Abstract
Prolidase (PEPD) is the only hydrolase that cleaves the dipeptides containing C-terminal proline or hydroxyproline-the rate-limiting step in collagen biosynthesis. However, the molecular regulation of prolidase expression remains largely unknown. In this study, we have identified overlapping binding sites for the transcription factors Krüppel-like factor 6 (KLF6) and Specificity protein 1 (Sp1) in the PEPD promoter and demonstrate that KLF6/Sp1 transcriptionally regulate prolidase expression. By cloning the PEPD promoter into a luciferase reporter and through site-directed deletion, we pinpointed the minimal sequences required for KLF6 and Sp1-mediated PEPD promoter-driven transcription. Interestingly, Sp1 inhibition abrogated KLF6-mediated PEPD promoter activity, suggesting that Sp1 is required for the basal expression of prolidase. We further studied the regulation of PEPD by KLF6 and Sp1 during transforming growth factor β1 (TGF-β1) signaling, since both KLF6 and Sp1 are key players in TGF-β1 mediated collagen biosynthesis. Mouse and human fibroblasts exposed to TGF-β1 resulted in the induction of PEPD transcription and prolidase expression. Inhibition of TGF-β1 signaling abrogated PEPD promoter-driven transcriptional activity of KLF6 and Sp1. Knock-down of KLF6 as well as Sp1 inhibition also reduced prolidase expression. Chromatin immunoprecipitation assay supported direct binding of KLF6 and Sp1 to the PEPD promoter and this binding was enriched by TGF-β1 treatment. Finally, immunofluorescence studies showed that KLF6 co-operates with Sp1 in the nucleus to activate prolidase expression and enhance collagen biosynthesis. Collectively, our results identify functional elements of the PEPD promoter for KLF6 and Sp1-mediated transcriptional activation and describe the molecular mechanism of prolidase expression.
Keywords: prolidase, transcriptional regulation, Kruppel-like factor 6 (KLF6), Specificity protein 1 (Sp1), collagen, transforming growth factor beta (TGF-β)
Prolidase is a metalloproteinase and is the only enzyme that hydrolyzes dipeptides containing C-terminal proline or hydroxyproline (1, 2). Encoded by the PEPD gene, prolidase is also known as peptidase D, Xaa-Pro dipeptidase, X-Pro dipeptidase, and proline dipeptidase (1, 2). The enzymatic activity of prolidase releases proline or hydroxyproline during the final stages of the catabolism of endogenous and dietary proteins, most notably, collagen-the predominant component of the extracellular matrix (ECM) (3). Collagen is the prevalent structural protein in the human body with proline and hydroxyproline constituting more than 20% of its total amino acids (4). The degradation of extracellular collagen is initiated by specific metalloproteinases (5), the products of which are further degraded by non-specific proteases (6). The resulting short fragments are internalized into cells and are degraded into individual amino acids in the lysosomes, except the imido-dipeptides such as glycyl-proline and glycyl-hydroxyproline (7, 8). The cytoplasmic prolidase is the only enzyme that degrades these imido-dipeptides to complete collagen degradation (9). Prolidase activity recovers ∼90% of the free proline (10) that are recycled to synthesize more collagen and other proteins (9, 11, 12). Therefore, alterations in prolidase activity are linked to dysfunction in collagen metabolism and ECM remodeling (13). Prolidase also plays key roles in a number of other physiological and pathological processes including carcinogenesis, fibrosis, cell proliferation, and wound healing (14). Notably, prolidase deficiency-a rare genetic disorder impairs skeletal formation and wound healing; both of which rely on collagen turnover (15). A number of reports also show increased prolidase mRNA levels and higher enzymatic activity in wound fluid and scar tissues (16, 17). Furthermore, a recent study showed that recombinant human prolidase induces cell growth, proliferation, migration, and collagen biosynthesis in human fibroblasts (18). However, the mechanisms and pathways that regulate prolidase expression are poorly understood.
Interestingly, there is evidence that mechanisms and pathways that regulate collagen and ECM interactions are also important in the regulation of prolidase activity (19, 20). For instance, insulin-like growth factor-1 (IGF1) signaling has been implicated in the regulation of prolidase (21, 22, 23). Phosphorylation of prolidase has been recognized as a potential post-transcriptional mechanism for regulating its enzymatic activity (24, 25). Inflammatory stimuli such as nitric oxide, which is induced during wound healing, induce serine/threonine phosphorylation of prolidase in fibroblasts (26). Previous studies from our laboratory have demonstrated that nitric oxide production induces prolidase phosphorylation and activity (25). In addition, products of prolidase activity; proline and hydroxyproline increase nuclear hypoxia-inducible factor levels (HIF-1α) by inhibiting HIF-1α degradation during wound healing (12). Importantly, there is evidence of the non-enzymatic function of prolidase. For example, prolidase serves as a ligand of ErbB1 (EGFR) and ErbB2 (HER2) receptors to stimulate downstream signaling in the EGFR and Src pathways, respectively (27, 28). Prolidase has also been reported as a novel regulator of p53 tumor suppressor (29) and shown to bind to p53 to suppress both transcription-dependent and transcription-independent activities of p53. In addition, a unique role of prolidase in regulating type I interferon (IFN-I) immune response during flavivirus infection has been reported (30). Despite these critical roles in various cellular processes and diseased conditions, the mechanisms of prolidase expression at the transcriptional and translational levels remain largely unknown.
In this study, we have characterized the PEPD promoter and have identified key functional elements that are involved in the transcriptional regulation of prolidase. Our results demonstrate that the transcription factor Sp1 is essential for the basal expression of prolidase. Importantly, KLF6 cooperates with Sp1 to induce PEPD transcription. We also observed that the ECM remodeling stimuli- TGF-β1 induced KLF6-mediated prolidase expression in a Sp1-dependent manner. Interestingly, KLF6/Sp1 mediated transcriptional activation of PEPD promoter also enhanced expression of collagen, type I, alpha 1, (ColA1) implying the functional relevance of this pathway in collagen biosynthesis. Collectively, these results provide novel insights into the mechanisms driving PEPD transcriptional regulatory pathway and its functional relevance in collagen biosynthesis.
Results
Identification of putative transcriptional regulatory elements of PEPD promoter
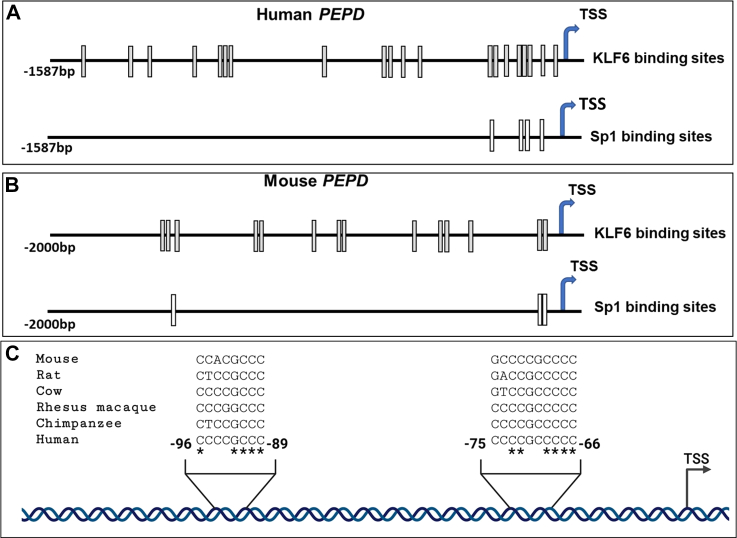
Human prolidase is encoded by the PEPD gene, located on the long arm of chromosome 19 at position 13.11 (Gene ID: 5184). The 130 kilobase (kb) gene contains 15 exons to encode three mRNA transcript variants (31). Transcript variant one is the most abundant and longest isoform that yields the canonical 493 amino acid prolidase protein. For identifying the putative transcriptional regulatory elements in the PEPD promoter, first, we carried out in silico analyses of the 5′ untranslated region (UTR) of sequences ranging from −1587 bp to +50 bp of the PEPD gene (+1 bp representing the transcription start site-TSS) using TRANScription FACtor (TRANSFAC) (32, 33). Interestingly, this analysis revealed that the PEPD promoter lacks a canonical TATA box and contains putative binding sites for a number of transcription factors (data not shown). Among the transcription factors, the KLF6 binding sites were the most abundant and distributed throughout the 5′-UTR of the PEPD promoter (Fig. 1A). Notably, a cluster of KLF6 binding sites was situated proximal to the TSS and overlapped with another cluster of Sp1 binding sites (Fig. 1A). TRANSFAC analyses of the mouse PEPD promoter (−2000 bp to +50 bp) also identified overlapping KLF6 and Sp1 binding sites in the 5′-UTR near the TSS (Fig. 1B). Strikingly, the overlapping KLF6 and Sp1 binding sites near the TSS were conserved across multiple mammalian species, implying the functional significance of these sites in the transcriptional regulation of PEPD (Fig. 1C). In fact, there is evidence that both KLF6 and Sp1 play important roles in collagen biosynthesis (34). Since prolidase catalyzes the rate-limiting step of collagen degradation, the overlapping KLF6 and Sp1 binding sites within the PEPD promoter provided a strong basis to test the role of these two transcription factors in PEPD transcriptional regulation.
Figure 1.
In silicoanalyses ofPEPDpromoter. Schematic illustration of (A) human and (B) mouse proximal PEPD promoter region- TRANSFAC based in silico analysis of −1587 bp of human PEPD promoter and −2000 bp of mouse PEPD promoter relative to TSS (+1 bp). The putative KLF6 and Sp1 binding sites are represented by gray/white boxes. C, sequence alignment of overlapping KLF6 and Sp1 binding sites in the human PEPD promoter between −96 bp to −89 bp and −75 bp to −66 bp relative to TSS among different mammalian species.
PEPD promoter-driven transcription is regulated by Sp1 and KLF6
To study whether the overlapping binding sites of KLF6 and Sp1 on the PEPD promoter (Fig. 1) are transcriptionally functional, we constructed a luciferase reporter construct (PEPD-Luc) by cloning the human PEPD promoter region containing sequences from −1537 bp to +50 bp. Then, increasing amounts of PEPD-Luc or the empty vector (Null-Luc) were transfected into HEK293T cells, and lysates were prepared for luciferase activity measurements (Fig. 2A). A dose-dependent increase in luciferase activity was detected after transfection of increasing amounts of PEPD-Luc relative to the Null-Luc (Fig. 2A). These results establish that the PEPD-Luc reporter construct is functional, and thus was used to identify the minimal sequences required for the PEPD promoter-driven transcription.
Figure 2.
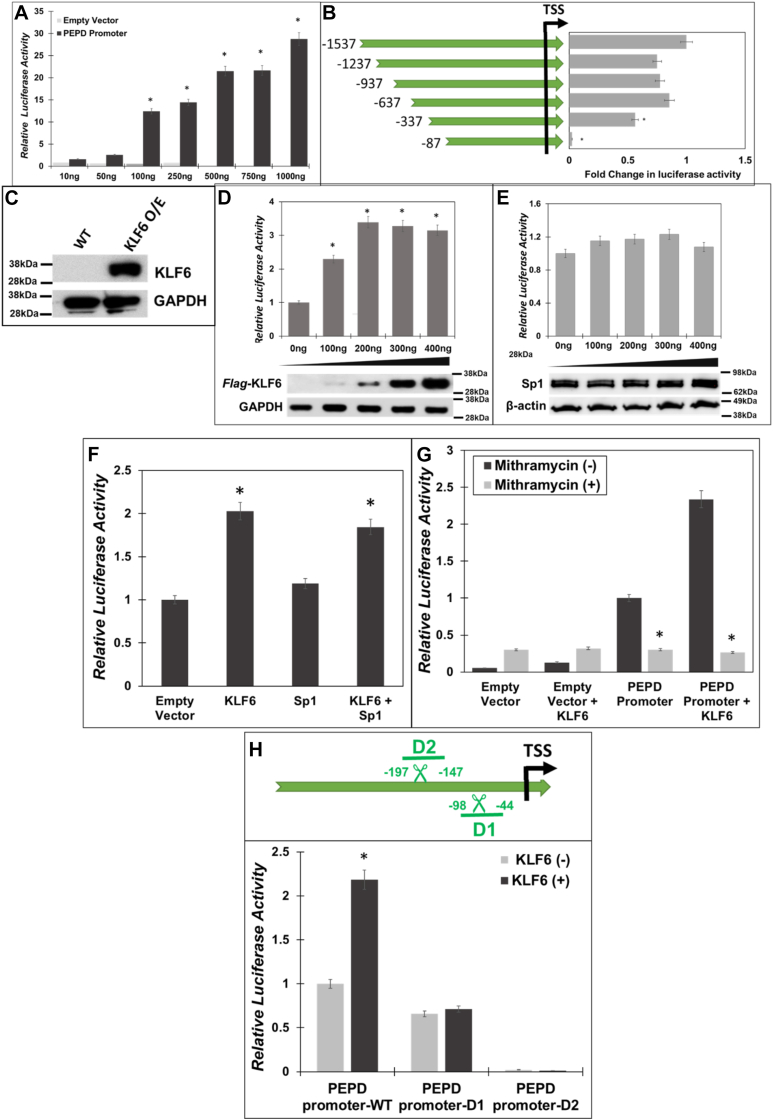
Sp1 and KLF6 drive PEPD promoter activity.A, the human PEPD promoter region (−1537 bp to +50 bp relative to the TSS) was cloned into the pGL3-basic luciferase reporter (PEPD-Luc). HEK293T cells were co-transfected with increasing amounts of PEPD-Luc or PEPD-null and Renilla Luciferase (pRL-null). Post transfection (24 h), the firefly luciferase activity was measured and normalized to Renilla luciferase activity as an internal control. B, mapping of minimal PEPD promoter: Schematic representation of sequential deletions of PEPD promoter from the 5′-end. Several truncated PEPD-Luc constructs were generated and transfected along with pRLnull into HEK293T cells. Luciferase activity was normalized relative to untruncated PEPD-Luc. C, expression of KLF6 in HEK293T cells without and with the overexpression construct. D–F, HEK293T cells were co-transfected with PEPD-Luc and increasing amounts of either human Flag-KLF6 or Sp1 expression plasmids or both. Then luciferase activity was measured in the cellular lysates and were plotted relative to the control cells. G, HEK293T cells were pretreated with 1 μM of Mith A or vehicle (DMSO) for 1 h, following which the cells were co-transfected with PEPD-Luc with and without human Flag-KLF6. 24 h post transfection the luciferase activity was measured in the cellular lysates and were plotted relative to controls. H, schematic of the two deletion constructs that contained the overlapping KLF6/Sp1 binding sites (upper panel). deletions between – 98 bp and −44 bp (delta 1-D1) and −197 bp to −147 bp (delta 2-D2) relative to TSS of PEPD promoter construct were introduced. These constructs were co-transfected in HEK293T cells with and without human Flag-KLF6. 24 h post transfection the luciferase activity was measured in the cellular lysates and the normalized fold change in luciferase activity was determined. Data are mean values of three independent experiments with error bars representing SEM. ∗ represents a p value of < 0.05 for the statistical comparison of (A and B) empty vector vs PEPD-Luc, (D) empty vector vs PEPD-Luc and KLF6 expression construct, (E) empty vector vs PEPD-Luc and Sp1 expression construct (F) empty vector vs KLF6 or KLF6/Sp1 expression constructs, (G) Untreated vs Mith A treated, and (H) control vs KLF6 expression construct.
To identify elements of PEPD promoter that drive transcriptional activity, we introduced a series of 5′-deletions in the PEPD-Luc construct by sequentially removing 300 bp starting from −1537 bp sequences of the promoter (Fig. 2B, left panel). These deletions were introduced by PCR-based site-directed mutagenesis and were verified by sequencing (data not shown). Each of these deletion mutant PEPD-Luc reporters were then transfected into HEK293T cells to test promoter-driven transcriptional activity. We observed that step-wise deletion of the PEPD promoter sequences starting from −1537 bp up to −337 bp marginally reduced luciferase activity when compared to the full-length reporter (Fig. 2B). However, the deletion of sequences beyond −337 bp rendered the PEPD promoter non-functional in driving the transcription of the luciferase reporter. These luciferase-reporter studies of the deletion mutants strongly suggested that the sequences between positions −337 to +50 bp of the PEPD promoter are critically required for transcriptional activity. Interestingly, the overlapping binding sites of KLF6 and Sp1 were also located within the PEPD promoter sequences between positions −337 to +50 bp (Fig. 1). Therefore, we carried out transfection studies using expression constructs of KLF6 and Sp1 in HEK293T cells to assess the individual effects of these transcription factors on PEPD promoter-driven transcriptional activity. We used HEK293T cells for these co-transfection studies since these cells lack endogenous KLF6 (Fig. 2C) but express Sp1 (Fig. 2D, bottom panel). Transfection of KLF6 expression construct resulted in a dose-dependent increase in KLF6 protein levels as detected by Western blot (Fig. 2D, bottom panel). Interestingly, co-transfection of KLF6 and PEPD-Luc significantly stimulated PEPD promoter activity in a dose-dependent manner (Fig. 2D, top panel). Particularly, a maximal increase in luciferase activity (∼3 to 4-fold) was obtained with 200 ng of the KLF6 construct when compared to the empty vector control (Fig. 2D). Surprisingly, transfection of Sp1 expression construct did not increase the basal promoter activity of PEPD-Luc reporter (Fig. 2E). It should be noted that endogenous Sp1 levels is robust in these cells and transfection of the Sp1 expression construct did not increase the protein levels significantly (Fig. 2E, bottom panel). Thus, it is likely that endogenous levels of Sp1 in HEK293T cells is sufficient for transcriptional activation of the PEPD promoter.
Thereafter, we carried out co-transfection of KLF6 and Sp1 expression constructs along with PEPD-Luc reporter to test the combined effects of KLF6 and Sp1 on PEPD promoter activity (Fig. 2F). As expected, expression of KLF6 significantly increased PEPD promoter-driven luciferase activity, whereas transfection of Sp1 alone showed minimal effect (Fig. 2F). Interestingly, co-transfection of both KLF6 and Sp1 constructs did not show measurable enhancement in luciferase activity over the KLF-6 transfected cells (Fig. 2F). To further probe the role of KLF6 and Sp1 in PEPD promoter activity, we utilized the selective Sp1 inhibitor; Mithramycin A (Mith A) in our co-transfection studies. Mith A inhibits Sp1 binding to target promoters and blocks Sp1-mediated transcriptional activity (35). As expected, Mith A pretreatment followed by transfection with PEPD-Luc constructs significantly repressed PEPD promoter-driven luciferase activity (Fig. 2G). Interestingly, Mith A pre-treatment followed by co-transfection of KLF6 and PEPD-Luc also significantly inhibited luciferase activity compared to untreated cells (Fig. 2G). These results provide further support to our hypothesis that endogenous Sp1 optimally activates PEPD promoter-driven transcription. Most importantly, these observations also indicate that activation of PEPD promoter-driven transcription by KLF6 is dependent on Sp1.
PEPD promoter sequences between −197 to −147 bp are required for KLF6/Sp1-driven transcription
Our results indicated that the sequences up to −337 bp from the TSS are critical for PEPD promoter-driven transcription (Fig. 2B) and contain a number of highly conserved and overlapping KLF6/Sp1 binding sites (Fig. 1). To further probe the functional role of these putative binding sites during KLF6/Sp1-driven PEPD promoter activity, we constructed a specific deletion mutant that removed these overlapping binding sites (D1 with deletions of −98 bp till −44 bp) (Fig. 2H). We also designed a deletion mutant (D2) where a deletion was created outside of these binding sites at sequences spanning from −197 to −147 bp. Each of these two PEPD promoter deletion constructs was individually transfected into HEK293T cells without or with the KLF6 expression construct, and then luciferase activity was assessed as a measure of PEPD promoter activity (Fig. 2H). We observed that without KLF6 PEPD-Luc-driven basal promoter activity was marginally reduced by the D1 mutant when compared to the control (PEPD-Luc full length). However, deletion of the D2 region completely abrogated the basal promoter activity, suggesting that sequences spanning from −197 to −147 bp are critical for PEPD promoter-driven transcription. As expected, co-transfection of the KLF6 construct activated the full-length PEPD-Luc. Interestingly, KLF6 expression failed to activate the PEPD promoter activity of constructs lacking the D1 or D2 regions. These results suggested that the regions between −197 and −147 bp of the PEPD promoter (D2 region) are critical for both basal and KLF6-driven transcriptional activity (Fig. 2H).
KLF6 and Sp1 directly bind to the PEPD promoter to regulate prolidase expression
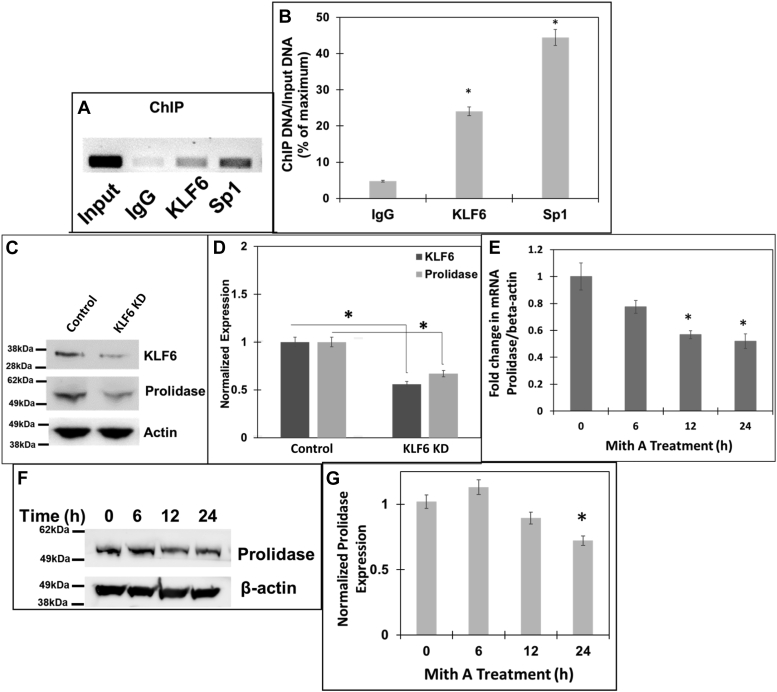
Our luciferase reporter studies in HEK293T cells revealed that PEPD promoter-driven transcription is controlled by Sp1 at the basal level, whereas KLF6 expression further stimulates Sp1-mediated promoter activation (Figs. 1 and 2). To better understand the role of KLF6 and Sp1 in PEPD promoter activity in a physiologically relevant system, we utilized mouse embryonic fibroblast (NIH3T3) cell line since fibroblasts are primarily responsible for collagen biosynthesis and turnover (36). Additionally, our in silico analyses revealed a similar pattern of overlapping KLF6 and Sp1 binding sites in both human and mouse PEPD promoter near the TSS (Fig. 1). First, we carried out chromatin immunoprecipitation (ChIP) assay to probe whether KLF6 and Sp1 directly bind to the PEPD promoter sequences. DNA fragments prepared from NIH3T3 cell pellets were subjected to ChIP with specific antibodies of Sp1, KLF6 or IgG control. The extracted ChIP-DNA was used as a template for PCR amplification using specific primers complementary to the PEPD promoter regions containing Sp1 and KLF6 overlapping binding sites. The results of the PCR assay showed that chromatin fragments containing KLF6/Sp1-binding sites in the PEPD promoter were precipitated and amplified when pulled down by the antibodies of Sp1 as well as KLF6 (Fig. 3A). Since, ChIP with the isotype IgG control did not amplify the PEPD promoter sequences, these results provided strong evidence that Sp1 and KLF6 directly bind to the PEPD promoter in these fibroblast cell lines. Interestingly, densitometric analysis of the ChIP assay revealed a stronger binding by Sp1 when compared to KLF6 (Fig. 3B). These results suggested that Sp1 most likely binds to the PEPD promoter at a higher affinity than KLF6.
Figure 3.
Role of Sp1 and KLF6 in driving expression of prolidase.A and B, chromatin immunoprecipitation (ChIP) assay of PEPD promoter showing binding of Sp1 and KLF6. Soluble chromatin from (NIH3T3 cells was subjected to ChIP using either KLF6 or Sp1 antibodies or IgG control antibody. Immuno-precipitates were analyzed by PCR with specific primers for the PEPD promoter region encompassing the overlapping Sp1/KLF6 binding sites. A, the PCR products were detected by agarose gel electrophoresis. The input represented the DNA in cell extracts prior to immunoprecipitation. B, densitometric analysis of the immunoprecipitated DNA pulled down with the respective antibodies and are normalized to input from three independent experiments. C and D, NIH3T3 cells were transfected with either non-targeting or KLF6 targeting ASO and then 24 h post transfection, cellular lysates were prepared for Western blot. C, Representative blot (n = 3) showing expression of KLF6, prolidase and β-actin, in KLF6 knocked-down and control cells. D, densitometric analysis of the immunoblot of KLF6 and prolidase that are normalized to β-actin levels. E–G, NIH3T3 cells were treated with 1 μM Mith A or vehicle (DMSO) in a time-dependent manner. E, prolidase mRNA expression in these cells was measured by qPCR and normalized to β-actin mRNA. Prolidase mRNA expression is represented as fold change in Mith A-treated vs untreated cells based on ΔΔCt values. F, equal protein amounts of cell lysates were subjected to immunoblot to measure prolidase protein expression. Representative immunoblot (F) and densitometry analyses (G) of prolidase expression normalized to β-actin protein levels. Data are mean values of three independent experiments with error bars representing SEM. ∗ represents a p value of < 0.05 for the statistical comparison of (B) IgG control vs KLF6 or Sp1, (D) Non-target siRNA control vs KLF6 siRNA, (E and G) untreated vs Mith A treated samples.
To solidify the transcriptional regulation of prolidase by KLF6 and Sp1, we next employed genetic and pharmacological studies. First, we carried out a knock-down of KLF6 in NIH3T3 cells using siRNA-based methods. We were able to reduce the levels of KLF6 to a significant level (∼50% lower) by using AUMsilenceTM FANA Antisense Oligos (FANA ASOs) in these cells (Fig. 3, C and D). Then, prolidase protein levels were measured in the KLF6 knocked-down cells by Western blot. Interestingly, prolidase expression was significantly reduced in cells with reduced levels of KLF6 compared to the non-target control cells (Fig. 3, C and D). Unfortunately, our siRNA-based knock-down of Sp1 in NIH3T3 cells was unsuccessful. Therefore, we measured prolidase expression in the NIH3T3 cells in response to Mith A (1 μM) treatment in a time-dependent manner. Since activation of PEPD promoter by KLF6 required Sp1 (Fig. 2) and KLF6 knock-down reduced prolidase levels (Fig. 3, C and D), we predicted that blocking Sp1 binding to the promoter would affect prolidase expression. As expected, these fibroblast cell lines express prolidase mRNA and protein (Fig. 3, C–E). Interestingly, Mith A treatment decreased both the mRNA and protein levels of prolidase supporting that Sp1-inhibition suppresses prolidase expression. Particularly, prolidase mRNA levels were significantly decreased after 12 to 24 h of Mith A treatment, whereas the protein levels were significantly decreased by 24 h. These results establish that KLF6 and Sp1 bind to the PEPD promoter and blocking Sp1-binding to the promoter or reducing KLF6 expression lowers prolidase expression likely by inhibiting PEPD promoter-driven transcription.
TGF-β1 induces KLF6 and Sp1 to drive prolidase expression
Our results in Figure 1, Figure 2, Figure 3 provided strong evidence that KLF6 and Sp1 are directly involved in the molecular regulation of prolidase via transcriptional activation of the PEPD promoter. To test the physiological relevance of activation of PEPD promoter by KLF6/Sp1, we probed the effects of TGF-β1 signaling on KLF6- and Sp1-mediated expression of prolidase. TGF-β1 is a cytokine that regulates collagen metabolism, fibroblast differentiation, and wound healing (37, 38). Particularly, TGF-β1 stimulates collagen biosynthesis/turnover by activating the transcriptional activity of KLF6 and Sp1 (34). Since prolidase carries out the final step of collagen degradation (39) and KLF6/Sp1 binds to the PEPD promoter, we probed the role of TGF-β1 signaling in the regulation of prolidase expression.
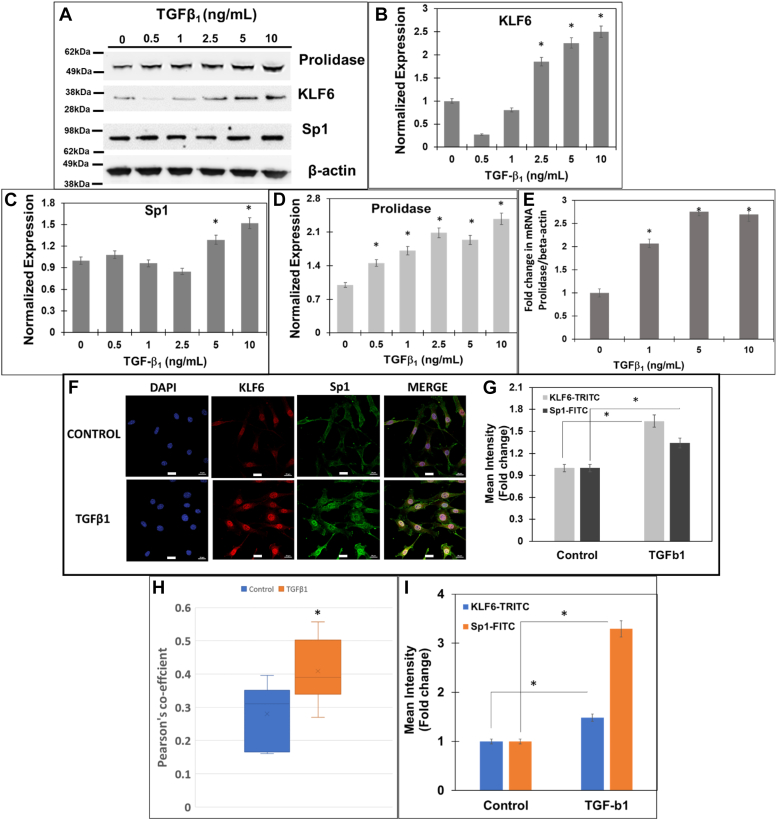
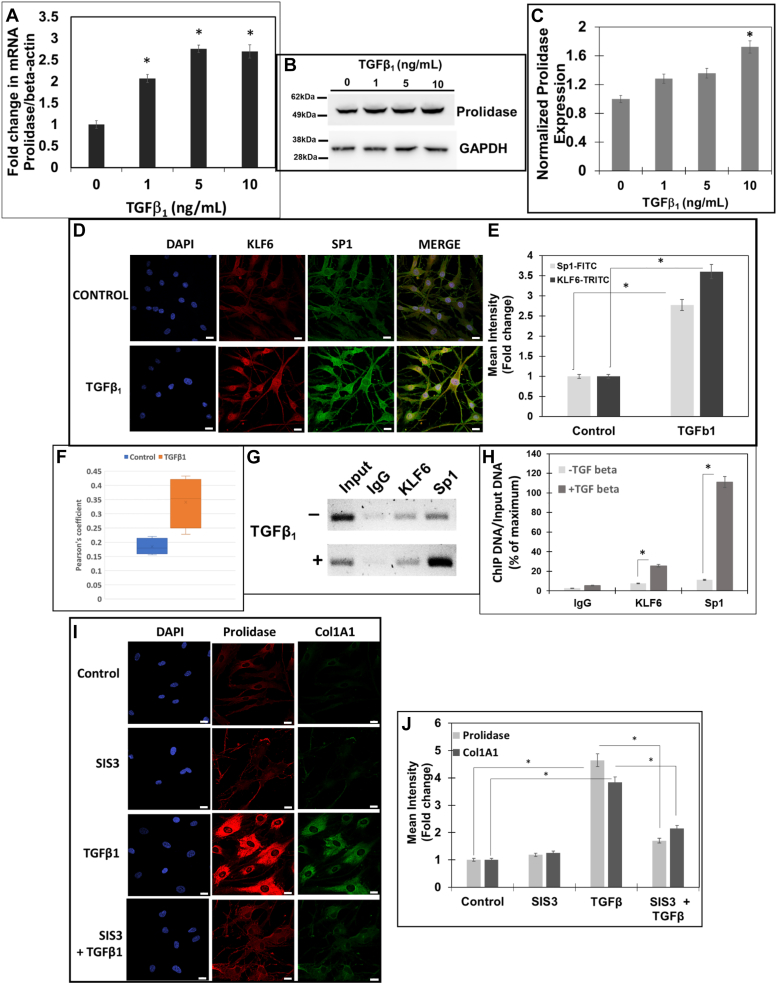
First, we treated NIH3T3 cells with increasing concentrations of TGF-β1 (0–10 ng/ml) and measured KLF6 and Sp1 expression by Western blot (Fig. 4). Initially, we carried out TGF-β1 treatments in a time-dependent manner and observed optimal effects on KLF6 and Sp1 at 6 h (data not shown). Thus, we treated the cells with TGF-β1 for 6 h and harvested cells for protein and RNA extraction. Western blot analysis showed increased levels of KLF6 with TGF-β1 treatment in a dose-dependent manner (Fig. 4, A and B). For instance, KLF6 expression was significantly increased with 2.5 to 10 ng/ml of TGF-β1 treatment, with a maximum of ∼2.5-fold increase at 10 ng/ml. Interestingly, Sp1 level was also increased with TGF-β1 treatment (∼1.5-fold) only with treatments above 5 ng/ml. Interestingly, a dose-dependent increase in prolidase expression was also detected in the TGF-β1 treated cells concurrent with the higher levels of KLF6 and Sp1 (Fig. 4, A and D). Densitometry analyses showed a maximum induction of ∼2 to 2.5-fold increase in prolidase levels in cells treated with 2.5 to 10 ng/ml of TGF-β1 relative to vehicle-treated cells (Fig. 4D). Together, these results demonstrated that TGF-β1 treatment upregulates KLF6 and Sp1 expression concurrent with higher levels of prolidase. To verify whether the increased prolidase levels is a consequence of transcriptional activation, we measured the PEPD mRNA levels by qPCR (Fig. 4E). A dose-dependent increase in prolidase mRNA levels was also observed in NIH3T3 cells after the treatment with increasing amounts of TGF-β1 (Fig. 4E). Interestingly, a maximum increase of ∼2 to 2.5-fold in mRNA levels was measured relative to the untreated controls with 5 to 10 ng/ml of TGF-β1 treatment, similar to the increase in the prolidase protein expression at these conditions (Fig. 4D). These results strongly suggest that TGF-β1 treatment induces KLF6 and Sp1 expression in mouse fibroblast cell lines concurrent with significantly higher mRNA and protein levels of prolidase.
Figure 4.
TGF-β1increases expression of prolidase concurrent with increased KLF6 and Sp1 in mouse fibroblasts. NIH3T3 cells were serum-starved for 6 h and treated with TGF-β1 in a concentration and time-dependent manner. After treatment total RNA was isolated for mRNA quantification and cell lysates were prepared for protein measurements. Prolidase mRNA expression was measured by qPCR and normalized to β-actin. Equal protein amounts of cell lysates were subjected to immunoblot analyses to measure protein expression. A, representative Immunoblot showing expression of prolidase, KLF6, Sp1, and β-actin. Densitometry analyses of (B) KLF6, (C) Sp1, and (D) prolidase expression normalized to β-actin. E, the fold change in prolidase mRNA expression with increasing concentration of TGF-β1 treatment is represented as treated vs untreated cells based on ΔΔCt values. F–I, confocal microscopy analysis of KLF6 and Sp1 in NIH3T3 cells after treatment with TGF-β1. Following treatment cells were fixed, permeabilzed, and stained with blue staining of nuclei with DAPI, anti-KLF6 (red), and anti-Sp1 (green). F, representative confocal images showing the expression of KLF6 and Sp1 and their colocalization (merge). Scale bars: 20 μm. G, the calculated mean fluorescence intensity values show the increase in expression of both KLF6 and Sp1 following TGF-β1 treatment as compared to control cells. H, Pearson’s coefficient shows the co-localization of KLF6 and Sp1 following TGF-β1 treatment as compared to control cells. I, nuclear co-localization of KLF6 and Sp1 following TGF-β1 treatment as compared to control cells. Data are mean values of three independent experiments with error bars representing SEM. ∗ represents a p value of < 0.05 for the statistical comparison of (B–E and G–I) untreated vs TGF-β1 treated samples.
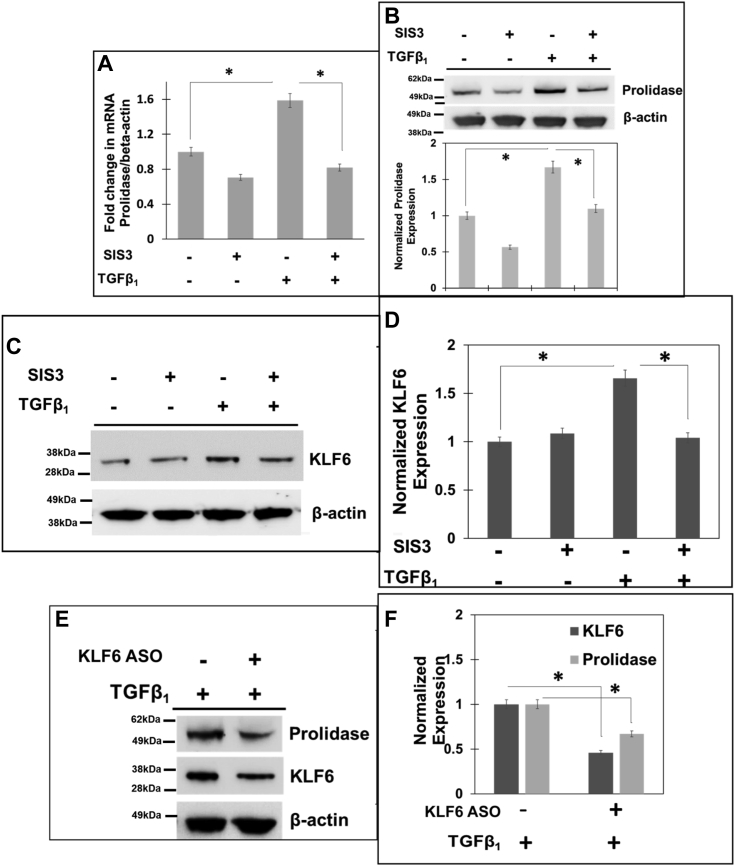
Then, we conducted confocal microscopy to further solidify the effects of TGF-β1 on KLF6 and Sp1 expression in these cells (Fig. 4, F and G). TGF-β1 treatment increased the expression of KLF6 and Sp1 as measured by the mean fluorescent intensities. Furthermore, we observed a strong co-localization between KLF6 and Sp1 in the nucleus upon treatment with TGF-β1. For instance, calculation of the Pearson’s coefficient indicated a significant increase in the co-localization of KLF6 and Sp1 in these cells (Fig. 4H), strongly supporting the interaction between these two transcription factors during TGF-β1 treatment. The data in Figure 4, G and H indicated that the colocalization of KLF6 and Sp1 is more robust than the induction of Sp1 at the protein level (∼1.5 fold) (Fig. 4G). Therefore, we compared the levels of nuclear localization of KLF6 and Sp1 in both untreated and TGF-β1 treated cells (Fig. 4I). Interestingly, the levels of nuclear localization of KLF6 were increased in cells treated with TGF-β1 to a level comparable to the increased protein levels (Fig. 4G). However, there was a robust (∼3-fold higher) Sp1 nuclear localization in TGF-β1 treated cells when compared to the untreated cells (Fig. 4I), although the levels of Sp1 protein only increased modestly (Fig. 4G). These results strongly suggested that TGF-β1 treatment increases nuclear localization of both Sp1 and KLF6 concurrent with higher co-localization between these two proteins in the nucleus. We predict that these phenotypes support Sp1/KLF6 mediated activation of prolidase expression during TGF-β1 signaling. The canonical TGF-β1 signaling pathway activates phosphorylation of the Smad2/3 family of transcription factors to activate genes involved in collagen synthesis, collagen turnover, and ECM remodeling (40). Thus, to further probe the role TGF-β1 signaling on prolidase expression, we utilized the specific inhibitor of Smad3 (SIS3) that inhibits Smad3 phosphorylation and abrogates collagen synthesis (41). To test this, serum-starved NIH3T3 cells were pretreated with SIS3 (10 μM) for 1 h followed by treatment with either TGF-β1 (5 ng/ml) or vehicle. Then, the cells were harvested for RNA and protein extraction to measure prolidase mRNA and protein levels. Our qPCR results revealed that SIS3 treatment alone reduced the levels of prolidase mRNA (Fig. 5A) and protein expression (Fig. 5B). As expected, TGF-β1 treatment induced both the mRNA and protein levels of prolidase (Fig. 5, A and B). Interestingly, pre-treatment of the cells with SIS3 abrogated upregulation of prolidase mRNA and protein expression in TGF-β1 treated cells (Fig. 5, A and B). These results confirmed that prolidase expression is induced during TGF-β1 signaling.
Figure 5.
Inhibition of TGF-β1signaling inhibits prolidase expression in a KLF6-dependentmanner. A–D, NIH3T3 cells were serum-starved for 6 h and treated with either vehicle (DMSO) or 10 μM of SIS3 for 1 h. Then, the cells were stimulated with TGF-β1 for 6 h. After treatment, total RNA and cell lysates were analyzed. A, Prolidase mRNA expression was measured by qPCR and normalized to β-actin mRNA. Prolidase mRNA expression is plotted based on ΔΔCt values and as fold change in SIS3-treated vs untreated cells in the presence and absence of TGF-β1. B–D, equal protein amounts of cell lysates were subjected to immunoblot analyses to measure (B) prolidase protein expression (representative immunoblot-upper panel and densitometry analyses-lower panel) and (C and D) KLF6 protein expression (C-representative immunoblot and D-densitometry analyses) as normalized to β-actin. E and F, KLF6 was knocked-down in NIH3T3 cells with ASO and then 24 h post transfection, cellular lysates were prepared for Western blot. E, representative blots (n = 3) showing expression of KLF6, prolidase and β-actin, in KLF6 knocked-down and control cells. F, Densitometric analysis of the immunoblots in E of KLF6 and prolidase that are normalized to β-actin levels. Data are mean values of three independent experiments with error bars representing SEM. ∗ p value of < 0.05 for the statistical comparison of untreated vs TGF-β1 treated samples and TGF-β1 treated vs TGF-β1 + SIS three treated samples.
Our results in Figure 2, Figure 3 demonstrated that Sp1 drives basal expression of prolidase, whereas KLF6 further increases prolidase expression. Therefore, we probed the effects of SIS3 on KLF6 expression with TGF-β1 treatment. We observed that SIS3 treatment alone minimally affected KLF6 expression (Fig. 5, C and D). As expected, KLF6 expression was upregulated in these cells upon TGF-β1 treatment. However, SIS3 treatment abrogated TGF-β1-induced KLF6 expression. To solidify that upregulation of prolidase during TGF-β1 signaling is mediated by KLF6, we also probed prolidase expression in KLF6 knocked-down cells after TGF-β1 treatment. Similar to data presented in Figure 3C, KLF6 level was significantly reduced in siRNA knocked-down cells compared to the control cells (Fig. 5, E and F). Interestingly, when these cells were treated with TGF-β1, prolidase expression remained lower in KLF6 knocked-down cells when compared to the control cells (Fig. 5, E and F). Collectively, these results indicate that KLF6 is critical for prolidase expression during TGF-β1 signaling.
TGF-β1 -mediated prolidase expression is induced by the binding of KLF6 with Sp1
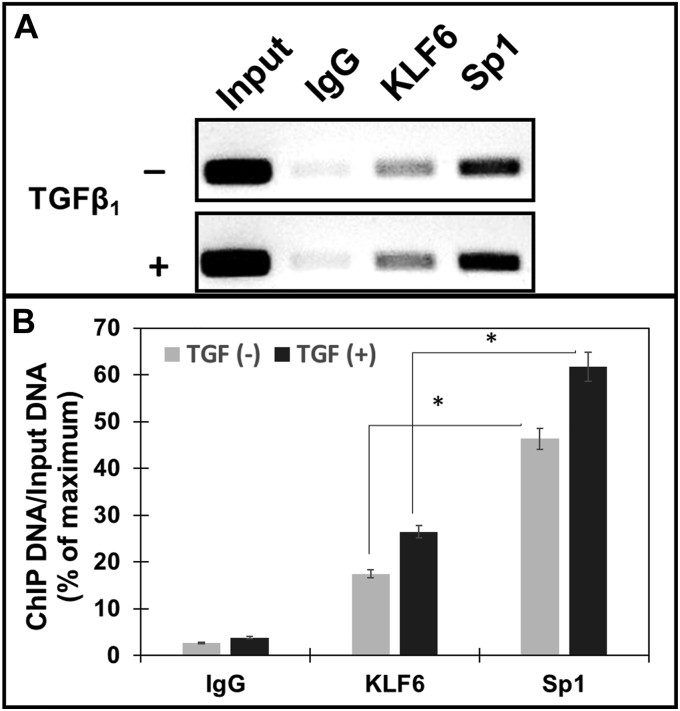
Our results in Figures 4 and 5 demonstrated that TGF-β1 treatment induced prolidase expression concurrent with the upregulation of KLF6 and Sp1 in NIH3T3 cells. Additionally, our observations demonstrated that KLF6 activates Sp1-driven prolidase expression. To test whether higher levels of KLF6 and Sp1 transactivate PEPD promoter to increase prolidase levels, we carried out ChIP assay. NIH3T3 cells treated with and without TGF-β1 were used for DNA isolation. Isolated DNA fragments were subjected to ChIP with antibodies of Sp1, KLF6, and IgG control, and the precipitated DNA was analyzed by PCR (Fig. 6A). Our results clearly showed that PEPD promoter sequences containing KLF6 and Sp1-binding sites were enriched when compared to the isotype IgG controls (Fig. 6A). We also observed that both KLF6 and Sp1 bind to the endogenous PEPD promoter in both untreated and TGF-β1 treated cells. Notably, the binding of both KLF6 and Sp1 to the PEPD promoter was further increased upon TGF-β1 treatment (Fig. 6A). Densitometric analysis revealed a stronger pull-down by Sp1 as compared to KLF6 both in the untreated and treated cells (Fig. 6B), suggesting that Sp1 most likely binds to the PEPD promoter at higher and/or stronger affinity compared to KLF6. Given that our results in Figures 4 and 5 demonstrated that TGF-β1 induced upregulation of both KLF6 and Sp1, these binding assay results provide strong evidence that TGF-β1 signaling activates KLF6/Sp1 for the transactivation of PEPD promoter. Together, these results suggest that PEPD transcriptional activation by KLF6 and Sp1 is mediated by the binding of these transcription factors to the promoter sequences.
Figure 6.
TGF-β1increases interaction of Sp1 and KLF6 on prolidase promoter: NIH3T3 cells plated in 6-well plates were serum-starved for 6 h were stimulated further with TGF-β1 for 6 h. Following treatment, the cells were subjected to ChIP analyses to probe the binding of Sp1 and KLF6 to the PEPD promoter using anti-KLF6, anti-Sp1 and IgG (control) antibodies. Co-precipitated chromatin fragments were purified and analyzed by PCR amplification using primers (Table 1) against the mouse PEPD proximal promoter region. PCR products were separated by agarose gel electrophoresis. Results were normalized to total input chromatin. A, representative blot and (B) Densitometry analyses of PCR amplified DNA from ChIP assay. Data in (B) are mean values of three independent experiments with error bars representing SEM. ∗ p value of < 0.05 for the statistical comparison of untreated vs TGF-β1 treated samples.
Prolidase expression induced by TGF-β1 is associated with increased collagen type 1 expression
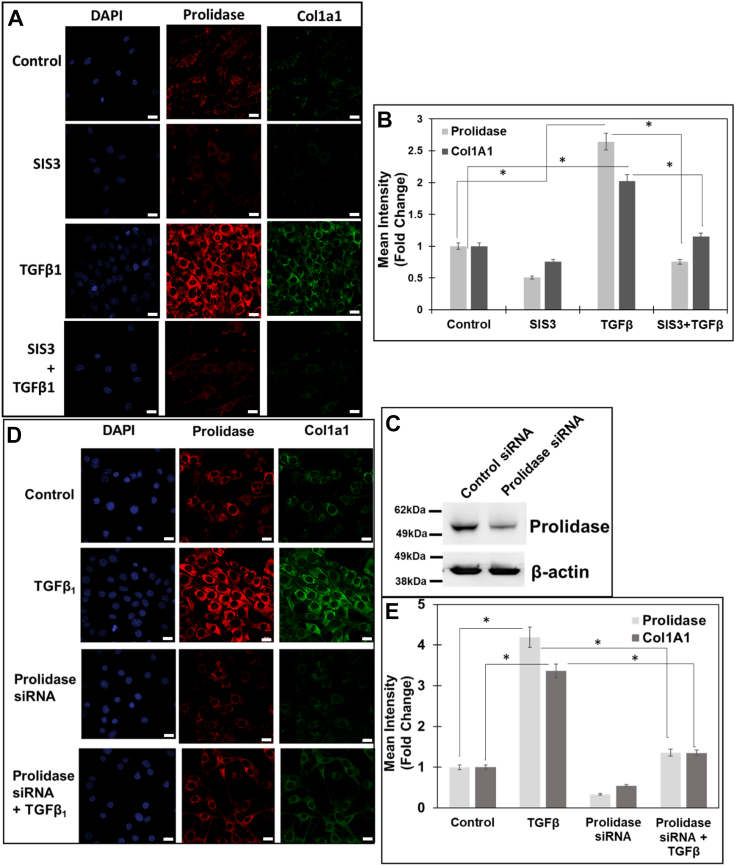
Next, we investigated whether KLF6 and Sp1-mediated upregulation of prolidase is functionally linked to collagen biosynthesis. Fibroblasts activated by TGF-β1 facilitate collagen synthesis that begins as rope-like procollagen molecules each comprising of three chains (42). Type I collagen is composed of two pro-α1(I) chains (produced from the COL1A1 gene) and one pro-α2(I) chain (encoded by the COL1A2 gene) (43). To probe the functional link between prolidase expression and collagen synthesis, we performed immunofluorescence studies and measured the effects of TGF-β1 on prolidase and Col1A1 expression in NIH3T3 cell lines (Fig. 7). NIH3T3 cells were treated with TGF-β1 in the presence and absence of SIS3 and the expression of Col1A1 was measured after 6h. As expected, TGF-β1 treatment resulted in a marked increase in Col1A1 expression concurrent with higher prolidase expression (Fig. 7, A and B). Inhibition of TGF-β1 signaling with the pre-treatment with SIS3 attenuated the TGF-β1-induced expression of both prolidase and Col1A1. To confirm the functional link between prolidase and Col1A1 expression, we carried out knocked-down studies of prolidase in these cells by siRNA-based method (Fig. 7, C–E). Western blot analysis confirmed significantly reduced levels of prolidase in siRNA transfected cells compared to the control cells (Fig. 7C). Then, confocal microscopy analysis showed that reduced prolidase expression is associated with lower levels of Col1A1 in NIH 3T3 cells. Interestingly, reduced levels of Col1A1 were observed in cells untreated or treated with TGF-β1 (Fig. 7, D and E). These observations establish the functional contribution of prolidase in ColA1 expression during TGF-β1 signaling.
Figure 7.
TGF-β1 induced prolidase-dependent increase in collagen type 1 expression in NIH3T3 fibroblasts.A and B, NIH3T3 fibroblasts were plated in 8-well chamber slides and serum-starved for 6 h and were treated with either vehicle (DMSO) or SIS3 (10 μM) 1 h prior to stimulation with TGF-β1 (5 ng/ml) for 6 h. A, representative confocal images of cells stained for prolidase (red) and Col1A1 (green) and the nuclei (blue). Scale bar, 20 μm. B, the graph represents the mean fluorescence intensity of prolidase and Col1A1 in SIS3-treated vs untreated cells in the presence and absence of TGF-β1. C–E, NIH3T3 cells were plated in 8-well chamber slides and transfected with either control siRNA or prolidase specific siRNA for 48 h. Following which the cells were serum starved and stimulated with TGF-β1 (5 ng/ml) for 6 h. After treatment the samples were prepared for confocal microscopy. C, representative immunoblot showing the knockdown of prolidase expression with siRNA, using β-actin as loading control. D, representative confocal images of prolidase and Col1A1 expression in NIH3T3 cells after staining. Scale bar, 20 μm. E, the graph represents the mean fluorescence intensity of prolidase and Col1A1 in siRNA-transfected vs control-siRNA-transfected cells in the presence and absence of TGF-β1. Data in (B and E) are mean values of three independent experiments with error bars representing SEM. ∗ p value of < 0.05 for the statistical comparison of (B) untreated control vs TGF-β1 treated samples and TGF-β1 treated vs TGF-β1 + SIS3 treated samples, and (E) untreated control vs TGF-β1 treated samples and TGF-β1 treated vs TGF-β1 + Prolidase siRNA samples.
TGF-β1 activates KLF6/Sp1-mediated prolidase expression to enhance Col1A1 levels in human fibroblasts
Our studies of HEK293T cells and NIH3T3 mouse fibroblast cells established that KLF6 and Sp1 upregulated by TGF-β1 induces prolidase expression via transcriptional activation (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6). Our results also indicated that increased levels of prolidase were associated with higher synthesis of Col1A1 in mouse fibroblast cell lines (Fig. 7). To determine whether this regulatory pathway is also activated in human fibroblasts, we investigated the effects of TGF-β1 on KLF6, Sp1, prolidase and Col1A1 in primary human dermal fibroblast (HDF). HDFs were cultured and treated with TGF-β1 in a dose-dependent manner and prolidase mRNA and protein levels were measured by qPCR and Western blot, respectively (Fig. 8A). TGF-β1 treatment increased the mRNA levels of prolidase up to two to 3-fold similar to the data obtained with NIH3T3 cells (Fig. 4). Prolidase protein levels were also increased in these cells upon TGF-β1 treatment (Fig. 8, B and C). Notably, confocal imaging studies revealed that the expression of KLF6 and Sp1 was also upregulated in HDFs after TGF-β1 treatment when compared to the untreated control cells (Fig. 8, D and E). Calculation of Pearson’s coefficient demonstrated strong co-localization of KLF6 and Sp1 in these cells after TGF-β1 treatment (Fig. 8F). We also carried out a ChIP assay to probe binding of KLF6 and Sp1 to the PEPD promoter sequences in these HDFs. Results from these analyses confirmed that both Sp1 and KLF6 bind to the PEPD promoter compared to the isotype IgG control (Fig. 8, G and H). Similar to the mouse fibroblast cells, strong binding of Sp1 to the PEPD promoter was detected when compared to the binding of KLF6. Interestingly, the binding of both Sp1 and KLF6 to the promoter was significantly increased in TGF-β1 treated HDFs (Fig. 8, G and H). Finally, we also observed increased levels of prolidase protein concurrent with higher expression of Col1A1 in TGF-β1 treated HDFs compared to the untreated control cells (Fig. 8, I and J). Notably, inhibition of the TGF-β1 pathway by SIS3 significantly reduced the levels of prolidase and Col1A1. Collectively, these results using HDFs provide strong evidence that PEPD transcriptional activation by KLF6 and Sp1 is mediated by the direct binding of these transcription factors to the promoter sequences and the ensuing production of Col1A1-a key component of collagen.
Figure 8.
TGF-β1 induced KLF6/SP1/prolidase axes increase Col1A1 expression in primary human dermal fibroblasts. Serum-starved primary HDFs were treated with increasing concentrations of TGF-β1 (0–10 ng/ml). A, prolidase mRNA levels normalized to GAPDH analyzed by qPCR. B and C, Prolidase protein expression was analyzed by Western blot using GAPDH as a loading control. B, a representative blot, and (C) Densitometric analyses of prolidase expression normalized to GAPDH. D and E, confocal microscopy of KLF6 and Sp1 expression. HDFs were treated with TGF-β1 (10 ng/ml), were fixed/permeabilized and stained with DAPI, anti-KLF6 (red) and anti- Sp1 (green). D, representative images showing KLF6 and Sp1 and colocalization (merge). Scale bars: 20 μm. E, calculated mean fluorescence intensity showing expression of both KLF6 and Sp1 following TGF-β1 treatment as compared to control cells. F, Pearson’s coefficient showing the co-localization of KLF6 and Sp1 following TGF-β1 treatment as compared to control cells. G and H, ChIP was performed using anti-KLF6, anti-Sp1 and IgG (control) antibodies. Co-precipitated chromatin fragments were purified and analyzed by PCR using primers (Table 1) against the human PEPD promoter. PCR products were resolved by agarose gel electrophoresis. Results were normalized to total input chromatin. G, representative blot and (H) Densitometry analyses of PCR amplified DNA. (I and J) HDFs were treated with either vehicle (DMSO) or SIS3 (10 μM) 1 h prior to stimulation with TGF-β1 (10 ng/ml) for 6 h. I, representative confocal images of cells stained for prolidase (red) and Col1A1 (green) and the nuclei (blue). Scale bar, 20 μm. J, the graph represents the mean fluorescence intensity of prolidase and Col1A1 in SIS3-treated vs untreated cells in the presence and absence of TGF-β1. Data are mean values of three independent experiments with error bars representing SEM. ∗ p value of < 0.05 for the statistical comparison of (A–C, E–F, and G) untreated vs TGF-β1 treated samples and (E) untreated control vs TGF-β1 treated samples and TGF-β1 treated vs TGF-β1 + SIS3 treated samples.
Discussion
Prolidase catalyzes the rate-limiting step in collagen degradation, turn-over and synthesis (14, 39, 44). Importantly, proline released by the enzymatic activity of prolidase is utilized for protein synthesis (including collagen) to support cell growth and proliferation (14, 39, 44). Therefore, alterations in prolidase activity is associated with a number of pathological conditions such as cancer, liver disease & wound healing (39, 44). Dysregulation in prolidase activity is also associated with prolidase deficiency (PD)- a rare genetic disease that manifests imidodipeptiduria, hard-to-heal wounds, mental retardation and impaired immune system (39, 44). Prolidase also regulates other cellular processes, including EGFR- and HER2-dependent signaling pathways, p53 activity, and interferon response, a function that is not dependent on its enzymatic activity (27, 29, 30, 45, 46, 47). Despite of these critical roles, the molecular and cellular mechanisms that regulate prolidase expression during either normal or pathological conditions remain largely unknown. In this study, we provide evidence that the transcriptional factors, KLF6 and Sp1, directly bind to and transcriptionally activate the PEPD promoter. We also demonstrate that during TGF-β1 signaling KLF6 activates Sp1-driven PEPD promoter transcription to increase prolidase expression and enhance collagen synthesis.
Cellular protein expression is regulated at transcriptional, post-transcriptional, translational, and post-translational levels. However, the underlying role of these fundamental regulatory mechanisms in prolidase expression remains largely unknown. There are only few studies that describe the involvement of regulatory mechanisms for prolidase. For instance, there is evidence that post-translational mechanisms such as phosphorylation regulate prolidase expression (25, 26). An early study by Tanoue et al. (1990) reported that PEPD gene lacks a TATA-box (31), a feature similar to other housekeeping genes that are stimulated during cell growth and proliferation. Like many TATA-less promoters, the proximal 5'-flanking region of the human and mouse prolidase promoter has numerous GC-rich elements (48). They also described that PEPD promoter contains a canonical "CAAT" box and putative Sp1 binding sites (31). However, to the best of our knowledge experimental evidence that supports transcriptional regulation of PEPD by Sp1 or any other transcription factors is lacking. Interestingly, our in silico analyses of the PEPD promoter sequences from −1537 bp to +50 bp (+1 bp being the TSS) identified overlapping and evolutionarily conserved bindings sites for Sp1 and KLF6 (Fig. 1). The KLF6 binding sites were abundant and distributed across the entire PEPD promoter region, while the Sp1 sites were relatively few and were concentrated proximal to the TSS, in accordance with the study by Tanoue et al. (31). Identification of the overlapping Sp1 and KLF6 binding sites in the PEPD promoter coupled with the reported role of these two transcription factors in collagen biosynthesis (34) and the critical role of prolidase in collagen turnover (39) provided a strong scientific rationale to study transcriptional regulation of the PEPD promoter.
First, we combined luciferase reporter assay with site-directed promoter deletion studies to demonstrate transcriptional regulation of PEPD promoter. This approach identified that the 5′ upstream sequences spanning from −337 bp to the TSS are required for PEPD promoter-driven transcriptional activity (Fig. 2, A and B). Our results also pinpointed that the overlapping Sp1 and KLF6 binding sites are clustered within the −337 bp region and deletion of these sequences abrogated PEPD promoter activity (Fig. 2B). Additionally, the KLF6 and Sp1 binding sites located 5′ upstream of the PEPD promoter sequences from −1537 to −337 bp were mostly dispensable for promoter activity, since deletion of these sequences marginally reduced PEPD promoter activity (Fig. 2B). These key results established that the sequences from −337 bp to the TSS are functionally critical for PEPD promoter-driven transcription. Next, we probed whether Sp1 and KLF6 functionally regulate PEPD promoter-driven transcription. Luciferase reporter-based PEPD promoter studies in HEK293T cells revealed that Sp1 is responsible for the basal transcription of PEPD promoter (Fig. 2E). This is in accordance with the well-recognized role of Sp1 as a ubiquitous transcription factor that drives basal transcription of a wide variety of housekeeping genes (49). Interestingly, Sp1-driven PEPD promoter-driven transcription was significantly stimulated by KLF6 (Fig. 2, D and F). Our ChIP-based binding assays confirmed that transcriptional activation of the PEPD by Sp1 and KLF6 was a consequence of the direct binding of these transcriptional factors to specific sequences in the promoter (Fig. 3, A and B). Particularly, Sp1 showed strong binding to the PEPD promoter when compared to the binding of KLF6. Inhibition of Sp1 binding abrogated PEPD promoter activity, demonstrating that Sp1 is required for transcriptional activation (Fig. 2G). Notably, when Sp1 binding was inhibited, KLF6 failed to activate PEPD promoter-driven transcription (Fig. 2F), thus demonstrating that KLF6-mediated regulation of PEPD promoter activity is dependent on Sp1. Accordingly, both inhibition of Sp1 and knock-down of KLF6 reduced prolidase expression (Fig. 3, C–G). It is noteworthy that the cooperative function of KLF6 and Sp1 in transcriptional regulation has been reported for a number of genes (34, 50). Collectively, our molecular, genetic, and biochemical studies support a direct and functional role of KLF6 and Sp1 in the transcriptional activation of PEPD promoter and the expression of prolidase.
Fibroblasts, the most abundant type of cells in connective tissues are required for the biosynthesis, degradation, and remodeling of ECM (36). The TGF-β super-family of growth factors plays critical roles during the synthesis and degradation of ECM components (38). Among the three TGF-β isoforms (TGF-β1, TGF-β2, and TGF-β3); TGF-β1 is the most important regulator of collagen biosynthesis in fibroblasts. Since prolidase catalyzes the rate-limiting step in collagen biosynthesis and TGF-β1 is a master regulator of collagen synthesis/turnover during ECM remodeling, we evaluated the effects of TGF-β1 on prolidase expression in both mouse and human fibroblasts. We also probed whether KLF6 and Sp1-mediated transcriptional regulation of PEPD promoter is activated during TGF-β1 signaling. Interestingly, in both mouse and human fibroblasts, TGF-β1 treatment significantly upregulated prolidase expression both at the transcriptional (mRNA) and translational (protein) level (Figs. 4 and 8). It is well-known that TGF-β1 exerts its effects by activating the serine/threonine kinase receptor complex comprising of type I and II TGF-β1 receptors and the subsequent activation of the canonical SMAD signaling pathway (40). Particularly, the binding of TGF-β1 activates both receptor-associated SMADs (SMAD2 and SMAD3) by phosphorylation. Activated SMAD2 and SMAD3 hetero-oligomerize with other SMADs to form complexes that translocate to the nucleus and regulate the expression of several genes important in ECM remodeling. Thus, to confirm the role of TGF-β1 signaling in prolidase expression, we used a specific SMAD3 inhibitor “SIS3” that inhibits SMAD3 phosphorylation, prevents its binding to SMAD4, and inhibits the downstream signaling cascade (37, 38, 41).
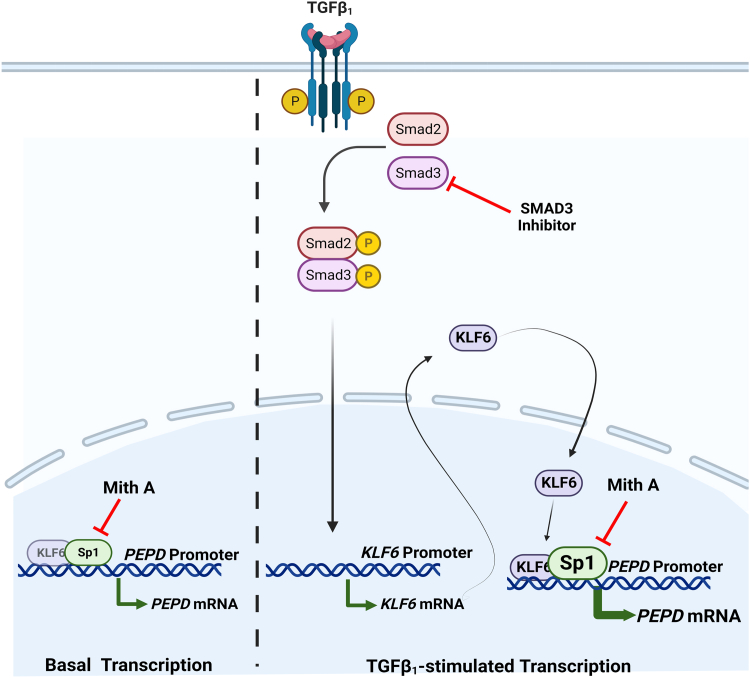
SIS3 treatment significantly diminished TGF-β1-induced prolidase expression both at mRNA and protein levels (Fig. 5, A and B) suggesting that the SMAD pathway is involved in the molecular regulation of prolidase. Interestingly, SIS3 treatment also abrogated the effects of TGF-β1-mediated upregulation of KLF6 and Sp1 (Figs. 4 and 8). The effects of TGF-β1 on KLF6 and Sp1 expression is not surprising, since TGF-β1 has been reported to regulate KLF6 expression and promote its cooperative interaction with Sp1 to drive target gene expression (37, 38). Accordingly, in both mouse and human fibroblasts, TGF-β1 treatment-induced KLF6 and Sp1 overexpression also increased binding of KLF6 and Sp1 to the PEPD promoter (Figs. 6 and 8, G and H). Even though the molecular and biochemical mechanisms underlying the cooperative function of KLF6 and Sp1 are not fully understood, there is evidence that KLF6 and Sp1 physically interact to transcriptionally regulate target genes (51, 52, 53). For instance, KLF6 binding to Sp1 has been reported to stabilize the binding of Sp1 to target gene promoters, thereby promoting the ability of Sp1 to transactivate (54). Our immunofluorescence studies demonstrated that KLF6 and Sp1 colocalize in the nucleus of the mouse and human fibroblasts (Figs. 4FI and 8, D–F). The nuclear colocalization of KLF6 and Sp1 was further enhanced when these fibroblasts were treated with TGF-β1. These results provide further support for the interaction between KLF6 and Sp1 in fibroblasts in response to TGF-β1 signaling. The regulation of prolidase at the transcriptional level by KLF6 and Sp1 is a novel finding highlighting the functional link between prolidase and the Sp/KLF families of transcriptional regulators and their target genes related to collagen turnover/synthesis. Therefore, we predict that TGF-β1 signaling in fibroblasts stimulates prolidase expression by a) upregulating KLF6 and Sp1 expression, b) increasing nuclear localization of KLF6 and Sp1, c) enhancing binding of these two transcription factors to specific sequences of PEPD promoter, and d) activating KLF6 and Sp1-driven PEPD promoter transcription (Fig. 9).
Figure 9.
Hypothetical model describing regulation of prolidase expression by TGF-β1-Sp1-KLF6 axes. The schematic model shows the expression of prolidase under basal and TGF-β1 stimulated conditions. Left Panel- We propose that the basal expression of prolidase is primarily regulated at the transcriptional step by Sp1, since inhibition studies of Sp1 using Mith A abrogated prolidase expression. Right panel- We predict that TGF-β1 activates the SMAD pathway to induce the expression of KLF6. Higher levels of KLF6 enhance Sp1-mediated PEPD promoter activity. The cooperative interaction of KLF6 and Sp1 at the overlapping binding sites within the PEPD promoter upregulates prolidase expression. Inhibition of TGF-β1 signaling by an SMAD3 inhibitor suppresses KLF6/Sp1-mediated activation of PEPD promoter-driven transcription. Finally, upregulation of prolidase during TGF-β1 signaling catalyzes the rate-limiting step in collagen biosynthesis to produce higher levels of collagen such as Col1A1. Collectively, this hypothetical model describes a mechanism for the molecular regulation of prolidase at the transcriptional level.
Finally, our studies demonstrate the functional significance of prolidase expression to collagen biosynthesis, given that prolidase catalyzes the rate-limiting step in collagen degradation, turn-over, and synthesis (14). Fibroblasts activated by TGF-β1 facilitate collagen biosynthesis that starts with the procollagen molecules of three chains (38). Type I collagen is composed of two pro-α1(I) chains encoded by the COL1A1 gene and one pro-α2(I) chain encoded by the COL1A2 gene. Therefore, as a proof of concept, we studied the expression of ColA1 in fibroblasts in response to TGF-β1 stimuli, since type I collagen is the most abundant among the various types of collagens (55). Our immunofluorescence studies of both mouse and human fibroblasts showed that TGF-β1 treatment resulted in higher levels of prolidase expression concurrent with increased ColA1 expression (Figs. 7 and 8). Interestingly, reduced levels of prolidase, by siRNA-mediated knock-down, resulted in a lower level of Col1A1 expression in these fibroblasts (Figs. 7 and 8). Moreover, a similar reduction in ColA1 expression concurrent with lower expression of prolidase, was also obtained when TGF-β1 signaling was inhibited by SIS3 (Figs. 7 and 8). These data strongly support that induction of prolidase expression by TGF-β1 signaling is functionally associated with increased collagen biosynthesis. Interestingly, it has been reported that inhibitors of prolidase catalytic activity decrease the expression of TGF-β1 and its receptor, while supplementation of proline and hydroxyproline, the products of prolidase catalytic activity resulted in an increased expression of TGF-β1 and its receptor (56). Therefore, the stimulating effects of TGF-β1 on prolidase expression imply the presence of a positive regulatory feedback loop that may be activated in response to mediate collagen synthesis, turnover, and ECM remodeling.
In summary, in this report we provide firsthand evidence for the transcriptional regulation of prolidase by KLF6 and Sp1 (Fig. 9). Our hypothetical model predicts that basal expression of prolidase is transcriptionally regulated by Sp1. Notably, upon induction of KLF6 by TGF-β1 stimuli, prolidase expression is further stimulated via cooperative transcriptional activation by KLF6 and Sp1. Therefore, we propose that higher levels of prolidase during TGF-β1 signaling enhances collagen synthesis. Finally, our findings demonstrate that prolidase is a target of TGFβ1 signaling. We predict that TGFβ1-induced prolidase expression has significant biological implications due to the necessity of prolidase in collagen turnover/biosynthesis and the wound healing process.
Experimental procedures
Reagents
Recombinant human Transforming Growth Factor β1 (TGF-β1) was obtained from Shenandoah Biotechnology and Genscript and resuspended/stored as per the instructions provided by the manufacturer. Mithramycin A (MithA) was purchased from Active Motiff, and Smad3 Inhibitor (SIS3) was purchased from MedChemExpress. The wild-type flag-hKLF6 (1006) was a gift from Scott Friedman (Addgene plasmid # 49488; http://n2t.net/addgene:49,488; RRID: Addgene_49488) and pN3-Sp1FL was a gift from Guntram Suske (Addgene plasmid # 24543; http://n2t.net/addgene:24,543; RRID: Addgene_24543). pGL3-basic, pRL-null, and Dual-Luciferase Reporter Assay System obtained from Promega. The primary antibodies used were as follows: anti-Sp1 (cat# PA5-29165), anti-collagen A1 (cat# PA5-29569) and anti-prolidase (cat# PA5-53335) were purchased from Thermofisher, anti-KLF6 (cat# MABN119) was purchased from EMD Millipore, anti-GAPDH was obtained from Sigma-Aldrich and anti-β-actin was obtained from Proteintech. The secondary antibodies used were goat anti-rabbit or goat anti-mouse purchased from BioRad laboratories.
Cell culture
HEK293T and NIH3T3 mouse embryonic fibroblasts were obtained from American Type Culture Collection (ATCC) and maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 2 mM glutamine and 1% antibiotics (penicillin-streptomycin) at 37 °C/5% CO2. HEK293T cells were supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Gibco) while NIH3T3 cells were supplemented with 10% (vol/vol) heat-inactivated calf bovine serum (Gibco). Normal Human Dermal Fibroblasts – Adult (HDF) were obtained from Lifeline Cell Technology and maintained as per the manufacturer's protocol at 37 °C/5% CO2. NIH3T3 and HDF cells were refed serum-free medium and stimulated with TGF-β1 with a dose-dependent and time-dependent manner with or without the pre-treatment of Mith A (1 μM) or SIS3 (10 μM).
Mapping, in silico analyses, and promoter cloning
Prolidase promoter sequences upstream of the transcription start site (human, mouse, rat, cow, rhesus macaque, chimpanzee) were retrieved as a FASTA file from NCBI Genome Browse and ENSEMBL. These sequences were aligned using Clustal W. Human and mouse prolidase promoter sequences were analyzed using TRANSFAC to identify putative transcription factors. The human prolidase promoter − 1537 to + 50 bp relative to the Transcription start site (TSS) was amplified from genomic DNA. The primers used are listed in Table 1. The conditions for PCR amplification were: denaturation at 98 °C for 30 s, 72 °C for 1 min, |40 cycles, and final extension at 72 °C for 5 min. The amplified PCR product was digested and cloned into the XhoI and HindIII (NEB) restriction sites of the pGL3 vector (Promega) to generate the PEPD promoter-luciferase reporter construct (PEPD-Luc) and confirmed by sequencing.
Table 1.
List of primers
| Name | (5′→ 3′) |
|---|---|
| Forward primer for prolidase promoter | TATACTCGAGCTGGCAGCTTTGGTCTC |
| Reverse primer for prolidase promoter | TATAAAGCTTGCCAGCGGGAAAGAG |
| Mouse promoter ChIP forward | AGCTGCAGGACTCCTCCACTTAAGG |
| Mouse promoter ChIP reverse | ACAGTGGACGCCATGTTCACTCGG |
| Human promoter ChIP forward | AACCCGACCTACTGTCTGCAGC |
| Human promoter ChIP reverse | TCACGTGAAGTGCGGCGTCAG |
| pGL3-hPEPDpro-1237 bp/+50 bp forward Q5 | TATAGGATCCAAACAAGAAGCATCCCCC |
| pGL3-hPEPDpro −937 bp/+50 bp forward Q5 | TATAGGATCCTTAAAGATAGTCTCGACCTCATAG |
| pGL3-hPEPDpro −637 bp/+50 bp forward Q5 | TATAGGATCCACCTAGAAAAGATCCTACCAG |
| pGL3-hPEPDpro −337 bp/+50 bp forward Q5 | TATAGGATCCCAGAATCAAGATGTCCTCTGC |
| pGL3-hPEPDpro −37/+50 bp forward Q5 | TATAGGATCCGCCGCACTTCACGTG |
| Delta 1 deletion forward | TCAGCTGACGCCGCACTT |
| Delta 1 deletion reverse | ATGGAGTCCTTGACCCACC |
| Delta 2 deletion forward | CCCGTCCCGTGGAATTTG |
| Delta 2 deletion reverse primer | AGATTGGGGGCCCAAGAG |
| Human/Mouse prolidase cDNA forward | GCTGGGGAATGAAACCCTGA |
| Human/Mouse prolidase cDNA reverse | CGCCCAGTGAAAGAAGGACT |
| Human/Mouse KLF6 cDNA forward | CACAGGAGAAAAGCCTTACAGATGC |
| Human/Mouse KLF6 cDNA reverse | AGGTGCCTCTTCATGTGCAGGGC |
| Human/Mouse Sp1 cDNA forward | TCAAATACAGATCATACCAGGTGCAAACC |
| Human/Mouse Sp1 cDNA reverse | TTGACAGGTAGCAAGGTGATGTTCC |
| GAPDH cDNA forward | GAAGGTGAAGGTCGGAGTC |
| GAPDH cDNA reverse | GAAGATGGTGATGGGATTTC |
| Actin cDNA forward | GCTCGTCGTCGACAACGGCTC |
| Actin cDNA reverse | CAAACATGATCTGGGTCATCTTCTC |
Promoter mutagenesis
Using the PEPD-Luc construct several constructs of varying length (−1237 bp/+50 bp, −937 bp/+50 bp, −637 bp/+50 bp, −337 bp/+50 bp, −37/+50 bp) of the PEPD-Luc plasmid were also generated to determine the minimal promoter by PCR using primers as described in Table 1. In addition, deletion constructs of the PEPD-Luc plasmid were generated using the Q5 site-directed mutagenesis kit as per the instructions provided by the manufacturer (NEB) and primers listed in Table 1. Two constructs with deletions in the PEPD promoter regions −44 to −98 (delta 1-D1) and −147 to −197 (delta 2-D2) containing Sp1/KLF6 transcription factor binding sites were generated and confirmed by sequencing.
Dual-luciferase reporter assay
PEPD transcriptional activity was measured using the Dual-Luciferase Reporter Assay (Promega) according to the manufacturer's protocol. HEK293T cells were grown in 48-well plates to approximately 70% to 80% confluence before transfection with reporter plasmids. The cells were co-transfected with the PEPD-Luc construct and pRL-null, a Renilla construct for normalizing transfection efficiency. Transfection was performed using polyethylenimine (PEI) as per the manufacturer’s instructions. To determine the effect of KLF6/Sp1 on PEPD promoter activity, equivalent amounts of KLF6/Sp1 or pcDNA3.1 empty vector plasmid were transfected along with the reporter constructs. Following which, the transfected cells were lysed, and luciferase activity was measured with equal amounts of cell extract using a plate reader (BioTek). Samples were assayed in triplicate.
Quantitative Real-time PCR
Total RNA was isolated from NIH3T3 or primary HDFs using Quick-RNA Miniprep (Zymo Research). cDNA was synthesized using OneScript Plus cDNA Synthesis Kit (Abmgood). qPCR assay was performed by subjecting 100 ng of cDNA to iTaq Universal SYBR Green chemistry (Bio-Rad) in a C1000 Touch CFX96 Real time System (Bio-Rad). Expression data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and/or Actin as an internal control, and the relative expression was calculated using the ΔΔCt method. Primers are listed in Table 1. Relative expression of mRNA is expressed as 2-delta Ct values as described previously and fold change in expression was calculated by comparing the 2-delta Ct values of the treated sample with that of untreated control. All samples were analyzed in triplicate.
Western blot analyses
After treatment cell lysates were prepared and protein concentrations were quantified by Bicinchoninic acid assay (BCA) (Pierce). Equal amounts of protein were electrophoresed on SDS-polyacrylamide gels and transferred to nitrocellulose membranes using a semi-dry blotter (Bio-Rad). Membranes were blocked with 5% (w/v) nonfat milk in Tris Buffered Saline with Tween 20. (TBST), pH 8.0 (Sigma) and then probed with the primary antibody in blocking buffer. Subsequently the blot was incubated with a secondary antibody conjugated to horseradish peroxidase (1:2000). All blots were washed in TBST and developed using the enhanced chemiluminescence (ECL) procedure using BioRad ChemiDoc Imaging System. Blots were stripped using Restore Plus stripping buffer (Pierce) and re-probed with anti-GAPDH/anti-β-actin monoclonal antibodies to serve as loading controls. Densitometry analyses were performed using ImageJ software (National Institutes of Health). Data were normalized to levels of β-actin or GAPDH.
ChIP assay
NIH3T3 and HDF cells were grown in T75 and T182 flasks, respectively, to approximately 70% to 80% confluency and treated with or without TGF-β1. Chromatin immunoprecipitation was carried out using the Active Motif ChIP-IT Express Enzymatic Kit as per the manufacturer’s instructions. Cells were cross-linked using 1% paraformaldehyde and incubated at room temperature for 15 min, and fixing was stopped using glycine provided by the manufacturer. After washing and scraping, cells were lysed, and chromatin was enzymatically sheared. The enzymatic reaction was stopped using 0.5 M EDTA. Chromatin amount equivalent to 40 μg was used from pulldown with anti-KLF6, anti-Sp1, and IgG overnight at 4 °C. Pulled-down chromatin-bead complexes were washed, eluted, reverse cross-linked, and proteinase K treated. Equal amounts of DNA were used for endpoint PCR using MegaFi Pro Fidelity DNA Polymerase with primers listed in Table 1.
Knockdown studies
Prolidase-specific siRNAs and non-specific scrambled controls were purchased from Santa Cruz Biotechnology. NIH3T3 (2 × 105 cells/well) grown in 6-well culture plates were transfected with 100 to 300 PM of prolidase-specific siRNAs or scrambled controls using INTERFERIN (Polypus) as per the manufacturer’s protocol. Post transfection, cells were incubated for 36 to 48 h at 37 °C/5% CO2, washed with PBS (1×) and harvested by gentle scraping for protein isolation. Knock-down of prolidase was confirmed by immunoblot analysis.
For KLF6 knock-down studies, AUMsilenceTM FANA Antisense Oligo (FANA ASOs) targeting the mouse KLF6 (5′-AATGAATTTGGTCCACAGGTC-3′) was designed and synthesized by AUM LifeTech, LLC. The day before ASO addition, NIH-3T3 cells were seeded at 50 to 60% cell density in 12-well plate. The KLF6 ASO or scrambled ASO were gymnotically delivered to the cells twice at 24-h interval and at a final concentration of 5 μM. 24 h post second round ASO delivery, cells were collected for further analysis. For TGF-β1 treatment, cells were serum starved for 1 h before treatment with 5 ng/ml of TGF-β1 for 6 h. Kncok-down of KLF6 was analyzed by immunoblot analysis.
Confocal microscopy
NIH3T3 and HDF cells were seeded on polylysine-coated chamber slides for treatment. After treatment cells were fixed using 3.7% (wt/vol) paraformaldehyde (PFA) for 5 min at room temperature. The fixing solution was removed, and the cells were washed three times with PBS. Following this the cells were permeabilized and blocked using 0.5% Triton X-100 with 10% FBS in PBS for 30 min at 4C. Thereafter, the cells were washed three times with PBS and incubated at 4 °C overnight with appropriately labeled primary antibodies. The next day, the primary antibody was removed, and the samples were carefully washed three times for 5 min with PBS. The chamber on the slides were removed and mounted with coverslip slides using Diamond antifade mounting medium with 49,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific) and allowed to dry at room temperature overnight. Imaging was performed using a Nikon A1R confocal laser scanning microscope. The excitation/emission wavelengths were set at 405/425 to 475 nm for DAPI, 488/500 to 550 nm for green fluorescence, and 561/570 to 620 nm for red fluorescence. Regions of interest (ROIs) were drawn to determine the localization and colocalization with DAPI via Pearson’s correlation coefficients using Nikon Elements Advanced Research imaging software and GraphPad Prism (GraphPad Software).
Statistical analysis
Data were expressed as mean ± SEM obtained from three independent experiments. The significance of differences between control and treated samples was determined by either a One-way ANOVA or Two-way ANOVA based on the number of treatment groups followed by appropriate post-hoc analyses (Tukey test). A p-value of < 0.05 was considered statistically significant.
Data availability
All the data generated in this study are included in the manuscript.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Author contributions
I. E-A., M. B., C. D., and J. P. conceptualization, I. E-A., O. K., J. S. G., M. B., C. D., and J. P. methodology, I. E-A. software, I. E-A., Z. M. L., F. I., O. K., J. S. G. investigation, I. E-A., C. D. and J. P. data curation, I. E-A., C. D., and J. P. writing–original draft preparation; M. B., C. D., and J. P. supervision; C. D. and J. P. resources; C. D. and J. P. formal analysis; C. D. and J. P. writing–reviewing and editing; C. D. and J. P. visualization; C. D. and J. P. project administration; C. D. and J. P. funding acquisition.
Funding and additional information
This work was supported by the Research Centers in Minority Institutions (RCMI) grant U54MD007586 to J. P. and C. D., National Institutes of Health Grants R01AI136740, R56AI122960. I. E. was in part supported by the NIH 5R25GM059994 (RISE). This work is also in part supported by the Meharry Translational Research Center (MeTRC) grant 5U54MD007593 to 10 and Tennessee CFAR Grant P30AI110527 from the National Institutes of Health to C. D.
Reviewed by members of the JBC Editorial Board. Edited by Brian D. Strahl
Contributor Information
Chandravanu Dash, Email: cdash@mmc.edu.
Jui Pandhare, Email: jpandhare@mmc.edu.
References
- 1.Endo F., Tanoue A., Nakai H., Hata A., Indo Y., Titani K., et al. Primary structure and gene localization of human prolidase. J. Biol. Chem. 1989;264:4476–4481. [PubMed] [Google Scholar]
- 2.Kitchener R.L., Grunden A.M. Prolidase function in proline metabolism and its medical and biotechnological applications. J. Appl. Microbiol. 2012;113:233–247. doi: 10.1111/j.1365-2672.2012.05310.x. [DOI] [PubMed] [Google Scholar]
- 3.Kular J.K., Basu S., Sharma R.I. The extracellular matrix: structure, composition, age-related differences, tools for analysis and applications for tissue engineering. J. Tissue Eng. 2014;5 doi: 10.1177/2041731414557112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramshaw J.A.M., Shah N.K., Brodsky B. Gly-X-Y tripeptide frequencies in collagen: a context for host-guest triple-helical peptides. J. Struct. Biol. 1998;122:86–91. doi: 10.1006/jsbi.1998.3977. [DOI] [PubMed] [Google Scholar]
- 5.Cui N., Hu M., Khalil R.A. Biochemical and biological attributes of matrix metalloproteinases. Prog. Mol. Biol. Transl. Sci. 2017;147:1–73. doi: 10.1016/bs.pmbts.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krane S.M. The importance of proline residues in the structure, stability and susceptibility to proteolytic degradation of collagens. Amino Acids. 2008;35:703–710. doi: 10.1007/s00726-008-0073-2. [DOI] [PubMed] [Google Scholar]
- 7.Laurent G.J. Dynamic state of collagen: pathways of collagen degradation in vivo and their possible role in regulation of collagen mass. Am. J. Physiol. 1987;252:C1–C9. doi: 10.1152/ajpcell.1987.252.1.C1. [DOI] [PubMed] [Google Scholar]
- 8.McAnulty R.J., Laurent G.J. Collagen synthesis and degradation in vivo. Evidence for rapid rates of collagen turnover with extensive degradation of newly synthesized collagen in tissues of the adult rat. Coll. Relat. Res. 1987;7:93–104. doi: 10.1016/s0174-173x(87)80001-8. [DOI] [PubMed] [Google Scholar]
- 9.Surazynski A., Miltyk W., Palka J., Phang J.M. Prolidase-dependent regulation of collagen biosynthesis. Amino Acids. 2008;35:731–738. doi: 10.1007/s00726-008-0051-8. [DOI] [PubMed] [Google Scholar]
- 10.Jackson S.H., Dennis A.W., Greenberg M. Iminodipeptiduria: a genetic defect in recycling collagen; a method for determining prolidase in erythrocytes. Can Med. Assoc. J. 1975;113:762–763. [PMC free article] [PubMed] [Google Scholar]
- 11.Duong H.S., Zhang Q.-Z., Le A.D., Kelly A.P., Kamdar R., Messadi D.V. Elevated prolidase activity in keloids: correlation with type I collagen turnover. Br. J. Dermatol. 2006;154:820–828. doi: 10.1111/j.1365-2133.2006.07167.x. [DOI] [PubMed] [Google Scholar]
- 12.Surazynski A., Donald S.P., Cooper S.K., Whiteside M.A., Salnikow K., Liu Y., et al. Extracellular matrix and HIF-1 signaling: the role of prolidase. Int. J. Cancer. 2008;122:1435–1440. doi: 10.1002/ijc.23263. [DOI] [PubMed] [Google Scholar]
- 13.Cechowska-Pasko M., Pałka J., Wojtukiewicz M.Z. Enhanced prolidase activity and decreased collagen content in breast cancer tissue. Int. J. Exp. Pathol. 2006;87:289–296. doi: 10.1111/j.1365-2613.2006.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misiura M., Miltyk W. Current understanding of the emerging role of prolidase in cellular metabolism. Int. J. Mol. Sci. 2020;21:5906. doi: 10.3390/ijms21165906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez A.C., Costa T.F., Andrade Z.A., Medrado A.R. Wound healing - a literature review. Bras Dermatol. 2016;91:614–620. doi: 10.1590/abd1806-4841.20164741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senboshi Y., Oono T., Arata J. Localization of prolidase gene expression in scar tissue using in situ hybridization. J. Dermatol. Sci. 1996;12:163–171. doi: 10.1016/0923-1811(95)00505-6. [DOI] [PubMed] [Google Scholar]
- 17.Oono T., Fujiwara Y., Yoshioka T., Arata J. Prolidase activity in chronic wound and blister fluids. J. Dermatol. 1997;24:626–629. doi: 10.1111/j.1346-8138.1997.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 18.Baszanowska W., Niziol M., Oscilowska I., Czyrko-Horczak J., Miltyk W., Palka J. Recombinant human prolidase (rhPEPD) induces wound healing in experimental model of inflammation through activation of EGFR signalling in fibroblasts. Molecules. 2023;28:851. doi: 10.3390/molecules28020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palka J.A., Phang J.M. Prolidase activity in fibroblasts is regulated by interaction of extracellular matrix with cell surface integrin receptors. J. Cell. Biochem. 1997;67:166–175. doi: 10.1002/(sici)1097-4644(19971101)67:2<166::aid-jcb2>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 20.Surazyński A., Pałka J., Wołczyński S. Acetylsalicylic acid-dependent inhibition of collagen biosynthesis and beta1-integrin signaling in cultured fibroblasts. Med. Sci. Monit. 2004;10:BR175–BR179. [PubMed] [Google Scholar]
- 21.Miltyk W., Karna E., Wołczyński S., Pałka J. Insulin-like growth factor I-dependent regulation of prolidase activity in cultured human skin fibroblasts. Mol. Cell. Biochem. 1998;189:177–183. doi: 10.1023/a:1006958116586. [DOI] [PubMed] [Google Scholar]
- 22.Blackstock C.D., Higashi Y., Sukhanov S., Shai S.Y., Stefanovic B., Tabony A.M., et al. Insulin-like growth factor-1 increases synthesis of collagen type I via induction of the mRNA-binding protein LARP6 expression and binding to the 5′ stem-loop of COL1a1 and COL1a2 mRNA. J. Biol. Chem. 2014;289:7264–7274. doi: 10.1074/jbc.M113.518951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sienkiewicz P., Pałka M., Pałka J. Oxidative stress induces IGF-I receptor signaling disturbances in cultured human dermal fibroblasts. A possible mechanism for collagen biosynthesis inhibition. Cell Mol. Biol. Lett. 2004;9:643–650. [PubMed] [Google Scholar]
- 24.Surazyński A., Pałka J., Wołczyński S. Phosphorylation of prolidase increases the enzyme activity. Mol. Cell. Biochem. 2001;220:95–101. doi: 10.1023/a:1010849100540. [DOI] [PubMed] [Google Scholar]
- 25.Ysrayl B.b., Balasubramaniam M., Albert I., Villalta F., Pandhare J., Dash C. A novel role of prolidase in cocaine-mediated breach in the barrier of brain microvascular endothelial cells. Sci. Rep. 2019;9:2567. doi: 10.1038/s41598-018-37495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surazynski A., Liu Y., Miltyk W., Phang J.M. Nitric oxide regulates prolidase activity by serine/threonine phosphorylation. J. Cell Biochem. 2005;96:1086–1094. doi: 10.1002/jcb.20631. [DOI] [PubMed] [Google Scholar]
- 27.Yang L., Li Y., Ding Y., Choi K.-S., Kazim A.L., Zhang Y. Prolidase directly binds and activates epidermal growth factor receptor and stimulates downstream signaling. J. Biol. Chem. 2013;288:2365–2375. doi: 10.1074/jbc.M112.429159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi H., Press M.F. The Breast: Comprehensive Management of Benign and Malignant Diseases. 5th ed. Elsevier; Amsterdam, The Netherlands: 2018. Molecular oncology of breast cancer; pp. 282–307. [Google Scholar]
- 29.Yang L., Li Y., Bhattacharya A., Zhang Y. PEPD is a pivotal regulator of p53 tumor suppressor. Nat. Commun. 2017;8:2052. doi: 10.1038/s41467-017-02097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubick K.J., Robertson S.J., McNally K.L., Freedman B.A., Rasmussen A.L., Taylor R.T., et al. Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell Host Microbe. 2015;18:61–74. doi: 10.1016/j.chom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanoue A., Endo F., Matsuda I. Structural organization of the gene for human prolidase (peptidase D) and demonstration of a partial gene deletion in a patient with prolidase deficiency. J. Biol. Chem. 1990;265:11306–11311. [PubMed] [Google Scholar]
- 32.Wingender E., Dietze P., Karas H., Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Botella L.M., Sanz-Rodriguez F., Komi Y., Fernandez L.A., Varela E., Garrido-Martin E.M., et al. TGF-beta regulates the expression of transcription factor KLF6 and its splice variants and promotes co-operative transactivation of common target genes through a Smad3-Sp1-KLF6 interaction. Biochem. J. 2009;419:485–495. doi: 10.1042/BJ20081434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sleiman S.F., Langley B.C., Basso M., Berlin J., Xia L., Payappilly J.B., et al. Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J. Neurosci. 2011;31:6858–6870. doi: 10.1523/JNEUROSCI.0710-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kendall R.T., Feghali-Bostwick C.A. Fibroblasts in fibrosis: novel roles and mediators. Front. Pharmacol. 2014;5:123. doi: 10.3389/fphar.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ismaeel A., Kim J.S., Kirk J.S., Smith R.S., Bohannon W.T., Koutakis P. Role of transforming growth factor-beta in skeletal muscle fibrosis: a review. Int. J. Mol. Sci. 2019;20:2446. doi: 10.3390/ijms20102446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frangogiannis N. Transforming growth factor-beta in tissue fibrosis. J. Exp. Med. 2020;217 doi: 10.1084/jem.20190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eni-Aganga I., Lanaghan Z.M., Balasubramaniam M., Dash C., Pandhare J. PROLIDASE: a review from discovery to its role in Health and disease. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.723003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu F., Liu C., Zhou D., Zhang L. TGF-beta/SMAD pathway and its regulation in hepatic fibrosis. J. Histochem. Cytochem. 2016;64:157–167. doi: 10.1369/0022155415627681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jinnin M., Ihn H., Tamaki K. Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-beta1-induced extracellular matrix expression. Mol. Pharmacol. 2006;69:597–607. doi: 10.1124/mol.105.017483. [DOI] [PubMed] [Google Scholar]
- 42.Kim K.K., Sheppard D., Chapman H.A. TGF-beta1 signaling and tissue fibrosis. Cold Spring Harb. Perspect. Biol. 2018;10:a022293. doi: 10.1101/cshperspect.a022293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S.J., Yuan W., Mori Y., Levenson A., Trojanowska M., Varga J. Stimulation of type I collagen transcription in human skin fibroblasts by TGF-beta: involvement of Smad 3. J. Invest. Dermatol. 1999;112:49–57. doi: 10.1046/j.1523-1747.1999.00477.x. [DOI] [PubMed] [Google Scholar]
- 44.Rossignol F., Wang H., Ferreira C. In: GeneReviews((R)) Adam M.P., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., et al., editors. University of Washington; Seattle (WA): 1993. Prolidase deficiency. [Google Scholar]
- 45.Yang L., Li Y., Zhang Y. Identification of prolidase as a high affinity ligand of the ErbB2 receptor and its regulation of ErbB2 signaling and cell growth. Cell Death Dis. 2014;5:e1211. doi: 10.1038/cddis.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L., Li Y., Bhattacharya A., Zhang Y. Inhibition of ERBB2-overexpressing tumors by recombinant human prolidase and its enzymatically inactive mutant. EBioMedicine. 2015;2:396–405. doi: 10.1016/j.ebiom.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L., Li Y., Bhattacharya A., Zhang Y. Dual inhibition of ErbB1 and ErbB2 in cancer by recombinant human prolidase mutant hPEPD-G278D. Oncotarget. 2016;7:42340–42352. doi: 10.18632/oncotarget.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang C., Bolotin E., Jiang T., Sladek F.M., Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Connor L., Gilmour J., Bonifer C. The role of the ubiquitously expressed transcription factor Sp1 in tissue-specific transcriptional regulation and in disease. Yale J. Biol. Med. 2016;89:513–525. [PMC free article] [PubMed] [Google Scholar]
- 50.Botella L.M., Sanchez-Elsner T., Sanz-Rodriguez F., Kojima S., Shimada J., Guerrero-Esteo M., et al. Transcriptional activation of endoglin and transforming growth factor-beta signaling components by cooperative interaction between Sp1 and KLF6: their potential role in the response to vascular injury. Blood. 2002;100:4001–4010. doi: 10.1182/blood.V100.12.4001. [DOI] [PubMed] [Google Scholar]
- 51.Gong M., Yu W., Pei F., You J., Cui X., McNutt M.A., et al. KLF6/Sp1 initiates transcription of the tmsg-1 gene in human prostate carcinoma cells: an exon involved mechanism. J. Cell Biochem. 2012;113:329–339. doi: 10.1002/jcb.23359. [DOI] [PubMed] [Google Scholar]
- 52.Gong M.Z., You J.F., Pei F., Cui X.L., Li G., Zheng J. Transcriptional activation of TMSG-1 by complex of KLF6 and Sp1. Zhonghua Bing Li Xue Za Zhi. 2011;40:542–548. [PubMed] [Google Scholar]
- 53.Kong L.M., Yao L., Lu N., Dong Y.L., Zhang J., Wang Y.Q., et al. Interaction of KLF6 and Sp1 regulates basigin-2 expression mediated proliferation, invasion and metastasis in hepatocellular carcinoma. Oncotarget. 2016;7:27975–27987. doi: 10.18632/oncotarget.8564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rubinstein M., Idelman G., Plymate S.R., Narla G., Friedman S.L., Werner H. Transcriptional activation of the insulin-like growth factor I receptor gene by the Kruppel-like factor 6 (KLF6) tumor suppressor protein: potential interactions between KLF6 and p53. Endocrinology. 2004;145:3769–3777. doi: 10.1210/en.2004-0173. [DOI] [PubMed] [Google Scholar]
- 55.Al-Habeeb F., Aloufi N., Traboulsi H., Liu X., Nair P., Haston C., et al. Human antigen R promotes lung fibroblast differentiation to myofibroblasts and increases extracellular matrix production. J. Cell Physiol. 2021;236:6836–6851. doi: 10.1002/jcp.30380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Surazynski A., Miltyk W., Prokop I., Palka J. Prolidase-dependent regulation of TGF beta (corrected) and TGF beta receptor expressions in human skin fibroblasts. Eur. J. Pharmacol. 2010;649:115–119. doi: 10.1016/j.ejphar.2010.09.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data generated in this study are included in the manuscript.