Summary
Background
Previous metabolic profiling of liver cancer has mostly used untargeted metabolomic approaches and was unable to quantitate the absolute concentrations of metabolites. In this study, we examined the association between the concentrations of 186 targeted metabolites and liver cancer risk using prediagnostic plasma samples collected up to 14 years prior to the clinical diagnosis of liver cancer.
Methods
We conducted a nested case–control study (n = 322 liver cancer cases, n = 322 matched controls) within the Shanghai Men's Health Study. Conditional logistic regression models adjusted for demographics, lifestyle factors, dietary habits, and related medical histories were used to estimate the odds ratios. Restricted cubic spline functions were used to characterise the dose–response relationships between metabolite concentrations and liver cancer risk.
Findings
After adjusting for potential confounders and correcting for multiple testing, 28 metabolites were associated with liver cancer risk. Significant non-linear relationships were observed for 22 metabolites. The primary bile acid biosynthesis and phenylalanine, tyrosine and tryptophan biosynthesis were found to be important pathways involved in the aetiology of liver cancer. A metabolic score consisting of 10 metabolites significantly improved the predictive ability of traditional epidemiological risk factors for liver cancer, with an optimism-corrected AUC increased from 0.84 (95% CI: 0.81–0.87) to 0.89 (95% CI: 0.86–0.91).
Interpretation
This study characterised the dose–response relationships between metabolites and liver cancer risk, providing insights into the complex metabolic perturbations prior to the clinical diagnosis of liver cancer. The metabolic score may serve as a candidate risk predictor for liver cancer.
Funding
National Key Project of Research and Development Program of China [2021YFC2500404, 2021YFC2500405]; US National Institutes of Health [subcontract of UM1 CA173640].
Keywords: Liver cancer, Metabolomics, Mass spectrometry, Prospective study, Plasma, Biomarker
Research in context.
Evidence before this study
Metabolic alterations associated with liver cancer have been characterised by many hospital-based studies, yet these studies are susceptible to reverse causation. Reports of prospective epidemiological studies remain scarce. To our knowledge, six studies have to date investigated the association between circulating metabolite profiles and liver cancer risk in a prospective manner. However, these studies used untargeted metabolomic approaches that quantify metabolites on a relative scale. Findings from these untargeted metabolomic studies warranted further confirmation.
Added value of this study
The application of targeted metabolomics technology in this study allowed the absolute quantification of metabolites, thereby strengthening the credibility of our findings and enabling confirmation of associations previously reported by untargeted metabolomics. Moreover, we examined potential non-linear relationships and visualised the dose–response curves between plasma metabolites and liver cancer risk. This provided additional insights into the complex metabolic perturbations prior to the clinical diagnosis of liver cancer.
Implications of all the available evidence
Our study offers insights into the aetiology of liver cancer, suggesting that abnormal metabolism might be present long before diagnosis and the association between several plasma metabolite concentrations and liver cancer risk may not be linear.
Introduction
Liver cancer ranks as the sixth most commonly diagnosed cancer and the third leading cause of cancer death worldwide, with approximately 906,000 new cases and 830,000 deaths occurring in 2020.1 Most cases of HCC, the predominant form of liver cancer, can be attributed to infections with HBV and HCV, accounting for about 44% and 21%. Meanwhile, lifestyle risk factors contribute to approximately 26% (alcohol consumption), 13% (tobacco smoking), 9% (obesity), and 7% (type 2 diabetes mellitus, T2DM) of the cases.2 Although the understanding of risk factors has greatly improved, the underlying molecular mechanisms linking them to liver cancer warrant further investigation.
Due to the asymptomatic nature of early-stage liver cancer, the majority of patients are diagnosed at an advanced stage, resulting in poor prognoses. Between 2000 and 2014, the 5-year relative survival for liver cancer was less than 30% in most countries and regions.3,4 Although widely used for the early detection of liver cancer, the efficacy of ultrasonography is highly operator-dependent and influenced by patient-related factors such as obesity and cirrhosis, while the sensitivity and specificity of serum alpha-fetoprotein (AFP) are insufficient.5, 6, 7 Novel biomarkers, especially those capable of indicating metabolic alterations well before the onset of symptoms, are currently attracting significant attention for risk assessment in liver cancer.
Metabolomics technology allows for a comprehensive characterisation of both endogenous and exogenous molecules present in biospecimens, offering a robust tool to dynamically monitor metabolic alterations at the molecular level in disease processes.8 The integration of metabolomic technology into epidemiological studies constitutes an emerging field of research, possessing the potential to enhance the understanding of disease aetiology and to identify early disease markers.9
Substantial efforts have been made to characterise the metabolic alterations associated with liver cancer.10, 11, 12, 13, 14 Several metabolites and biochemical pathways have been identified, such as bile acid biosynthesis, biosynthesis of amino acids and proteins, lipid metabolism, etc.15,16 However, the majority of existing studies, conducted in hospital settings and including prevalent patients with cancer, may be susceptible to reverse causation. Additionally, these studies often have relatively small sample sizes, potentially compromising their power to detect weak associations. Several population-based studies have prospectively investigated the association between circulating metabolites and liver cancer risk.17, 18, 19, 20, 21, 22 Given the limited number of existing studies and the significant disparities in study design, metabolomics platforms, and statistical analyses among them, it is necessary to provide additional evidence for the role of circulating metabolites in the development of liver cancer in a prospective setting, where study participants are recruited from the general population, and biospecimens are collected prior to disease diagnosis.
In this research, we conducted a 1:1 individual matched case–control study nested within a prospective cohort study and quantitated the plasma metabolites in study participants. The objective of this study was to examine the associations between the concentrations of plasma metabolites and liver cancer risk, to elucidate the biochemical pathways implicated in the aetiology of liver cancer, and to identify a panel of metabolites that may function as risk predictors for liver cancer.
Methods
Study design
The present nested case–control study was conducted based on the Shanghai Men's Health Study (SMHS), the rationale for which has been published elsewhere.23, 24, 25 Briefly, the SMHS is a prospective and population-based cohort initiated by the Shanghai Cancer Institute and Vanderbilt University in 2002–2006. SMHS included 61,469 men aged 40–74 years from an urban district in Shanghai, China. In-person interviews were conducted by trained staff at baseline to collect information on demographic background, medical history, family history of cancer, dietary habits, physical activity, and other lifestyle factors of the study participants. The food frequency questionnaire and physical activity questionnaire have been proven to have reasonable reproducibility and validity.24,25 Anthropometrics were also measured at study enrolment according to a standard protocol. A 10-mL blood sample was collected from each of the 46,244 willing participants (75.1%). All samples were kept at 4 °C, processed within 6 h, and stored at −75 °C. Information on the date and time of sample collection, time of last meal, intake of selected foods, smoking, and medication use in the past 24 h and the previous week was collected at the time of biospecimen collection.23 HBV infection status was determined by quantitating the HBsAg levels using a chemiluminescent microparticle immunoassay (CMIA) with the Architect HBsAg Reagent Kit and Architect i2000 analyser from Abbott Diagnostics (Abbott Park, IL, USA). According to the manufacturer's instructions, the seronegative of HBsAg was defined as a titre of less than 0.05 IU/mL.26
A combination of database linkage and active follow-up was utilised to track new cancer events in study participants. Incident cancer cases and deaths were updated annually by linking the Shanghai Cancer Registry and the Shanghai Vital Statistics Registry.27 Three-round of follow-ups have been conducted, all with high response rates (2004–2008: 97.6%; 2008–2011: 93.7% and 2012–2017: 93.6%). Possible cancer diagnoses were verified through home visits and review of medical records by clinical specialists and pathologists.
By the end of 2016, SMHS had identified 444 incident liver cancer cases (ICD-9: 155). The present nested case–control study was conducted based on 326 cases that had adequate blood samples. A density sampling method was used to select appropriate controls. Cohort members who had adequate blood samples and were free of cancer at the index time served as potential controls. For each case, we randomly selected one control, matching in terms of age (±two years), date of blood collection (±30 days), time of blood collection (morning/afternoon), and antibiotics use during the preceding week (yes/no). After excluding four cases that lacked suitable controls, the nested case–control study comprised 322 liver cancer cases along with their respective controls (Supplementary Figure S1).
Quantitation of plasma metabolites
The metabolomics analysis was performed by a Q300 Kit from Metabo-Profile Biotechnology (Shanghai, China) Co., Ltd.28 Sample preparation, instrument settings, and quality control (QC) were carried out according to the manufacturer's protocol. Details on the source of instruments and reagents have been described in detail elsewhere.28 Briefly, an ultraperformance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) system (ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA, USA) was used to quantitate all targeted metabolites. The raw data files generated by UPLC-MS/MS were processed using TMBQ software (v1.0, Human Metabolomics Institute, Shenzhen, Guangdong, China) for peak integration, calibration, and quantitation of each metabolite. We examined the reproducibility and reliability of the metabolite quantitation using pooled QC samples, which were prepared by mixing aliquots of the study samples. Principal component analysis (PCA) were conducted based on 644 study samples and 58 pooled QC samples. The multivariate control chart and the PCA score plot based on the first two principal component were depicted. The Spearman's rank correlation matrix for the pooled QC samples were also characterised. Inter-batch coefficient of variances (CVs) and the average of intra-batch CVs for each metabolites were calculated to assess the analytical performance. Results of these analysis suggested that the metabolite quantitation was robust and suitable for further analysis (Supplementary Figure S2, Supplementary Table S1).
Categorisation of the metabolite concentrations
After a standard QC procedure, the absolute concentrations (μmol/L) of 186 known metabolites were successfully quantitated and used for further analysis (Supplementary Table S1, Supplementary Figure S3). Values below the lower limit of quantitation were imputed with half of the minimum observed concentration for that metabolite. Metabolite concentrations were considered in both categorical and continuous ways. For the categorical manner, the metabolites were categorised into four groups based on corresponding quartile cutoffs of the controls, with the lowest quartiles as reference categories. Categorical analysis enhance the understanding of the relationship between different metabolite levels and the risk of liver cancer, and capture the possible nonlinear relationship. For the continuous manner, a log2-transformation was adopted to address the skewed distribution of plasma metabolite concentrations. Compared with other transformation method such as generalised log transformation and cube root transformation, log2 transformation allows for an intuitive interpretation. A 1-unit increase could be interpreted as a doubling in the metabolite concentrations.
Selection of covariates
Potential confounders were selected a priori based on their known associations with liver cancer and their potential to affect plasma metabolite concentrations. We created a directed acyclic graph using the DAGitty web application to determine the minimal sufficient adjustment sets (Supplementary Figure S4).29 Based on the causal diagram, the following covariates were included in the multivariable-adjusted models: age (years old, continuous), cigarette smoking (0 and tertiles of pack-year), alcohol drinking (0 and tertiles of drinks/d), body mass index (BMI) (<18.5 kg/m2, 18.5–23.9 kg/m2, 24–27.9 kg/m2, ≥28 kg/m2), Chinese Food Pagoda (CHFP) score (0–45 points, tertiles), total physical activity (MET-hour/week, tertiles), medical history of chronic hepatitis and cirrhosis (yes, no), medical history of cholelithiasis (yes, no) and medical history of T2DM (yes, no). The CHFP score indicated adherence to the Chinese dietary guidelines and the calculation has been reported in our previous publications.30, 31, 32 Considering the potential collinearity of seropositive HBsAg, medical history of chronic hepatitis, and medical history of cirrhosis, we combined the three variables into one (medical history of chronic hepatitis and cirrhosis) to indicate potential liver damage at blood collection.
Statistical analyses
Baseline characteristics of the study participants were described as frequency with proportion for categorical variables, as mean with SD for continuous variables with approximate normal distribution, and as median with 25th and 75th percentile for skewed variables. Categorical variables were compared using conditional logistic regression model. Continuous variables were compared using Student's paired t-test or Wilcoxon signed-rank test, as appropriate. The concentrations of plasma metabolites were presented as geometric means with geometric SDs and compared using geometric mean ratios with 95% CIs.
The associations between plasma metabolites and liver cancer risk were examined by conditional logistic regression models. The ORs and corresponding 95% CIs were calculated based on two models. Model 1 was conditioned on matching factors. Model 2 further adjusted for potential confounders mentioned above. Since only three continuous variables (pack-year, drinks/day, and CHFP score) have missing values and the missing proportions were low (0.2%, 0.8%, and 2.0%), we imputed missing values with corresponding median values. To account for multiple comparisons, the Benjamini-Hochberg false discovery rate (FDR) was computed.33 The Bonferroni-corrected P values (raw P value/186) were also presented. The S value [-log2 (P value)] was calculated to show the compatibility of data with hypotheses.34,35 The S-value has an intuitive interpretation in a physical experimental coin tossing. For example, an S value of 3 could seem about as surprising as seeing 3 heads in a row from fair coin-tossing.
The restricted cubic spline (RCS) function was used to investigate potential non-linear relationships and to characterise the dose–response curves flexibly.36 Log2-transformed metabolite concentrations fitted with three knots (10th, 50th, and 90th percentile) were included in the multivariable-adjusted models as a three-knot RCS function is much more flexible and would be more powerful in detecting departure from linearity.37 Nonlinearity was evaluated by Wald χ2 tests. Dose–response curves for metabolites with a Pnon-linearity of less than 0.05 were characterised. To best capture the dose–response relationships of these metabolites and liver cancer risk, RCS function with four (5th, 35th, 65th, and 95th percentile) and five (5th, 25th, 50th, 75th and 95th percentile) knots were further fitted for them and the Akaike information criterion was used to select the best-fitted model.38
We estimated the pairwise correlations of liver cancer-associated metabolites by calculating the raw and partial (adjusted for age and fasting time) Spearman rank correlation coefficients (Spearman's ρ). To avoid potential collider bias, the correlation analyses were performed only in controls.
To identify the most relevant biochemical pathways associated with the development of liver cancer, we conducted a metabolic pathway analysis based on metabolites with an FDR of <0.05. The Human Metabolome Database (HMDB) IDs were used as standard metabolite names.39 The pathway enrichment analysis and pathway topology analysis were conducted using the latest version of the R package provided by MetaboAnalyst.40 The Kyoto Encyclopedia of Genes and Genomes (KEGG) Homo sapiens database was selected as the pathway library.41 We employed the hypergeometric test as the method for over-representation analysis and relative betweenness centrality as the measure of node importance in topology analysis. GHCA (HMDB0240607) did not match with any compounds in the pathway library and was excluded from subsequent pathway analysis.
To examine the robustness of the observed associations, the following sensitivity analyses were conducted. First, we excluded participants with a follow-up time of less than two years to rule out possible reverse causation. Second, taking into account the impact of prevalent hepatobiliary diseases on plasma metabolite concentrations, we excluded participants with a medical history of chronic hepatitis, cirrhosis, or cholelithiasis or who were seropositive for HBsAg. Unconditional logistic regression models were employed in the above analyses as the matched case–control pairs had been split. Third, we replicated the observed associations by using a bootstrap resampling method (accounted for matched pairs) with 2000 repetitions to examine the robustness of the risk estimates. The bias-corrected and accelerated (BCa) method was used to calculate the 95% CIs of the bootstrapped ORs.42 Fourth, a multivariable-adjusted Cox regression model was fitted to offer insights into the temporal relationships between metabolites and liver cancer risk.
To extract the most informative and least redundant panel of metabolites that could discriminate liver cancer cases from healthy controls, a penalised least absolute shrinkage selection operator (LASSO) logistic regression analysis was performed.43 The optimal tuning parameter (λ) was determined by a tenfold cross-validation procedure. We chose the most parsimonious model whose deviance is no more than one SE above the deviance of the best model.44 All the metabolites with nonzero coefficients in the model were obtained and used to construct a liver cancer-associated metabolic score. The metabolic score was calculated as the weighted sum of the selected metabolites with weights equal to the coefficients from the LASSO logistic regression model. We used a multivariable-adjusted conditional logistic regression model incorporating RCS function terms (three knots: 10th, 50th, and 90th percentile) to characterise the dose–response relationship between the metabolic score and liver cancer risk.
To assess the predictive value of the metabolic score, we developed three logistic regression models, including a model with established risk factors for liver cancer that are available in our datasets (age, BMI, HBsAg, family history of liver cancer, cigarette smoking, and alcohol drinking), a model with LASSO-derived metabolic score alone, and a model with both established risk factors and the metabolic score. The Brier scores were calculated to evaluate the overall performance of the three models. Discrimination of the models was evaluated by receiver operating characteristics (ROCs) and AUCs. ROC curves were compared using DeLong's test.45 Calibration was examined by the Hosmer–Lemeshow goodness-of-fit test. The calibration slope, intercept, as well as the unreliability index (U) and its P value were also calculated to test unreliability (H0: intercept = 0, slope = 1) of the calibration curves. Apparent and bias-corrected calibration curves using the loess algorithm were also constructed to visualise the agreement between observed outcomes and predicted probabilities. To account for overfitting, the bootstrap technique was used for internal validation of the prediction models. We calculated the optimism-corrected Brier scores and AUCs and their 95% CIs with 2000 bootstrap repetitions.
Statistical analyses were conducted in SAS 9.4 (SAS Inc., Cary, N.C., USA) and R 4.3.0 (R Core Team, Vienna, Austria). A 2-sided P value of less than 0.05 was considered statistically significant. P values were reported with two significant digits. All raw data and statistical programs are permanently stored on servers at the Shanghai Cancer Institute.
Ethical approval
This study was conducted in accordance with both the Declarations of Helsinki and Istanbul. Approval was granted by the Ethics Committee of the Renji Hospital Ethics Committee of Shanghai Jiao Tong University School of Medicine (KY2021-029). Informed consent was obtained from all individual participants included in the study.
Role of the funding source
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Results
The descriptive characteristics of the cases and controls are presented in Table 1. The median age at blood collection was 57.57 years old [Q1 = 50.66, Q3 = 68.31] for cases and 57.01 years old (Q1 = 50.66, Q3 = 68.59) for controls. The median age at diagnosis for liver cancer cases was 65.03 years old (Q1 = 55.85, Q3 = 73.80) and the median time between sample collection and liver cancer diagnosis was 5.89 years (Q1 = 3.02, Q3 = 8.90). Compared to their respective controls, liver cancer cases were more likely to have a seropositive HBsAg, a medical history of chronic hepatitis, cirrhosis, and cholelithiasis, as well as a family history of liver cancer. The distributions of education, personal income, and cigarette smoking were also significantly different between the two groups.
Table 1.
Baseline characteristics of the study participants.a
| Cases (N = 322) | Controls (N = 322) | P valueb | |
|---|---|---|---|
| Age at blood collection, years old, median (Q1, Q3) | 57.57 (50.66, 68.31) | 57.01 (50.66, 68.59) | 0.26 |
| Fasting time, hours, n (%) | 0.90 | ||
| <3 | 79 (24.5) | 76 (23.6) | |
| 3–6 | 165 (51.2) | 166 (51.6) | |
| ≥6 | 78 (24.2) | 80 (24.8) | |
| Education, n (%) | 0.040 | ||
| Elementary school or less | 34 (10.6) | 35 (10.9) | |
| Middle school | 243 (75.5) | 219 (68.0) | |
| College or above | 45 (14.0) | 68 (21.1) | |
| Personal income, Yuan/month, n (%) | 0.0058 | ||
| <1000 | 213 (66.2) | 175 (54.4) | |
| 1000–2999 | 84 (26.1) | 115 (35.7) | |
| ≥3000 | 25 (7.8) | 32 (9.9) | |
| Cigarette smoking, pack-year, n (%)c | 0.043 | ||
| 0 | 90 (28.0) | 119 (37.0) | |
| ≤16.73 | 87 (27.0) | 67 (20.8) | |
| ≤30.00 | 74 (23.0) | 66 (20.5) | |
| >30.00 | 71 (22.1) | 70 (21.7) | |
| Alcohol drinking, drinks/day, n (%)c | 0.89 | ||
| 0 | 213 (66.2) | 217 (67.4) | |
| ≤1.19 | 32 (9.9) | 34 (10.6) | |
| ≤2.27 | 34 (10.6) | 35 (10.9) | |
| >2.27 | 43 (13.4) | 36 (11.2) | |
| HBV infection, n (%)d | 205 (63.7) | 20 (6.2) | <0.0001 |
| History of chronic hepatitis, n (%) | 85 (26.4) | 11 (3.4) | <0.0001 |
| History of cirrhosis, n (%) | 41 (12.7) | 1 (0.3) | <0.0001 |
| History of cholelithiasis, n (%) | 46 (14.3) | 21 (6.5) | 0.0014 |
| History of type 2 diabetes mellitus, n (%) | 33 (10.3) | 27 (8.4) | 0.42 |
| Family history of liver cancer, n (%) | 30 (9.3) | 10 (3.1) | 0.0022 |
| Body mass index, kg/m2, mean (SD) | 23.79 (3.44) | 23.53 (3.02) | 0.29 |
| CHFP score, mean (SD)e | 30.23 (5.06) | 30.53 (5.11) | 0.46 |
| Total physical activity, MET-hour/week, median (Q1, Q3) | 58.13 (36.70, 83.41) | 58.07 (35.43, 85.43) | 0.83 |
Categorical variables were compared using conditional logistic regression analyses.
Continuous variables were compared using Student's paired t-test or Wilcoxon signed-rank tests, as appropriate.
Never smoker/drinkers = 0; ever smoker/drinkers were categorised by tertiles distributions.
Six participants lacked this information. Their self-reported history of chronic hepatitis was used as a proxy.
Chinese Food Pagoda score.
A total of 186 types of metabolites, which involved amino acids, benzenoids, benzoic acids, bile acids, carbohydrates, carnitines, fatty acids, indoles, nucleotides, organic acids, phenols, phenylpropanoic acids, pyridines, and short-chain fatty acids (SCFAs), were quantitated in our study (Supplementary Figure S3, Supplementary Table S1). The geometric means, geometric SDs, and the ratios of geometric means of the plasma metabolite concentrations are shown in Supplementary Table S2.
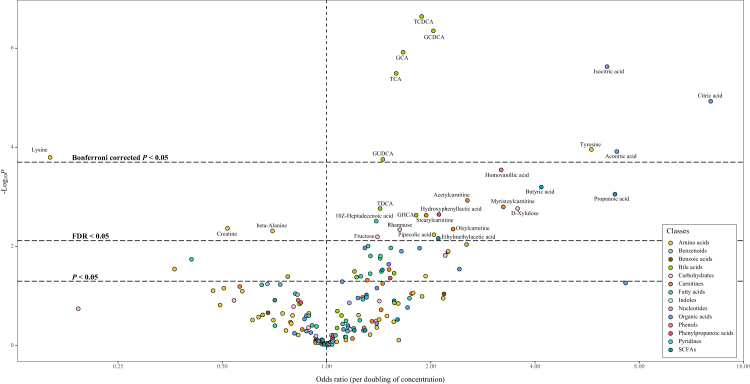
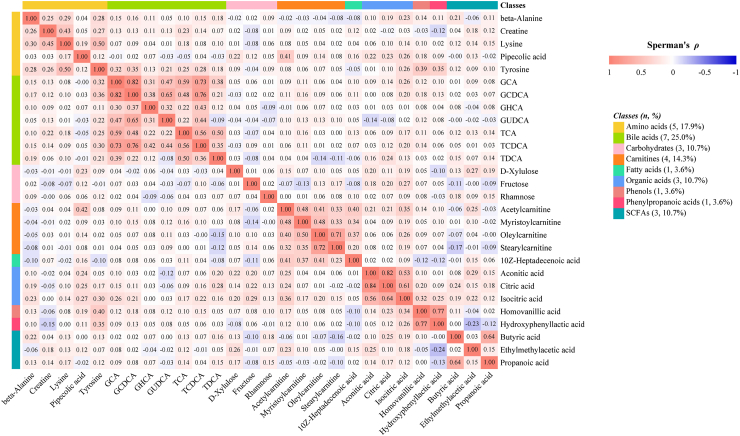
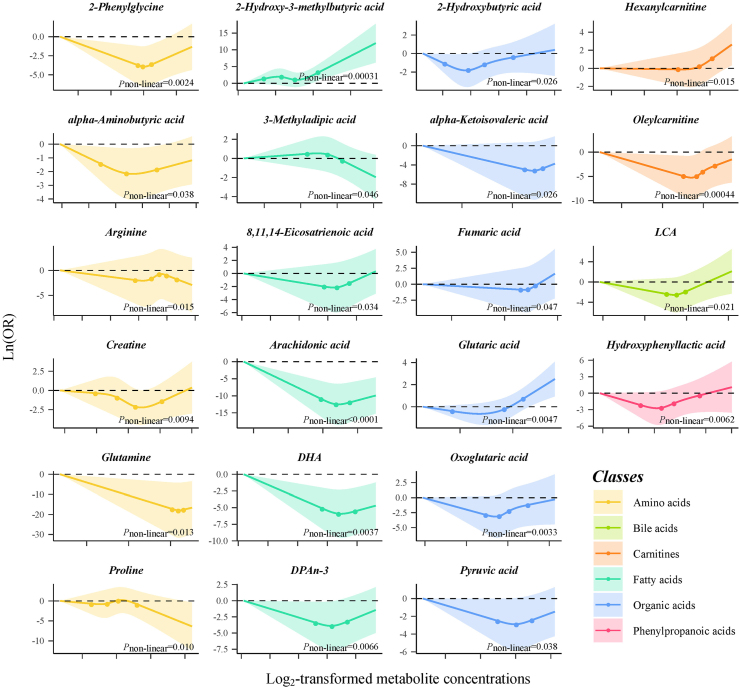
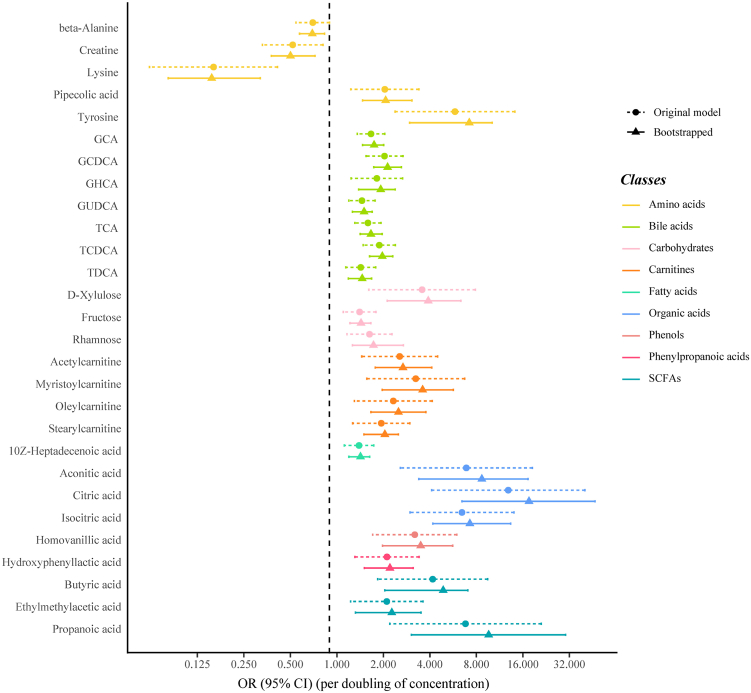
Results from the conditional logistic regression models showed that the plasma concentrations of 89 metabolites were significantly associated with liver cancer risk (FDR<0.05) conditioned on matching factors (Model 1) (Supplementary Table S3). Totally, 28 metabolites had an FDR of less than 0.05 after further adjusted for multiple potential confounders (Model 2) (Table 2, Fig. 1), predominantly consisting of bile acids (n = 7, 25%), amino acids (n = 5, 18%) and carnitines (n = 4, 14%) (Fig. 2). The ORs (95% CI), P values, and S values for all the metabolites are shown in Supplementary Table S3 (Model 1) and Supplementary Table S4 (Model 2). Citric acid and Lysine had the strongest association with liver cancer risk based on effect estimates (OR per doubling = 12.86, 95% CI: 4.10–40.30 and 0.16, 95% CI: 0.06, 0.41). Potential non-linear relationships were observed for 22 metabolites and liver cancer risk, with a Pnon-linearity of less than 0.05 (Supplementary Table S4). The dose–response curves for these metabolites are presented in Fig. 3. Arachidonic acid had an FDRnon-linear of less than 0.05 and the dose–response curve suggested a “v” shape between the log2-transformed concentrations of arachidonic acid and liver cancer risk (Fig. 3). Pairwise correlations between liver cancer-associated metabolites varied substantially (Fig. 2). The highest correlation was observed in citric acid and aconitic acid (ρraw = 0.84, ρpartial = 0.82).
Table 2.
Associations between selected metabolites and liver cancer risk.a
| Metabolite | Class | Categorial analysis |
Continuous analysis |
|||||
|---|---|---|---|---|---|---|---|---|
| Q1 | ORQ2 (95% CI) | ORQ3 (95% CI) | ORQ4 (95% CI) | FDRnon-linearb | ORdoubling (95% CI) | FDR | ||
| beta-Alanine | Amino acids | Ref. | 0.51 (0.24, 1.08) | 0.61 (0.27, 1.37) | 0.44 (0.19, 1.00) | 0.62 | 0.70 (0.54, 0.90) | 0.036 |
| Creatine | Amino acids | Ref. | 0.61 (0.30, 1.24) | 0.30 (0.13, 0.67) | 0.46 (0.20, 1.06) | 0.35 | 0.52 (0.33, 0.81) | 0.035 |
| Lysine | Amino acids | Ref. | 0.56 (0.25, 1.26) | 0.16 (0.06, 0.42) | 0.32 (0.14, 0.76) | 0.59 | 0.16 (0.06, 0.41) | 0.0033 |
| Pipecolic acid | Amino acids | Ref. | 2.68 (1.12, 6.40) | 2.92 (1.16, 7.37) | 4.06 (1.63, 10.10) | 0.70 | 2.04 (1.23, 3.39) | 0.041 |
| Tyrosine | Amino acids | Ref. | 1.08 (0.38, 3.05) | 3.22 (1.14, 9.08) | 5.38 (1.97, 14.70) | 0.74 | 5.82 (2.38, 14.20) | 0.0028 |
| GCA | Bile acids | Ref. | 3.39 (1.15, 9.94) | 5.15 (1.62, 16.41) | 12.74 (4.17, 38.89) | 0.62 | 1.66 (1.35, 2.04) | <0.0001 |
| GCDCA | Bile acids | Ref. | 2.42 (0.77, 7.57) | 5.25 (1.85, 14.86) | 14.97 (4.92, 45.53) | 0.47 | 2.03 (1.54, 2.68) | <0.0001 |
| GHCA | Bile acids | Ref. | 1.10 (0.38, 3.21) | 2.67 (0.81, 8.85) | 2.95 (0.98, 8.88) | 0.64 | 1.81 (1.24, 2.66) | 0.022 |
| GUDCA | Bile acids | Ref. | 1.25 (0.52, 2.99) | 1.86 (0.73, 4.75) | 3.53 (1.48, 8.45) | 0.74 | 1.45 (1.20, 1.77) | 0.0033 |
| TCA | Bile acids | Ref. | 4.28 (1.33, 13.78) | 2.63 (0.82, 8.45) | 13.36 (4.35, 41.03) | 0.67 | 1.59 (1.31, 1.93) | 0.00012 |
| TCDCA | Bile acids | Ref. | 2.94 (0.88, 9.82) | 3.79 (1.26, 11.41) | 19.06 (5.91, 61.49) | 0.82 | 1.88 (1.48, 2.39) | <0.0001 |
| TDCA | Bile acids | Ref. | 2.25 (0.87, 5.82) | 0.92 (0.33, 2.54) | 4.95 (1.88, 13.01) | 0.85 | 1.43 (1.14, 1.78) | 0.019 |
| D-Xylulose | Carbohydrates | Ref. | 2.35 (0.90, 6.15) | 2.94 (1.09, 7.98) | 3.61 (1.41, 9.26) | 0.85 | 3.56 (1.61, 7.88) | 0.019 |
| Fructose | Carbohydrates | Ref. | 1.36 (0.59, 3.11) | 1.16 (0.49, 2.76) | 2.34 (1.01, 5.44) | 0.52 | 1.40 (1.10, 1.79) | 0.044 |
| Rhamnose | Carbohydrates | Ref. | 0.63 (0.25, 1.62) | 1.48 (0.58, 3.77) | 2.60 (1.03, 6.59) | 0.85 | 1.63 (1.16, 2.28) | 0.035 |
| Acetylcarnitine | Carnitines | Ref. | 1.93 (0.85, 4.38) | 2.17 (0.93, 5.11) | 2.82 (1.26, 6.28) | 0.74 | 2.55 (1.45, 4.49) | 0.016 |
| Myristoylcarnitine | Carnitines | Ref. | 1.13 (0.42, 3.05) | 2.76 (0.94, 8.10) | 4.11 (1.60, 10.60) | 0.85 | 3.24 (1.56, 6.72) | 0.019 |
| Oleylcarnitine | Carnitines | Ref. | 0.84 (0.35, 2.04) | 2.47 (0.95, 6.40) | 4.14 (1.60, 10.69) | 0.13 | 2.32 (1.30, 4.14) | 0.035 |
| Stearylcarnitine | Carnitines | Ref. | 1.05 (0.46, 2.39) | 1.60 (0.71, 3.60) | 3.37 (1.43, 7.97) | 0.62 | 1.94 (1.27, 2.97) | 0.022 |
| 10Z-Heptadecenoic acid | Fatty acids | Ref. | 1.46 (0.62, 3.44) | 1.31 (0.58, 3.00) | 3.35 (1.49, 7.51) | 0.71 | 1.39 (1.12, 1.73) | 0.027 |
| Arachidonic acid | Fatty acids | Ref. | 0.44 (0.22, 0.90) | 0.52 (0.23, 1.19) | 0.66 (0.31, 1.45) | 0.016 | 0.65 (0.42, 1.02) | 0.18 |
| Aconitic acid | Organic acids | Ref. | 1.14 (0.40, 3.26) | 3.53 (1.22, 10.22) | 3.74 (1.26, 11.06) | 0.85 | 6.89 (2.57, 18.43) | 0.0028 |
| Citric acid | Organic acids | Ref. | 1.10 (0.43, 2.83) | 1.70 (0.60, 4.80) | 5.33 (1.94, 14.66) | 0.62 | 12.86 (4.10, 40.30) | 0.00036 |
| Isocitric acid | Organic acids | Ref. | 2.36 (0.77, 7.24) | 3.10 (1.11, 8.67) | 9.22 (3.06, 27.83) | 0.92 | 6.45 (2.98, 14.00) | 0.00011 |
| Homovanillic acid | Phenols | Ref. | 1.32 (0.54, 3.23) | 1.88 (0.68, 5.21) | 3.78 (1.48, 9.71) | 0.56 | 3.19 (1.71, 5.98) | 0.0048 |
| Hydroxyphenyllactic acid | Phenylpropanoic acids | Ref. | 0.28 (0.10, 0.83) | 0.75 (0.31, 1.85) | 1.68 (0.69, 4.10) | 0.12 | 2.11 (1.31, 3.41) | 0.022 |
| Butyric acid | SCFAs | Ref. | 2.76 (1.03, 7.43) | 6.79 (1.95, 23.66) | 6.49 (1.59, 26.48) | 0.87 | 4.17 (1.84, 9.46) | 0.0098 |
| Ethylmethylacetic acid | SCFAs | Ref. | 2.87 (1.13, 7.28) | 2.87 (1.05, 7.85) | 3.48 (1.24, 9.78) | 0.83 | 2.10 (1.23, 3.60) | 0.046 |
| Propanoic acid | SCFAs | Ref. | 1.44 (0.58, 3.53) | 3.47 (1.33, 9.01) | 5.98 (2.01, 17.79) | 0.52 | 6.80 (2.20, 21.05) | 0.013 |
Conditioned on matching factors and adjusted for age, cigarette smoking, alcohol drinking, BMI, physical activity, CHFP score, medical history of hepatitis and cirrhosis, medical history of cholelithiasis and medical history of T2DM.
Log2-transformed metabolite concentrations was fitted with a 3-knots restricted cubic spline (10th, 50th, and 90th percentile).
Fig. 1.
Associations between plasma metabolite concentrations and male liver cancer risk (Logistic regression models conditioned on matching factors and adjusted for age, cigarette smoking, alcohol drinking, BMI, physical activity, CHFP score, medical history of hepatitis and cirrhosis, medical history of cholelithiasis and medical history of T2DM were used. The horizontal dashed lines represent thresholds for statistical significance. Metabolites located above the lines have reached significance levels at the corresponding thresholds annotated).
Fig. 2.
Correlation matrix for liver cancer-associated metabolites among controls (The numbers within the squares represent Spearman's ρ. The lower left section displays the raw correlation coefficients, while the upper right section shows the partial correlation coefficients adjusted for age and fasting time).
Fig. 3.
Dose–response relationships between selected metabolites and liver cancer risk (Logistic regression models conditioned on matching factors and adjusted for age, cigarette smoking, alcohol drinking, BMI, physical activity, CHFP score, medical history of hepatitis and cirrhosis, medical history of cholelithiasis and medical history of T2DM were used. The solid lines represent the estimated dose–response curves, the dots indicate the knots used by RCS function, and the shaded areas denote the 95% CIs).
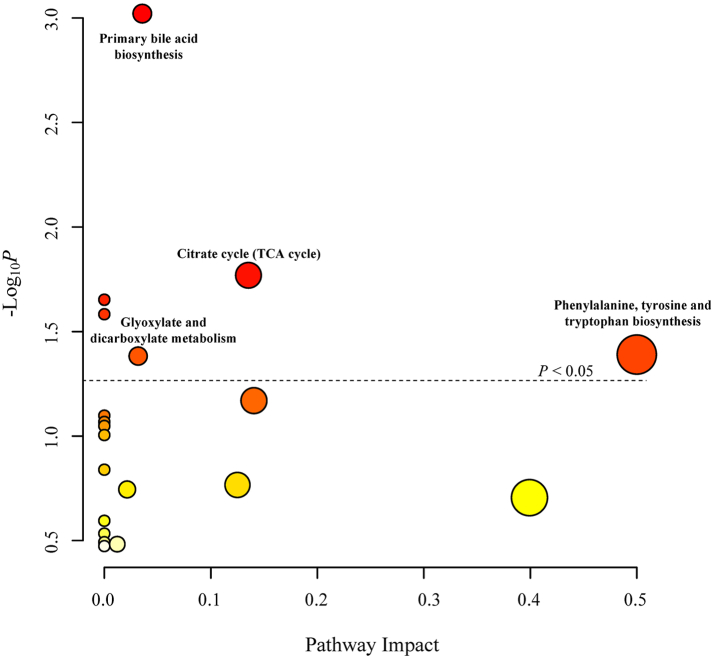
As shown in Fig. 4, four pathways were significantly associated with liver cancer risk, among which the primary bile acid biosynthesis had the lowest P value and the phenylalanine, tyrosine and tryptophan biosynthesis had the highest impact. The inclusion of arachidonic acid, whose FDRnon-linear was less than 0.05, did not alter the results of pathway analysis.
Fig. 4.
Pathway analysis based on liver cancer-associated metabolites (Metabolites with an FDR of <0.05 were included in the analysis. Pathways that lie above the dashed line surpass the significance threshold of P < 0.05).
The results of the sensitivity analysis substantially supported the main findings. The exclusion of participants with a follow-up time of less than two years had a minor impact on most significant associations (Supplementary Table S5). When excluding participants with hepatobiliary diseases, the magnitude of some observed associations was moderately attenuated (Supplementary Table S6) and did not reach statistical significance. All of the significant associations were replicated using the bootstrapping method (Fig. 5). The non-linear relationship between arachidonic acid and liver cancer risk remained stable after excluding participants whose follow-up time was less than two years or with hepatobiliary diseases (Supplementary Figure S5). Results of the Cox regression models also supported the main findings (Supplementary Table S7).
Fig. 5.
Replication of the observed metabolites-liver cancer associations using a bootstrapping method with 2000 repetitions (Logistic regression models conditioned on matching factors and adjusted for age, cigarette smoking, alcohol drinking, BMI, physical activity, CHFP score, medical history of hepatitis and cirrhosis, medical history of cholelithiasis and medical history of T2DM were used. The error bar refers to 95% CI).
The penalised LASSO logistic regression model derived a panel of 10 metabolites (creatine, glutamine, tyrosine, TCA, TCDCA, rhamnose, AMP, glutaric acid, isocitric acid, and homovanillic acid) most significantly associated with liver cancer risk while robust to collinearity. Seven metabolites were associated with liver cancer risk at the FDR<0.05 level (Table 2). The distribution frequency of the LASSO-derived metabolic score varied substantially between liver cancer cases and controls (Supplementary Figure S6). An increased level of metabolic score was significantly associated with liver cancer risk after adjusting for potential confounders (Supplementary Figure S6).
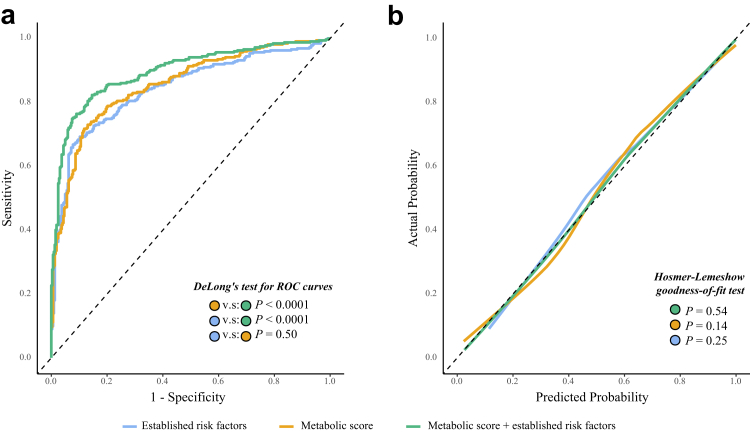
The performance of the three predictive models was shown in Fig. 6 and Table 3. Overall, the metabolic score exhibited discrimination comparable to that of established risk factors [the optimism-corrected AUCs = 0.86 (95% CI: 0.82–0.88) and 0.84 (95% CI: 0.81–0.87), respectively; DeLong's test P = 0.50]. Incorporating the metabolic score into the model with established risk factors improved the overall performance of the model. The optimism-corrected Brier score decreased from 0.15 (95% CI: 0.14–0.17) to 0.13 (95% CI: 0.11–0.14). The combined model yielded an optimism-corrected AUC of 0.89 (95% CI: 0.86–0.91), which was significantly higher than that of either metabolites or epidemiological variables alone (DeLong's tests P < 0.0001). The calibration plots demonstrated good predictive accuracy between observed and predicted probabilities across all three models (Fig. 6, Supplementary Figure S7). Test for calibration slope and intercept showed that all the models are well calibrated (U = −0.003, P > 0.999 for three apparent calibration curves). The discrimination of the individual metabolites used to construct the metabolic score was presented in Supplementary Figure S8, among which TCDCA showed the best discriminative ability (AUC = 0.82, 95% CI: 0.78–0.85).
Fig. 6.
Discrimination and calibration of models with LASSO-derived metabolic score and established risk factors on male liver cancer (Panel (a) represents the ROC curves for three models. Panel (b) shows the loess-smoothed apparent calibration curves for the models).
Table 3.
Brier scores and AUCs for three prediction models.
| Apparent | Optimism-correcteda | |
|---|---|---|
| Brier score (95% CI) | ||
| Established risk factorsb | 0.15 (0.14, 0.17) | 0.15 (0.14, 0.17) |
| Metabolic score | 0.15 (0.14, 0.17) | 0.15 (0.14, 0.17) |
| Metabolic score & established risk factors | 0.12 (0.11, 0.14) | 0.13 (0.11, 0.14) |
| AUC (95% CI) | ||
| Established risk factorsb | 0.84 (0.81, 0.87) | 0.84 (0.81, 0.87) |
| Metabolic score | 0.86 (0.83, 0.88) | 0.86 (0.82, 0.88) |
| Metabolic score & established risk factors | 0.90 (0.87, 0.92) | 0.89 (0.86, 0.91) |
Bootstrapping method with 2000 repetitions.
Including age, BMI, HBsAg, family history of liver cancer, cigarette smoking and alcohol drinking.
Discussion
Based on a case–control study nested within a prospective cohort study, we examined the associations between the prediagnostic plasma concentrations of 186 metabolites and the risk of liver cancer. We found 28 metabolites were associated with liver cancer risk after adjusting for potential confounders and correcting for multiple testing. These associations remained stable when excluding participants with a short follow-up time or prevalent hepatobiliary diseases. Our study highlighted the non-linear relationships between 22 metabolites and liver cancer risk. The primary bile acid biosynthesis and phenylalanine, tyrosine and tryptophan biosynthesis were identified as important biochemical pathways in the development of liver cancer. Furthermore, we derived a metabolic score consisting of 10 metabolites that significantly improved the predictive ability of established risk factors for liver cancer risk.
Understanding the early metabolic alterations in liver cancer can provide insights into the aetiology and aid in the search for novel biomarkers for risk assessment of liver cancer. To our knowledge, six studies have to date investigated the association between circulating metabolite profiles and liver cancer risk in a prospective manner.17, 18, 19, 20, 21, 22 In addition, several studies paid particular attention to specific categories of metabolites including bile acids, amino acids, and one-carbon metabolites.46, 47, 48, 49, 50, 51 Consistent with the Alpha-Tocopherol, Beta-Carotene Cancer Prevention (ATBC) study,19 the prospective Korean Cancer Prevention Study-II (KCPS-II),18 two cohort studies in Chinese population,20,22 and three reports of the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort,17,21,51 we found that elevated levels of circulating tyrosine were associated with an increased risk of liver cancer. The associations between beta-alanine, creatine, and lysine and liver cancer risk were also reported by Hang et al. and two reports of EPIC.22,47,51 Additionally, our study suggested that elevated levels of pipecolic acid was associated with liver cancer risk. The liver plays a critical role in the transport, signaling, and metabolism of amino acids.52 Altered amino acid profiles have been reported to be associated with established risk factors for liver cancer, such as obesity, excessive alcohol intake, T2DM, and metabolic syndrome.53, 54, 55, 56, 57, 58, 59 The alterations in circulating amino acid levels may act as potential mediators linking these risk factors to liver cancer.
Our study highlighted the importance of bile acid perturbations in the aetiology of liver cancer. We found that elevated concentrations of seven bile acids were associated with an increased risk of liver cancer, which was consistent with previous publications.18,22,48, 49, 50, 51 Similar to our study, Jee et al. suggested that dysregulation of primary bile acid biosynthesis plays an important role in the development of liver cancer.18 Bile acids are detergent molecules that can be highly toxic if accumulated in the liver and other tissues.60 Increased levels of bile acids represent early indicators of liver dysfunction.60 Except for liver cancer, alterations in bile acid homeostasis could also be led by other hepatobiliary diseases, such as liver fibrosis, cirrhosis, and NAFLD.11,61,62 Nevertheless, we observed similar associations in participants without prevalent hepatobiliary diseases. This suggested that the associations between circulating bile acids and liver cancer risk might not be solely attributed to these diseases.
Previous studies have seldom focused on the association between organic acids and liver cancer risk. Only two prospective studies reported a significant association between citrate and liver cancer risk.17,20 We found that elevated plasma concentrations of three organic acids were associated with an increased risk of liver cancer. Additionally, the citrate cycle was identified as one of the most abundant pathways, with three liver cancer-associated organic acids included in the pathway. The citrate cycle, also referred to as the tricarboxylic acid cycle or the Krebs cycle, is a crucial process of energy production in cells. In this process, Acetyl-CoA derived from the breakdown of sugars, fats, and proteins were oxidised in cells to produce the high-energy molecule ATP.63 The citrate cycle also generates important intermediates used in the synthesis of fatty acids, cholesterol, and other biomolecules. A hospital-based study suggested that oxaloacetate, a citrate cycle-related metabolite, could be used as a potential biomarker to discriminate patients with HCC from patients with liver cirrhosis as well as healthy volunteers.11 They suggested that the alterations of metabolites in the citrate cycle revealed high energy demand as well as altered enzyme activities in association with rapid tumour growth.11 Our study indicated that abnormal energy metabolism may be present long before the diagnosis of liver cancer.
An interesting observation in the present study was the significantly non-linear relationship between arachidonic acid (AA) and liver cancer risk, which was inconsistent with the previous publication of the prospective KCPS-II20 and a hospital-based cross-sectional study.64 The previous studies indicated that the level of AA was higher in the HCC group than in the cancer-free control group, while the ORs were consistently less than one in our study. AA is a long-chain polyunsaturated fatty acid present in human phospholipid cell membranes and can be obtained from animal food sources, particularly meat, fish, and eggs, or derived from linoleic acid, etc.65 Experimental studies suggested that AA metabolism yields eicosanoids, like prostaglandins, thromboxanes, and leukotrienes via cyclooxygenase and lipoxygenase pathways, which are implicated in inflammation and cancer progression by influencing tumour cell dynamics and angiogenesis.66 However, Mendelian randomisation did not support the causal relationship between circulating AA levels and most common cancers including liver cancer.65,67 Chen et al. suggested that genetically predicted higher plasma phospholipid AA levels were associated with an increased risk of NAFLD and cirrhosis but not with liver cancer.68 The combined OR for per SD increase in AA levels was 0.99 (95% CI: 0.94–1.05; P = 0.77) for liver cancer. Given the inconsistent results of existing studies, the association between arachidonic acid and liver cancer needs further investigation. This example indicated the complexity of the metabolic alterations in the development of liver cancer.
Several studies have examined the possibility of circulating metabolites serving as risk predictors for liver cancer. For example, Huang et al. derived a metabolic score consisting of tyrosine, glutamate, and citrate that improved the c-statistic of a clinical model predicting future HCC (age, HBeAg, viral load, and alanine aminotransferase) from 0.70 to 0.79.20 Using the orthogonal partial least-square analysis, a metabolic signature was derived from the EPIC study, presenting an AUC of 0.85.17 Furthermore, Hang et al. utilised LASSO regression to select independent metabolites and built a logistic regression model to assess the performance of the predictors. They obtained a combination of 18 metabolites that showed the potential to predict HCC risk with AUCs of 0.87 (95% CI: 0.82–0.92) and 0.86 (95% CI: 0.80–0.93) in the training and validation sets, respectively.22 Similar to Hang et al., we derived a panel of 10 metabolites that are robust to collinearity through LASSO. A metabolic score consisting of these metabolites produced a similar predictive performance with a combination of established risk factors for liver cancer. Considering the high cost of circulating metabolite assays, easily available epidemiological risk factors may still be a better choice for practical applications.
The key strength of this study lies in its prospective design, in which blood samples were collected up to 14 years prior to the clinical diagnosis of primary liver cancer, thereby reducing the possibility of reverse causation. Detailed information on risk factors for liver cancer, such as HBsAg, lifestyle factors, dietary habits, and medical histories allows for comprehensive control of confounders and improves the reliability of the results. Besides, the 1:1 individual matching design significantly reduced the variance and improved the statistical efficiency. In addition, compared to the untargeted approach that quantifies metabolites on a relative scale, targeted metabolomics provides absolute concentrations of identified compounds. This is achieved by utilizing chemical standards to construct calibration curves for each metabolite. To our knowledge, most of the existing studies used an untargeted metabolomic approach17, 18, 19, 20, 21, 22 and were unable to quantitate the absolute concentrations of metabolites as in our study (in μmol/L). The application of targeted metabolomics technology in this study assured the precision of metabolite identification and quantitation, thereby strengthening the credibility of our findings and enabling confirmation of associations previously reported by untargeted metabolomics. Moreover, we examined potential non-linear relationships and visualised the dose–response curves between plasma metabolites and liver cancer risk. This provided additional insights into the complex metabolic perturbations prior to the clinical diagnosis of liver cancer.
Several limitations pertinent to the present study warrant acknowledgment. First, our study lacked biochemical markers that indicate liver function, such as alanine aminotransferase, aspartate aminotransferase, and total bilirubin. Potential liver damage might affect the concentrations of plasma metabolites, possibly resulting in biased risk estimates. To address this, we utilised self-reported medical histories of chronic hepatitis, cirrhosis, and seropositive HBsAg as proxies for underlying liver dysfunction and included them as covariates in the regression models. We also performed sensitivity analyses by excluding participants with these conditions and the results remained stable. Still, future metabolomics studies should assess liver function markers concurrently and give full consideration to this issue. Second, as is typical of most previous publications, metabolomic profiling was conducted only once in our study. Although the reproducibility of many circulating metabolites over time has been demonstrated,68, 69, 70 longitudinal studies with repeated measurements are necessitated to account for within-person variations and to assess the dynamic changes of metabolites in relation to liver cancer risk. Third, the concentration of plasma metabolites would be influenced by diet and medication. To improve the compliance of the study participants, fasting was not required before blood collection. Although we considered sampling time and use of antibiotic drugs in the study, there will inevitably be deviations through individual subjective traceability. While a sensitivity analysis in a fasted subset could be informative, it was not feasible in our study due to the very low proportion of fasting samples (<10%). Nevertheless, such non-differential misclassification was more likely to result in an underestimation of the effect. Fourth, residual confounding from strong liver cancer risk factors such as cigarette smoking and alcohol drinking, and unmeasured confounding cannot be fully eliminated. Fifth, the definition of liver cancer cases in our study was based on ICD-9 code 155, which encompasses not only HCC but also intrahepatic cholangiocarcinoma and other rare histological subtypes. In this case, our findings primarily reflect the metabolic alterations in the aetiology of HCC (comprising 75%–85% of the cases).1 Given the potential differences in the aetiology of liver cancer among various histological subtypes, analysing each type separately would offer a more comprehensive understanding of the metabolic differences in liver cancer. However, the current study cannot accommodate this analysis because the histological subtypes of the liver cancer cases were unavailable. The SWHS was established at an earlier time, when the design did not facilitate the precise collection of detailed histological subtype data. Last but not least, participants in the present study are middle-aged Chinese men from an urban area whose genetic background, lifestyles, and dietary habits may differ from those of the other populations. Therefore, our findings should be interpreted and extrapolated with caution. Further investigations in women and other ethnic groups are needed. We also acknowledge the importance of conducting validation in similar source populations in future studies, which could substantially strengthen the generalizability of our findings.
In conclusion, we conducted a prospective metabolomics investigation to examine the associations between concentrations of plasma metabolites and liver cancer risk and search for potential risk predictors for the disease. This study provided insights into the aetiology of liver cancer and offers potential candidate biomarkers for risk prediction, suggesting that abnormal metabolism might be present long before diagnosis and the association between several plasma metabolites and liver cancer risk may not be linear. Future studies are warranted to confirm our findings in other populations and to further investigate the underlying molecular mechanisms.
Contributors
Y-BX designed research and obtained funding; Z-YL, Q-MS, J-W, J-YT, Y-TT, H-LL, and Y-BX conducted the study; Z-YL, Q-MS, and Y-BX analysed the data and interpreted the results; Z-YL, and Q-MS verified the underlying data; Z-YL and Y-BX prepared and wrote the first draft; All authors reviewed, edited and approved the final version of the paper; and Y-BX has primary responsibility for final content.
Data sharing statement
Due to privacy and confidentiality concerns, raw data of the current study will not be uploaded to public databases. But de-identified datasets and statistical programs may be available on request pending approval by the scientific committee of the relevant institutes.
Declaration of interests
The authors have no relevant financial or non-financial interests to disclose.
Acknowledgements
We would like to thank all participants and staff from the Shanghai Men's Health Study for their contribution to this research. This work was supported by the National Key Project of Research and Development Program of China [2021YFC2500404, 2021YFC2500405 to Yong-Bing Xiang]; and the parent cohort was supported by a subcontract from the grant of the US National Institutes of Health [subcontract of UM1 CA173640 to Yong-Bing Xiang].
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.104990.
Appendix A. Supplementary data
S1–S7
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Baecker A., Liu X., La Vecchia C., Zhang Z.F. Worldwide incidence of hepatocellular carcinoma cases attributable to major risk factors. Eur J Cancer Prev. 2018;27(3):205–212. doi: 10.1097/CEJ.0000000000000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allemani C., Matsuda T., Di Carlo V., et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y.F., Li Z.Y., Ji X.W., Shen Q.M., Tuo J.Y., Yuan H.Y. Global pattern and trend of liver cancer survival: a systematic review of population-based studies. Hepatoma Res. 2020;6(52):1–13. [Google Scholar]
- 5.Piñero F., Dirchwolf M., Pessôa M.G. Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells. 2020;9(6):1370. doi: 10.3390/cells9061370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel M., Shariff M.I.F., Ladep N.G., et al. Hepatocellular carcinoma: diagnostics and screening. J Eval Clin Pract. 2012;18(2):335–342. doi: 10.1111/j.1365-2753.2010.01599.x. [DOI] [PubMed] [Google Scholar]
- 7.Thun M.J., Linet M.S., Cerhan J.R., Haiman C., Schottenfeld D., editors. Cancer epidemiology and prevention. 4th ed. Oxford University Press; New York, NY: 2018. [Google Scholar]
- 8.Goodacre R., Vaidyanathan S., Dunn W.B., Harrigan G.G., Kell D.B. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22(5):245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Su L.J., Fiehn O., Maruvada P., et al. The use of metabolomics in population-based research. Adv Nutr. 2014;5(6):785–788. doi: 10.3945/an.114.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadei Gardini A., Del Coco L., Marisi G., et al. 1H-NMR based serum metabolomics highlights different specific biomarkers between early and advanced hepatocellular carcinoma stages. Cancers. 2020;12(1):241. doi: 10.3390/cancers12010241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y., Hong Z., Tan G., et al. NMR and LC/MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int J Cancer. 2014;135(3):658–668. doi: 10.1002/ijc.28706. [DOI] [PubMed] [Google Scholar]
- 12.Luo P., Yin P., Hua R., et al. A large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2018;67(2):662–675. doi: 10.1002/hep.29561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T., Zheng X., Yen Y., Liu P., Jia W. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics. 2011;10(7) doi: 10.1074/mcp.M110.004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao J.F., Varghese R.S., Zhou B., et al. LC-MS based serum metabolomics for identification of hepatocellular carcinoma biomarkers in Egyptian cohort. J Proteome Res. 2012;11(12):5914–5923. doi: 10.1021/pr300673x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X., Wang Z., Luo L., Shu D., Wang K. Metabolomics in hepatocellular carcinoma: from biomarker discovery to precision medicine. Front Med Technol. 2022;4 doi: 10.3389/fmedt.2022.1065506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng N., Yu F., Yu F., et al. Metabolomic biomarkers for hepatocellular carcinoma: a systematic review. Medicine. 2022;101(3) doi: 10.1097/MD.0000000000028510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fages A., Duarte Salles T., Stepien M., et al. Metabolomic profiles of hepatocellular carcinoma in a European prospective cohort. BMC Med. 2015;13:242. doi: 10.1186/s12916-015-0462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jee S.H., Kim M., Kim M., et al. Metabolomics profiles of hepatocellular carcinoma in a Korean prospective cohort: the Korean cancer prevention study-II. Cancer Prev Res. 2018;11(5):303–312. doi: 10.1158/1940-6207.CAPR-17-0249. [DOI] [PubMed] [Google Scholar]
- 19.Loftfield E., Rothwell J.A., Sinha R., et al. Prospective investigation of serum metabolites, coffee drinking, liver cancer incidence, and liver disease mortality. J Natl Cancer Inst. 2020;112(3):286–294. doi: 10.1093/jnci/djz122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang B.Y., Tsai M.R., Hsu J.K., et al. Longitudinal change of metabolite profile and its relation to multiple risk factors for the risk of developing hepatitis B-related hepatocellular carcinoma. Mol Carcinog. 2020;59(11):1269–1279. doi: 10.1002/mc.23255. [DOI] [PubMed] [Google Scholar]
- 21.Stepien M., Keski Rahkonen P., Kiss A., et al. Metabolic perturbations prior to hepatocellular carcinoma diagnosis: findings from a prospective observational cohort study. Int J Cancer. 2021;148(3):609–625. doi: 10.1002/ijc.33236. [DOI] [PubMed] [Google Scholar]
- 22.Hang D., Yang X., Lu J., et al. Untargeted plasma metabolomics for risk prediction of hepatocellular carcinoma: a prospective study in two Chinese cohorts. Int J Cancer. 2022;151(12):2144–2154. doi: 10.1002/ijc.34229. [DOI] [PubMed] [Google Scholar]
- 23.Shu X.O., Li H., Yang G., et al. Cohort profile: the Shanghai Men's health study. Int J Epidemiol. 2015;44(3):810–818. doi: 10.1093/ije/dyv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurj A.L., Wen W., Xiang Y.B., et al. Reproducibility and validity of the Shanghai Men's Health Study physical activity questionnaire. Am J Epidemiol. 2007;165(10):1124–1133. doi: 10.1093/aje/kwk119. [DOI] [PubMed] [Google Scholar]
- 25.Villegas R., Yang G., Liu D., et al. Validity and reproducibility of the food-frequency questionnaire used in the Shanghai Men's Health Study. Br J Nutr. 2007;97(5):993–1000. doi: 10.1017/S0007114507669189. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y., Gao J., Li H.L., et al. Dose-response association between hepatitis B surface antigen levels and liver cancer risk in Chinese men and women. Int J Cancer. 2016;139(2):355–362. doi: 10.1002/ijc.30086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao C., Yang G., Hu J., Ma J., Xia W., Lopez A.D. Validation of cause-of-death statistics in urban China. Int J Epidemiol. 2007;36(3):642–651. doi: 10.1093/ije/dym003. [DOI] [PubMed] [Google Scholar]
- 28.Xie G., Wang L., Chen T., et al. A metabolite array technology for precision medicine. Anal Chem. 2021;93(14):5709–5717. doi: 10.1021/acs.analchem.0c04686. [DOI] [PubMed] [Google Scholar]
- 29.Textor J., van der Zander B., Gilthorpe M.S., Liskiewicz M., Ellison G.T. Robust causal inference using directed acyclic graphs: the R package 'dagitty'. Int J Epidemiol. 2016;45(6):1887–1894. doi: 10.1093/ije/dyw341. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen S., Li H., Yu D., et al. Adherence to dietary recommendations and colorectal cancer risk: results from two prospective cohort studies. Int J Epidemiol. 2020;49(1):270–280. doi: 10.1093/ije/dyz118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu D.X., Zhang X.L., Xiang Y.B., et al. Adherence to dietary guidelines and mortality: a report from prospective cohort studies of 134,000 Chinese adults in urban Shanghai. Am J Clin Nutr. 2014;100(2):693–700. doi: 10.3945/ajcn.113.079194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q.L., Zhao L.G., Zhang W., et al. Combined impact of known lifestyle factors on total and cause-specific mortality among Chinese men: a prospective cohort study. Sci Rep. 2017;7(1):5293. doi: 10.1038/s41598-017-05079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300. [Google Scholar]
- 34.Mansournia M.A., Nazemipour M., Etminan M. P-value, compatibility, and S-value. Glob Epidemiol. 2022;4 doi: 10.1016/j.gloepi.2022.100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole S.R., Edwards J.K., Greenland S. Surprise. Am J Epidemiol. 2021;190(2):191–193. doi: 10.1093/aje/kwaa136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durrleman S., Simon R. Flexible regression models with cubic splines. Stat Med. 1989;8(5):551–561. doi: 10.1002/sim.4780080504. [DOI] [PubMed] [Google Scholar]
- 37.Desquilbet L., Mariotti F. Dose-response analyses using restricted cubic spline functions in public health research. Stat Med. 2010;29(9):1037–1057. doi: 10.1002/sim.3841. [DOI] [PubMed] [Google Scholar]
- 38.Harrell F.E. Springer; 2015. Regression modeling strategies: with applications to linear models, logistic and ordinal regression, and survival analysis. [Google Scholar]
- 39.Wishart D.S., Guo A., Oler E., et al. HMDB 5.0: the human Metabolome database for 2022. Nucleic Acids Res. 2022;50(D1):D622–D631. doi: 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang Z., Chong J., Zhou G., et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49(W1):W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DiCiccio T.J., Efron B. Bootstrap confidence intervals. Stat Sci. 1996;11(3):189–228. [Google Scholar]
- 43.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B Methodol. 1996;58(1):267–288. [Google Scholar]
- 44.Hastie T., Tibshirani R., Friedman J.H., Friedman J.H. Vol. 2. Springer; 2009. (The elements of statistical learning: data mining, inference, and prediction). [Google Scholar]
- 45.DeLong E.R., DeLong D.M., Clarke Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 46.Butler L.M., Arning E., Wang R., et al. Prediagnostic levels of serum one-carbon metabolites and risk of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2013;22(10):1884–1893. doi: 10.1158/1055-9965.EPI-13-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stepien M., Duarte Salles T., Fedirko V., et al. Alteration of amino acid and biogenic amine metabolism in hepatobiliary cancers: findings from a prospective cohort study. Int J Cancer. 2016;138(2):348–360. doi: 10.1002/ijc.29718. [DOI] [PubMed] [Google Scholar]
- 48.Petrick J.L., Florio A.A., Koshiol J., et al. Prediagnostic concentrations of circulating bile acids and hepatocellular carcinoma risk: REVEAL-HBV and HCV studies. Int J Cancer. 2020;147(10):2743–2753. doi: 10.1002/ijc.33051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas C.E., Luu H.N., Wang R., et al. Association between pre-diagnostic serum bile acids and hepatocellular carcinoma: the Singapore Chinese Health Study. Cancers. 2021;13(11):2648. doi: 10.3390/cancers13112648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farhat Z., Freedman N.D., Sampson J.N., et al. A prospective investigation of serum bile acids with risk of liver cancer, fatal liver disease, and biliary tract cancer. Hepatol Commun. 2022;6(9):2391–2399. doi: 10.1002/hep4.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stepien M., Lopez Nogueroles M., Lahoz A., et al. Prediagnostic alterations in circulating bile acid profiles in the development of hepatocellular carcinoma. Int J Cancer. 2022;150(8):1255–1268. doi: 10.1002/ijc.33885. [DOI] [PubMed] [Google Scholar]
- 52.Paulusma C.C., Lamers W.H., Broer S., van de Graaf S.F.J. Amino acid metabolism, transport and signalling in the liver revisited. Biochem Pharmacol. 2022;201 doi: 10.1016/j.bcp.2022.115074. [DOI] [PubMed] [Google Scholar]
- 53.Moore S.C., Matthews C.E., Sampson J.N., et al. Human metabolic correlates of body mass index. Metabolomics. 2014;10(2):259–269. doi: 10.1007/s11306-013-0574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carayol M., Leitzmann M.F., Ferrari P., et al. Blood metabolic signatures of body mass index: a targeted metabolomics study in the EPIC cohort. J Proteome Res. 2017;16(9):3137–3146. doi: 10.1021/acs.jproteome.6b01062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Roekel E.H., Trijsburg L., Assi N., et al. Circulating metabolites associated with alcohol intake in the European Prospective Investigation into Cancer and Nutrition cohort. Nutrients. 2018;10(5):654. doi: 10.3390/nu10050654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taghizadeh H., Emamgholipour S., Hosseinkhani S., et al. The association between acylcarnitine and amino acids profile and metabolic syndrome and its components in Iranian adults: data from STEPs 2016. Front Endocrinol. 2023;14 doi: 10.3389/fendo.2023.1058952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun S., He D., Luo C., et al. Metabolic syndrome and its components are associated with altered amino acid profile in Chinese Han population. Front Endocrinol. 2021;12 doi: 10.3389/fendo.2021.795044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramzan I., Ardavani A., Vanweert F., Mellett A., Atherton P.J., Idris I. The association between circulating branched chain amino acids and the temporal risk of developing type 2 diabetes mellitus: a systematic review & meta-analysis. Nutrients. 2022;14(20):4411. doi: 10.3390/nu14204411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calvani R., Rodriguez Mañas L., Picca A., et al. Identification of a circulating amino acid signature in frail older persons with type 2 diabetes mellitus: results from the Metabofrail study. Nutrients. 2020;12(1):199. doi: 10.3390/nu12010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiang J.Y.L., Ferrell J.M. Bile acid metabolism in liver pathobiology. Gene Expr. 2018;18(2):71–87. doi: 10.3727/105221618X15156018385515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bing H., Li Y.L. The role of bile acid metabolism in the occurrence and development of NAFLD. Front Mol Biosci. 2022;9 doi: 10.3389/fmolb.2022.1089359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu A.N., Xu C.F., Liu Y.R., et al. Secondary bile acids improve risk prediction for non-invasive identification of mild liver fibrosis in nonalcoholic fatty liver disease. Aliment Pharmacol Ther. 2023;57(8):872–885. doi: 10.1111/apt.17362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verschueren K., Blanchet C., Felix J., et al. Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle. Nature. 2019;568(7753):571–575. doi: 10.1038/s41586-019-1095-5. [DOI] [PubMed] [Google Scholar]
- 64.Zhou L., Ding L., Yin P., et al. Serum metabolic profiling study of hepatocellular carcinoma infected with hepatitis B or hepatitis C virus by using liquid chromatography-mass spectrometry. J Proteome Res. 2012;11(11):5433–5442. doi: 10.1021/pr300683a. [DOI] [PubMed] [Google Scholar]
- 65.Larsson S.C., Carter P., Vithayathil M., et al. Genetically predicted plasma phospholipid arachidonic acid concentrations and 10 site-specific cancers in UK biobank and genetic consortia participants: a mendelian randomization study. Clin Nutr. 2021;40(5):3332–3337. doi: 10.1016/j.clnu.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang D., Dubois R.N. Eicosanoids and cancer. Nat Rev Cancer. 2010;10(3):181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen J., Ruan X., Sun Y., Li X., Yuan S., Larsson S.C. Plasma phospholipid arachidonic acid in relation to non-alcoholic fatty liver disease: mendelian randomization study. Nutrition. 2023;106 doi: 10.1016/j.nut.2022.111910. [DOI] [PubMed] [Google Scholar]
- 68.Floegel A., Drogan D., Wang Sattler R., et al. Reliability of serum metabolite concentrations over a 4-month period using a targeted metabolomic approach. PLoS One. 2011;6(6) doi: 10.1371/journal.pone.0021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Townsend M.K., Clish C.B., Kraft P., et al. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin Chem. 2013;59(11):1657–1667. doi: 10.1373/clinchem.2012.199133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carayol M., Licaj I., Achaintre D., et al. Reliability of serum metabolites over a two-year period: a targeted metabolomic approach in fasting and non-fasting samples from EPIC. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0135437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1–S7