Abstract
Severe cold, defined as a damaging cold beyond acclimation temperatures, has unique responses, but the signaling and evolution of these responses are not well understood. Production of oligogalactolipids, which is triggered by cytosolic acidification in Arabidopsis (Arabidopsis thaliana), contributes to survival in severe cold. Here, we investigated oligogalactolipid production in species from bryophytes to angiosperms. Production of oligogalactolipids differed within each clade, suggesting multiple evolutionary origins of severe cold tolerance. We also observed greater oligogalactolipid production in control samples than in temperature-challenged samples of some species. Further examination of representative species revealed a tight association between temperature, damage, and oligogalactolipid production that scaled with the cold tolerance of each species. Based on oligogalactolipid production and transcript changes, multiple angiosperm species share a signal of oligogalactolipid production initially described in Arabidopsis, namely cytosolic acidification. Together, these data suggest that oligogalactolipid production is a severe cold response that originated from an ancestral damage response that remains in many land plant lineages and that cytosolic acidification may be a common signaling mechanism for its activation.
Keywords: Abiotic stress, acidification, angiosperms, damage, membrane, oligogalactolipid, phylogeny, severe cold
Oligogalactolipid production is a response to severe cold in many land plant lineages. It occurs during times of membrane damage and can be reproduced in multiple species by cytosolic acidification.
Introduction
As the climate changes, weather extremes are becoming more common. One such extreme phenomenon is temperature stress, which affects the production of many worldwide staple crops. However, losses caused by temperature stress are difficult to predict because they occur in a non-linear fashion (Schlenker and Roberts, 2009). Cold stress has a large influence on crops originating from tropical environments and grown in temperate climates, such as maize (Zea mays L.). For many years, studies have been conducted on such crops to better understand how they survive cold temperatures (Sellschop and Salmon, 1928). Cold stress includes both chilling stress (temperatures >0 °C) and freezing stress (temperatures <0 °C), with adaptation to each of these stresses being species specific (Lyons, 1973).
One adaptative strategy used by plants in response to low-temperature stress relies on lipid remodeling of membranes (Uemura et al., 1995; Welti et al., 2002; Li et al., 2008; Moellering et al., 2010). Glycerolipids constitute the bulk of the membrane lipids, consisting of a polar head group attached to two fatty acid ‘tails’ through a glycerol backbone. Head group size, in addition to the level of saturation in the fatty acid tails, affects how a lipid bonds within the membrane, thus changing the physical properties of the entire membrane (Melser et al., 2011). In response to stress, lipid remodeling targets both the fatty acid tails and the lipid head group. The saturation level of fatty acid tails changes in response to low-temperature stress, with fatty acids becoming more unsaturated after low-temperature exposure (Hugly and Somerville, 1992; Miquel et al., 1993; Steponkus, 1996). Unsaturation weakens hydrophobic interactions of the hydrocarbon tail group, ultimately decreasing the temperature at which the membrane transitions to a gel phase. Changes in lipid head groups have also been observed (Raju et al., 2018). For example, SENSITIVE TO FREEZING 2 (SFR2) and ACYLATED GALACTOLIPID- ASSOCIATED PHOSPHOLIPASE 1 (AGAP1) in Arabidopsis (Arabidopsis thaliana) both convert monogalactolipids into other lipids during freezing challenge (Moellering et al., 2010; Nilsson et al., 2015; Barnes et al., 2016). Head groups influence membrane permeability, and the extent of physical space needed by head and tail groups modulates the effectiveness of weak hydrophobic interactions between lipids, thus modifying membrane flexibility and temperatures at which the membrane undergoes phase transition (Melser et al., 2011).
SFR2 resides on the outer envelope of the chloroplast (Fourrier et al., 2008), where it is required for remodeling lipid head groups in response to severe cold (Moellering et al., 2010), which we define as a damaging cold beyond acclimation temperatures. SFR2 converts monogalactosyldiacylglycerol (MGDG) to oligogalactolipids, including digalactosyldiacylglycerol (DGDG) and trigalactosyldiacylglycerol (TGDG). Lipids with three or more galactose head groups result uniquely from SFR2 activity and are not biosynthesized through any other known mechanism. The reaction involves removal of a galactose from one MGDG and subsequent linkage to another galactolipid, leading to the release of a diacylglycerol (Moellering et al., 2010; Roston et al., 2014). This diacylglycerol is then converted into triacylglycerol (TAG) through a multistep pathway. Fatty acids derived from MGDG are found in lipid droplets of TAG released after SFR2 activation (Barnes et al., 2016). SFR2 is constitutively present in Arabidopsis, but the accumulation of the unique TGDG lipid product is only observed following SFR2 activation after exposure to sub-zero freezing stress (severe cold) (Thorlby et al., 2004; Barnes et al., 2016).
The activity responsible for the production of TGDG has previously been studied under the name galactolipid-galactolipid galactosyltransferase (GGGT) (Dorne et al., 1982; Heemskerk et al., 1983) and was later attributed to SFR2. Much of the initial work was performed in isolated chloroplasts from spinach (Spinacia oleracea), where TGDG accumulates in response to the chloroplast isolation procedure and to ozone fumigation (Dorne et al., 1982; Heemskerk et al., 1983, 1986; Sakaki et al., 1985, 1990). TGDG accumulation is also associated with the response to other processing stresses and environmental cues, including protoplast isolation in common bean (Phaseolus vulgaris) and Arabidopsis (Webb and Williams, 1984; Barnes et al., 2019) and chloroplast isolation and wounding in Arabidopsis (Vu et al., 2015). Increased amounts of TGDG and a concomitant reduction in MGDG are observed in the desiccation plants rock violet (Boea hygroscopica) and blue gem (Craterostigma plantagineum) during the transition to the desiccated form (Navari-Izzo et al., 1994; Sgherri et al., 1994; Gasulla et al., 2013). Moreover, drought and salinity stress cause TGDG accumulation in tomato (Solanum lycopersicum), which was directly attributed to SFR2 through the use of mutant lines, as in studies of severe cold in Arabidopsis (Wang et al., 2016).
Cold is a stress that is relative to each species and has multiple definitions. Chilling stress is a type of cold stress that is non-damaging. It induces physiological changes that then allow acclimation to additional cold. Sensing of initial chilling is well characterized and has been reviewed extensively (Thomashow, 1999, 2001, 2010). Briefly, initial chilling is sensed at the plasma membrane through a flux of calcium ions (Knight et al., 1991, 1996); the resulting calcium spike causes changes in gene expression, including that of the C-REPEAT/DRE BINDING FACTOR (CBF) and COLD-REGULATED (COR) transcription factor genes (Fowler and Thomashow, 2002; Catala et al., 2003; Chinnusamy et al., 2007; Doherty et al., 2009). These transcriptional changes produce alterations in cellular membranes (Zhao et al., 2016), the accumulation of solutes within the cell (McKown et al., 1996), and metabolic rebalancing (Schulze et al., 2012). Exposure to lower temperatures then induces severe cold stress, from a damaging level of cold. In freezing-tolerant plants, this includes freezing stress. Studies of severe low-temperature stress across multiple species have shown that sub-zero acclimation, consisting of a period of below-zero non-lethal temperature, is distinct from initial chilling sensing and prepares plants for additional damaging temperatures (Castonguay et al., 1993; Monroy et al., 1993; Livingston, 1996; Herman et al., 2006; Le et al., 2008, 2015; Espevig et al., 2011; Takahashi et al., 2019). Indeed, many of the changes that occur during sub-zero acclimation are different from those observed during initial chilling sensing and involve modulation of numerous genes not induced during initial cold treatment, as well as proteome- and cell wall-specific changes (Takahashi et al., 2019). Little is known about how severe cold is sensed or transduced. Our previous work identified cytosolic acidification as one signaling mechanism employed by Arabidopsis that results in TGDG accumulation (Barnes et al., 2016), but it is unknown whether other species use the same mechanism.
Studies of closely related species that grow in both temperate and tropical climates have allowed a better understanding of how the evolutionary origin of low-temperature tolerance shapes species distribution and defines their effective growth areas. Investigation of temperature tolerance in the Pooideae subfamily of the grasses indicated that tolerance to initial cold is potentially more ancient than tolerance to drought (McKeown et al., 2016; Das et al., 2021). Furthermore, severe cold tolerance appears to have evolved more recently than tolerance to initial cold (McKeown et al., 2016; Das et al., 2021). Differences from severe cold tolerance even exist at the subspecies level in maize, when highland and lowland maize landraces are compared (Barnes et al., 2022). An improved understanding of the evolution of severe cold signals will allow the engineering of increased tolerance in crops.
To explore the evolution of severe cold responses, we used TGDG production as a marker and investigated its activation through cellular acidification. We examined TGDG production across bryophytes, gymnosperms, and angiosperms under both routine growth conditions and severe cold. We show here that TGDG production correlates with temperature and cellular acidity in multiple species and compare transcriptional changes induced by severe cold and artificial acidification treatments.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana [Columbia-0 (Col-0)] plants were grown on a mixture of Sungrow Propagation Mix soil and Turface at 22 °C under a 16 h light/8 h dark photoperiod. Plants were grown for 3–4 weeks before cold acclimation at 6 °C under a 12 h light/12 h dark photoperiod for 1 week. Garden peas (Pisum sativum ‘Little Marvel’) were grown in vermiculite at 22 °C under a 16 h light/8 h dark photoperiod. Pea plants were grown for 2–3 weeks under normal conditions before acclimation at 6 °C under a 12 h light/12 h dark photoperiod for 1 week. Maize (Zea mays ‘B73’) and sorghum (Sorghum bicolor ‘BTx623’) were both grown in chambers under a 16 h light/8 h dark photoperiod with 29 °C during the day and 22 °C at night on standard greenhouse soil mix [8:8:3:1 (w/w/w/w) peat moss:vermiculite:sand:screened topsoil, with 7.5:1:1:1 (w/w/w/w) Waukesha fine lime, Micromax, Aquagro, and Green Guard per 0.764 m2]. Acclimation was carried out at 16 °C for 1 week for both maize and sorghum, and maize was additionally acclimated for 3 d at 6 °C. Wheat (Triticum aestivum ‘Overland’) was also grown in standard greenhouse soil at 21 °C under a 16 h light/8 h dark photoperiod before cold acclimation at 6 °C under a 12 h light/12 h dark photoperiod for 1 week. All plants for all treatments were moved into low-temperature stress at the end of their respective day for treatment in the dark.
Plants for phylogeny tests, excluding trees, were grown under greenhouse conditions at 24 °C on standard greenhouse soil mix, as described above, under a 12 h light/12 h dark photoperiod. Physcomitrium patens was grown on PpNH4 Moss Medium (Caisson Laboratories, Smithfield, UT, USA), and Spirodela polyrhiza was grown on Schenk and Hildebrandt (Sigma Aldrich, Inc., St. Louis, MO, USA) basal salt medium with pH adjusted to 5.8 using potassium phosphate. The moss and duckweed were grown at 22 °C under a 16 h light/8 h dark photoperiod in a growth chamber. Tree samples were collected from the University of Nebraska-Lincoln campus; leaves were sampled in the early autumn before any nights approaching or below freezing had occurred. Plants were obtained from greenhouses dedicated to teaching plant diversity at the University of Nebraska-Lincoln. Species that were not already grown by the University of Nebraska-Lincoln were obtained through cuttings or seeds from the USDA-GRIN database.
Lipid analysis
Lipids were extracted from plant tissues using a modified Bligh and Dyer method and TLC analysis as described (Wang and Benning, 2011). Lipids were loaded onto EMD60 plates (Millipore, Burlington, MA, USA) and subsequently resolved in a solvent system of chloroform:methanol:acetic acid:water (85:20:10:4, v/v/v/v) as described (Barnes et al., 2016). Samples frozen in water to match ion leakage low-temperature treatments were allowed 30 min of thawing so that the water could be removed, and the lipids extracted. α-Naphthol stain was used to detect galactolipids on the chromatogram as described (Wang and Benning, 2011).
TGDG contents were calculated relative to DGDG, as DGDG is not responsive to severe cold temperatures and makes up ~25% of most plant membranes. ImageJ with the FIJI plug-in densitometry function (Schindelin et al., 2012, 2015; Schneider et al., 2012) was used for quantification of DGDG and TGDG contents, with fractional TGDG/DGDG ratios based on the gray value of each TLC spot measured (Rouser et al., 1966).
Phylogenetic analysis of TGDG accumulation
SFR2-like proteins were identified using Phytozome and NCBI Blast. Relationships in the cladogram were based on version 13 of the Angiosperm Phylogeny Website (http://www.mobot.org/mobot/research/apweb/) and other available phylogenomic analyses (Wickett et al., 2014; Gitzendanner et al., 2018).
Plant material was sampled by taking six leaf discs of 8 mm in diameter. For plants with irregular leaves, such as needles or small fronds, tissue samples equivalent to the weight of six leaf discs were collected with a razor blade. Corresponding fresh and frozen samples were collected, with samples of each equivalent to six discs. Lipids from fresh samples (controls) were immediately extracted for lipid analysis as described above. ‘Severe cold-challenged’ samples were obtained from plants cooled gradually over 24 h from 6 °C to –20 °C in a refrigerated circulator (Fig. 1A). Several plant varieties were sourced from the U.S. National Plant Germplasm System.
Fig. 1.
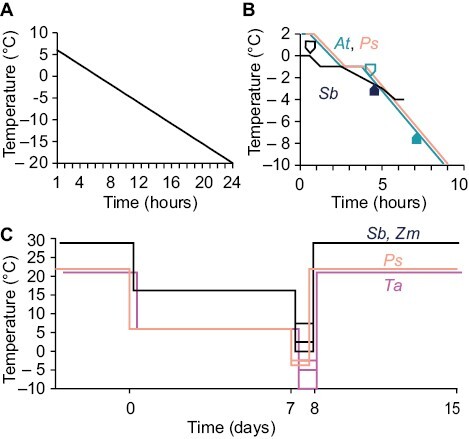
Schematics of low-temperature treatments. (A) Temperatures applied to samples for the plant phylogeny described in Fig. 2. All species received the same treatment. (B) Temperatures applied to Arabidopsis (At), pea (Ps), and sorghum (Sb) during ion leakage described in Figs 3 and 6. The line describing pea has been shifted to the right for clarity. Black arrows indicate sample treatment in At or Sb for comparison with artificial acidification in Fig. 6. (C) Temperature profiles applied to pea (Ps), sorghum (Sb), maize (Zm), and wheat (Ta) during whole-plant stresses described in Fig. 4. Lines for Sb, Zm, and Ta are shifted to the right for clarity.
Ion leakage
Plants used for ion leakage were grown as described above, and all plants were cold acclimated for 1 week at the appropriate temperature. Ion leakage was determined using a refrigerated circulator (AP15R-40, VWR, Radnor, PA, USA) with leaf pieces or punches floated onto 3 ml of ddH2O. For Arabidopsis, an entire leaf was used. For pea, a leaflet was used after removing the mid-vein. For sorghum, three leaf punches of 8 mm in diameter were used. Samples were collected from the second true leaf, except for Arabidopsis, where rosette leaves were sampled, with care taken to use older, expanded leaves and avoid cotyledons. Different plants were then chilled to temperatures sufficient to induce stress in the respective species. Stress was imposed on Arabidopsis as previously described (Warren et al., 1996), and similar conditions were used for pea. Briefly, samples were exposed to an initial equilibration at 2 °C for 30 min, nucleation was initiated with a ddH2O ice chip at –1 °C for 1 h, and subsequent chilling occurred at a rate of –2 °C h–1 (Fig. 1B). Samples for sorghum were collected at temperatures from 0 °C to –4 °C. Following a 30 min equilibration at 0 °C, samples were cooled from 0 °C to –1 °C at a rate of –2 °C h–1, and ice nucleation was initiated at –1 °C and held for 1 h before subsequent chilling at a rate of –1 °C h–1 from –1 °C to –3 °C and then –2 °C h–1 from –3 °C to –4 °C (Fig. 1B). The slower chilling between –1 °C and –3 °C allowed for sampling in 0.5 °C steps.
After chilling, leaf samples were incubated at 4 °C overnight. The temperature was then raised to room temperature, and the samples were shaken at 250 rpm for 15 min (Warren et al., 1996). This set of samples constituted the initial conductivity measurement, which was performed using an Accumet AB200 (Fisher Scientific, Hampton, NH, USA). For total conductivity measurements, samples were heated to 65 °C for 30 min to release all electrolytes, then cooled down to room temperature and shaken at 250 rpm for 15 min. To calculate leakage, the percentage relative to total ions was calculated, plotted, and fit to a sigmoidal curve (Warren et al., 1996). At each temperature point, an equivalent sample for lipid analysis and fractional TGDG accumulation was collected for all species. For Arabidopsis and sorghum, an additional and equivalent sample was also taken for transcriptome analysis.
Whole-plant freezing tests
Cold-acclimated soil-grown plants were treated as indicated in Fig. 1C. After cold acclimation, each species had multiple plants treated overnight at varying temperatures (pea, 0, −2, and −4°C; maize, 2 °C and 0 °C; sorghum, 2 °C and 0 °C; wheat, 0, −2, −5, and −10 °C). For all tests, ice chips were added on the soil surface to induce ice nucleation 1–2 h after being introduced to the treatment temperature if it was at or below 0°C. Leaf lipid samples for TGDG accumulation were collected from the second true leaf after 16 h of freezing by an extra-sharp double-sided razor blade directly into extraction buffer, extracted as described above, and analyzed via TLC. Any samples noted as damaged by pressure during sampling from evidence of green seepage from the leaf were discarded. Post-freezing recovery was performed by returning plants to a greenhouse at 24 °C for 1 week before photographs were taken.
Cytosolic acidification
All experiments were performed on excised leaves. For Arabidopsis, the leaf was placed into a cup of acid solution or water after having removed the leaf from the rosette of a full-sized 3-week-old plant. For pea, a leaflet was removed, gently scored with a razor blade on its abaxial side, and placed in a cup as above. A 20 mM acetic acid solution, adjusted to pH 5 with concentrated KOH, was used for acidification treatment along with water as a control, as described (Barnes et al., 2016). Treatments were conducted for 3 h. Leaves were patted dry before extracting lipids. For sorghum, the sorghum stalk above the soil surface was cut using a new razor blade for each plant and shoots were inserted into a tube containing 20 mM 2,4-dinitrophenol, pH 5, in 18.2% (v/v) methanol, adjusted with KOH, or into 18.2% (v/v) methanol/water as a control. These samples were immediately placed into a humidity chamber for 3 h with a minimum relative humidity of 84%. Leaf punches were taken from the second true leaf to mimic the samples used in ion leakage tests.
RNA-seq data generation and processing
Total RNA was isolated for each sample using a Zymo Quick-RNA Plant Mini-prep Kit (Zymo Research Corp, Irvine, CA, USA), and RNA-seq libraries were prepared according to Illumina TruSeq Sample Preparation V2 using 1 μg of starting total RNA. Libraries were sequenced using a 75 bp paired-end Illumina Miseq instrument at the University of Nebraska Medical Center. Raw reads were deposited in the NCBI SRA (Sequence Read Archive) database under the BioProject ID PRJNA894306. Trimmomatic 0.36 (Bolger et al., 2014) was used to remove low-quality reads and adapters using default parameters. The resulting clean reads from Arabidopsis and sorghum samples were aligned to the A. thaliana TAIR10 and S. bicolor v3.1 genomes, respectively (retrieved from Phytozome v12.0), using GSNAP (2018-03-25) (Wu and Nacu, 2010). Alignment files were converted to bam format using Samtools (v1.9) (Li et al., 2009) and used as input to HTSeq (0.6.1) (Anders et al., 2015) for generation of raw counts per gene.
Differentially expressed genes
The formula design ~=Replicate+Condition in DESeq2 was used to identify differentially expressed genes (DEGs) for each species for both artificial acidification and severe cold treatment using DESeq2 (Love et al., 2014). Two and three biological replicates were employed for the identification of DEGs during temporal and chemical treatment. Any gene in the condition factor (control versus treatment) with an adjusted P-value <0.05 and absolute log2 fold change >1 were classified as DEGs. Overall, four gene categories in each species were generated: up-regulated by chemical treatment; up-regulated by temperature treatment; down-regulated by chemical treatment; and down-regulated by temperature treatment. To identify differentially expressed orthologs between Arabidopsis and sorghum, a list of corresponding orthologs between each sorghum gene model and the best hit Arabidopsis gene models was retrieved from Phytozome v12.0. To compare with randomly overlapping orthologs in either treatment, only genes with 1:1 orthologs between sorghum and Arabidopsis were considered as background. Sorghum genes in each category were assigned to the best hit Arabidopsis gene models. To determine the significance of co-up-regulated or co-down-regulated orthologs between Arabidopsis and sorghum, we randomly picked the equal number of up-regulated or down-regulated genes in Arabidopsis, respectively, and tested the number of genes with orthologs in sorghum. The F-test was used to determine significance between real overlapping ortholog numbers and permutated overlapping ortholog numbers. In total, 100 permutations were performed.
Gene Ontology analysis
Lists of genes obtained from DEG analysis were analyzed via the Gene Ontology resource (Ashburner et al., 2000; Mi et al., 2019). GO enrichment analysis was performed, and significantly enriched categories were reported (Gene Ontology Consortium, 2021).
Results
TGDG production has multiple patterns in land plants
In Arabidopsis, SFR2 is always present (Thorlby et al., 2004; Barnes et al., 2016), and its activation leads to TGDG production in response to low-temperature stress. To better understand the similarity of severe cold responses across a set of diverse plant species, we analyzed TGDG accumulation in 43 species after identical low-temperature exposure (Fig. 1). Species were chosen for their economic importance, availability of sequenced genomes, or phylogenetic origin, and all 43 species are represented in a phylogenetic tree based on plant evolution (Fig. 2A).
Fig. 2.
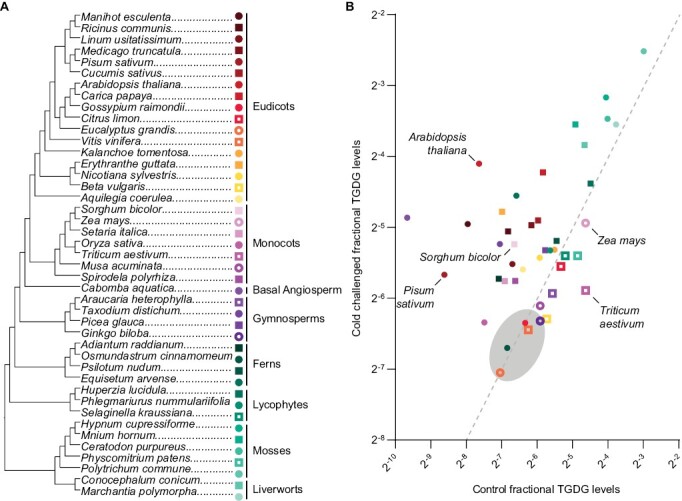
Most land plants accumulate TGDG, including species with high cold susceptibility. (A) Phylogenetic tree of plant evolution based on version 13 of the Angiosperm Phylogeny website and published phylogenetic trees. Colors indicate to which plant group each species belongs: red, orange, and yellow, eudicots; magenta, monocots; violet, basal angiosperms; plum, gymnosperms; green, ferns; teal, lycophytes; turquoise, mosses; aqua, liverworts. Symbols alternate circle and square for ease of visibility. Open symbols denote species with more fractional TGDG in control samples, and filled symbols denote species with more fractional TGDG in cold-challenged samples. (B) TGDG/DGDG ratio plotted in logarithmic scale for each species described in (A). Lipids were quantified in freshly sampled leaves (control, x-axis) or leaves after 24 h of cold challenge defined in Fig. 1 (cold stressed, y-axis). A gray oval indicates no observable TGDG; values in this area indicate the limit of detection in each species. The same color scheme is used in both panels.
Quantification of TGDG amounts allowed observation of four patterns of TGDG accumulation (Fig. 2B). Production of TGDG was below the detection limit for both control and cold-challenged samples in some species (Fig. 2B, shaded oval). These species did not appear to group clearly across phyla or relative cold hardiness. The second pattern of TGDG accumulation is from species tending to have low TGDG content in control samples and a substantial increase in TGDG content in cold-challenged samples (Fig. 2B). This pattern of TGDG accumulation appeared in some species of eudicots, monocots, basal angiosperms, ferns, and lycophytes. In the third pattern, TGDG contents were higher in control samples than in those challenged with cold (Fig. 2B, open symbols). The amount of TGDG detected under control conditions in these species, which included wheat (T. aestivum) and maize, may be caused by wounding damage of the leaf punch during sample collection, masking any increase in TGDG during low-temperature challenge. The species having this pattern of TGDG accumulation had no discernible similarities across phylogeny or cold hardiness. The final pattern of TGDG accumulation was characterized by high amounts of TGDG in both control and cold-challenged samples. Many species exhibiting this pattern were the most anciently diverged lineages of land plants such as mosses and liverworts (Fig. 2B). Among all phylogenetic groups, the angiosperms (eudicots, monocots, and basal angiosperms) appear to have the strongest differences in TGDG accumulation patterns between species (Fig. 2, red/orange/yellow/pink/purple).
Total cellular damage corresponds to TGDG accumulation
The presence of TGDG in control samples in Fig. 2 suggested that TGDG may accumulate in response to cellular damage caused by the mechanical wounding from taking a leaf punch. We hypothesized that severe cold stress might also cause sufficient cellular damage to trigger TGDG accumulation, rather than directly inducing the formation of this class of lipids. To assess the association between cellular damage and severe cold stress, we measured ion leakage, a quantitative measure of cell damage (Demidchik et al., 2014), in parallel with TGDG content in Arabidopsis, pea, and sorghum. Prior to severe cold stress, plants were cold acclimated at a temperature appropriate for each species. We then transferred leaf pieces or discs of cold-acclimated plants to a continuous ramp of severe cold in a refrigerated circulator and determined ion leakage and lipid contents at each temperature (Fig. 3).
Fig. 3.
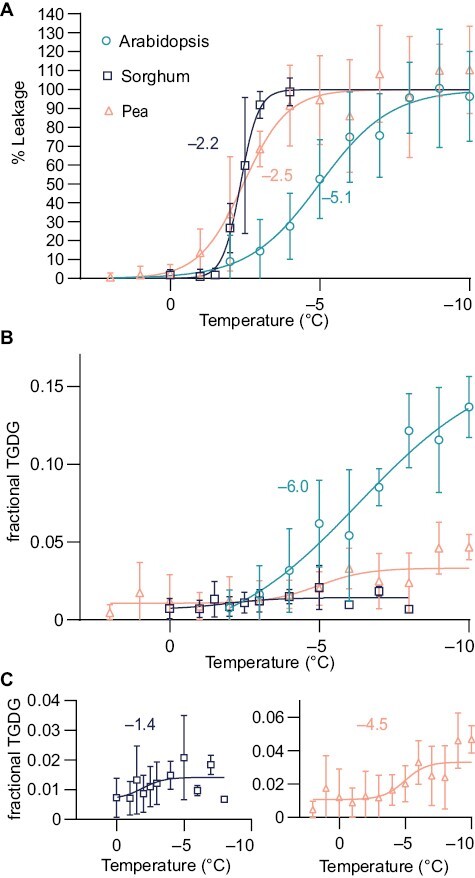
Severe low-temperature damage corresponds with TGDG accumulation. (A) Ion leakage curves for Arabidopsis, sorghum, and pea. Data are shown as means ±SD. Arabidopsis, n=8; pea, n=4–7; sorghum, n=6. The inflection point of each curve fit is noted. (B) Fractional TGDG/DGDG ratios for leaf samples treated as in (A). Data are shown as means ±SD. The inflection point of the Arabidopsis curve fit is shown. (C) A reproduction of the data shown in (B) of sorghum and pea fractional TGDG/DGDG ratios with adjusted y-axis values and the corresponding inflection point for each species. Arabidopsis, n=6; pea, n=5–15; sorghum, n=5. Additional temperature samples for line fit of sorghum were taken from –5 °C to –8 °C and are shown (n=2). Additional samples (n=8) were taken at –20 °C for pea to improve curve fitting; these are not shown.
The temperature at which 50% cellular leakage occurs, termed LT50, varied for each species and reflected their low-temperature tolerance (Fig. 3A). Arabidopsis results were similar to previously published outcomes (Warren et al., 1996), showing partial leakage from temperatures ranging from –2 °C to –10 °C (Fig. 3A) and an LT50 of –5.1 °C. In pea, all temperatures below freezing induced some cellular damage, as in Arabidopsis (Fig. 3A), but LT50 was reached earlier at around –2.5 °C. Sorghum leakage occurred rapidly, and exhibited ion leakage values close to total ion leakage at the range of temperatures tested (Fig. 3A), resulting in an LT50 of –2.2 °C.
The speed of the ion leakage assay (Fig. 1) provided an opportunity for direct comparisons between TGDG production and the extent of membrane damage, which we assessed at each temperature point. Final TGDG amounts were highest in Arabidopsis, while in sorghum and pea, accumulation occurred more modestly, with the amounts of TGDG nearly 10-fold lower than those observed in Arabidopsis (Fig. 3B). The amount of TGDG occurred in a similar order to the relative cold tolerance levels of the species, and all species accumulated TGDG within the narrow time frame of the experiment. The inflection points of TGDG accumulation were near the inflection points of ion leakage. The inflection point of TGDG accumulation was –6.0 °C (Fig. 3B) in Arabidopsis, –1.4 °C in sorghum (Fig. 3C), and –4.5 °C in pea (Fig. 3C). These results indicate a tight association between membrane damage and TGDG accumulation, as all species induced TGDG accumulation in the same temperature and time range as membrane damage occurred.
Whole-plant low-temperature treatments suggest that TGDG accumulation in freshly collected excised leaves results from wounding
Because damage may have a role in the severe cold response, we wished to use whole plants to ask if species with TGDG levels higher in control samples than in those challenged with cold in Fig. 2 (open symbolss) accumulate TGDG in response to severe cold. To accurately encompass the variety and divergence of TGDG accumulation patterns illustrated in Fig. 2B, we selected pea (P. sativum), maize (Z. mays), sorghum (S. bicolor), and wheat (T. aestivum). Pea had one of the highest relative accumulations of TGDG in the cold. Maize and sorghum are closely related and had similar TGDG contents, with one accumulating more TGDG in cold-challenged samples and one in control samples. Wheat was selected as it had one of the highest accumulations of TGDG in control samples. Each of these species is also of economic importance and has established growth conditions.
Plants were cold-acclimated prior to freezing at temperatures appropriate for their low-temperature tolerance: 6 °C for pea and wheat and 16 °C for maize and sorghum. Each freezing schedule was also adjusted to match the cold tolerance of the species. Pea, maize, and sorghum each died at temperatures 1 °C lower than shown (Fig. 4A). Wheat died at –10 °C. We exposed plants to cold stress at the end of the light cycle, which is when plants normally exhibit their maximal cold tolerance levels (Raju et al., 2018), thus mirroring the overnight conditions when freezing would be experienced in a field. Pea and wheat were the most tolerant of freezing of the four species, as they both survived exposure to below-freezing temperatures (Fig. 4A), despite being the most disparate in TGDG accumulation in their control and cold-challenged samples (Fig. 2). In all plants, TGDG accumulation increased in response to cold challenge (Fig. 4B,C). We confirmed that wheat, maize, and sorghum were very sensitive to wounding stress and that the sample collection method was important. Even when using whole plants, we detected higher amounts of TGDG in samples collected during control growth conditions if there was any leaf damage during lipid sampling. As seen in Fig. 3, TGDG contents also scaled with low-temperature tolerance. Wheat was the most tolerant species assayed and had the most TGDG accumulation. Sorghum and maize were the least tolerant species and accumulated less TGDG.
Fig. 4.
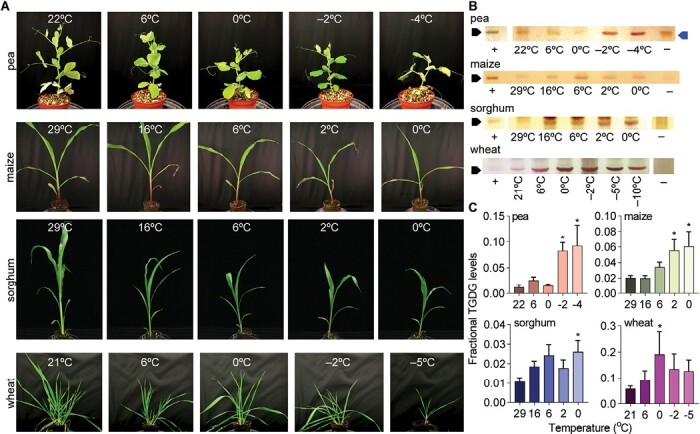
TGDG accumulates in response to low-temperature stress of whole plants. (A) Representative photographs of plants grown at the indicated temperatures and (B) corresponding thin-layer chromatograms stained with α-naphthol for galactose visualization for pea, maize, sorghum, and wheat. Black arrows indicate the position of TGDG; the blue arrow indicates a pigment that is not TGDG; +, positive control of frozen Arabidopsis lipids; –, negative control of freshly sampled Arabidopsis lipids without TGDG accumulation. (C) Fractional TGDG/DGDG amounts are quantified for each species with n≥3 biological replicates for each species. Averages are indicated with SEM error bars; asterisks indicate a difference between control temperatures by ANOVA corrected for multiple comparisons with Bonferroni’s method.
Cytosolic acidification mimics a freezing response in Arabidopsis, pea, and sorghum
Next, we asked whether TGDG accumulation is activated by similar mechanisms in TGDG-accumulating angiosperm species that differ in their ability to tolerate cold. We previously developed a protocol that artificially acidified the cytosol to mimic the lipid response and pH change that occurs during an overnight freezing test (Barnes et al., 2016). We thus excised whole leaves from pea and leaf pieces from sorghum, and treated them with low levels of organic acids, processing Arabidopsis plate-grown rosettes concomitantly as a control. In both Arabidopsis and pea, TGDG accumulated in response to acidification within 120 min (Fig. 5A, B, D).
Fig. 5.
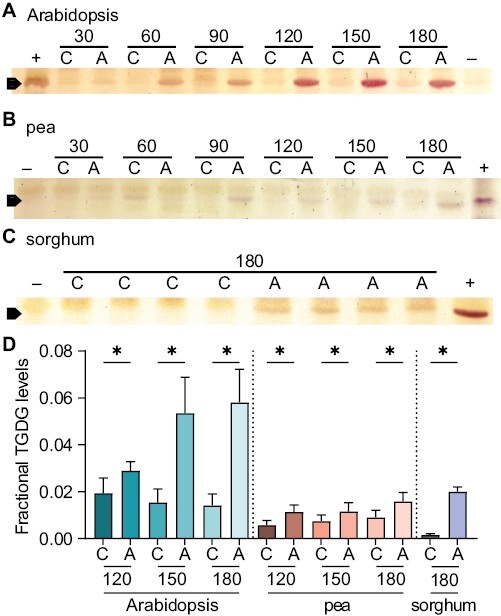
TGDG accumulates in Arabidopsis, pea, and sorghum upon acidification. Thin-layer chromatograms stained with α-naphthol to identify galactose head groups. (A) Arabidopsis, (B) pea, and (C) sorghum leaves were treated and collected at the number of minutes indicated above. ‘A’ denotes acidification by treatment with 20 mM acetic acid, pH 5 for Arabidopsis and pea or 20 mM 2,4-dinitrophenol, pH 5 in 18.2% methanol for sorghum; ‘C’ denotes control treatment of water for Arabidopsis and pea and 18.2% methanol for sorghum; +, positive control of frozen Arabidopsis lipids; –, negative control of freshly sampled Arabidopsis lipids without TGDG accumulation. (D) Quantification of the fractional TGDG/DGDG levels of the data shown and replicates, n≥6. Averages are indicated with SEM error bars, and asterisks indicating statistical significance in acidified versus control conditions by t-test with P<0.05. Times below 120 min were not statistically significant in pea or sorghum and are not shown.
In sorghum, sampling the leaves by razor blade or leaf punch caused TGDG accumulation in control samples after just 30 min. We reasoned that the considerable amounts of wax on sorghum leaves (Traore et al., 1989) might prevent them from taking up acid or water. We then transitioned to use whole excised shoots placed vertically to treat the sorghum, and a more hydrophobic organic acid, 2,4-dinitrophenol. After 180 min, we routinely observed TGDG accumulation in the acid-treated sorghum and none in the control (Fig. 5C, D). Thus, cytosolic acidification appears to activate TGDG accumulation in multiple species.
Severe cold and acidification treatments invoke overlapping transcriptome responses
We used our new assay for sorghum acidification to ask if sorghum and Arabidopsis would have any similarities in non-TGDG responses to the stimuli of severe cold or acidification. We already knew that lipid changes induced by low-temperature and acid treatments are similar in Arabidopsis (Barnes et al., 2016) and wanted to separate the response to severe cold challenge and response to acid treatment to identify any overlap between treatments or between species. Thus, we exposed Arabidopsis to 0 °C or –7 °C and sorghum to 0 °C or –2.5 °C through a quick ramp in temperature as for the ion leakage samples (Fig. 1). In parallel, we exposed plants grown simultaneously to the acidification and control treatments defined above. We then collected samples for RNA-seq analysis. We identified significant DEGs as any gene in the treatment group with an adjusted P-value <0.05 and an absolute log2 fold change >1 relative to the control group. We first defined eight groups: up-regulated or down-regulated in response to severe cold or when acidified in Arabidopsis (Fig. 6A), and the same four groups in sorghum (Fig. 6B). There was a statistically significant set of genes that were enriched in response to severe cold and acidification when either up- or down-regulated in each species.
Fig. 6.
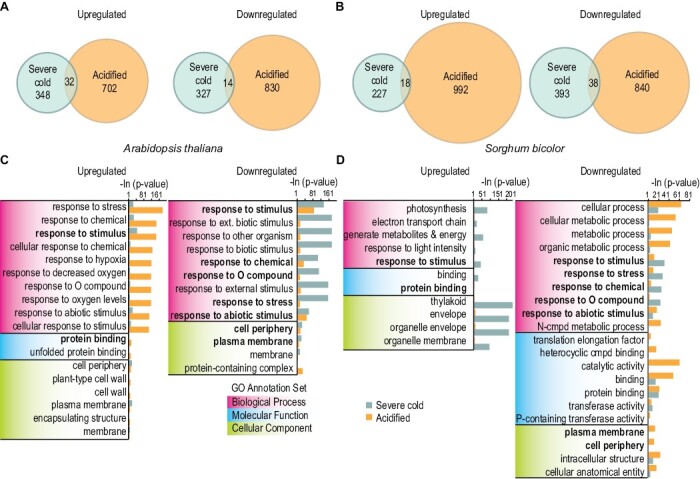
Severe cold treatment and acidification have overlapping responses in Arabidopsis and sorghum. Venn diagrams representing the number of DEGs separated into direction of regulation in response to severe cold (blue) or acid (orange) treatments and the overlap between the two for (A) Arabidopsis and (B) sorghum. DEGs were tested for GO enrichment in biological process, molecular function, or cellular component ontologies separately for both (C) Arabidopsis and (D) sorghum. The top 10 GO terms in each ontology with statistically relevant enrichments for both severe cold and acidification are reported. P-values are given as the negative natural log. GO categories that are enriched for in both species in response to severe cold and acidification are indicated in bold.
We attempted to define any genes enriched in response to both cold and acidification in both species using orthologous gene sets. There were overlaps between all single stress categories (i.e. up-regulation in response to cold in Arabidopsis and sorghum); however, no overlap was detected between the two species’ responses to both stresses, probably due to the small numbers of genes and the difficulty in identifying homologs between sorghum and Arabidopsis. Thus we investigated the categories of genes affected in both stresses and both species. To do so, we further separated the DEGs into eight groups by up-regulation and down-regulation in response to treatment. The eight groups were then used to identify GO terms of enrichment for each species (Fig. 6C, D). Complete lists of the GO term categories can be found in Supplementary Tables S1–S8. We identified multiple GO terms that were significantly enriched in both Arabidopsis and sorghum, under both severe cold and acid treatments (Fig. 6C, D, bold), and many more that were species specific. Importantly, we detected no significant enrichment for GO term categories in biological processes that were related to initial chilling responses such as response to cold, cold acclimation, and response to temperature stimulus among DEGs (Barah et al., 2013). Instead, response to stresses, oxygen-containing compounds, stimulus, and chemicals were significantly enriched in response to both treatments; the same GO categories are enriched in response to wounding and other stresses that cause cellular damage (Reymond et al., 2000; Ding et al., 2013; Mata-Pérez et al., 2015). GO terms for the molecular function and cellular component ontologies, protein binding, cellular periphery, and plasma membrane were significantly enriched in both species. This indicates the importance of membrane dynamics in response to cellular damage and stress. To summarize, there are similarities in non-TGDG responses to the stimuli of severe cold or acidification in multiple species.
Discussion
SFR2 is ubiquitous across the plant kingdom (Fourrier et al., 2008), raising the possibility that its product TGDG might also be. Our phylogenetic analysis of TGDG production in leaf discs indicated that most plants synthesize and accumulate TGDG (Fig. 2), with the most ancestrally diverged species displaying the highest contents of TGDG. It is possible that TGDG production is as ubiquitous as the presence of SFR2, though in some phylogenetically diverse species it remains below the detection limit in control and severe temperature challenge. Other species, including maize and wheat, appeared to accumulate higher amounts of TGDG under control conditions compared with severe cold challenge treatment, which prompted us to test the role of tissue damage, which probably occurred during sample preparation. We compared TGDG production with the extent of membrane damage through ion leakage assays in sorghum, pea, and Arabidopsis (Fig. 3). These assays showed tight associations between membrane damage and accumulation of TGDG. We then asked if a subset of species from the initial screen with high accumulations of TGDG in control conditions also accumulated TGDG in response to whole-plant cold challenge (Fig. 4). They did, and the scale of response matched well with the level of the species’ cold tolerance. This implied that a similar mechanism of activation may be used in all species. Tests of pea, sorghum, and Arabidopsis all showed that acidification can trigger TGDG accumulation independently of temperature (Fig. 5) (Barnes et al., 2016). Finally, we detected significant overlaps between DEGs and their associated pathways after acidifying or severe cold treating Arabidopsis and sorghum (Fig. 6). Many of these DEGs are not related to initial cold responses and instead are associated with more general cellular stresses and membrane dynamics. Together, this work suggests that the TGDG severe cold response has evolved from an ancestral response, the strength of the response has been modified multiple times in each phylum, and acidification of the cytosol plays a role in sensing severe cold.
Multiple studies have harnessed plant diversity and phylogeny to understand the loss and gain of tolerance mechanisms to abiotic stresses such as drought and salinity (Bromham et al., 2020; Marks et al., 2021). Costa and colleagues reported that drought stress imposed on vegetative tissue co-opts and reprograms some of the mechanisms used during seed desiccation (Costa et al., 2017). Drought tolerance of vegetative tissues has arisen separately multiple times, suggesting that this trait may emerge from the modification of regulatory regions of existing genes (VanBuren, 2017). An examination of the phylogeny of salt tolerance reveals a unique pattern. Rather than clusters of salt-tolerant species within families, these species are often located at the tips of a phylogeny, with few common relatives also exhibiting salt tolerance (Bromham et al., 2020). The authors suggest that such a pattern may originate due to one of three potential reasons: (i) a recent environmental change caused the gain of salt tolerance; (ii) salt tolerance is a highly labile trait with frequent loss and frequent gain; or (iii) the trait is quick to arise and has a high extinction rate. Currently, our understanding of severe low-temperature tolerance is consistent with each of these hypotheses. The angiosperms (Fig. 2, eudicots, monocots, and basal angiosperms) include species that produce high amounts of TGDG in response to cold (e.g. A. thaliana, Cabomba aquatica) and no detectable TGDG (Gossypium raimondii, Ginko biloba), as do the ferns (high, Equisetum arvense, low, Osmundastrum cinnamomeum). This is consistent with multiple separate evolutions or losses of severe cold tolerance, and may parallel the hypothesis of drought tolerance emergence from modification of existing genes involved in damage. Similarly, TGDG accumulations in response to cold varied widely between species within each phyla (Fig. 2), an observation that was confirmed within angiosperms by whole-plant cold challenge assays (Fig. 4). This is consistent with the salt tolerance hypotheses for trait lability. Future work could dissect patterns of response further within phyla, as we show that there are differences in TGDG accumulation in each.
The results presented here expand our initial understanding of TGDG accumulation in response to severe cold by demonstrating that TGDG content broadly scales with plant species’ cold tolerance levels and coincides with cellular damage. We previously reported in Arabidopsis that TGDG can accumulate in response to severe cold (Barnes et al., 2016). In Fig. 2, we tested a wide range of species, finding that some species accumulate more TGDG in control samples than in response to cold challenge. Figure 3 established a reduced time scale experiment, within which multiple species, Arabidopsis, sorghum, and pea, accumulate TGDG as membranes are damaged. Under control, whole-plant, growth conditions, and species-specific cold challenges, pea, maize, sorghum, and wheat all accumulated TGDG in response to cold challenge (Fig. 4). The temperature at which TGDG began to accumulate matched the cold tolerance limits of the species, with maize and sorghum accumulating TGDG at temperatures that would not be considered severe cold by Arabidopsis or pea plants. Wheat and maize had higher contents of TGDG in control treatments (Fig. 4C), which further increased when cold challenged. This finding contrasted with results shown in Fig. 2B, in which wheat and maize accumulated more TGDG in control than in cold challenge. Experimental differences between these two sets of cold challenges (Fig. 1) explain the accumulation of TGDG in response to cold. The tight association between damage and TGDG production (Fig. 3) suggests that the difference in TGDG accumulation in the control samples (Figs 2, 4) is probably in response to damage caused by leaf punching. The central vacuole and the apoplastic spaces of plant leaves are approximately pH 6 (Gao et al., 2004; Martinière et al., 2013), making them both reservoirs of acidity. Other work supports that when tissues are damaged by pathogens, acidification occurs (Lebrun-Garcia et al., 1999; Roos et al., 2006). This idea is supported by our previous observation that wounding changes cytosolic pH and initiates TGDG accumulation (Vu et al., 2015). Together, these data suggest that a component of the severe cold response is a response to membrane damage, both of which promote TGDG accumulation.
Membrane damage and cytosolic acidification seem to be consistent factors uniting the multiple stresses that activate SFR2 in angiosperms accumulating TGDG. Previous data from multiple reports have indicated that TGDG accumulates in response to various membrane-damaging stresses such as ozone treatment of spinach (Sakaki et al., 1985, 1990), salt and drought stress of tomato (Wang et al., 2016), and protoplast isolation from fava bean (Vicia faba) or Arabidopsis (Webb and Williams, 1984; Barnes et al., 2019). Our previous investigation revealed that TGDG accumulation promoted by protoplast isolation is pH dependent (Barnes et al., 2019). Membrane damage occurs in each of these stresses, suggesting a broad similarity in mechanisms across species. Many of the species we tested here showed an increase in TGDG amounts in response to severe cold, and at least a subset of these increases appeared to be associated with damage (Fig. 2B). Based on this information, we suggest that TGDG accumulation after a membrane-damaging event is the ancestral state.
The specifics of how membrane damage and cytosolic acidification are linked to TGDG accumulation remain unknown. The abundance of SFR2, the enzyme that produces TGDG, does not increase in response to severe cold and is not induced by either temperature or pH when tested in a heterologous system (Roston et al., 2014); therefore, post-transcriptional mechanisms must connect damage and cytosolic acidification to TGDG accumulation. The same mechanisms may underlie the overlapping DEGs responding to severe cold or acidification of Arabidopsis and sorghum (Fig. 6A, B). Interestingly, many of the gene categories were associated with response to multiple types of stress, but not cold specifically (Fig. 6C, D). This observation suggests a core response to these two stresses.
We used the prevalence of oligogalactolipid accumulation as a tool to better understand the evolution of plant cold tolerance. We linked cytosolic acidification with activation of a severe cold response including TGDG accumulation (Fig. 5) and multiple transcriptional changes (Fig. 6). We correlated TGDG accumulation with membrane damage over a short time window (Fig. 3). We also showed that TGDG accumulation in response to severe low temperature varies in each phyla investigated, probably originating from an ancestral state in which TGDG production was relatively higher (Fig. 2). Our results expand our understanding of severe low temperature tolerance across plant species, setting the stage for pursuing many more fundamental questions in stress signaling.
Supplementary data
The following supplementary data are available at JXB online.
Table S1. GO terms for Arabidopsis DEGs down-regulated in response to acidification.
Table S2. GO terms for Arabidopsis DEGs up-regulated in response to acidification.
Table S3. GO terms for Arabidopsis DEGs down-regulated in response to cold.
Table S4. GO terms for Arabidopsis DEGs up-regulated in response to cold.
Table S5. GO terms for sorghum DEGs down-regulated in response to acidification.
Table S6. GO terms for sorghum DEGs up-regulated in response to acidification.
Table S7. GO terms for sorghum DEGs down-regulated in response to cold.
Table S8. GO terms for sorghum DEGs up-regulated in response to cold.
Acknowledgments
We would like to acknowledge Samantha Link and Kandy Hanthorn for care and maintenance of the plants used in these studies. We further acknolwedge Zachery D. Shomo and Ngoc Pham Thien Thao for useful discussion.
Glossary
Abbreviations
- DEG
differentially expressed gene
- DGDG
digalactosyldiacylglycerol
- GO
Gene Ontology
- MGDG
monogalactosyldiacylglycerol
- SFR2
SENSITIVE TO FREEZING 2
- TGDG
trigalactosyldiacylglycerol
Contributor Information
Allison C Barnes, Department of Biochemistry, University of Nebraska-Lincoln, Lincoln, NE, USA.
Jennifer L Myers, Department of Biochemistry, University of Nebraska-Lincoln, Lincoln, NE, USA; Department of Horticulture, North Carolina State University, Raleigh, NC, USA.
Samantha M Surber, Department of Biochemistry, University of Nebraska-Lincoln, Lincoln, NE, USA.
Zhikai Liang, Department of Agronomy and Horticulture, University of Nebraska-Lincoln, Lincoln, NE, USA.
Jeffrey P Mower, Department of Agronomy and Horticulture, University of Nebraska-Lincoln, Lincoln, NE, USA.
James C Schnable, Department of Agronomy and Horticulture, University of Nebraska-Lincoln, Lincoln, NE, USA.
Rebecca L Roston, Department of Biochemistry, University of Nebraska-Lincoln, Lincoln, NE, USA.
Yuki Nakamura, RIKEN Center for Sustainable Resource Science, Japan.
Author contributions
ACB and RLR: design; ACB, SMS, and JLM: carrying out the experiments; all authors analyzed the results; ACB and RLR: writing the manuscript; all authors edited the manuscript.
Conflict of interest
JCS has equity interests in Data2Bio, LLC; Dryland Genetics Co; and EnGeniousAg LLC. He is a member of the scientific advisory board of GeneSeek. We have no other conflicts to disclose.
Funding
This project was supported by an National Science Foundation (NSF) IOS-1845175 grant to RR and United States Department of Agriculture (USDA) NIFA Predoctoral Fellowship 2018-67011-28008 and NSF-PGRP Fellowship 2010703 to ACB. It was partially supported by the Nebraska Agricultural Experiment Station with funding from the Hatch Multistate Research capacity funding program (accession number NC1200) from the USDA National Institute of Food and Agriculture. We would also like to acknowledge support from the University of Nebraska DNA Sequencing Core from the National Institute for General Medical Science (NIGMS) INBRE P20GM103427-14 and COBRE 1P30GM110768-01 grants, as well as The Fred & Pamela Buffett Cancer Center Support Grant P30CA036727 which supported the transcriptional analysis.
Data availability
The sequencing data that support the findings of this study are available in the NCBI SRA (Sequence Read Archive) database under the BioProject ID PRJNA894306. The full GO term analyses are available in the supplementary data.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barah P, Jayavelu ND, Rasmussen S, Nielsen HB, Mundy J, Bones AM. 2013. Genome-scale cold stress response regulatory networks in ten Arabidopsis thaliana ecotypes. BMC Genomics 14, 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AC, Benning C, Roston RL. 2016. Chloroplast membrane remodeling during freezing stress is accompanied by cytoplasmic acidification activating SENSITIVE TO FREEZING2. Plant Physiology 171, 2140–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AC, Elowsky CG, Roston RL. 2019. An Arabidopsis protoplast isolation method reduces cytosolic acidification and activation of the chloroplast stress sensor SENSITIVE TO FREEZING 2. Plant Signaling & Behavior 14, 1629270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AC, Rodríguez-Zapata F, Juárez-Núñez KA, et al. 2022. An adaptive teosinte mexicana introgression modulates phosphatidylcholine levels and is associated with maize flowering time. Proceedings of the National Academy of Sciences, USA 119, e2100036119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromham L, Hua X, Cardillo M. 2020. Macroevolutionary and macroecological approaches to understanding the evolution of stress tolerance in plants. Plant, Cell & Environment 43, 2832–2846. [DOI] [PubMed] [Google Scholar]
- Castonguay Y, Nadeau P, Laberge S. 1993. Freezing tolerance and alteration of translatable mRNAs in alfalfa (Medicago sativa L.) hardened at subzero temperatures. Plant and Cell Physiology 34, 31–38. [Google Scholar]
- Catala R, Santos E, Alonso JM, Ecker JR, Martinez-Zapater JM, Salinas J. 2003. Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. The Plant Cell 15, 2940–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu J-K. 2007. Cold stress regulation of gene expression in plants. Trends in Plant Science 12, 444–451. [DOI] [PubMed] [Google Scholar]
- Costa M-CD, Artur MAS, Maia J, et al. 2017. A footprint of desiccation tolerance in the genome of Xerophyta viscosa. Nature Plants 3, 17038. [DOI] [PubMed] [Google Scholar]
- Das A, Prakash A, Dedon N, Doty A, Siddiqui M, Preston JC. 2021. Variation in climatic tolerance, but not stomatal traits, partially explains Pooideae grass species distributions. Annals of Botany 128, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Straltsova D, Medvedev SS, Pozhvanov GA, Sokolik A, Yurin V. 2014. Stress-induced electrolyte leakage: the role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. Journal of Experimental Botany 65, 1259–1270. [DOI] [PubMed] [Google Scholar]
- Ding Y, Liu N, Virlouvet L, Riethoven J-J, Fromm M, Avramova Z. 2013. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biology 13, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. 2009. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. The Plant Cell 21, 972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorne A-J, Block MA, Joyard J, Douce R. 1982. The galactolipid:galactolipid galactosyltransferase is located on the outer surface of the outer membrane of the chloroplast envelope. FEBS Letters 145, 30–34. [Google Scholar]
- Espevig T, DaCosta M, Hoffman L, Aamlid TS, Tronsmo AM, Clarke BB, Huang B. 2011. Freezing tolerance and carbohydrate changes of two Agrostis species during cold acclimation. Crop Science 51, 1188–1197. [Google Scholar]
- Fourrier N, Bédard J, Lopez-Juez E, Barbrook A, Bowyer J, Jarvis P, Warren G, Thorlby G. 2008. A role for SENSITIVE TO FREEZING2 in protecting chloroplasts against freeze-induced damage in Arabidopsis. The Plant Journal 55, 734–745. [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. 2002. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. The Plant Cell 14, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Knight MR, Trewavas AJ, Sattelmacher B, Plieth C. 2004. Self-reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiology 134, 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasulla F, vom Dorp K, Dombrink I, Zähringer U, Gisch N, Dörmann P, Bartels D. 2013. The role of lipid metabolism in the acquisition of desiccation tolerance in Craterostigma plantagineum: a comparative approach. The Plant Journal 75, 726–741. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. 2021. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Research 49, D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitzendanner MA, Soltis PS, Wong GKS, Ruhfel BR, Soltis DE. 2018. Plastid phylogenomic analysis of green plants: a billion years of evolutionary history. American Journal of Botany 105, 291–301. [DOI] [PubMed] [Google Scholar]
- Heemskerk JW, Bögemann G, Wintermans JFGM. 1983. Turnover of galactolipids incorporated into chloroplast envelopes: an assay for galactolipid:galactolipid galactosyltransferase. Biochimica et Biophysica Acta 754, 181–189. [Google Scholar]
- Heemskerk JWM, Wintermans JFGM, Joyard J, Block MA, Dorne A-J, Douce R. 1986. Localization of galactolipid:galactolipid galactosyltransferase and acyltransferase in outer envelope membrane of spinach chloroplasts. Biochimica et Biophysica Acta 877, 281–289. [Google Scholar]
- Herman EM, Rotter K, Premakumar R, Elwinger G, Bae H, Ehler-King L, Chen S, Livingston DP 3rd. 2006. Additional freeze hardiness in wheat acquired by exposure to –3 °C is associated with extensive physiological, morphological, and molecular changes. Journal of Experimental Botany 57, 3601–3618. [DOI] [PubMed] [Google Scholar]
- Hugly S, Somerville C. 1992. A role for membrane lipid polyunsaturation in chloroplast biogenesis at low temperature. Plant Physiology 99, 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ. 1991. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352, 524–526. [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR. 1996. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. The Plant Cell 8, 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MQ, Engelsberger WR, Hincha DK. 2008. Natural genetic variation in acclimation capacity at sub-zero temperatures after cold acclimation at 4 °C in different Arabidopsis thaliana accessions. Cryobiology 57, 104–112. [DOI] [PubMed] [Google Scholar]
- Le MQ, Pagter M, Hincha DK. 2015. Global changes in gene expression, assayed by microarray hybridization and quantitative RT-PCR, during acclimation of three Arabidopsis thaliana accessions to sub-zero temperatures after cold acclimation. Plant Molecular Biology 87, 1–15. [DOI] [PubMed] [Google Scholar]
- Lebrun-Garcia A, Bourque S, Binet MN, Ouaked F, Wendehenne D, Chiltz A, Schaffner A, Pugin A. 1999. Involvement of plasma membrane proteins in plant defense responses. Analysis of the cryptogein signal transduction in tobacco. Biochimie 81, 663–668. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang R, Li M, Li L, Wang C, Welti R, Wang X. 2008. Differential degradation of extraplastidic and plastidic lipids during freezing and post-freezing recovery in Arabidopsis thaliana. Journal of Biological Chemistry 283, 461–468. [DOI] [PubMed] [Google Scholar]
- Livingston DP III. 1996. The second phase of cold hardening: freezing tolerance and fructan isomer changes in winter cereal crowns. Crop Science 36, 1568–1573. [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons JM. 1973. Chilling injury in plants. Annual Review of Plant Physiology 24, 445–466. [Google Scholar]
- Marks RA, Farrant JM, Nicholas McLetchie D, VanBuren R. 2021. Unexplored dimensions of variability in vegetative desiccation tolerance. American Journal of Botany 108, 346–358. [DOI] [PubMed] [Google Scholar]
- Martinière A, Bassil E, Jublanc E, Alcon C, Reguera M, Sentenac H, Blumwald E, Paris N. 2013. In vivo intracellular pH measurements in tobacco and Arabidopsis reveal an unexpected pH gradient in the endomembrane system. The Plant Cell 25, 4028–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Pérez C, Sánchez-Calvo B, Begara-Morales JC, et al. 2015. Transcriptomic profiling of linolenic acid-responsive genes in ROS signaling from RNA-seq data in Arabidopsis. Frontiers in Plant Science 6, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown M, Schubert M, Marcussen T, Fjellheim S, Preston JC. 2016. Evidence for an early origin of vernalization responsiveness in temperate pooideae grasses. Plant Physiology 172, 416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown R, Kuroki G, Warren G. 1996. Cold responses of Arabidopsis mutants impaired in freezing tolerance. Journal of Experimental Botany 47, 1919–1925. [Google Scholar]
- Melser S, Molino D, Batailler B, Peypelut M, Laloi M, Wattelet-Boyer V, Bellec Y, Faure J-D, Moreau P. 2011. Erratum to: Links between lipid homeostasis, organelle morphodynamics and protein trafficking in eukaryotic and plant secretory pathways. Plant Cell Reports 30, 675–676. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Research 47, D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, James D Jr, Dooner H, Browse J. 1993. Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proceedings of the National Academy of Sciences, USA 90, 6208–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Muthan B, Benning C. 2010. Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330, 226–228. [DOI] [PubMed] [Google Scholar]
- Monroy AF, Castonguay Y, Laberge S, Sarhan F, Vezina LP, Dhindsa RS. 1993. A new cold-induced alfalfa gene is associated with enhanced hardening at subzero temperature. Plant Physiology 102, 873–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navari-Izzo F, Pinzino C, Quartacci MF, Sgherri CLM, Izzo R. 1994. Intracellular membranes: kinetics of superoxide production and changes in thylakoids of resurrection plants upon dehydration and rehydration. Proceedings of the Royal Society of Edinburgh Section B: Biological Sciences 102, 187–191. [Google Scholar]
- Nilsson AK, Johansson ON, Fahlberg P, et al. 2015. Acylated monogalactosyl diacylglycerol: prevalence in the plant kingdom and identification of an enzyme catalyzing galactolipid head group acylation in Arabidopsis thaliana. The Plant Journal 84, 1152–1166. [DOI] [PubMed] [Google Scholar]
- Raju SKK, Barnes AC, Schnable JC, Roston RL. 2018. Low-temperature tolerance in land plants: are transcript and membrane responses conserved? Plant Science 276, 73–86. [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. 2000. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. The Plant Cell 12, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos W, Viehweger K, Dordschbal B, Schumann B, Evers S, Steighardt J, Schwartze W. 2006. Intracellular pH signals in the induction of secondary pathways—the case of Eschscholzia californica. Journal of Plant Physiology 163, 369–381. [DOI] [PubMed] [Google Scholar]
- Roston RL, Wang K, Kuhn LA, Benning C. 2014. Structural determinants allowing transferase activity in SENSITIVE TO FREEZING 2, classified as a family I glycosyl hydrolase. Journal of Biological Chemistry 289, 26089–26106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouser G, Slakotos AN, Fleischer S. 1966. Quantitative analysis of phospholipids by thin layer chromatography and phosphorus analysis of spots. Lipids 1, 85–86. [DOI] [PubMed] [Google Scholar]
- Sakaki T, Ohnishi J-I, Kondo N, Yamada M. 1985. Polar and neutral lipid changes in spinach leaves with ozone fumigation: triacyiglycerol synthesis from polar lipids. Plant and Cell Physiology 26, 253–262. [Google Scholar]
- Sakaki T, Saito K, Kawaguchi A, Kondo N, Yamada M, Abrams SR, Covello PS, Taylor DC, Savage L, Milcamps A. 1990. Conversion of monogalactosyldiacylglycerols to triacylglycerols in ozone-fumigated spinach leaves. Plant Physiology 94, 766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. 2015. The ImageJ ecosystem: an open platform for biomedical image analysis. Molecular Reproduction and Development 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenker W, Roberts MJ. 2009. Nonlinear temperature effects indicate severe damages to U.S. crop yields under climate change. Proceedings of the National Academy of Sciences, USA 106, 15594–15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze WX, Schneider T, Starck S, Martinoia E, Trentmann O. 2012. Cold acclimation induces changes in Arabidopsis tonoplast protein abundance and activity and alters phosphorylation of tonoplast monosaccharide transporters. The Plant Journal 69, 529–541. [DOI] [PubMed] [Google Scholar]
- Sellschop JP, Salmon SC. 1928. The influence of chilling, above the freezing point, on certain crop plants. Journal of Agricultrual Research 37, 315–338. [Google Scholar]
- Sgherri CLM, Quartacci MF, Bochicchio A, Navari-Izzo F. 1994. Defence mechanisms against production of free radicals in cells of ‘resurrection’ plants. Proceedings of the Royal Society of Edinburgh Section B: Biological Sciences 102, 291–294. [Google Scholar]
- Steponkus PL, ed. 1996. Advances in low-temperature biology. Amsterdam: Elsevier. [Google Scholar]
- Takahashi D, Gorka M, Erban A, Graf A, Kopka J, Zuther E, Hincha DK. 2019. Both cold and sub-zero acclimation induce cell wall modification and changes in the extracellular proteome in Arabidopsis thaliana. Scientific Reports 9, 2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. 2001. So what’s new in the field of plant cold acclimation? Lots! Plant Physiology 125, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. 2010. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiology 154, 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorlby G, Fourrier N, Warren G. 2004. The SENSITIVE TO FREEZING2 gene, required for freezing tolerance in Arabidopsis thaliana, encodes a β-glucosidase. The Plant Cell 16, 2192–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traore M, Sullivan CY, Rozowski JR, Lee KW. 1989. Comparative leaf surface morphology and the glossy characteristic of sorghum, maize, and pearl millet. Annals of Botany 64, 447–453. [Google Scholar]
- Uemura M, Joseph RA, Steponkus PL. 1995. Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions). Plant Physiology 109, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R. 2017. Desiccation tolerance: seedy origins of resurrection. Nature Plants 3, 17046. [DOI] [PubMed] [Google Scholar]
- Vu HS, Roston R, Shiva S, Hur M, Wurtele ES, Wang X, Shah J, Welti R. 2015. Modifications of membrane lipids in response to wounding of Arabidopsis thaliana leaves. Plant Signaling & Behavior 10, e1056422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Benning C. 2011. Arabidopsis thaliana polar glycerolipid profiling by thin layer chromatography (TLC) coupled with gas-liquid chromatography (GLC). Journal of Visualized Experiments (49), 2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Hersh HL, Benning C. 2016. SENSITIVE TO FREEZING2 aids in resilience to salt and drought in freezing-sensitive tomato. Plant Physiology 172, 1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, McKown R, Marin AL, Teutonico R. 1996. Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiology 111, 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MS, Williams JP. 1984. Changes in the lipid and fatty acid composition of Vicia faba mesophyll protoplasts induced by isolation. Plant and Cell Physiology 25, 1541–1550. [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H-E, Rajashekar CB, Williams TD, Wang X. 2002. Profiling membrane lipids in plant stress responses: role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. Journal of Biological Chemistry 277, 31994–32002. [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proceedings of the National Academy of Sciences, USA 111, E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhang Z, Xie S, Si T, Li Y, Zhu J-K. 2016. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiology 171, 2744–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data that support the findings of this study are available in the NCBI SRA (Sequence Read Archive) database under the BioProject ID PRJNA894306. The full GO term analyses are available in the supplementary data.


