization) was 68.9%.34 Currently, ICSI is a widely used assisted reproductive technology and is the indicated choice for couples with male-factor infertility.35 Previous studies found that in two patients with DNAH6 variants, one still did not achieve pregnancy after two cycles, and the other patient acquired pregnancy after transferring three embryos. However, no fetal heartbeat was detected on the ultrasound 28 days later (Table 1).13,24 In the present study, one patient with a DNAH6 mutation received ICSI treatment, and the outcomes were compared with those in other patients with MMAF;36 we found that only the high-quality blastocyst rate was lower than that in others. This could be affected by DNAH6 variants or other factors, but as the result was only observed in one patient, it should be further confirmed by the outcomes of other patients with DNAH6 variants.
We obtained semen samples only from patient AY0334 with the DNAH6 mutation. When we acquired sperm morphology slices from patient AY0159, we found that the patient had malformations in both the head and flagella of the sperm. In addition, we will continue to follow the results of ICSI treatment in patients AY0334 and AY0283.
In conclusion, we identified novel mutant sites in DNAH6 in patients with MMAF, and all patients presented with sperm flagella morphological defects that are consistent with the MMAF phenotype. In addition, sperm heads displayed various abnormalities. As a result, our findings expanded the genetic spectrum of men with MMAF carrying DNAH6 variants, provided a better understanding of the sperm phenotypes of patients with these variants, and demonstrated that ICSI treatment could be recommended for patients with asthenoteratozoospermia associated with DNAH6 variants.
AUTHOR CONTRIBUTIONS
ZMS, YTZ, MRL, and YPX participated in the data integration, and writing and modification of the manuscript. ZMS, SCG, HY, and MG performed the experiments. KKL analyzed the data. DDT contributed to the sample collection. KKL, DDT, YPX, and MRL provided important guidance for the study. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
Immunofluorescence and Western blotting analysis of SPAG6, RSPH1 and AKAP4 in patients with DNAH6 mutation are demonstrated. (a) Detection of variants and normal spermatozoa using the center pair of related protein SPAG6 (red), DNA was stained using Hoechst (blue). It was demonstrated that SPAG6 expression was dramatically reduced in patients with DNAH6 mutation AY0283. (b) Both normal and mutant spermatozoa were stained with radial spoke-associated protein RSPH1 (red) and monoclonal anti-tubulin acetylated antibody (green) together, and DNA was stained with Hoechst (blue). Radial spoke-associated protein RSPH1 expression was revealed to be reduced in expression in the mutant patients. (c) Immunofluorescence staining was performed on normal and mutant patient sperm with the fibrous sheath component AKAP4 (red) and monoclonal anti-tubule acetylation antibody (green), and Hoechst (blue) was used for staining of nuclei. The expression of AKAP4 showed a slightly reduced in patient AY0283. Scale bar represents 10 µm. (d) Expression changes of SPAG6, RSPH1 and AKAP4 proteins in variants AY0334 were analyzed by Western blot. β-actin was used as internal reference.
Illustration of immunofluorescence and protein analysis of acrosome-associated proteins ACTL7A and acrosin from patients with DNAH6 variants. The normal control and mutant spermatozoa were separately stained with the acrosome-associated proteins (a) acrosin (red) and (b) ACTL7A (red), and the nuclei were stained with Hoechst (blue). ACTL7A and acrosin are localized to the acrosome in normal individuals, whereas most of the acrosomes of spermatozoa from mutant patients showed deletion. Scale bar=10 µm. (c) Protein levels of ACTL7A and acrosin were measured in sperm from patient AY0334 using Western blot. The levels of the acrosome-associated proteins ACTL7A and acrosin were found to be decreased in mutant sperm compared to normal sperm. β-actin was used as internal reference.
ACKNOWLEDGMENTS
We are grateful to the patients and their families, as well as to the fertile men who provided semen samples for this study. We would also like to thank Zhen-Hai Tang and Hai-Jian Cai at the Center for Scientific Research of Anhui Medical University (Hefei, China) for their assistance with the SEM and TEM. This work was supported by the National Natural Science Foundation of China (No. 82071705), University Outstanding Youth Program of Anhui Provincial Education Department (2022AH030113), University Outstanding Young Talents Support Program (gxyq2021174), and Postgraduate Innovation Research and Practice Program of Anhui Medical University (No. YJS20210327).
REFERENCES
- 1.Fainberg J, Kashanian JA. Recent advances in understanding and managing male infertility. F1000Res. 2019;8 doi: 10.12688/f1000research.17076.1. F1000 Faculty Rev–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal A, Baskaran S, Parekh N, Cho CL, Henkel R, et al. Male infertility. Lancet. 2021;397:319–33. doi: 10.1016/S0140-6736(20)32667-2. [DOI] [PubMed] [Google Scholar]
- 3.Halder A, Kumar P, Jain M, Kalsi AK. Genomics: tool to predict and prevent male infertility. Front Biosci (Schol Ed) 2017;9:448–508. doi: 10.2741/s496. [DOI] [PubMed] [Google Scholar]
- 4.Joseph S, Mahale SD. Male infertility knowledgebase: decoding the genetic and disease landscape. Database (Oxford) 2021. 2021:baab049. doi: 10.1093/database/baab049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coutton C, Escoffier J, Martinez G, Arnoult C, Ray PF. Teratozoospermia: spotlight on the main genetic actors in the human. Hum Reprod Update. 2015;21:455–85. doi: 10.1093/humupd/dmv020. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen. 6th ed. Geneva: World Health Organization; 2021. [Google Scholar]
- 7.Ben Khelifa M, Coutton C, Zouari R, Karaouzène T, Rendu J, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shahrokhi SZ, Salehi P, Alyasin A, Taghiyar S, Deemeh MR. Asthenozoospermia: cellular and molecular contributing factors and treatment strategies. Andrologia. 2020;52:e13463. doi: 10.1111/and.13463. [DOI] [PubMed] [Google Scholar]
- 9.Toure A, Martinez G, Kherraf ZE, Cazin C, Beurois J, et al. The genetic architecture of morphological abnormalities of the sperm tail. Hum Genet. 2021;140:21–42. doi: 10.1007/s00439-020-02113-x. [DOI] [PubMed] [Google Scholar]
- 10.Inaba K. Molecular basis of sperm flagellar axonemes: structural and evolutionary aspects. Ann N Y Acad Sci. 2007;1101:506–26. doi: 10.1196/annals.1389.017. [DOI] [PubMed] [Google Scholar]
- 11.Lv M, Liu W, Chi W, Ni X, Wang J, et al. Homozygous mutations in DZIP1 can induce asthenoteratospermia with severe MMAF. J Med Genet. 2020;57:445–53. doi: 10.1136/jmedgenet-2019-106479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Q, Guo Q, Guo W, Song S, Wang N, et al. Loss of CEP70 function affects acrosome biogenesis and flagella formation during spermiogenesis. Cell Death Dis. 2021;12:478. doi: 10.1038/s41419-021-03755-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu C, Nie H, Meng L, Yuan S, He W, et al. Identification of DNAH6 mutations in infertile men with multiple morphological abnormalities of the sperm flagella. Sci Rep. 2019;9:15864. doi: 10.1038/s41598-019-52436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu C, Cong J, Zhang Q, He X, Zheng R, et al. Bi-allelic mutations of DNAH10 cause primary male infertility with asthenoteratozoospermia in humans and mice. Am J Hum Genet. 2021;108:1466–77. doi: 10.1016/j.ajhg.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan C, Meng L, Lv M, He X, Sha Y, et al. Bi-allelic variants in DNHD1 cause flagellar axoneme defects and asthenoteratozoospermia in humans and mice. Am J Hum Genet. 2022;109:157–71. doi: 10.1016/j.ajhg.2021.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Liu Y, Li Y, Li K, Xu C, et al. DNALI1 deficiency causes male infertility with severe asthenozoospermia in humans and mice by disrupting the assembly of the flagellar inner dynein arms and fibrous sheath. Cell Death Dis. 2023;14:127. doi: 10.1038/s41419-023-05653-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W, He X, Yang S, Zouari R, Wang J, et al. Bi-allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am J Hum Genet. 2019;104:738–48. doi: 10.1016/j.ajhg.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong J, Wang X, Amiri-Yekta A, Wang L, Kherraf ZE, et al. Homozygous mutations in CCDC34 cause male infertility with oligoasthenoteratozoospermia in humans and mice. J Med Genet. 2022;59:710–8. doi: 10.1136/jmedgenet-2021-107919. [DOI] [PubMed] [Google Scholar]
- 19.Liu C, Lv M, He X, Zhu Y, Amiri-Yekta A, et al. Homozygous mutations in SPEF2 induce multiple morphological abnormalities of the sperm flagella and male infertility. J Med Genet. 2020;57:31–7. doi: 10.1136/jmedgenet-2019-106011. [DOI] [PubMed] [Google Scholar]
- 20.He X, Li W, Wu H, Lv M, Liu W, et al. Novel homozygous CFAP69 mutations in humans and mice cause severe asthenoteratospermia with multiple morphological abnormalities of the sperm flagella. J Med Genet. 2019;56:96–103. doi: 10.1136/jmedgenet-2018-105486. [DOI] [PubMed] [Google Scholar]
- 21.Tang S, Wang X, Li W, Yang X, Li Z, et al. Biallelic mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2017;100:854–64. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coutton C, Martinez G, Kherraf ZE, Amiri-Yekta A, Boguenet M, et al. Bi-allelic mutations in ARMC2 lead to severe astheno-teratozoospermia due to sperm flagellum malformations in humans and mice. Am J Hum Genet. 2019;104:331–40. doi: 10.1016/j.ajhg.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Yagi H, Onuoha EO, Damerla RR, Francis R, et al. DNAH6 and its interactions with PCD genes in heterotaxy and primary ciliary dyskinesia. PLoS Genet. 2016;12:e1005821. doi: 10.1371/journal.pgen.1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Sha YW, Xu X, Mei LB, Qiu PP, et al. DNAH6 is a novel candidate gene associated with sperm head anomaly. Andrologia. 2018;50:e12953. doi: 10.1111/and.12953. [DOI] [PubMed] [Google Scholar]
- 25.Gershoni M, Hauser R, Yogev L, Lehavi O, Azem F, et al. A familial study of azoospermic men identifies three novel causative mutations in three new human azoospermia genes. Genet Med. 2017;19:998–1006. doi: 10.1038/gim.2016.225. [DOI] [PubMed] [Google Scholar]
- 26.Liu YJ, Zhuang XJ, An JT, Jiang H, Li R, et al. Identification of risk genes in Chinese nonobstructive azoospermia patients based on whole-exome sequencing. Asian J Androl. 2023;25:66–72. doi: 10.4103/aja202275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li K, Wang G, Lv M, Wang J, Gao Y, et al. Bi-allelic variants in DNAH10 cause asthenoteratozoospermia and male infertility. J Assist Reprod Genet. 2022;39:251–9. doi: 10.1007/s10815-021-02306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu F, Liu C, Wang F, Yang X, Zhang J, et al. Mutations in PMFBP1 cause acephalic spermatozoa syndrome. Am J Hum Genet. 2018;103:188–99. doi: 10.1016/j.ajhg.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2000;73:1155–8. doi: 10.1016/s0015-0282(00)00518-5. [DOI] [PubMed] [Google Scholar]
- 30.Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26:1270–83. doi: 10.1093/humrep/der037. [DOI] [PubMed] [Google Scholar]
- 31.Gao Y, Tian S, Sha Y, Zha X, Cheng H, et al. Novel bi-allelic variants in DNAH2 cause severe asthenoteratozoospermia with multiple morphological abnormalities of the flagella. Reprod Biomed Online. 2021;42:963–72. doi: 10.1016/j.rbmo.2021.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Gao Y, Liu L, Shen Q, Fu F, Xu C, et al. Loss of function mutation in DNAH7 induces male infertility associated with abnormalities of the sperm flagella and mitochondria in human. Clin Genet. 2022;102:130–5. doi: 10.1111/cge.14146. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa T. Axoneme structure from motile cilia. Cold Spring Harb Perspect Biol. 2017;9:a028076. doi: 10.1101/cshperspect.a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banker M, Dyer S, Chambers GM, Ishihara O, Kupka M, et al. International Committee for Monitoring Assisted Reproductive Technologies (ICMART): world report on assisted reproductive technologies, 2013. Fertil Steril. 2021;116:741–56. doi: 10.1016/j.fertnstert.2021.03.039. [DOI] [PubMed] [Google Scholar]
- 35.Haddad M, Stewart J, Xie P, Cheung S, Trout A, et al. Thoughts on the popularity of ICSI. J Assist Reprod Genet. 2021;38:101–23. doi: 10.1007/s10815-020-01987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wambergue C, Zouari R, Fourati Ben Mustapha S, Martinez G, Devillard F, et al. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum Reprod. 2016;31:1164–72. doi: 10.1093/humrep/dew083. [DOI] [PubMed] [Google Scholar]
- 37.Auger J, Jouannet P, Eustache F. Another look at human sperm morphology. Hum Reprod. 2016;31:10–23. doi: 10.1093/humrep/dev251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunofluorescence and Western blotting analysis of SPAG6, RSPH1 and AKAP4 in patients with DNAH6 mutation are demonstrated. (a) Detection of variants and normal spermatozoa using the center pair of related protein SPAG6 (red), DNA was stained using Hoechst (blue). It was demonstrated that SPAG6 expression was dramatically reduced in patients with DNAH6 mutation AY0283. (b) Both normal and mutant spermatozoa were stained with radial spoke-associated protein RSPH1 (red) and monoclonal anti-tubulin acetylated antibody (green) together, and DNA was stained with Hoechst (blue). Radial spoke-associated protein RSPH1 expression was revealed to be reduced in expression in the mutant patients. (c) Immunofluorescence staining was performed on normal and mutant patient sperm with the fibrous sheath component AKAP4 (red) and monoclonal anti-tubule acetylation antibody (green), and Hoechst (blue) was used for staining of nuclei. The expression of AKAP4 showed a slightly reduced in patient AY0283. Scale bar represents 10 µm. (d) Expression changes of SPAG6, RSPH1 and AKAP4 proteins in variants AY0334 were analyzed by Western blot. β-actin was used as internal reference.
Illustration of immunofluorescence and protein analysis of acrosome-associated proteins ACTL7A and acrosin from patients with DNAH6 variants. The normal control and mutant spermatozoa were separately stained with the acrosome-associated proteins (a) acrosin (red) and (b) ACTL7A (red), and the nuclei were stained with Hoechst (blue). ACTL7A and acrosin are localized to the acrosome in normal individuals, whereas most of the acrosomes of spermatozoa from mutant patients showed deletion. Scale bar=10 µm. (c) Protein levels of ACTL7A and acrosin were measured in sperm from patient AY0334 using Western blot. The levels of the acrosome-associated proteins ACTL7A and acrosin were found to be decreased in mutant sperm compared to normal sperm. β-actin was used as internal reference.


