Abstract
APS patients exhibit a wide clinical heterogeneity in terms of the disease’s origin and progression. This diversity can be attributed to consistent aPL profiles and other genetic and acquired risk factors. Therefore, understanding the pathophysiology of APS requires the identification of specific molecular signatures that can explain the pro-atherosclerotic, pro-thrombotic and inflammatory states observed in this autoimmune disorder. In recent years, significant progress has been made in uncovering gene profiles and understanding the intricate epigenetic mechanisms and microRNA changes that regulate their expression. These advancements have highlighted the crucial role played by these regulators in influencing various clinical aspects of APS. This review delves into the recent advancements in genomic and epigenetic approaches used to uncover the mechanisms contributing to vascular and obstetric involvement in APS. Furthermore, we discuss the implementation of novel bioinformatics tools that facilitate the investigation of these mechanisms and pave the way for personalized medicine in APS.
Keywords: antiphospholipid syndrome, transcriptomics, microRNA, epigenetics, bioinformatics
Rheumatology key messages.
Genomic and epigenomic research in APS has grown, driven by novel technologies identifying new potential signatures.
Limited multi-omics, advanced tools and small APS cohorts hinder comprehensive profiling of cell types.
Integrating clinical and molecular data using high-throughput technologies and AI can advance precision medicine for APS patients.
Introduction
APS is an autoimmune thrombophilia marked by circulating aPL antibodies, pivotal in vascular thrombosis and obstetric complications. In the past decade, extensive research has led to the development of new ACR/EULAR criteria for diagnosing APS, which were very recently published. These criteria require the presence of at least one positive aPL antibody test within 3 years of identifying an aPL-associated clinical criterion as an entry requirement. Subsequently, additional weighted criteria, ranging from 1 to 7 points each, are grouped into two laboratory domains [LA functional coagulation assays and ELISAs for IgG/IgM aCL and/or IgG/IgM anti-β2-glycoprotein I (GPI) antibodies] and six clinical domains (macrovascular venous thromboembolism, macrovascular arterial thrombosis, microvascular, obstetric, cardiac valve and haematologic). Patients who accumulate at least 3 points from both the laboratory and clinical domains are categorized as having APS [1, 2]. Also, it is linked to other manifestations beyond these new diagnostic criteria: atherosclerosis, neurologic disorders and pulmonary hypertension [3–5].
Most of the positive results in LA, aCL and anti-β2GPI assays can be attributed to antibodies targeting β2GPI. However, antibodies against the phosphatidylserine–prothrombin (PS-PT) complex can also contribute to LA effects, either alone or in combination with anti-β2GPI antibodies in samples that test positive for LA [6]. The presence of LA alone carries greater predictive power for clinical manifestations compared with isolated positive results in aCL or anti-β2GPI assays, particularly when LA is accompanied by positivity for anti-PS-PT antibodies. These antibodies seem to enhance overall predictability when combined with the three formal classification laboratory assays, a combination referred to as ‘tetra-positive’ patients [7].
aPL antibodies not only serve as markers of the disease, but also play a crucial role in driving the pathogenesis of APS. Initially, aPL antibodies were thought to mainly interfere with the anticoagulant systems involved in regulating coagulation and fibrinolysis. Subsequent research revealed their additional role in activating various blood and innate immune cells, resulting in pro-coagulant and pro-inflammatory effects. Thus it has been demonstrated that APS serves as a valuable model for understanding thrombo-inflammation, the intricate process linking inflammation and thrombosis, contributing to cardiovascular disease (CVD) development [8].
There is significant variability in APS symptoms among individuals, with each person presenting a unique combination of manifestations. This heterogeneity may be attributed to consistent aPL profiles alongside other genetic and acquired risk factors. Therefore, to gain a comprehensive understanding of APS pathophysiology, it is imperative to identify APS-specific gene signatures that can explain the pro-atherosclerotic, pro-thrombotic and inflammatory states observed in this autoimmune disorder.
Over the past few years, remarkable progress has been made in uncovering gene profiles as well as understanding the intricate workings of epigenetic mechanisms and microRNA (miRNA) changes that govern their expression. These advancements have shed light on the pivotal role played by these regulators in influencing various clinical aspects of APS [9].
This review explores the recent advancements in genomic and epigenetic approaches utilized to uncover the mechanisms contributing to vascular and obstetric involvement in APS. It seeks to elucidate the clinical heterogeneity observed among APS patients concerning the disease’s origin and progression. Additionally, we discuss the implementation of novel bioinformatics tools that enable the investigation of these mechanisms and pave the way for personalized medicine in APS.
Genomic and epigenetic changes associated with aPLs and clinical profiles in APS patients: previous studies
Numerous studies have identified key genes associated with thrombosis, inflammation and endothelial dysfunction in APS patients, including TF, VEGF, Flt1, IL8, PAR1, PAR2, TLR2 and TLR4, which are highly expressed in immune and vascular cells and promote thrombin production. Research has explored various aspects, such as the impact of anti-β2GPI antibodies on gene expression in human umbilical vein endothelial cells (HUVECs), unique molecular pathways in monocytes exposed to aPLs and genomic patterns in monocytes associated with atherosclerosis and thrombosis. A systematic review identified genetic risk factors for thrombotic APS, crucial for immunity and coagulation [10–18].
It is widely acknowledged that the genome produces a vast array of regulatory non-protein-coding RNAs, including miRNAs, which play crucial roles as major transcriptional and post-transcriptional regulators of gene expression. Previous studies showed that miRNAs like miR-19b and miR-20a play regulatory roles in APS, with cardiovascular miRNAs decreasing in APS monocytes and neutrophils. Additionally, plasma miRNA signatures correlated with clinical features. Lastly, epigenetic analysis revealed genome-wide DNA methylation signatures in neutrophils, potentially impacting foetal health and development [19–24] (Table 1).
Table 1.
Previous studies showing genomic and epigenetic changes associated with aPLs and clinical profiles in APS patients.
| Source/type of study | Featured molecules | Biomarkers | Main findings | References |
|---|---|---|---|---|
| Monocytes (in vivo) | Genes and proteins | TF | Monocyte TF expression is overexpressed in APS monocytes and involved in its thrombotic complications | Cuadrado et al. [10] |
| Monocytes (in vivo/in vitro) | Genes and proteins | VEGF and Flt-1 | Monocytes from primary APS patients show increased expression of VEGF and Flt-1, further induced by aPL, through the p38 MAPK pathway | Cuadrado et al. [11] |
| Human umbilical vein endothelial cells (in vitro) | Genes (Affymetrix array) | Chemokines (CCL20, CXCL3, CX3CL1, etc); Tenascin C, OLR1, and IL-18-R1; growth factors (CSF2, CSF3 IL-6, IL-1β, FGF18) and signalling molecules (ID2) | Anti-β2GPI promotes in HUVEC an altered gene profile (including 101 genes upregulated and 14 genes downregulated) involving several genes related to APS vasculopathy | Hamid et al. [12] |
| Monocytes (in vivo/in vitro) | Genes and proteins | PAR1, PAR-2 | First demonstration of increased expression of PARs 1 and 2 in APS monocytes, further induced by aPL | López-Pedrera et al. [13] |
| PBMCs (in vivo/in vitro) | Genes | TLR-2 and TLR-4 | aPLs induce TLR pathway activation in PBMCs, promoting increased TLR-2 and TLR-4 expression | Benhamou et al. [14] |
| Monocytes (in vivo and in vitro) | Genes | Genes linked to mitochondrial biogenesis and function, the regulation of oxidative stress and antioxidant defence-related mechanisms | This study identified distinctive genetic patterns in monocytes from APS, SLE and APS + SLE patients, associated with atherosclerosis and thrombosis and further induced by aPLs | Perez-Sanchez et al. [15] |
| Meta-analysis (in vivo) | Genes | PF4V1, SELP, TLR2, TLR4, SERPINE1, APOH, ITGA2, GP1BA, F2R, F2RL1, TFPI, F3, VEGFA, FLT1, TNF and F2 | The 16 genes identified as deregulated in APS are expressed in 32 different organs, indicating the high risk of developing thrombosis anywhere in the body of primary APS patients | Islam et al. [16] |
| Monocytes (in vitro) | Genes | Genes involved in cell response to stress, MAPK signalling modulation, cell adhesion, extracellular matrix dynamics and embryonic and skeletal development | This study identified, by genome array, novel pathways differentially modulated in vitro in monocytes by isolated aPL-IgG from APS patients with either pregnancy morbidity or vascular thrombosis | Ripoll et al. [17] |
| Neutrophils (in vivo) | Genes | 17 hypomethylated and 25 hypermethylated CpG | Hypomethylation of ETS1 and EMP2 in APS neutrophils suggests trophoblast dysregulation, possibly contributing to foetal morbidity | Weeding et al. [18] |
| Monocytes (in vivo) | miRNAs | miR-19b, miR-20a | miR-19b and miR-20a are downregulated in APS and inversely correlated with levels of TF and procoagulant activity in monocytes | Teruel et al. [19] |
| Endothelial cells (in vitro) | miRNAs | mIR-126, mIR-100, mIR-1185-1, mIR-10b, mIR-576, mIR-1251, mIR-543, mIR-26A1, mIR-32, mIR-365b, mIR-339 | Treating endothelial cells with anti-β2GPI antibodies led to the release of extracellular vesicles containing a specific miRNA signature further implicated in the activation of ECs through autocrine or paracrine mechanisms | Wu et al. [20] |
| Human first trimester extra-villous trophoblast cell line (HTR8) (in vivo/in vitro) | miRNAs | miR-146a-5p, miR-146a-3p, miR-155, miR-210 | APS patients with adverse pregnancy outcomes present elevated miR-146a-3p levels. Trophoblasts exposed to aPLs show increased miR-146a-5p, miR-146a-3p, miR-155 and miR-210, all associated with TLR signalling | Gysler et al. [21] |
| Monocytes and neutrophils (in vivo/in vitro) | miRNAs | miR-124a-3p, miR-125a-5p, miR-125b-5p, miR-146a-5p, miR-155-5p, miR-222-3p | Several miRNAs related to CVD are altered in APS monocytes and neutrophils by effect of aPL antibodies | Perez-Sanchez et al. [22] |
| Plasmacytoid dendritic cells (in vivo) | miRNAs | miR-361-5p, miR-128-3p, miR-181-2-3p | Lower miRNA expression in pDCs, shared between SLE, SLE + APS and APS, is related to the IFN signature | van den Hoogen et al. [23] |
| Plasma (in vivo/in vitro) | miRNAs | miRNAs 34a-5p, 15a-5p, 133b-3p, 145a-5p, 124-3p, 20a-5p, 19b-3p, 210-3p, 206, 296-5p, 374a-5p | Circulating miRNAs, deregulated in plasma from APS patients by effect of aPLs, are associated with both foetal loss and type of thrombosis | Perez-Sanchez et al. [24] |
TF: tissue factor; VEGF: vascular endothelial growth factor; Flt-1: VEGF receptor 1; PAR-1–4: protease activator receptor 1–4; TLR: toll-like receptor; CCL20: macrophage inflammatory protein-3α; CXCL3: chemokine (C-X-C motif) ligand 3; CX3CL1: fractalkine; CXCL5: epithelial neutrophil activating peptide; CXCL2: chemokine (C-X-C motif) ligand 2; CXCL1: chemokine (C-X-C motif) ligand 1; OLR1: oxidized low-density lipoprotein receptor 1; CSF2: colony-stimulating factor 2; CSF3: colony-stimulating factor 3; FGF18: fibroblast growth factor 18; ID2: inhibitor of DNA binding 2; MAPK: mitogen-activated protein kinase; VT: vascular thrombosis; PM: pregnancy morbidity; PAI-1: plasminogen activator inhibitor-1; VEGFA: vascular endothelial growth factor A; VEGF-R1: vascular endothelial growth factor receptor 1; MCP-1: monocyte chemoattractant protein-1; qRT-PCR: quantitative RT-PCR; PTPN2: protein tyrosine phosphatase non-receptor type 2; ETS1: ETS proto-oncogene 1; pDCs: plasmacytoid dendritic cells.
Genomic and epigenetic changes associated with aPLs and clinical profiles in APS patients: update from the last 5 years
Transcriptomics and miRNA regulators in APS
Recent studies have focused on understanding the pathological effects of aPL in various contexts at the transcriptomic level, leading to valuable insights.
Patsouras et al. [25] employed RNAseq analysis to investigate alterations in the transcriptome of endothelial cells (HUVECs) treated in vitro with purified total IgG from APS patients’ sera (1 mg/ml), along with human native β2GPI (20 μg/ml). Their sequencing analysis revealed significant changes in gene expression, with 906 genes displaying differential expression after 6 h of treatment: 395 genes were upregulated, while 511 genes were downregulated. Among the upregulated genes, several were associated with immune response and inflammation, including key transcription factors and signal transduction molecules (NF-κB, YAP1, SMAD, MAP2K5, MAP2K3), as well as chemokines, adhesion molecules and growth factors (IL-8, E-selectin, VCAM-1, ICAM-1, TGFβ-2), among others. Pathway enrichment analysis of the altered genes highlighted a significant impact on biological processes related to MAPK38 signalling, TGF-β signalling, TNF-α signalling and Hippo pathways. To further validate these findings at the protein level, immunostaining analyses were conducted on the same cells. The results confirmed elevated protein expression of important pro-inflammatory markers such as IL-6, IL-8 and NF-κB in HUVECs treated with aPLs. Notably, the parallel examination of placenta biopsies from two APS patients and five healthy donors corroborated the upregulation of several inflammatory protein mediators, including IL-8, IL-6, TNF-α, NF-κB1, E-selectin, P-selectin, ICAM-1, VCAM-1 and TGF-β2. Overall, these findings strongly support the notion that aPLs induce endothelial cell activation in line with the upregulation of classical inflammatory pathways.
In another recent study conducted by Plunde et al. [26], the role of aPL at the transcriptomic level was examined in a different context. The researchers aimed to investigate the prevalence and impact of aPL in individuals with calcific aortic valve stenosis (CAVS) within the general population. First, they observed a significantly higher prevalence of aPL IgG in CAVS patients (n = 223) compared with age- and sex-matched controls (n = 124), with an odds ratio of 8.3. Moving forward, the molecular profile was analysed in aortic valve tissue from CAVS patients with IgG/IgM aPL (n = 5) and CAVS patients without aPL (n = 5) using the Human Transcriptome Arrays 2.0 from Affymetrix (Santa Clara, CA, USA). The CAVS patients positive for aPL exhibited differential expression of 100 genes compared with the negative group, with 46 genes being upregulated and 65 genes downregulated. Functional analysis of these genes indicated their association with antigen processing and IFN signalling. Furthermore, through machine learning analysis, they demonstrated that these 100 deregulated genes could accurately predict the occurrence of thickening, resilience and calcification in valves within an independent cohort of 64 patients. Taken together, these findings strongly suggest that aPL plays a pathological role in the development of CAVS by perturbing specific transcriptomic signatures.
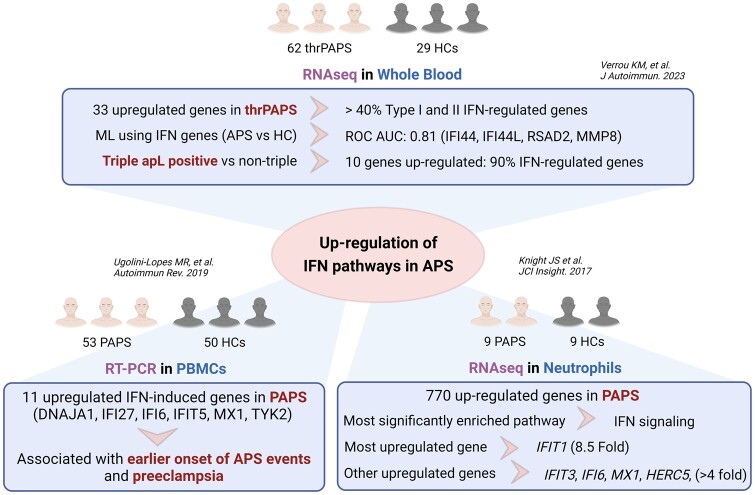
Recent studies have focused on identifying altered transcriptome signatures directly in samples from APS patients. In a pioneering study by Verrou et al. [27], high-throughput RNA sequencing was employed to analyse the transcriptome of 62 primary APS patients with thrombosis (thrPAPS) and 29 age- and sex-matched healthy controls (HCs) using whole blood samples. Although unsupervised clustering analysis did not effectively differentiate thrPAPS from HCs based on the entire transcriptome, a specific signature of 34 genes exhibiting differential expression between the two groups was identified. This signature comprised one downregulated gene, MYOM2, and 14 upregulated genes associated with type I and II IFN pathways. Notably, machine learning analysis utilizing these IFN-regulated genes (IRGs) successfully distinguished APS patients from HCs with considerable accuracy (area under the curve 0.811), with IFI44L, IFI44, RSAD2 and MMP8 displaying the highest information scores. When comparing patients with venous and arterial thrombosis, five genes were found to be downregulated (NECTIN2, RAP1GAP, AC104389.5, G0S2, SMIM1), while 11 genes, including nine IRGs, were upregulated in the venous thrombosis group. Moreover, altered expression of IRGs was also observed in triple-aPL-positive APS patients compared with non-triple-aPL patients. Additionally, patients with recurrent thrombosis exhibited significant changes in the expression of several genes (MYOM2, MMP8, CAMP, OLFM4, CRISP3, CEACAM6, CEACAM8, DEFA3, DEFA4, LCN2 and LTF) when compared with those with non-recurrent thrombosis. In general, this study suggests that the IFN pathway may contribute to the pathogenesis of APS patients with worse clinical outcomes. These findings corroborate previous observations presented by Knight et al. [28] in a study utilizing RNAseq analysis in neutrophils from a small cohort of nine APS patients and nine HCs, where the upregulation of genes associated with IFN signalling was observed. In line with all these findings, Ugolini-Lopes et al. [29] performed a comprehensive analysis using RT-PCR to investigate a panel of 41 IFN-induced genes in peripheral blood mononuclear cells (PBMCs) obtained from 53 primary APS patients and 50 HCs. The analysis revealed a distinct signature comprising 11 IFN-induced genes that exhibited significant upregulation in primary APS patients. Notable genes in this signature included IFI5, IFI6, IFIT27, DNAJA1, TYK2 and MX1. The presence of this IFN signature was found to be associated with pre-eclampsia and an earlier onset of APS events, further highlighting the significance of IFN-related pathways in the pathogenesis of primary APS (Fig. 1).
Figure 1.
Alteration of the IFN pathway in APS: consistent findings across independent studies. Recent independent studies have shed light on alteration of the IFN pathway in APS. Notably, three distinct investigations have consistently reported the upregulation of IFN-related genes in patients with APS, demonstrating their significant association with various clinical features such as autoantibody profile, thrombosis and pre-eclampsia. Despite the diversity in study cohorts, sample types (whole blood, PBMCs and neutrophils) and methodologies (RNAseq and RT-PCR), the consistent finding across all studies is upregulation of the IFN pathway. ML: machine learning; ROC AUC: area under the receiver operating characteristics curve
The identified transcriptomic alterations in APS patients, particularly the upregulation of genes associated with IFN pathways, hold significant clinical implications. First, these findings suggest the potential for targeted therapeutic interventions aimed at modulating the IFN signalling pathway in APS patients. Additionally, the distinct gene signatures associated with different thrombosis types and disease manifestations hint at the possibility of tailored treatment strategies based on the patient’s thrombotic profile. Moreover, the observed correlation between IFN and clinical outcomes, like pre-eclampsia and early onset of APS events, underscores the importance of monitoring and managing IFN-related pathways to mitigate the severity and progression of APS, offering a pathway towards personalized and more effective clinical management.
Our research group has been dedicated to unravelling the pathogenesis of APS through innovative approaches, including analysis of the splicing process in purified cell types and the integration of different omics in purified monocytes. In a recent study, we provided novel insights by demonstrating that the regulation of splicing, a crucial biological process involved in the generation of mature mRNAs, is impaired in leucocyte subsets of APS and SLE patients [30]. To investigate this, we examined the expression of 45 splicing machinery components (SMCs) using a quantitative PCR microfluidic array in purified monocytes, lymphocytes and neutrophils obtained from 80 patients (22 APS, 23 APS + SLE and 35 SLE). Overall, we observed a reduction in the expression of several SMCs, which exhibited cell-type and pathology-specific alterations. Specifically, in APS, we identified differential expression of 20 genes in monocytes, 16 genes in lymphocytes and 10 genes in neutrophils. Some SMCs were simultaneously altered in multiple cell types, including MAGOH, PRPF8, RBM22, KHDRBS1, SND1, SRSF-10, SRSF-5, SRSF-9, TRA2B and U4atac. Most of them were involved in autoimmune and inflammatory processes that characterize this disease, thus suggesting their relevance in autoimmunity [31–33].
Accordingly, correlation studies revealed significant associations between the levels of altered SMCs and key clinical features such as autoantibody titres (anti-β2GPI and aCL), cardiovascular risk markers (hyperlipidaemia and arterial hypertension) and the adjusted Global Antiphospholipid Syndrome Score.
Furthermore, transfection studies involving the overexpression or silencing of selected SMCs demonstrated their direct impact on relevant immune functions, including monocyte chemotaxis, lymphocyte proliferation and NETosis. While our study offers valuable mechanistic insights into the regulation of SMCs in APS, it remains uncertain whether these SMC alterations are inherently ‘primary’ or rather a consequence of persistent inflammation or autoimmune-related mechanisms influencing gene transcription within the cells. This pivotal aspect warrants further investigation.
Our study sheds light on the intricate molecular mechanisms underlying APS and might open avenues for further research and potential therapeutic interventions.
In another innovative study, we showed for the first time in APS that the integration of two levels of molecular complexity—the transcriptome and its regulation by miRNA—allowed stratification of APS patients according to their CVD risk [15]. In an exploratory cohort of APS patients, we conducted microarray analysis of mRNA and miRNA in monocytes and found that 547 mRNAs and 22 miRNAs exhibited differential expression compared with HCs. These differentially expressed genes and miRNAs were closely related to inflammatory, cardiovascular and reproductive disorders. Leveraging bioinformatics tools based on sequence homology, we integrated the altered genes and miRNAs and discovered a specific signature comprising 17 interconnected genes (IL1A, TGFB2, OXR1, ALOX15B, PXDNL, GPX8, STAT1, MAP2K6, MAP3K4, VEGFA, SERPINB2, VCAM1, SELE, ITGA2, TNFRSF1A, CCL2, IFNG) that may be potentially modulated by 9 miRNAs (miR-26a-5p, -150-5p, -146b-5p, -145-5p, -30b-5p, -494-3p, -17-5p, -376c-3p, -199a-5p). To validate the significance of this signature, we assessed its expression in a cohort of 40 primary APS patients and 40 HCs. Notably, the altered expression of this signature remained stable over time and was specific to non-autoimmune thrombotic patients. Experimental validation through transfection and luciferase assays confirmed the potential interactions among the altered genes and miRNAs. Furthermore, in vitro studies demonstrated that aPL antibodies isolated from the serum of APS patients promoted the altered expression of the mRNA–miRNA signature. Importantly, employing unsupervised clustering analysis with this altered molecular signature allowed us to effectively stratify APS patients into distinctive prothrombotic risk groups. In conclusion, our findings highlight the significance of integrating different omics data in APS, as it can provide valuable insights into identifying distinctive clinical phenotypes and pave the way for the development of personalized and tailored treatments for APS patients.
These compelling findings carry significant clinical implications for the management of APS. The ability to stratify APS patients based on their CVD risk through molecular signatures offers a promising avenue for tailored treatment strategies. The identified interconnected mRNAs and miRNAs represent potential therapeutic targets, providing a foundation for the development of precision medicines that could mitigate CVD risk in APS patients. Moreover, the molecular signature’s stability and specificity to non-autoimmune thrombotic patients emphasize its potential as a valuable diagnostic and prognostic tool. Implementing such stratification approaches in clinical settings could enhance risk assessment and guide the selection of appropriate therapeutic interventions, ultimately improving patient outcomes in APS.
Potential relationship between COVID-19 and APS at the transcriptomic level
COVID-19, like APS, increases the thrombosis risk in blood vessels [34]. Investigations of COVID-19 patients show parallels with APS, including abnormal activation of neutrophils [35, 36], endothelial cells [37] and platelets [38]. Several studies have found conventional aPL and ‘non-criteria’ aPL in COVID-19 patients. It is unclear if these aPL antibodies are transient or persistent, indicating long-term thrombotic risk. Indeed, the association between aPL and macrovascular thrombotic events in COVID-19 is not well established. Nevertheless, the relationship between APS and COVID-19 is an emerging area of research [39, 40].
Recent investigations have been conducted to explore the potential relationship between COVID-19 and APS at the transcriptomic level. Zhang et al. [30] employed publicly available microarray datasets and bioinformatic tools to identify commonly altered genes. They utilized a previous dataset generated by our group (GSE50395), which consisted of monocytes from APS patients and HCs [15], as well as the COVID-19 dataset GSE164805, which focused on PBMCs. Through a comparison with HCs, a specific common signature of eight altered genes emerged: ITGB8, FGF1, TIMELESS, PPARGC1A, FGF2, FUBP1, SYCP2 and FERMT1. Pathway enrichment analysis of this gene signature revealed its association with various biological processes, such as endothelial cell chemotaxis towards fibroblast growth factor, regulation of the actin cytoskeleton pathway, lung development and the PI3K-Akt signalling pathway.
Latest epigenetics advances in APS
While there is limited new research specifically linking APS and DNA methylation, this and other epigenetic alterations have been implicated in various autoimmune diseases [2]. Recently, Barturen et al. [27] integrated transcriptome and methylome data of seven systemic autoimmune diseases, including APS, in four clusters characterized by ‘inflammation’ functions, ‘lymphoid’ functions, ‘interferon’ functions and ‘undefined’, which had no clearly defined functional modules. Curiously, when they compared those clusters with controls, the inflammatory cluster had the highest number of differentially expressed genes and differentially methylated CpGs, followed by the ‘interferon’ cluster, the ‘lymphoid’ cluster and finally the ‘undefined’ cluster, in which no CpGs were observed, being more similar to controls [27].
Furthermore, research has indicated that DNA methylation alterations in specific genes can influence the expression of pro-inflammatory cytokines and other immune-related factors. Patsouras et al. [28] observed a significant reduction in methylation of the IL-8 promoter and significantly increased methylation of the tissue factor (F3) gene in whole blood of APS patients compared with HCs. In vitro, they treated monocytes and endothelial cells with anti-β2GPI, β2GPI and CXCL4, which reduced relative methylation of the IL-8 promoter and in the first intron of the F3 gene, which increased their expression. These changes in DNA methylation may potentially contribute to the pro-inflammatory state and thrombotic tendency seen in APS.
On the other hand, Tang et al. [41] found that the C677T mutation of methylenetetrahydrofolate reductase is a risk factor for arterial thrombosis in patients with APS. This enzyme catalyses the irreversible conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which is the methyl donor for the remethylation of homocysteine to methionine, leading to an increase in plasma homocysteine levels and consequently to an increased risk of thrombosis.
Overall, although some advances have been made in this area in recent years, the relationship between APS and DNA methylation is still poorly understood and further research is needed to establish a more comprehensive understanding of the molecular mechanisms involved.
Histone post-translational modifications are chemical modifications that occur on histone proteins, the proteins that help package DNA into chromatin. These modifications, which include acetylation, methylation, phosphorylation, ubiquitination and others, play a crucial role in regulating gene expression and chromatin structure [42]. To date, the modifications most studied are histone acetylation and methylation, highlighting H3K4me3 (histone 3 lysine 4 tri-methylation) and H3K9ac (histone 3 lysine 9 acetylation), which are often located at the promoter regions of target genes [43, 44]; H3K4me1 and H3K27ac, which are markers of silent enhancers and correlate with transcription repression [43, 45]; H3K27me3 and H3K36me3, which are involved in extending transcription and maintaining genomic stability via DNA mismatch repair [46, 47]; and H4K20me3 and H3K9me3, which are post-translational modifications for blocking gene expression [48, 49].The relationship between APS and histone modifications is an area of ongoing research, and the exact mechanisms are not yet fully understood. However, research illustrates the involvement of epigenetic mechanisms and related processes in the pathogenesis of autoimmune disorders and vascular complications, which are relevant to APS. Several studies have shown that EZH2 levels, an epigenetic regulator that mediates H3K27me3 and modulates DNA methylation, are increased in CD4+ T cells from patients with lupus, and its overexpression causes hypomethylation of F11-R, which encodes junctional adhesion molecule A (JAM-A), resulting in increased expression of JAM-A associated with increased leucocyte adhesion and migration [50]; overexpression of IFN-stimulated genes, contributing to the overactivation of the IFN-I signalling pathway [51]; and favours the formation of germinal centres by B cells, increasing the humoral immune response [52]. Furthermore, Liu et al. [53] showed that downregulation of UHRF1 (ubiquitin-like with PHD and RING finger domains 1) leads to increased BCL6 expression, a master regulator of T follicular helper (Tfh) cells, by decreasing DNA methylation and H3K27me3 levels, promoting Tfh cell differentiation in vitro and in vivo. These findings reveal that EZH2 inhibition and UHRF1 regulation may be useful for the treatment of lupus and other autoimmune disorders, such as APS.
It has been demonstrated an increase in the expression of HDAC9 (histone deacetylase 9) in monocytes stimulated with specific aPL antibodies. HDAC9 can impact TLR signalling and innate immunity influencing the development of atherosclerosis and metabolic disease [54].
Additionally, several unfavourable histone modifications increase the adverse pregnancy outcomes in APS. Reduced peroxisome proliferator-activated receptor gamma (PPARγ) delays foetal growth via upregulating H3K4me3 and H3K9ac in the formation of trophoblasts. Consequently, the administration of the specific PPARγ agonist ciglitazone represses H3K4me3 and H3K9ac enrichment and protects against intrauterine growth retardation [55]. Interestingly, H3K9me2 levels are increased in peripheral blood during proper trophoblast differentiation, and blocking H3K9me2 through the inhibitor of histone deacetylase delays trophoblast differentiation [56].
Further research is needed to fully elucidate the specific histone modifications involved in APS and their functional consequences. Understanding the interplay between histone modifications, epigenetic regulation and the pathogenesis of APS may provide insights into potential therapeutic targets or interventions for management of the condition.
Prospects and emerging opportunities in APS research
Bioinformatics and high-throughput technologies play a significant role in modern science, combining biology, computer science and statistics to analyse complex biological data [57]. The generation of data across various omics disciplines, including genomics, transcriptomics, proteomics and metabolomics, within the context of APS, is profoundly enriching our understanding and furthering the advancement of knowledge about this pathology. Bioinformatics has played a crucial role in advancing our understanding of APS by facilitating the analysis and interpretation of large-scale biologic data. Despite the limited availability of extensive cohorts and datasets, it has contributed to the identification of genetic risk factors, the characterization of molecular pathways, the discovery of biomarkers and the development of predictive models for APS [17, 27, 58].
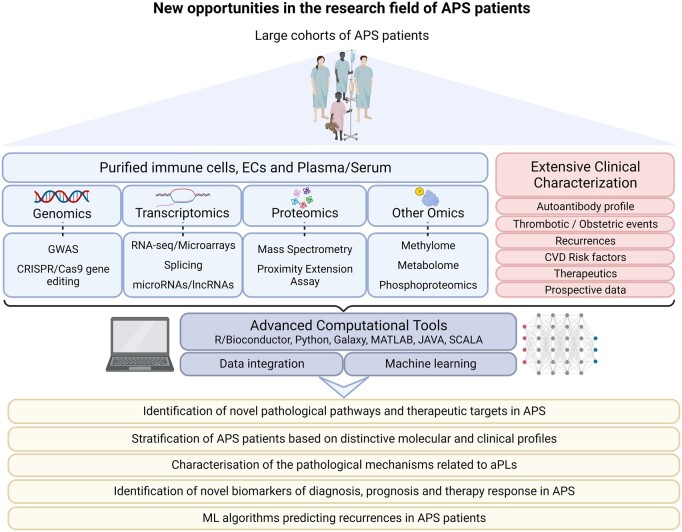
Machine learning has the potential to revolutionize biomedicine by improving disease diagnosis, accelerating drug discovery, enabling personalized medicine, advancing medical imaging analysis and enhancing healthcare decision making [59–63]. Through comprehensive integration of molecular data with thorough clinical characterization, these models possess the potential to address crucial clinical requirements in APS. This encompasses the identification of novel pathological pathways and therapeutic targets, patient stratification based on distinctive molecular and clinical profiles, characterization of the pathological mechanisms associated with aPLs and the discovery of novel biomarkers for diagnosis, prognosis and therapy response (Fig. 2).
Figure 2.
Uncovering new opportunities and perspectives in APS research. Despite the advancements in new technologies, the field of APS still faces a shortage of large-scale studies. However, with the emergence of multi-omics technologies and innovative computational tools, particularly machine learning methodologies, there is an unprecedented opportunity to transform APS research. Integrating diverse clinical and molecular data from extensive cohorts of patients using these cutting-edge approaches holds the potential to unlock novel pathological mechanisms underlying APS. Additionally, these methods offer the promise of identifying previously unknown biomarkers and therapeutic targets. As a result, the development of precision medicine in APS can be significantly accelerated, offering more tailored and effective treatments to patients
Further evolution in cutting-edge technologies, enhanced computational methodologies, refined data analysis algorithms and strengthened interdisciplinary collaborations will undoubtedly facilitate the revelation of previously hidden facets of this pathology. Consequently, this will play a pivotal role in laying the groundwork for the advancement of precision medicine in the context of APS.
Conclusions
The advancement of new technologies and bioinformatics tools has significantly enhanced our understanding of APS pathology. As a result, new therapeutic targets have emerged, such as the role of the IFN pathway in the disease. Moreover, integrating analysis across different molecular levels can prove effectiveness in stratifying patients based on key clinical features. While there is a current lack of epigenetics studies, there are some promising insights that suggest epigenetic mechanisms may play a crucial role in the development and progression of APS.
Numerous studies have demonstrated that aPL have a direct impact on immune system cells and vascular endothelium, both in in vivo contexts and in vitro experiments, contributing to various alterations. Nevertheless, it is possible that additional comorbidities or independent/interrelated pathological mechanisms may also influence the development of diverse clinical manifestations associated with this disease, and further research is needed to fully characterize these aspects.
To further improve our understanding and management of APS, utilizing novel omic technologies in large cohorts of patients with purified cell types as well as conducting independent validation studies will be essential. Additionally, employing integrated computational tools based on machine learning methodologies could open up new avenues for research and patient care in APS. By leveraging these approaches, the scientific community can potentially revolutionize how we approach the disease and improve patient outcomes.
Contributor Information
Chary López-Pedrera, Rheumatology Service, Maimonides Institute of Biomedical Research of Cordoba (IMIBIC), Reina Sofia University Hospital, University of Córdoba, Córdoba, Spain.
Tomás Cerdó, Rheumatology Service, Maimonides Institute of Biomedical Research of Cordoba (IMIBIC), Reina Sofia University Hospital, University of Córdoba, Córdoba, Spain; Centre for Rheumatology Research, Division of Medicine, University College London, London, UK.
Elizabeth C Jury, Centre for Rheumatology Research, Division of Medicine, University College London, London, UK.
Laura Muñoz-Barrera, Rheumatology Service, Maimonides Institute of Biomedical Research of Cordoba (IMIBIC), Reina Sofia University Hospital, University of Córdoba, Córdoba, Spain.
Alejandro Escudero-Contreras, Rheumatology Service, Maimonides Institute of Biomedical Research of Cordoba (IMIBIC), Reina Sofia University Hospital, University of Córdoba, Córdoba, Spain.
M A Aguirre, Rheumatology Service, Maimonides Institute of Biomedical Research of Cordoba (IMIBIC), Reina Sofia University Hospital, University of Córdoba, Córdoba, Spain.
Carlos Pérez-Sánchez, Rheumatology Service, Maimonides Institute of Biomedical Research of Cordoba (IMIBIC), Reina Sofia University Hospital, University of Córdoba, Córdoba, Spain; Department of Cell Biology, Immunology and Physiology, Agrifood Campus of International Excellence, University of Córdoba, ceiA3, Córdoba, Spain.
Data availability
No new data were generated or analysed in support of this research.
Authors’ contributions
C.L.P. and C.P.S. defined the scope and objectives of the review. C.L.P., C.P.S., T.C. and L.M.B. conducted literature review summarazing research findings. C.L.P., T.C. and C.P.S. drafted the initial version and developed the overall structure. E.C.J., A.E.C. and M.A.A. provided in-depth critiques of the manuscript and offered constructive feedback. T.C. and L.M.B. designed visual content like figures and tables. C.L.P. and C.P.S. supervised the final format.
Funding
This study was supported by grants PI21/00591, funded by Instituto de Salud Carlos III (ISCIII) and co-funded by the European Union, and RD21/0002/0033 Health Outcomes-Oriented Cooperative Research Networks, granted by the Instituto de Salud Carlos III (ISCIII) and funded by the European Union—NextGenerationEU via Mecanismo de Recuperación y Resiliencia and Plan de Recuperación, Transformación y Resiliencia. C.L.-P. was supported by a contract from the Spanish Junta de Andalucía (Nicolas Monardes program). T.C. was supported by the Sara Borrell program of the ISCIII (CD21/00187). L.M.B. was supported by a predoctoral grant from the ISCIII (FI22/00299). C.P.S. was financed by a contract from the MINECO Ramon y Cajal program (RYC2021-033828-I), cofounded by the European Union NextGenerationEU/PRTR.
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1. Miyakis S, Lockshin MD, Atsumi T. et al. International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS). J Thromb Haemost 2006;4:295–306. [DOI] [PubMed] [Google Scholar]
- 2. Barbhaiya M, Zuily S, Naden R. et al. ; ACR/EULAR APS Classification Criteria Collaborators. 2023 ACR/EULAR antiphospholipid syndrome classification criteria. 2023;82:1258–70. [DOI] [PubMed] [Google Scholar]
- 3. Cervera R, Serrano R, Pons-Estel G. et al. ; Euro-Phospholipid Project Group (European Forum on Antiphospholipid Antibodies). Morbidity and mortality in the antiphospholipid syndrome during a 10-year period: a multicentre prospective study of 1000 patients. 2015;74:1011–8. [DOI] [PubMed] [Google Scholar]
- 4. Linnemann B. Antiphospholipid syndrome–an update. Vasa 2018;47:451–64. [DOI] [PubMed] [Google Scholar]
- 5. Bertolaccini ML, Amengual O, Andreoli L. et al. ; 14th International Congress on Antiphospholipid Antibodies Task Force. Report on antiphospholipid syndrome laboratory diagnostics and trends. 2014;13:917–30. [DOI] [PubMed] [Google Scholar]
- 6. Meroni PL, MOJFii B.. Antiphospholipid antibody assays in 2021: looking for a predictive value in addition to a diagnostic one. 2021;12:726820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Savino S, Nicola B, Luigi MP. et al. Autoantibodies testing in autoimmunity: diagnostic, prognostic and classification value. Autoimmun Rev 2023;22:103356. [DOI] [PubMed] [Google Scholar]
- 8. Salet DM, Bekkering S, Middeldorp S, van den Hoogen L.. Targeting thromboinflammation in antiphospholipid síndrome. J Thromb Haemost 2023;21:744–57. [DOI] [PubMed] [Google Scholar]
- 9. Lopez-Pedrera C, Barbarroja N, Patiño-Trives AM. et al. New biomarkers for atherothrombosis in antiphospholipid syndrome: genomics and epigenetics approaches. 2019;10:764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cuadrado MJ, López‐Pedrera C, Khamashta MA. et al. Thrombosis in primary antiphospholipid syndrome. A pivotal role for monocyte tissue factor expression. 1997;40:834–41. [DOI] [PubMed] [Google Scholar]
- 11. Cuadrado M, Buendía P, Velasco F. et al. Vascular endothelial growth factor expression in monocytes from patients with primary antiphospholipid syndrome. 2006;4:2461–9. [DOI] [PubMed] [Google Scholar]
- 12. Hamid C, Norgate K, D'Cruz DP. et al. Anti-β2GPI-antibody-induced endothelial cell gene expression profiling reveals induction of novel pro-inflammatory genes potentially involved in primary antiphospholipid syndrome. 2007;66:1000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. López‐Pedrera C, Aguirre MA, Buendía P. et al. Differential expression of protease‐activated receptors in monocytes from patients with primary antiphospholipid syndrome. 2010;62:869–77. [DOI] [PubMed] [Google Scholar]
- 14. Benhamou Y, Bellien J, Armengol G. et al. Role of Toll‐like receptors 2 and 4 in mediating endothelial dysfunction and arterial remodeling in primary arterial antiphospholipid syndrome. 2014;66:3210–20. [DOI] [PubMed] [Google Scholar]
- 15. Perez-Sanchez C, Barbarroja N, Messineo S. et al. Gene profiling reveals specific molecular pathways in the pathogenesis of atherosclerosis and cardiovascular disease in antiphospholipid syndrome, systemic lupus erythematosus and antiphospholipid syndrome with lupus. 2015;74:1441–9. [DOI] [PubMed] [Google Scholar]
- 16. Islam MA, Khandker SS, Alam F, Kamal MA, Gan S.. Genetic risk factors in thrombotic primary antiphospholipid syndrome: a systematic review with bioinformatic analyses. 2018;17:226–43. [DOI] [PubMed] [Google Scholar]
- 17. Ripoll VM, Pregnolato F, Mazza S. et al. Gene expression profiling identifies distinct molecular signatures in thrombotic and obstetric antiphospholipid syndrome. 2018;93:114–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weeding E, Coit P, Yalavarthi S. et al. Genome-wide DNA methylation analysis in primary antiphospholipid syndrome neutrophils. 2018;196:110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teruel R, Perez‐Sanchez C, Corral J. et al. Identification of miRNAs as potential modulators of tissue factor expression in patients with systemic lupus erythematosus and antiphospholipid syndrome. 2011;9:1985–92. [DOI] [PubMed] [Google Scholar]
- 20. Wu M, Barnard J, Kundu S, McCrae K.. A novel pathway of cellular activation mediated by antiphospholipid antibody‐induced extracellular vesicles. J Thromb Haemost 2015;13:1928–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gysler SM, Mulla MJ, Guerra M. et al. Antiphospholipid antibody-induced miR-146a-3p drives trophoblast interleukin-8 secretion through activation of Toll-like receptor 8. 2016;22:465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pérez-Sánchez C, Aguirre M, Ruiz-Limón P. et al. Atherothrombosis-associated microRNAs in antiphospholipid syndrome and systemic lupus erythematosus patients. 2016;6:31375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van den Hoogen LL, Rossato M, Lopes AP. et al. microRNA downregulation in plasmacytoid dendritic cells in interferon-positive systemic lupus erythematosus and antiphospholipid syndrome. 2018;57:1669–74. [DOI] [PubMed] [Google Scholar]
- 24. Pérez-Sánchez C, Arias-de la Rosa I, Aguirre MÁ. et al. Circulating microRNAs as biomarkers of disease and typification of the atherothrombotic status in antiphospholipid syndrome. 2018;103:908–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patsouras M, Alexopoulou E, Foutadakis S. et al. Antiphospholipid antibodies induce proinflammatory and procoagulant pathways in endothelial cells. J Transl Autoimmun 2023;6:100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Plunde O, Svenungsson E, Ferrannini G, Franco-Cereceda A, Bäck MJR.. Antiphospholipid antibodies in patients with calcific aortic valve stenosis. 2023;62:1187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verrou K-M, Sfikakis PP, Tektonidou M.. Whole blood transcriptome identifies interferon-regulated genes as key drivers in thrombotic primary antiphospholipid syndrome. 2023;134:102978. [DOI] [PubMed] [Google Scholar]
- 28. Knight JS, Meng H, Coit P. et al. Activated signature of antiphospholipid syndrome neutrophils reveals potential therapeutic target. JCI Insight 2017;2:e93897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ugolini-Lopes MR, Torrezan GT, Gândara APR. et al. Enhanced type I interferon gene signature in primary antiphospholipid syndrome: association with earlier disease onset and preeclampsia. Autoimmun Rev 2019;18:393–8. [DOI] [PubMed] [Google Scholar]
- 30. Zhang W, Di L, Liu Z. et al. TIMELESS is a key gene mediating thrombogenesis in COVID-19 and antiphospholipid syndrome. 2022;12:17248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Cheng Y, Wu W. et al. SRSF10 regulates alternative splicing and is required for adipocyte differentiation. 2014;34:2198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dima A, Jurcut C, Baicus CJRI.. The impact of anti-U1-RNP positivity: systemic lupus erythematosus versus mixed connective tissue disease. 2018;38:1169–78. [DOI] [PubMed] [Google Scholar]
- 33. Sun W, Qin R, Wang R. et al. Sam68 promotes invasion, migration, and proliferation of fibroblast-like synoviocytes by enhancing the NF-κB/P65 pathway in rheumatoid arthritis. 2018;41:1661–70. [DOI] [PubMed] [Google Scholar]
- 34. Connors JM, Levy JHJB.. COVID-19 and its implications for thrombosis and anticoagulation . J Am Soc Hematol 2020;135:2033–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zuo Y, Yalavarthi S, Shi H. et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020;5:e138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi H, Zuo Y, Yalavarthi S. et al. Neutrophil calprotectin identifies severe pulmonary disease in COVID-19. 2021;109:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Varga Z, Flammer AJ, Steiger P. et al. Endothelial cell infection and endotheliitis in COVID-19. 2020;395:1417–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manne BK, Denorme F, Middleton EA. et al. Platelet gene expression and function in patients with COVID-19. 2020;136:1317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zuo Y, Estes SK, Ali RA. et al. Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. 2020;12:eabd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hollerbach A, Müller‐Calleja N, Pedrosa D. et al. Pathogenic lipid‐binding antiphospholipid antibodies are associated with severity of COVID‐19. 2021;19:2335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Undas A, Brozek J, Szczeklik A.. Homocysteine and thrombosis: from basic science to clinical evidence. J Thromb Haemost 2005;94:907–15. [DOI] [PubMed] [Google Scholar]
- 42. Joseph FM, Young NL.. Histone variant-specific post-translational modifications. Semin Cell Dev Biol2023;135:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beacon TH, Delcuve GP, López C.. The dynamic broad epigenetic (H3K4me3, H3K27ac) domain as a mark of essential genes. 2021;13:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li C, Xiong W, Liu X. et al. Hypomethylation at non-CpG/CpG sites in the promoter of HIF-1α gene combined with enhanced H3K9Ac modification contribute to maintain higher HIF-1α expression in breast cancer. 2019;8:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bae S, Lesch B, biology d. H3K4me1 distribution predicts transcription state and poising at promoters. 2020;8:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Raas MW, Zijlmans DW, Vermeulen M, Marks H.. There is another: H3K27me3-mediated genomic imprinting. 2022;38:82–96. [DOI] [PubMed] [Google Scholar]
- 47. Sun Z, Zhang Y, Jia J. et al. H3K36me3, message from chromatin to DNA damage repair. 2020;10:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu J, Kidder B.. H4K20me3 co-localizes with activating histone modifications at transcriptionally dynamic regions in embryonic stem cells. 2018;19:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nicetto D, Zaret KS, Zaret KSJCoig, development. Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. 2019;55:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsou PS, Coit P, Kilian NC, Sawalha AH.. EZH2 modulates the DNA methylome and controls T cell adhesion through junctional adhesion molecule A in lupus patients. J Rheumatology 2018;70:98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu L, Jiang X, Qi C. et al. EZH2 inhibition interferes with the activation of type I interferon signaling pathway and ameliorates lupus nephritis in NZB/NZW F1 mice. 2021;12:653989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhen Y, Smith RD, Finkelman FD, Shao W-H.. Ezh2-mediated epigenetic modification is required for allogeneic T cell-induced lupus disease. Arthritis Res Ther 2020;22:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu L, Hu L, Yang L.. UHRF1 downregulation promotes T follicular helper cell differentiation by increasing BCL6 expression in SLE. 2021;13:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Patsouras M, Karagianni P, Kogionou P, Vlachoyiannopoulos P.. Differential CpG methylation of the promoter of interleukin 8 and the first intron of tissue factor in Antiphospholipid syndrome. 2019;102:159–66. [DOI] [PubMed] [Google Scholar]
- 55. Meister S, Hahn L, Beyer S. et al. Regulation of epigenetic modifications in the placenta during preeclampsia: PPARγ influences H3K4me3 and H3K9ac in extravillous trophoblast cells. 2021;22:12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kafer GR, Tanaka Y, Rillo-Bohn R. et al. Sequential peripheral enrichment of H2A. Zac and H3K9me2 during trophoblast differentiation in human embryonic stem cells. 2020;133:jcs245282. [DOI] [PubMed] [Google Scholar]
- 57. Piatek P, Tarkowski M, Namiecinska M. et al. H3K4me3 Histone ChIP-Seq Analysis Reveals Molecular Mechanisms Responsible for Neutrophil Dysfunction in HIV-Infected Individuals. 2021;12:682094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pérez-Sánchez L, Patiño-Trives AM, Aguirre-Zamorano MÁ. et al. Characterization of antiphospholipid syndrome atherothrombotic risk by unsupervised integrated transcriptomic analyses. 2021;41:865–77. [DOI] [PubMed] [Google Scholar]
- 59. Shi T, Song E, Nie S. et al. Advances in targeted proteomics and applications to biomedical research. 2016;16:2160–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu T, Huang W, Qi J. et al. Research trends and frontiers on antiphospholipid syndrome: a 10-year bibliometric analysis (2012–2021). 2022;13:1035229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang L, Li Y, Lin X, Jia C, Yu XJPO.. Liquid chromatography/mass spectrometry based serum metabolomics study on recurrent abortion women with antiphospholipid syndrome. 2019;14:e0225463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Choi MY, Chen I, Clarke AE. et al. Machine learning identifies clusters of longitudinal autoantibody profiles predictive of systemic lupus erythematosus disease outcomes. 2023;82:927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kremers R, Zuily S, De Groot P. et al. Development of a neural network to predict thrombosis in the antiphospholipid syndrome with an accuracy of 87%. 2018;132:3796. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.