Abstract
A low-molecular-weight recombinant Brucella abortus protein reactive with antibodies from a variety of naturally and experimentally infected hosts and T lymphocytes from experimentally infected mice was identified and given the designation BA14K. The gene encoding BA14K was cloned and characterized, and the predicted amino acid sequence of this immunoreactive protein showed no significant homology with previously described proteins. Sequences homologous to the cloned fragment encoding BA14K were identified by Southern blot analysis of genomic DNAs from representatives of all of the currently recognized Brucella species. Studies employing BA14K should contribute to our efforts to better understand the antigenic specificity of protective immunity to brucellosis.
Brucella spp. are gram-negative, facultative intracellular bacterial pathogens which cause abortion and infertility in numerous domestic and wild mammals, as well as a disease known as undulant fever in humans (1). People become infected through direct contact with infected animals or animal products (16). Consequently, brucellosis in animals used for food is not only a serious economic problem but also a potential public health hazard. Attenuated live vaccines, such as Brucella abortus S19, which until recently was the vaccine used in cattle in the United States, and Brucella melitensis Rev1, which is used in sheep and goats in other parts of the world, have been used successfully in eradication and control programs (1). Although these vaccines have been invaluable components of eradication programs, there are significant problems associated with their use. These include the virulence of S19 and Rev1 for humans (24), the potential for abortion when these strains are used in pregnant animals (2, 17), and the development of agglutinating antibodies in animals vaccinated as adults which are indistinguishable from those elicited by natural infection (17). Clearly, the construction of brucellosis vaccines lacking these undesirable properties would be of great benefit to both veterinary medicine and human medicine.
Similarly to other infections caused by facultative intracellular pathogens, the induction of specific cell-mediated immunity is required for effective clearance of Brucella infections (4, 22). Unfortunately, the nature and antigenic specificity of protective cellular immunity against brucellosis are unclear (13). Therefore, the identification of Brucella cellular components which contribute to the induction of protective responses in the host will be an important step in designing improved vaccines. The cloning and characterization of genes encoding immunoreactive Brucella proteins will provide a useful source of antigen for immunologic assays and subunit immunization studies. It will also facilitate the construction of replicating antigen delivery systems such as those based on salmonellae (7) and vaccinia virus (15). Studies employing both purified subunit preparations and live, recombinant antigen delivery systems should allow a comprehensive evaluation of the relative importance of specific Brucella proteins in eliciting protective immunity.
In an attempt to identify Brucella proteins capable of inducing protective immune responses, a collection of recombinant Escherichia coli clones expressing Brucella proteins reactive in immunoassays with sera from a variety of experimentally and naturally infected hosts was assembled (18). One of these clones, which was designated IV-4, produced a recombinant Brucella protein with an apparent molecular mass of 14 kDa as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis. Restriction enzyme analysis of the plasmid encoding the recombinant 14-kDa protein, which was designated pBA44, revealed the presence of a 1.8-kb insert. For ease of communication, the recombinant Brucella protein produced by clone IV-4 was designated BA14K.
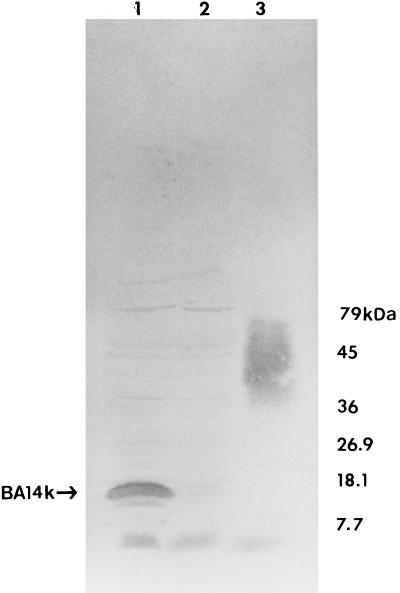
To further examine the extent of the reactivity of BA14K with sera from naturally and experimentally infected hosts, cell lysates of recombinant E. coli DH5α [F− φ80dlacZM15Δ (lacZYA-argF)U169 deoR recA1 endA1 phoA hsdR17 (rK− mK+) supE44 λ− thi-1 gyrA96 relA1] carrying pBA44 and pUC9 were prepared and subjected to SDS-PAGE and immunoblot analysis by previously described methods (19, 20). Affinity-purified, species-specific anti-immunoglobulin G (IgG) horseradish peroxidase conjugates (Sigma Chemical Co., St. Louis, Mo.) were employed in these experiments. IgG-type antibodies reactive with BA14K were detected in sera from naturally infected dogs (Fig. 1), cattle (data not shown), and humans (data not shown) and also in sera obtained from BALB/c mice and goats experimentally infected with B. abortus 2308 (data not shown). The detection of IgG-type antibodies specific for BA14K in sera from these hosts is relevant for several reasons. First, cattle, goats, dogs, and humans are important natural hosts for Brucella infections (1, 16), and mice represent a well-established model for both human (6, 25) and ruminant (14) brucellosis. Second, levels of IgG in infected hosts are directly correlated with the presence of active infections (8). Third, protective antibodies in the mouse model appear to be predominantly of the IgG class (9).
FIG. 1.
Reaction of recombinant E. coli DH5α producing BA14K in Western blots with serum from a dog naturally infected with Brucella canis. Lanes: 1, DH5α/pBA44; 2, DH5α/pUC9; and 3, B. abortus 2308 cell lysate.
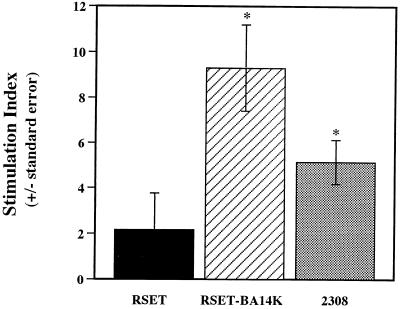
Lymphocyte proliferation assays were used to determine if BA14K-specific T lymphocytes were present in mice with chronic Brucella infections. Female BALB/c mice (Harlan Sprague Dawley, Indianapolis, Ind.) that were 8 to 10 weeks of age were infected with 5 × 104 CFU of B. abortus 2308, B. melitensis 16M, or B. abortus S19 via the intravenous route by previously described procedures (10, 14). Between 28 and 30 weeks postinfection, five mice from each experimental group were euthanized with a halothane overdose. Their spleens were aseptically removed, and lymphocyte transformation assays were performed on pooled, single-cell suspensions of T-lymphocyte-enriched splenocytes by previously described methods (13). Test antigens were added to the splenocytes at concentrations of 0.5, 1, and 2 μg of protein per well. A commercial T7 polymerase-based expression system (RSET; Invitrogen, San Diego, Calif.) was used for enhanced production of BA14K in recombinant E. coli. An overexpression plasmid (pRS44.8) was constructed by cloning the 1.8-kb fragment encoding BA14K into pRSETB (Invitrogen). This plasmid and pRSETB were independently introduced into E. coli BL21 (F− ompT r−B m−B) (DE3) by the procedures described by Hanahan (11). E. coli cell lysates and B. abortus 2308 whole killed cells were prepared as previously described (20). Concanavalin A (ConA) (Sigma Chemical Co.) was used as a positive control. Splenocytes were incubated with antigen or ConA for 48, 72, 96, or 120 h and pulsed with [3H]thymidine 18 h prior to harvesting. The results of the lymphocyte transformation assays were expressed as stimulation indices, which were calculated by dividing the counts per minute obtained with cells treated with antigen or mitogen by the counts per minute obtained with untreated cells. A stimulation index of ≥3 was considered a positive response. Statistical comparisons between experimental groups were performed by the two-tailed Student t test (23). At least four separate lymphocyte transformation assays were performed with T-cell-enriched splenocytes from mice infected with each of the Brucella strains used as immunogens. T lymphocytes reactive with BA14K were detected in BALB/c mice infected with virulent B. melitensis 16M (Fig. 2), B. abortus 2308 (data not shown), or the attenuated live vaccine strain 19 (data not shown) at 28 to 30 weeks postinfection. Because of the intracellular nature of Brucella infections, the induction of a cellular immune response is critical for effective host clearance of infection. Thus, detection of BA14K-reactive T lymphocytes in mice infected with both virulent and vaccine strains suggests that further investigation of the immunoreactive nature of this protein along with its capacity to induce protective immune responses is justified.
FIG. 2.
Proliferative responses of T lymphocyte-enriched splenocyte cultures from B. melitensis 16M-infected mice at 28 weeks postinfection. Lymphocytes were stimulated with antigen (1-μg protein concentration) for 96 h and pulsed with [3H]thymidine for 18 h. Antigens: RSET, E. coli BL21 (DE3)/pRSETB cell lysate; RSET-BA14K, E. coli BL21 (DE3)/pRS44.8; 2308, B. abortus 2308 whole killed cells. Stimulation indices were calculated as described in the text. ∗, P < 0.05 compared to RSET.
Western blot analysis showed that expression of BA14K was isopropyl-β-d-thiogalactopyranoside inducible in E. coli JM109 [Δ(lac-proAB) (F′ traD36 proAB lacIq ZΔM15) e14-(mcrA) recA1 endA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1], suggesting that the cloned gene encoding BA14K was expressed from the lac promoter resident in pUC9 (data not shown). By employing the dideoxynucleotide-based procedures described by Sanger et al. (21), nucleotide sequence analysis of the 1.8-kb insert in pBA44 confirmed that BA14K was produced in E. coli as a fusion composed of the C-terminal 133 amino acids of a native Brucella protein fused to the N-terminal 13 amino acids of the α-subunit of β-galactosidase encoded by pUC9.
The complete gene encoding the native Brucella protein was cloned in the following manner. Southern blot analysis of B. abortus 2308 genomic DNA employing a 1.5-kb HindIII fragment from pBA44 which encompassed the BA14K coding region and the HindIII site from the multiple cloning site of pUC9 was used to identify a HindIII fragment of approximately 4.6 kb in the genomic DNA preparation which likely contained the complete gene. Following NaCl gradient fractionation (12), HindIII fragments of 2308 genomic DNA ranging in size from 4 to 10 kb were cloned into the HindIII site of pBC/SK+ (Stratagene, La Jolla, Calif.), and the resulting recombinant plasmid bank was used to transform E. coli DH5α. Transformants were screened by colony blot hybridization with the 1.5-kb HindIII fragment from pBA44, and six reactive clones were identified. All six of these transformants contained recombinant plasmids containing 4.6-kb HindIII fragments that hybridized with the 1.5-kb HindIII probe derived from pBA44 by Southern blot analysis. One of these recombinant plasmids was selected for further evaluation and was given the designation pRLF6. Nucleotide sequence analysis with primers specific for the partial BA14K coding region in pBA44 revealed that the open reading frame encoding the complete Brucella protein was present in pRLF6 (Fig. 3). Computer-assisted analysis of this sequence indicated that it encoded a polypeptide of 147 amino acids with a predicted molecular weight of 16,822 and an isoelectric point of 11.47. An inverted repeat which may act as a rho-independent terminator is present 18 nucleotides downstream of the predicted stop codon for this coding region, and an open reading frame encoding the C-terminal 132 amino acids of a protein showing limited homology with the lactaldehyde reductase protein FucO of E. coli (5) (GenBank accession no. M31059) ends approximately 172 nucleotides upstream of the predicted start codon of the BA14K coding sequence, with no detectable consensus promoter elements in the intervening sequences. This organization suggests that the gene encoding the immunoreactive Brucella protein is the last component of an operon. Further characterization of the upstream nucleotide sequence in pRLF6 has not been performed.
FIG. 3.
Nucleotide sequence of the gene encoding the B. abortus immunoreactive protein BA14K and the deduced amino acid sequence of this protein. Underlining, putative ribosome-binding site; asterisks, stop codons; double dagger, fusion site between the Brucella DNA sequence and pUC9 in pBA44; open triangle, the predicted signal peptidase cleavage site; dashed arrows, inverted repeat, which may serve as a rho-independent terminator. A dot above every 10th nucleotide is present.
Although the unprocessed form of the immunoreactive Brucella protein has a predicted molecular mass of approximately 17 kDa, computer-assisted analysis of its amino acid sequence employing the Protein Analysis Toolbox suite of programs (MacVector 6.0; Oxford Molecular Group, Campbell, Calif.) suggests that the first 26 amino acids of this polypeptide form a potential signal sequence for export. Removal of the predicted leader sequence would result in the production of a protein of approximately 14,200 Da and with a pI of 11.25, which is consistent with the mobility of BA14K detected by SDS-PAGE and Western blot analysis. Therefore, we have retained the designation BA14K for this protein. Amino acids 83 to 98 of the unprocessed form of BA14K form a potential transmembrane domain; thus, it appears likely that this protein is associated with the bacterial cell envelope; however, further biochemical characterization will be required to confirm this subcellular localization. No homology to previously deposited protein sequences in the SWISS-PROT database could be detected when the predicted BA14K amino acid sequence was evaluated with the FSTPSCAN program from PCGENE (Intelligenetics, Mountain View, Calif.) or when this sequence was compared with multiple protein sequence databases by using the BLAST algorithm (3). This latter service was provided by the National Center for Biotechnology Information.
The results presented here indicate that the B. abortus protein BA14K is strongly immunoreactive, inducing both humoral and cellular immune responses in hosts during the course of infection. BA14K-specific humoral immune responses were detected in a variety of relevant natural and experimental hosts, which is consistent with the observation that nucleotide sequences homologous to the 1.8-kb cloned fragment encoding BA14K were detected by Southern blot analysis in genomic DNAs from strains representing all six of the currently recognized Brucella species (data not shown). BA14K-reactive T lymphocytes were also detected in experimentally infected BALB/c mice, which are an established model for both human (6, 25) and ruminant (14) brucellosis. Based on these findings, it appears that further investigation of the biological significance of BA14K-specific immune responses is warranted. For example, it would be interesting to determine if BA14K-reactive lymphocytes are present in naturally infected hosts such as cattle and goats or in infected humans. Information obtained from such studies may provide the basis for the design of improved vaccines and diagnostic reagents for brucellosis in humans and animals. The construction of relevant mutants may also provide insight into the biological function of BA14K.
Nucleotide sequence accession number.
The nucleotide sequence of the BA14K coding region has been deposited in GenBank under accession no. U62541.
Acknowledgments
This work was supported by grants from the National Institutes of Health (AI28867 to R.M.R. II and NS32464 to S.R.J.) and the LSUMC Center for Excellence in Cancer Research, Treatment and Education.
We thank Rebecca Freeland and Bryan Bellaire for technical support, and the assistance of the University of Florida DNA Sequencing Core Laboratory is also greatly appreciated.
REFERENCES
- 1.Acha P, Szyfres B. Zoonoses and communicable diseases common to man and animals. Washington, D.C: Pan American Health Organization; 1980. pp. 28–45. [Google Scholar]
- 2.Alton G G. Brucella melitensis. In: Nielsen K H, Duncan J R, editors. Animal brucellosis. 1990. pp. 383–409. p. 383–409. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 3.Altschul S, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Araya L N, Elzer P H, Rowe G E, Enright F M, Winter A J. Temporal development of protective cell-mediated and humoral immunity in BALB/c mice infected with Brucella abortus. J Immunol. 1989;143:3330–3337. [PubMed] [Google Scholar]
- 5.Chen Y-M, Lu Z, Lin E C C. Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on l-1,2-propanediol. J Bacteriol. 1989;171:6097–6105. doi: 10.1128/jb.171.11.6097-6105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford R M, Van De Berg L, Yuan L, Hadfield T L, Warren R L, Drazek E S, Houng H-S H, Hammack C, Sasala K, Polsinelli T, Thompson J, Hoover D L. Deletion of purE attenuates Brucella melitensis infection in mice. Infect Immun. 1996;64:2188–2192. doi: 10.1128/iai.64.6.2188-2192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtiss R, III, Kelley S M, Gulig P A, Gentry-Weeks C R, Galán J E. Avirulent salmonellae expressing virulence antigens from other pathogens for use as orally administered vaccines. In: Roth J R, editor. Virulence mechanisms of bacterial pathogens. Washington, D.C: American Society for Microbiology; 1988. pp. 311–328. [Google Scholar]
- 8.Díaz R, Moriyón I. Laboratory techniques in the diagnosis of human brucellosis. In: Young E J, Corbel M J, editors. Human brucellosis: clinical and laboratory aspects. Boca Raton, Fla: CRC Press; 1989. pp. 73–83. [Google Scholar]
- 9.Elzer P H, Jacobson R H, Jones S M, Nielsen K H, Douglas J T, Winter A J. Antibody-mediated protection against Brucella abortus in BALB/c mice at successive periods after infection: variation between virulent strain 2308 and attenuated strain 19. Immunology. 1994;82:651–658. [PMC free article] [PubMed] [Google Scholar]
- 10.Elzer P H, Phillips R W, Kovach M E, Peterson K M, Roop R M., II Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect Immun. 1994;62:4135–4139. doi: 10.1128/iai.62.10.4135-4139.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser K, Murray N E. The use of phage lambda replacement vectors in the construction of representative genomic DNA libraries. In: Glover D M, editor. DNA cloning, a practical approach. Vol. 1. Washington, D.C: IRL Press; 1985. pp. 1–47. [Google Scholar]
- 13.Lauderdale T-L, Jones S M, Winter A J. Response of murine T cells to antigens of Brucella abortus at sequential periods after infection. Immunol Infect Dis. 1991;1:59–66. [Google Scholar]
- 14.Montaraz J A, Winter A J. Comparison of living and nonliving vaccines for Brucella abortus in BALB/c mice. Infect Immun. 1986;53:245–251. doi: 10.1128/iai.53.2.245-251.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss B. Vaccinia virus: a tool for research and vaccine development. Science. 1991;252:1662–1667. doi: 10.1126/science.2047875. [DOI] [PubMed] [Google Scholar]
- 16.Nicoletti P L. Relationship between animal and human disease. In: Young E J, Corbel M J, editors. Brucellosis: clinical and laboratory aspects. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 41–51. [Google Scholar]
- 17.Nicoletti P L. Vaccination. In: Nielsen K H, Duncan J R, editors. Animal brucellosis. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 283–299. [Google Scholar]
- 18.Roop R M, Bagchi T, Sriranganathan N, Boyle S M, Schurig G G. Proceedings of the 69th Annual Conference of Research Workers in Animal Disease. Ames, Iowa: Iowa State Press; 1988. Expression of Brucella abortus immunogens in Escherichia coli, abstr. 199; p. 35. [Google Scholar]
- 19.Roop R M, II, Price M L, Dunn B E, Boyle S M, Sriranganathan N, Schurig G G. Molecular cloning and nucleotide sequence analysis of the gene encoding the immunoreactive Brucella abortus Hsp60 protein, BA60K. Microb Pathog. 1992;12:47–62. doi: 10.1016/0882-4010(92)90065-v. [DOI] [PubMed] [Google Scholar]
- 20.Roop R M, II, Fletcher T W, Sriranganathan N, Boyle S M, Schurig G G. Identification of an immunoreactive Brucella abortus HtrA stress response protein homolog. Infect Immun. 1994;62:1000–1007. doi: 10.1128/iai.62.3.1000-1007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith R., III . T lymphocyte-mediated mechanisms of acquired protective immunity against brucellosis in cattle. In: Adams L G, editor. Advances in brucellosis research: an international symposium. College Station, Tex: Texas A & M University Press; 1990. pp. 164–190. [Google Scholar]
- 23.Snedecor G W, Cochran W G. Statistical methods. Ames, Iowa: Iowa State University Press; 1985. [Google Scholar]
- 24.Spink W W, Hall III J W, Finstad J, Malle E. Immunization with viable Brucella organisms. Results of a safety test in humans. Bull W H O. 1962;26:409–419. [PMC free article] [PubMed] [Google Scholar]
- 25.Van de Berg L L, Hartman A B, Bhattacharjee A K, Tall B D, Yuan L, Sasala K, Hadfield T L, Zollinger W D, Hoover D L, Warren R L. Outer membrane protein of Neisseria meningitidis as a mucosal adjuvant for lipopolysaccharide of Brucella melitensis in mouse and guinea pig intranasal immunization models. Infect Immun. 1996;64:5263–5268. doi: 10.1128/iai.64.12.5263-5268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]