Summary
Angiogenesis, the growth of new blood vessels from pre-existing vessels, occurs during development, injury repair, and tumorigenesis to deliver oxygen, immune cells, and nutrients to tissues. Defects in angiogenesis in cardiovascular and inflammatory diseases, and chronic, non-healing wounds, yet treatment options are limited. Here, we provide a map of the early angiogenic niche by analyzing single-cell RNA sequencing of mouse skin wound healing. Our data implicate Langerhans cells (LCs), a phagocytic, skin-resident immune cell, in driving angiogenesis during skin repair. Using lineage-driven reporters, three-dimensional (3D) microscopy, and mouse genetics, we show that LCs are situated at the endothelial cell leading edge in mouse skin wounds and are necessary for angiogenesis during repair. These data provide additional future avenues for the control of angiogenesis to treat disease and chronic wounds and extend the function of LCs beyond their canonical role in antigen presentation and T-cell immunity.
eTOC
Angiogenesis repairs blood vessels after injury by creating new blood vessels from preexisting endothelial cell networks. Using scRNA sequencing, Wasko et al. provide a map of the predicted molecular and cellular cues that drive angiogenesis and identify Langerhans cells as a major regulator of angiogenesis in murine skin wounds.
Graphical Abstract
Introduction
Angiogenesis, the growth of new blood vessels from existing vasculature, is an essential feature of tissue development, tissue repair, and also occurs during tumorigenesis1. While defects in this process underlie diseases of the cardiovascular system and inflammation, tissue defects, and chronic non-healing wounds, the mechanisms that drive angiogenesis are not fully defined. Within tissues, angiogenesis is controlled by the combination of pro-and anti-angiogenic factors produced by tissue-resident cells to control endothelial cells (ECs) at the tips of blood vessels to proliferate and migrate to form a leading edge of newly sprouting vessels. Tip cell extension is supported by adjacent stalk cells that maintain connection to the originating vessel. The most well-studied angiogenic factor, vascular endothelial growth factor alpha (VEGF-A), induces collective migration, proliferation, and cellular rearrangement of ECs to form new blood vessels. Despite our in-depth understanding of VEGF signaling, clinical therapies involving VEGF have not been fruitful2,3, suggesting that additional factors control angiogenesis. Indeed, pro-angiogenic factors such as Notch signaling, integrins, and other guidance cues and growth factors have been shown to promote angiogenesis4. However, the angiogenic niche has not been fully defined and may reveal additional cell types and/or molecules that control EC biology.
Skin wound repair is an excellent model to study angiogenesis since the repair of blood vessels occurs within the injured tissue. Tissue repair occurs through a series of temporally-overlapping and tightly regulated steps of inflammation, proliferation, and remodeling5. The inflammatory phase involves the recruitment of blood-derived monocytes, which differentiate and activate into sequential waves of inflammatory and anti-inflammatory macrophages. During the proliferative phase of healing, macrophages, fibroblasts, and keratinocytes interact with ECs to induce angiogenesis. Indeed, deletion of VEGF-A in LysM-Cre expressing monocyte-derived cells and Keratin-5 (K5)-expressing keratinocytes leads to defects in angiogenesis during murine skin repair6,7. Activation of Notch signaling within ECs promotes skin repair-associated angiogenesis, while Hedgehog and the transcription factor Sox9 inhibit EC activation after skin injury8. Following angiogenesis, ECs and other cell types are pruned during tissue remodeling as the tissue attempts to return to a pre-injured state. Tissue repair mechanisms are compromised with age and disease, and defects in angiogenesis can lead to chronic non-healing wounds. Thus, identifying cellular and molecular mechanisms involved in angiogenesis could inform new clinical strategies for cardiovascular diseases, cancer, chronic inflammatory disorders, and defective wound repair.
Here, we provide a combinatorial map of the cells and their ligands that could drive angiogenesis within skin wounds. Using computational analysis of single-cell RNA sequencing (scRNA-seq) data, we identify multiple distinct ligands produced by fibroblasts, keratinocytes, and macrophages that likely activate receptors on ECs to drive gene expression changes during angiogenesis in skin wounds. Surprisingly, we find that, after injury, Langerhans cells (LCs), a specialized phagocytic immune cell in the skin, upregulate mRNAs associated with an angiogenic program and are predicted to activate signaling in skin wound ECs. After injury, LCs localize near ECs at the leading edge of sprouting vessels during wound-induced angiogenesis. Using mouse genetics, we demonstrate that LCs are essential for angiogenesis, particularly induction of EC proliferation during wound repair. Our data suggest that a combination of cells and angiogenic factors, including LC-derived ligands, are essential for proper angiogenesis during tissue repair.
Results:
Identification of angiogenic regulators in skin wounds
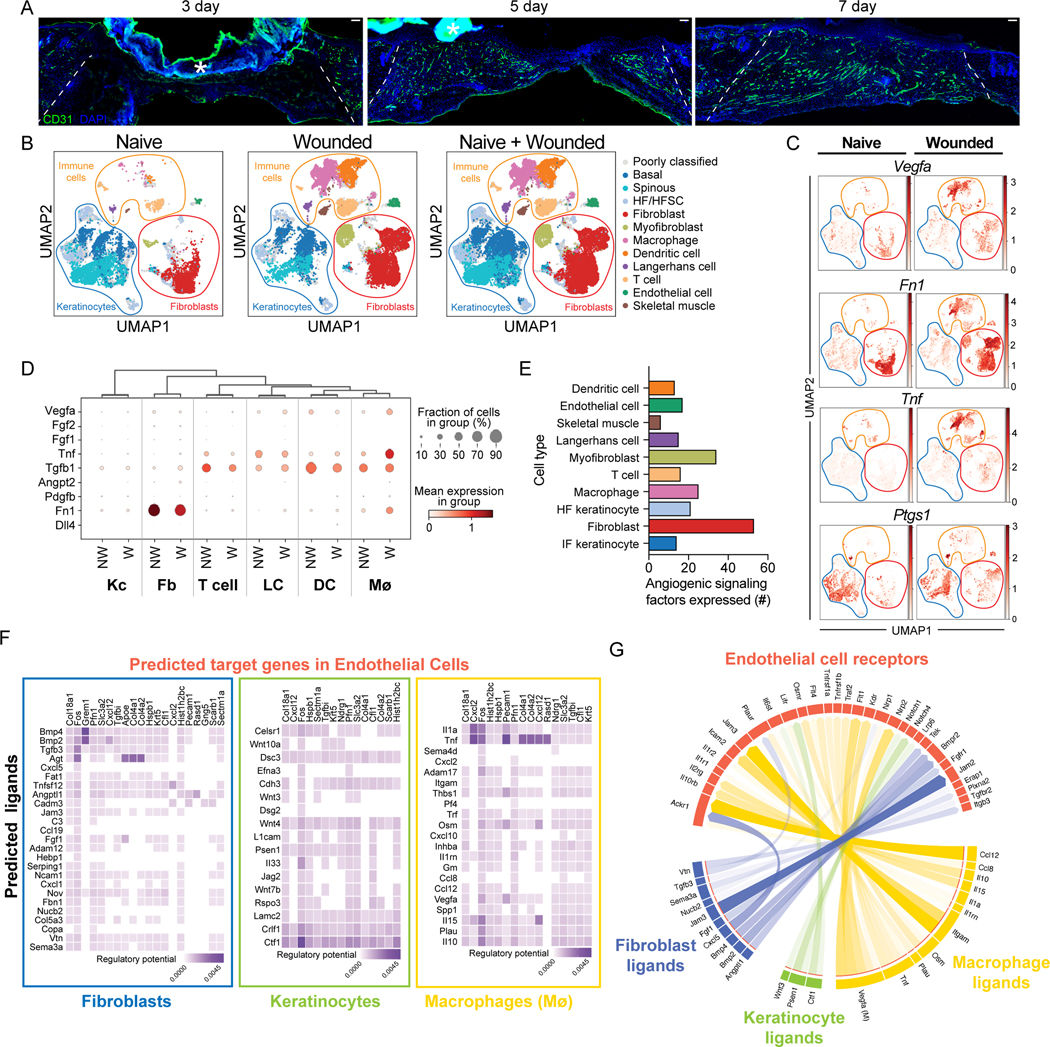
Angiogenesis is initiated at the beginning of the proliferation phase of wound healing as indicated by immunostaining with CD31 (platelet endothelial cell adhesion molecule, PECAM-1)9 antibodies of skin sections of wounds 3, 5, and 7 days after injury (Fig. 1A)10. To identify factors that contribute to the angiogenic niche within skin wounds, we analyzed scRNA-seq data from mouse skin wounds at the beginning of the proliferative phase of repair11. By training a neural network to identify cell types based on expression of established marker genes (following the approach from Kumar et al. 2018) (Figs. S1A–B), we classified 11 broad cell types in the scRNA-seq data11, including classes of immune cells, keratinocytes, fibroblasts, endothelial cells, and skeletal muscle cells (Figs. 1B and S1C). Next, we examined the expression angiogenic ligands known to be expressed in skin keratinocytes, macrophages, and fibroblasts (Vegfa, Tnf, Ptgs1, and Fn1)4,12–14 across the 11 cell clusters in wounded skin compared to non-wounded skin (Fig. 1C). We found that several known proangiogenic factors are expressed in non-wounded and wounded skin at varying levels, including VEGFA (Fig. 1D and S1D–E). Multiple cell types express Tnf, Vegfa, and Ptgs1, and fibroblasts predominantly express Fn1, indicating an unappreciated complexity to the early angiogenic niche in skin wounds.
Figure 1: Identification of angiogenic regulators in skin wounds.
(A) Images of CD31 immunostaining (green) and DAPI-stained nuclei (blue) in cross-sections of 3-, 5-, and 7-day wound beds. The white dashed lines delineate wound edges. Asterisks (*) indicate non-specific staining of scab. Scale bars, 100 μm.
(B) Uniform Manifold Approximation and Projection (UMAP) plots of scRNA-seq data (Haensel et al. (GSE142471)) from mouse skin: non-wounded (Naive), 4-day Wounded, and combined.
(C) Feature plots showing expression of VEGFA, Tnf, Ptgs1, and Fn1 in single cells of Naive and Wounded skin from (B).
(D) Bubble plot depicting average mRNA expression (color) of angiogenic signaling factors expressed by single keratinocytes (Kc), fibroblasts (Fb), T cells, Langerhans cells, dendritic cells (DC), and macrophages (Mø) in wounded (W) and nonwounded (NW) samples from (B). Bubble size indicates the percent of cells expressing that gene.
(E) Quantification of the number of angiogenic signaling factors with an average mRNA expression level ≥1 in each indicated cell type.
(F) Heatmaps showing potential links between ligands expressed by Fb, Kc, and Mø and EC downstream target genes. Fibroblast data includes fibroblast and myofibroblast populations.Keratinocyte data includes basal, spinous, and HF/HFSC populations.
(G) Chord diagram summarizing the top 50 ligand-receptor links during wound healing. Arrows represent ligands from Fb, Kc, and Mø binding to EC receptors. See also Figures S1 and S2.
To gain a comprehensive picture of the angiogenic niche within skin wounds, we utilized the NicheNet algorithm, which infers ligand binding from patterns of target gene expression in receiving cells15. Specifically, we sought to infer signaling to ECs from the other classes of cells identified in the scRNA-seq data. We defined target genes in ECs as genes that were differentially expressed (log2FC > 0.25 and adjusted p-value < 0.05) in ECs after wounding as compared to non-wounded conditions (Supplemental Table 1). Correlation of observed expression of these EC target genes against NicheNet’s prior model implicated 202 potential ligands (i.e., angiogenic factors) as driving patterns in EC gene expression after wounding (Fig. S2A). The predicted angiogenic factor mRNAs ranged from 53 (fibroblasts) to 8 (skeletal muscle cells) (Fig. 1E). Among the immune cell populations, macrophages expressed the most angiogenic mRNAs (Fig. 1E), which is consistent with their well-established role driving angiogenesis in skin wounds16. Surprisingly, we found that dendritic cells, LCs, and T cells expressed a similar number of angiogenic mRNAs as interfollicular keratinocytes (Fig. 1E), which have been implicated as a major regulator of angiogenesis during wound healing7.
To characterize the cellular interactions between endothelial cells and other cell types within skin wounds, we focused on ligands expressed by specific cell types and used NicheNet to predict the potential for these ligands to interact with receptors expressed by ECs (i.e., interaction potential), and their potential to activate EC downstream target genes (i.e., regulatory potential) within skin wounds (Fig. 1F). We found that fibroblasts (including fibroblast and myofibroblast subsets) upregulated several ligands that are predicted to interact with integrins, atypical chemokine receptors (ACKRs), bone morphogenetic protein receptors (BMPR), and fibroblast growth factor (FGF) receptors, and activate several genes in endothelial cells (Figs. 1F and S2B). Keratinocytes (including hair follicle [HF] and interfollicular [IF] subsets) expressed agonists for Wnt and Notch signaling, and cytokine receptors that are predicted to activate several signaling factors and extracellular matrix gene expression in endothelial cells (Fig. 1F). Macrophages upregulated multiple angiogenic cytokines (Tnf, Il10, Il1a) and additional ligands that are predicted to induce ECM and other signaling factors (Fig. 1F). Endothelial receptors were predicted to be activated by distinct ligands produced by each cell type (Fig. S2B), suggesting that each cell within the repairing wound contributes uniquely to the angiogenic niche. To strengthen our confidence that the NicheNet-predicted receptors are robustly expressed by ECs in our dataset, we examined the average mRNA expression of each receptor in each cell population and found that most of these receptors were robustly and predominantly expressed by endothelial cells (Fig. S2C). These receptors are predicted to activate several EC target genes to control angiogenesis including Fos, Cxcl12, Col4a2, and Col18a1 (Fig. 1F). Together, these data provide a map for the multiple interactions that may drive early events in angiogenesis within skin wounds (Fig. 1G). Furthermore, only a few mRNAs upregulated in ECs within skin wounds also are defined as ECspecific genes in other tissues17 (Table 1), consistent with a unique skin wound niche for angiogenesis.
Table 1. Comparison of EC specific genes in different cell types.
Top 50 marker genes of tissuespecific EC populations from Table S2 in Kaluck et al11 compared to the top 50 mRNAs expressed in skin wound ECs from GSE142471. mRNAs shared with skin wound ECs in each tissue are highlighted in yellow.
| Data from Kalucka et al., 2020 Table S2 | Data from Wasko et al. Table l Top 50 upregulate d genes in wounded ECs | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Top-50 marker genes of tissue- specific EC phenotypes | ||||||||||||||
|
| ||||||||||||||
| Brain-largeartery | Brain-capillary | Brain-largevein | Testis-largeartery | Testis-capillary | Liver-Largeartery | Liver-capillary | Liver-proliferating | Lung-artery | Lung-capillary 1 | Lunc-capillary 2 | Colon-artery | Colon-capillary 1 | Colon-capillary 2 | Skin wound EC |
| Mgp | Cxcl12 | Tmsb10 | Edn1 | Car4 | Sdc1 | Dnase1l3 | Stmn1 | Mgp | Car4 | Gpihbp1 | Clu | Car4 | Esm1 | Cst3 |
| Cytl1 | Spock2 | Vcam1 | Ifi27l2a | Ptn | Clu | Clec4g | Hmgb2 | Tm4sf1 | Igfbp7 | Sema3c | Gja4 | Rgcc | Plpp3 | Scarb1 |
| Fbln5 | Gm9946 | Slc38a5 | Sox17 | Slco1a4 | Ehd4 | Stab2 | H2afz | Cxcl12 | Emp2 | Gm12216 | Fbln5 | Plpp1 | Nrp1 | Lrat |
| Clu | Lrrn3 | Icam1 | Gadd45g | Bsg | Cyp1a1 | Ctsl | Cdkn1a | Plat | Pmp22 | Stmn1 | Glul | Aplnr | Mef2c | Pltp |
| Eln | Gstm7 | Tmem252 | Amd1 | Gpcpd1 | Plac8 | Bmp2 | Top2a | Plac8 | Fibin | Sox17 | Azin1 | Ramp3 | Cd24a | Bcam |
| Bgn | Palm | Lcn2 | Ssu2 | Slc3a2 | Col8a1 | Maf | Cdk1 | Gja4 | AW112010 | Ier3 | Igfbp4 | Col4a1 | Ehd4 | S100a11 |
| Igfbp4 | Tbx3 | Vwf | Ptprr | Ier3 | Lrg1 | Gatm | Cks2 | Mecom | Cyp4b1 | Plvap | Stmn2 | Sparc | Tmem176a | Ly6c1 |
| Cfh | Prdm16 | Ctsc | Crip1 | Ephb1 | Mgp | Kdr | Birc5 | Igfbp4 | Ptp4a3 | Socs3 | S100a6 | Nkx2−3 | Fam167b | Apoe |
| Lmcd1 | Rasgrp2 | Tgm2 | Ptma | Caskin2 | Mecom | Mt1 | Tuba1b | Atp13a3 | Sept4 | Wfdc1 | Edn1 | Hspg2 | Rbp7 | Rasa4 |
| Bpgm | Cd83 | Ackr1 | S100a4 | Tmem201 | Cd200 | Plpp3 | Tk1 | Apoe | Ednrb | Aplnr | Sox17 | AW112010 | Col13a1 | Pfn1 |
| Cmip | Pik3ip1 | Apod | Wfdc1 | Kazn | Fbln2 | Cyp4b1 | Hmgn2 | Gja5 | Cd34 | Sgk1 | Sema3g | Mgll | Oaz2 | Slc30a1 |
| Htra1 | Pmaip1 | Apoe | Cav1 | Cspp1 | Hspb1 | Gpr182 | Lgals1 | Fxyd5 | Prx | H2-Ab1 | Ace | Sparcl1 | Tmem176b | Lgals1 |
| Sat1 | Tnfrsf21 | Nfkbia | Ier2 | Usp32 | AU021092 | Mrc1 | Cks1b | Tsc22d1 | Chst1 | Cadm1 | Cst3 | Fmo2 | Smco4 | Cxcl2 |
| Bmx | Prkcb | Hs3st1 | Jun | Setdb1 | Ly6c1 | Fam167b | Nucks1 | Edn1 | Tbx3 | Casc4 | Tsc22d1 | Trp53i11 | Ces2e | Cxx1a |
| Reck | Plekhh2 | Ptn | Alox12 | Zdhhc9 | Cyr61 | Fcgr2b | Pcna | Efnb2 | Clu | Btg2 | Tspan8 | Col15a1 | Vwa1 | Marcksl1 |
| Emp3 | Gpr160 | Ch25h | Btg2 | Mcf2 | Cd63 | Adam23 | H2afx | Sparcl1 | Nrp1 | Sparc | Slc6a6 | Car8 | Jam3 | Serf2 |
| Vim | Rftn2 | Fth1 | Lmna | Tiam1 | Adgrg6 | Oit3 | Cdca8 | Htra1 | Kdr | Kit | Fbln2 | Lamb1 | Plscr2 | Thy1 |
| S100a11 | Cnnm2 | Ptgs2 | Tgfb2 | Aars2 | Plvap | Cldn5 | Spc24 | Fbln2 | Ly6c1 | Hilpda | Tm4sf1 | Sgk1 | Kdr | C1qtnf9 |
| Timp2 | Krt222 | Cxcl1 | Klf2 | Ndufaf1 | Gja5 | Apoe | Lockd | Hey1 | Bcam | Tubb2a | Heg1 | Enpp6 | Thbd | Gm694 |
| Ptgis | Helz | Net1 | Stmn1 | Mamstr | Crip1 | Lgmn | Smc4 | Fam3c | Tmcc2 | Lpl | Gja5 | Ivns1abp | Hspb1 | Col18a1 |
| Fhl1 | Nanos1 | Itih5 | Cdkn1c | Nol4l | Cst3 | Stab1 | Cdkn2c | Sulf1 | Tppp3 | Sox11 | Sat1 | Jun | Pltp | Cxcl12 |
| Ccnd2 | Naa30 | Rgs16 | Chd3 | Mfsd12 | Npr3 | Clec1b | Hjurp | Sema3g | Rgs12 | Plk2 | Gadd45g | Sox4 | Ncoa7 | Ndrg1 |
| Cp | Napg | Rnase4 | Klf4 | Alg6 | Sncg | Hpgd | Smc2 | Id1 | Phlda3 | Jund | Mast4 | Dusp6 | Flt1 | Gng5 |
| Emp1 | Ric1 | Mt1 | Eef1a1 | Atg4a | Ednrb | Cd300lg | Tyms | Mgst1 | Hspb1 | Atp1a1 | Hey1 | Mcam | Slc16a1 | Lgals7 |
| Id2 | Nt5dc2 | Csf1 | Cdk19 | Sdad1 | Ly6e | Aass | Tubb6 | Heg1 | Apln | H2-Q6 | Plat | Apold1 | Gm13889 | Pecam1 |
| Rhob | Vangl2 | Serpinb9 | Hspa1a | Tyw5 | Mgst1 | Akr1b8 | Racgap1 | Efna1 | Enho | Ncald | Fn1 | Ankrd37 | Plscr1 | Grem1 |
| Tm4sf1 | 2310040G24Rik | Rbp1 | Co18a1 | Hspb6 | Cd34 | Tfpi | Lmnb1 | Bmx | Pcdh1 | Ccr12 | Ssu2 | Ednrb | Cd200 | Fkbp1a |
| Fn1 | Htatip2 | Gm9844 | Zfp36 | 0610010F05Rik | Nrg1 | Col13a1 | Nrm | Sat1 | Nhlrc2 | Npr3 | Mecom | Rapgef4 | Exoc3l2 | Krt5 |
| Emp2 | Utp14b | Mt2 | Rsad2 | Galc | Vegfc | Mt2 | Fen1 | Crip1 | Ly6a | Ier2 | Eps8l2 | Thbs1 | Lrrc3b | Fam167b |
| Pam | Rbfa | Lrg1 | Bmp2 | Pign | Tm4sf1 | Dab2 | Fbxo5 | Cdh13 | Kitl | Adrb1 | ssu2 | Fos | Ptp4a3 | Rhoc |
| Cd82 | Adck2 | Nr2f2 | Hist1h1c | Gm17619 | Ldb2 | Lpar6 | Cdc20 | Mmrn2 | Pllp | Car2 | Syt15 | Itga6 | Itm2a | Col4a1 |
| Jag1 | Eif2ak4 | Gbp4 | Neb1 | Zcchc10 | Ier2 | Clec14a | Mcm5 | Ccnd2 | Cldn5 | Rdx | Hspala | Chrm2 | Pcdh17 | Fos |
| Spint2 | Med27 | Jun | Akap13 | Fbxw4 | Efna1 | Mertk | Cenpa | Slc6a6 | Tmem176a | Nckap5 | 8430408G22Rik | Apln | Tnfaip2 | Tmsb4x |
| Irf6 | 2700046A07Rik | Il1r1 | Med1 | Gba2 | Ddit4 | Manf | Gmfg | Rps28 | Rasgrp2 | H2-Eb1 | Crip1 | Rad51ap1 | Rsad2 | Myct1 |
| Kcnn4 | Pias3 | Tmem176b | Cit | Klhl17 | Dusp1 | Slc40a1 | Ccdc34 | Rbp7 | Mgll | Junb | Dusp1 | Selenbp1 | Id3 | Prelid1 |
| Atp2a3 | Fam136a | Foxf1 | Dbndd2 | Mms19 | Syt15 | Chst15 | Ncapd2 | Nrarp | Tbx2 | Plcb1 | 40787 | F2r13 | Crim1 | Rasd1 |
| Ace | 2610301B20Rik | Cpe | 2810403A07Rik | Marf1 | Fbln5 | Ftl1 | Tcf19 | 8430408G22Rik | Aard | Ifi47 | Ybx1 | Gbp2 | Actg1 | Col4a2 |
| Nuak1 | A630072M18Rik | Zfp36l2 | Ccnl1 | Zfp335 | Abcg2 | Sparc | Rfc3 | Camk2d | Rprml | Car14 | S100a4 | Iigp1 | Gas7 | Ctla2a |
| Gja4 | Nup37 | Serpinb6b | Rnf152 | Dgkq | Gm13889 | Cyp26b1Myo10 | Cxcl10 | Cdc42ep3 | Tspan8 | Tmem221 | Car7 | Hes1 | Serpinb1a | mt-Nd5 |
| Lmo1 | Dnase2a | Dusp23 | Ppp1r15a | Gxylt1 | Fxyd5 | Myo10 | Ltc4s | Psen2 | Stmn2 | Ada mts1 | Bmx | Hrct1 | Prdm1 | Csrp2 |
| Sdc1 | Zfp788 | Polk | Tmem243 | Cwc27 | Utrn | Mylip | Hsp90aa1 | Gata6 | Dhrs3 | Ldb2 | Klf4 | Arrdc3 | Efhd1 | Hspb1 |
| Synpo | Slc43a2 | Cd14 | Smarcd1 | Fam212b | Lmo7 | Adgrf5 | Lig1 | Rgs3 | Plaur | Pcdh17 | Htra1 | Chchd10 | Penk | Cf11 |
| Edn1 | Usp43 | Dnm3 | Il20rb | Traf3ip1 | Fos | Cpne2 | Usp1 | Tmem158 | Ccdc184 | Irf1 | Jag1 | Btg2 | Abcg2 | RP23-354H24.9 |
| Mmrn2 | Cbx8 | Il6st | Cd36 | Fbxl18 | 8430408G22Rik | SlcB9a8 | Bok | Mast4 | Icam1 | Cdc42ep4 | Pcsk5 | H1f0 | Actb | Sectm1a |
| Tspan7 | Nhlrc3 | Car14 | Adarb1 | Gm20045 | Ltbp4 | Ets2 | Raph1 | Fmo2 | Cd24a | Pxdc1 | Lima1 | Bpgm | Akr1c14 | Tgfbi |
| Phlda3 | Clk2 | Gm5127 | Klhdc8b | NrarP | Plet1os | Pold2 | Fads3 | Slc48a1 | Vgll4 | Lamb2 | Xpc | Nr5a2 | Slc3a2 | |
| Arl15 | Angptl4 | Wnt5a | RasgrP3 | Cmklr1 | Itpkb | Nav2 | Mcam | Grina | Ankrd44 | Efnb2 | Col4a3 | B2m | Myeov2 | |
| Dkk2 | BC0B70B4 | Sema6a | Tmbim1 | Cd9 | Hsp90b1 | Rpa2 | Itga6 | Scn7a | Cd1d1 | Atp13a3 | Ankrd44 | Tnfrsf11b | Bsg | |
| Fstl1 | Naa16 | Flrt2 | Rsrp1 | Ehd2 | Cd84 | Ezh2 | Lgals3bp | Serpine1 | Rapgef4 | Mal | Hic1 | Rcan2 | Sectm1b | |
| H2-Q6 | B4galt7 | Car2 | Ppp1r15a | Trim47 | Itga9 | Rrm1 | Ebf1 | Phlda1 | Tubb2b | Adamts1 | Serpine1 | Cdc14a | Hist1h2bc | |
Langerhans cells upregulate angiogenic mRNAs in skin wounds
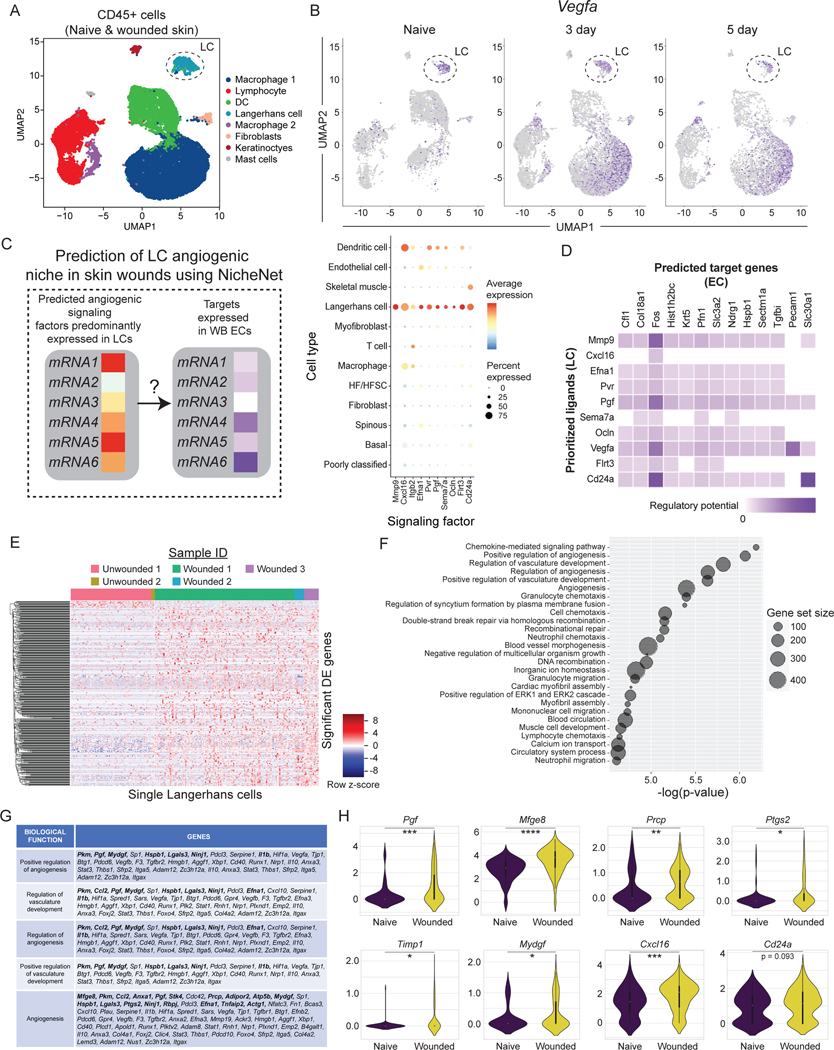
We were surprised by the expression of several angiogenic mRNAs in LCs, a subset of phagocytic immune cells resident in the epidermis (Fig. 1D)(Supplemental Table 2). Others have shown that depletion of all langerin+ cells, which includes LCs and a subpopulation of dermal dendritic cells, resulted in enhanced wound repair in mice18. CD11c+ dendritic cells have been implicated in the repair of burn wounds in studies that depleted CD11c+ cells but not LCs using genetics mouse models19. Thus, we sought to determine if LCs function in skin wound repair, and specifically in angiogenesis.
Consistent with LCs playing a unique role in wound repair compared to other DC populations19, LCs cluster distinctly from DCs, macrophages, and lymphocytes in unwounded and wounded samples after injury (Fig. 1B). CD207+ LCs comprised 0.56% of CD45+ cells in non-wounded samples and increased to 1.41% of CD45+ cells in skin wounds (Fig. 1B), which is consistent with prior reports showing an increase in LCs in skin wounds after injury20. These cells also express EpCAM, MHC-II genes, and CD11c, supporting that these cells express canonical LC genes (Fig. S3A)21. This distinct clustering of LCs compared to dermal DCs and other immune cells also occurred when immune cells were enriched via FACS purification based on CD45 expression (Figs. 2A, and S3B–D; GSE166950)22, and the expression of the angiogenic factor Vegfa was noted in LCs in non-wounded and wounded skin as well as in macrophages and in keratinocytes (Fig. 2B). Interestingly, we identified 10 angiogenic mRNAs expressed dominantly by LCs and not by other cell types in non-wounded and wounded skin, including mRNAs that function as cytokines such as Placental growth factor (Pgf)23 and Chemokine Ligand 16 (Cxcl16)24,25, the cytoskeletal modulator Matrix Metallo-protease9 (Mmp9)26, and cell adhesion molecules such as Integrin b2 (Itgb2)27 and Semaphorin 7a (Sema7a)28 (Fig. 2C; Key resource Table). During tissue repair, the LC-derived ligands are predicted to interact with several receptors and target genes that are upregulated by wound derived ECs (Fig. 2D). In particular, the LC expression of Cxcl16, Vegfa, and Pgf mRNAs are predicted to bind to multiple receptors upregulated by ECs in skin wounds (Fig. S3E). Further, LC ligands are infered to activate Fos, Profilin1 (Pfn1), and Pecam1 expression by ECs, which contribute to EC proliferation and migration (Fig. 2D)4,29–31.
Figure 2: Langerhans cells upregulate angiogenic mRNAs in skin wounds.
(A) UMAP plot of scRNA-seq data of FACS-purified CD45+ cells from nonwounded and wounded (3-day and 5-day post-injury) (GSE166950). Dashed circle identifies Langerhans cell (LC) cluster.
(B) Feature plots showing expression of VEGFA mRNA in scRNA-seq samples from non-wounded (Naive) skin, 3-day wounds, and 5-day wounds. Dashed circle highlights LC cluster.
(C and D) NicheNet analysis of predicted signaling from LCs to endothelial cells (EC) from GSE142471.
(C) Bubble plot depicting average mRNA expression (color) of angiogenic signaling factors expressed by LCs in wounded and nonwounded samples from GSE142471 (B). Bubble size indicates the percent of cells expressing that gene.
(D) Heatmaps showing potential links between LC ligands and EC downstream target genes predicted by NicheNet.
(E) Heatmap of differentially expressed genes in LCs from wounded and nonwounded skin.
(F) Plot of gene ontology (GO) terms associated with changes in LC gene expression during wound healing.
(G) Table of angiogenic genes differentially expressed in LCs after injury. Bolded genes have a padj value < 0.05. Non-bolded genes have a p-value < 0.05.
(H) Violin plots of LC mRNA expression of Pgf, Mfge8, Prcp, Ptgs2, Timp1, Mydgf, Cxcl16, and Cd24a transcripts in wounded and nonwounded samples. unpaired T-tests *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.00005. See also Figures S2 and S3.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC/eFluor 780 anti-mouse CD45 rat monoclonal | eBioscience | Cat# 47-0451-82; RRID: AB_1548781 (Clone 30-F11) |
| Alexa Fluor 700 anti-mouse CD11b rat monoclonal | eBioscience | Cat# 56-0112-82; RRID: AB_657585 (Clone M1/70) |
| eFluor 450 anti-mouse F4/80 rat monoclonal | eBioscience | Cat# 48-4801-82; RRID: AB_1548747 (Clone BM8) |
| PE/Cy7 anti-mouse Ly6G rat monoclonal (clone 1A8) | Biolegend | Cat# 127618; RRID: AB_1877261 |
| APC anti-mouse Ly6C rat monoclonal (clone HK1.4) | eBioscience | Cat# 17-5932; RRID: AB_1724155 |
| Alexa Fluor 488 anti-mouse CD206 rat monoclonal | Biolegend | Cat# 141710; RRID: AB_10900445 (clone C068C2) |
| PE anti-mouse MHCII rat monoclonal | eBioscience | Cat# 12-5321-82; AB_465928 (clone M5/114.15.2) |
| APC anti-mouse EpCam rat monoclonal (clone G8.8) | BD Biosciences | Cat# 563478; RRID: AB_2738234 |
| PerCp/Cy5.5 anti-mouse CD64 rat monoclonal | Biolegend | Cat# 139308; RRID: AB_2561963 (clone X54-5/7.1) |
| FITC anti-mouse CD3e Armenian hamster | eBioscience | Cat# 11-0031-82; RRID: AB_464882 monoclonal (clone 145-2C11) |
| PerCp anti-mouse CD4 rat monoclonal (clone GK1.5) | Biolegend | Cat# 100434; RRID: AB_893324 |
| APC anti-mouse CD8a rat monoclonal (clone 53-6.7) | eBioscience | Cat# 17-0081-83; RRID: AB_469336 |
| PE anti-mouse gd-TCR Armenian hamster | BD Biosciences | Cat# 553178; RRID: AB_394689 monoclonal (clone GL3) |
| Alexa Fluor 700 anti-mouse CD29 Armenian hamster | Biolegend | Cat# 102218; RRID: AB_493711 monoclonal (clone HMbeta1-1) |
| Brilliant Violet 421 antimouse CD34 rat monoclonal | Biolegend | Cat# 119321; RRID: AB_10900980 (clone MEC14.7) |
| APC-Fire750 anti-mouse CD31 rat monoclonal (390) | Biolegend | Cat# 102434; RRID: AB_2629683 |
| Anti-CD31 (PECAM-1) Armenian hamster monoclonal | Millipore | Cat# MAB1398Z, RRID:AB_94207 (clone 2H8) |
| Rat Anti-Mouse CD31, Clone MEC 13.3 (RUO) | BD Biosciences | Cat# 550274, RRID: AB_393571 |
| Anti-ER-TR7 rat monoclonal | Abcam | Cat# ab51824; RRID: AB_881651 |
| Anti-GFP chicken polyclonal | Abcam | Cat# ab13970; RRID: AB_300798 |
| Anti-mouse/human CD207 (Langerin) Antibody | BioLegend | Cat# 144202; RRID: AB_2562088 |
| Rabbit Anti-mouse Lyve1 antibody | Abcam | Cat# ab14917; RRID: AB_301509 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma | T5648 |
| EdU (5-ethynyl-2’-deoxyuridine) | Invitrogen | E10187 |
| Sytox Orange | Invitrogen | S34861 |
| Sytox Blue | Invitrogen | S34857 |
| Collagenase 1 | Worthington | LS004196 |
| Liberase TM | Roche | 5401127001 |
| Liberase TL | Roche | 5401020001 |
| Ethyl cinnamate | Sigma-Aldrich | 112372 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit | Invitrogen | C10419 |
| Click-iT EdU Alexa Fluor 647 Imaging Kit | Invitrogen | C10340 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | Charles River Laboratories | 027 |
| Mouse: B6.FVB-Tg(CD207-Dta)312Dhka/J | The Jackson Laboratory | 017949 |
| Mouse: Tg(CD207-cre/ERT2)1Dhka/J | The Jackson Laboratory | 028287 |
| Mouse: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | The Jackson Laboratory | 007676 |
| Deposited Data | ||
| Haensel et al. (2020) scRNA-seq dataset (raw) | GEO | GSE142471 |
| Konieczny et al. (2022) scRNA-seq dataset (processed) | GEO | GSE166950 |
| Analysis Code to reproduce analysis | Github depository | https://github.com/khbridges/waskolangerhans DOI: 10.5281/zenodo.7324227 |
| Software and Algorithms | ||
| FIJI (ImageJ) | NIH | https://fiji.sc |
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| FlowJo | FlowJo, LLC | https://www.flowjo.com |
| GraphPad Prism | GraphPad Software, Inc | https://www.graphpad.com |
| Scanpy | Python | https://github.com/theislab/scanpy |
| Tensorflow | Python | https://www.tensorflow.org |
| NicheNet | R | https://github.com/saeyslab/nichenetr |
| Circlize (for chord diagram visualization) | R | https://github.com/jokergoo/circlize |
| Seurat 3.0 | Stuart et al., 2019 | https://satijalab.org/seurat/ |
| Cell Ranger Single-Cell Software Suite | 10x Genomics | https://support.10xgenomics.com/singlecell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
LCs also significantly upregulated 577 genes in response to injury (Fig. 2E; p-adj value < 0.05), and gene ontology analysis of the changed genes in LCs during wound repair revealed that angiogenesis was a major category (Figs. 2F–G). Furthermore, several angiogenic genes were upregulated by LCs after injury (Fig. 2H). Mapping the top ligands predicted to interact with EC receptors during wound healing, we observed that LCs are a significant source of these interactions, along with fibroblasts, keratinocytes, and macrophages (Fig. S3F).
LCs localize at the leading edge of the angiogenic front in skin wounds
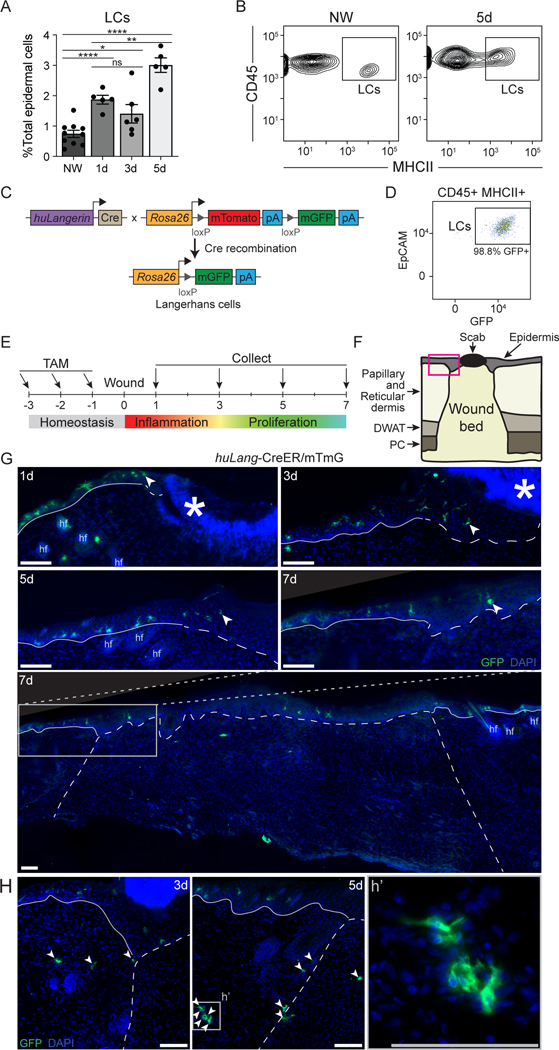
Langerhans cells are seeded in the skin during embryonic development32, and are replenished by low rates of local proliferation33, or by bone marrow-derived precursors that migrate into the epidermis and differentiate into LCs34. In consonance with prior analysis of the location of langerin+ in skin wounds18, we found a significant increase in MHCII+, CD45+ LCs within the healing epidermis in skin wounds from days 1 to 5 (Figs. 3A, 3B), which is consistent with the increase in LCs noted in scRNA sequencing data (Fig. 1B).
Figure 3: LCs localize at the epidermal and dermal edges of skin wounds.
(A) Flow cytometry quantification of LCs in nonwounded (NW) epidermis and epidermal edges of WT skin wounds 1, 3, and 5 days after injury. LCs quantified as a percent of total epidermal cells Data are 5–9 mice as indicated. Error bars indicate mean +/− SEM. two-way ANOVA, multiple comparisons with each timepoint, *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.00005.
(B) Representative flow cytometry plots of LCs (CD45+ MHCII+ epidermal cells) in nonwounded epidermis (NW, left) and at the edges of 5-day wounds (5d, right) in WT mice.
(C) Schematic summarizing genetic strategy to express membrane-associated GFP (mGFP) in mature LCs in huLang-CreER/mTmG (LC-iGFP) inducible fluorescent reporter mice.
(D) Representative flow cytometry plot demonstrating the efficiency of GFP labeling of mature LCs (CD45+ MHCII+ epidermal cells) in LC-iGFP mice.
(E) Schematic depicting tamoxifen treatment timeline and wound healing time points for histological analysis of LC-iGFP mice.
(F) Schematic of skin wound cross-section. Pink box outlines the epidermal wound edge. DWAT = dermal white adipose tissue, PC = panniculus carnosus.
(G) Fluorescent imaging of GFP+ LCs (green) and DAPI (blue) in epidermal wound edges 1, 3, 5, and 7 days after injury in LC-iGFP mice. Solid white lines trace the non-wounded epidermis, and the dashed white lines delineate the wound bed. White arrows label the LC closest to the wound center. Asterisks indicate non-specific labeling of scab, hair follicles (hf). Scale bars, 100 μm.
(H) Fluorescent imaging of GFP+ LCs in the dermis at wound edges of 3- and 5-day wounds. White arrows label LCs in dermis and wound bed. White dashed lines delineate wound edges. Scale bars, 100 μm. See also Figure S4.
To precisely and accurately define the spatial location of LCs in mouse wounds, we generated an inducible fluorescent reporter mouse in which membrane-associated GFP can be activated in LCs with high specificity by crossing huLangerin-CreER mice35 to mT/mG dual fluorescent reporter mice36 to generate huLangerin-CreER;mT/mG (LC-iGFP) (Fig. 3C). Low dose tamoxifen treatment of these mice induces the nuclear localization of Cre recombinase in LCs but not in langerin+ dermal dendritic cells (dDCs)35 (Fig. S4A). Indeed, when we treated LC-iGFP mice with tamoxifen (Fig. 3E), we noted that 98% of EpCAM+, CD45+, MHCII+ LCs were labeled (Fig. 3D), while Langerin+, EpCAM- dDCs were GFP (Fig. S4A). Thus, this mouse model labels LCs specifically, which is consistent with the specific activity of the huLangerin promoter in LCs21. Furthermore, this mouse model allows specific labelling of resident LCs by administering tamoxifen (which induces LC-specific GFP expression) prior to injury, thereby only labeling the pool of fully-differentiated LCs present in naive skin.
Given the specificity and high efficiency of LC labeling in LC-iGFP mice, we examined the location of GFP+ cells in skin wounds 1, 3, 5, and 7 days post-injury (Figs. 3E–G). At each time point, we observed LCs in the epidermis at the edges of skin wounds and in adjacent skin (Fig. 3G). At later stages of healing, 5 and 7 days after injury, we also observed LCs in the newly-regenerated epidermis (Fig. 3G). GFP+ LCs were also present in the dermis in wound-adjacent skin, at wound edges, and later, in the wound bed (Fig. 3H).
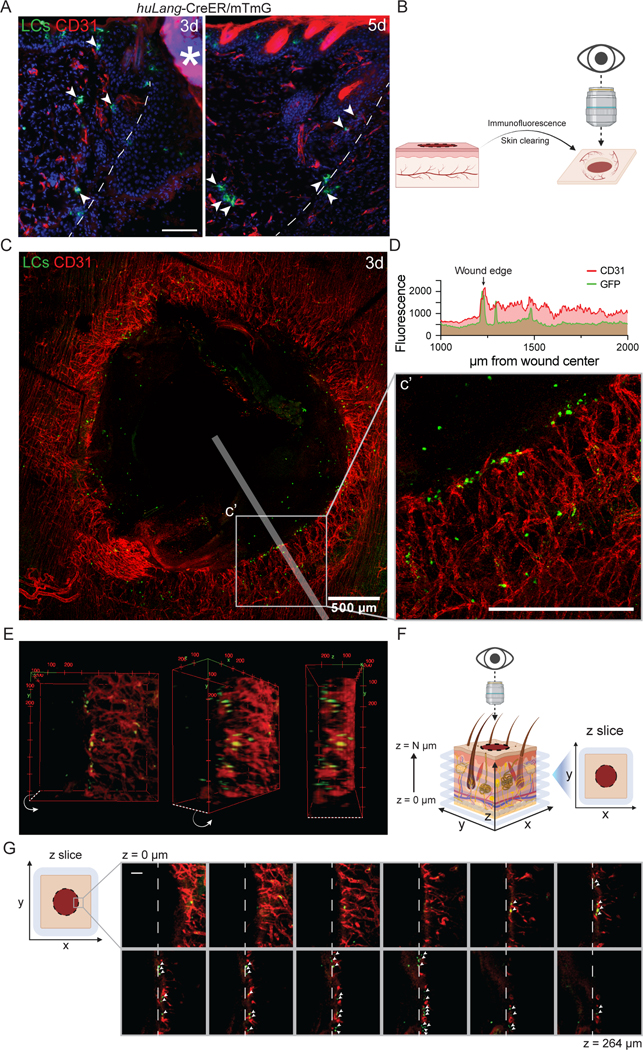
Since the main genes induced by LCs after injury were associated with angiogenesis (Fig. 2F), we sought to define the spatial relationship between LCs and ECs during skin repair. Using LC-iGFP mice, we injected tamoxifen daily for 3 days prior to wounding to induce GFP expression in LCs (Fig. 3E), and then co-stained wound beds for CD31+ endothelial cells. In cross sections of skin wounds, LCs in the dermis were near ECs 3 and 5 days after injury (Fig. 4A). To examine the spatial relationship between LCs and sprouting vessels in the three-dimensional tissue, we performed tissue clearing of 3-day wound beds of LC-iGFP mice and immunostained the whole mount tissue with antibodies against CD31 and GFP (Fig. 4B). Imaging throughout the depth of the skin tissue provided a view of the regenerating endothelial vessels as the tip cells enter the repairing dermal compartment of the wound bed (Fig. 4C). We noted that the middle of the wound bed contained GFP+ cells that were likely resident in the re-epithelizing keratinocyte layer (Fig. S4B). Interestingly, we found that many LCs were clustered at the leading tips of the repairing endothelial vessels (Figs. 4C–G). Additionally, a few LCs were also observed along the length of vessels (Figs. 4C–D). These data indicate that LCs are spatially poised to promote dermal angiogenesis during wound healing.
Figure 4: Langerhans cells localize near tips of regenerating blood vessels during wound healing.
(A) Imaging of GFP+ LCs (green) and immunostaining of CD31+ blood vessels (red) in cross-sections of 3- and 5-day wounds from LC-iGFP mice. Arrows label LCs close to blood vessels. White dashed lines delineate wound edges. Asterisks (*) indicates non-specific labeling of scab. Scale bars, 100 μm.
(B) Schematic depicting the orientation of 3-dimensional wound beds for whole mount confocal microscopy.
(C) Maximum intensity projection of confocal imaging of CD31+ blood vessels (red) and GFP+ LCs (green) in 3-day whole mount wounds of LC-iGFP mice. Transparent white line depicts line scan path quantified in D. Scale bar, 500 μm.
(D) Quantification of CD31 (red) and GFP (green) fluorescence in line scan (75 μm wide) along wound radius. Arrow indicates wound edge.
(E) 3-Dimensional volume rendering of CD31+ blood vessels (red) and GFP+ LCs (green) at the edges of a 3-day wounds in LC-iGFP mice.
(F) Schematic depicting the orientation of optical z-slices acquired from confocal microscopy of whole mount wound beds.
(G) Montage of z-slice images from deep (z = 0 μm) to superficial (z = 264 μm) depth of a 3-day LC-iGFP wound bed. White arrows indicate GFP+ LCs (green) close to CD31+ (red) blood vessels at the wound leading edge (white dashed lines). Scale bar, 100 μm.
Langerhans Cells are Necessary for Efficient Wound Healing
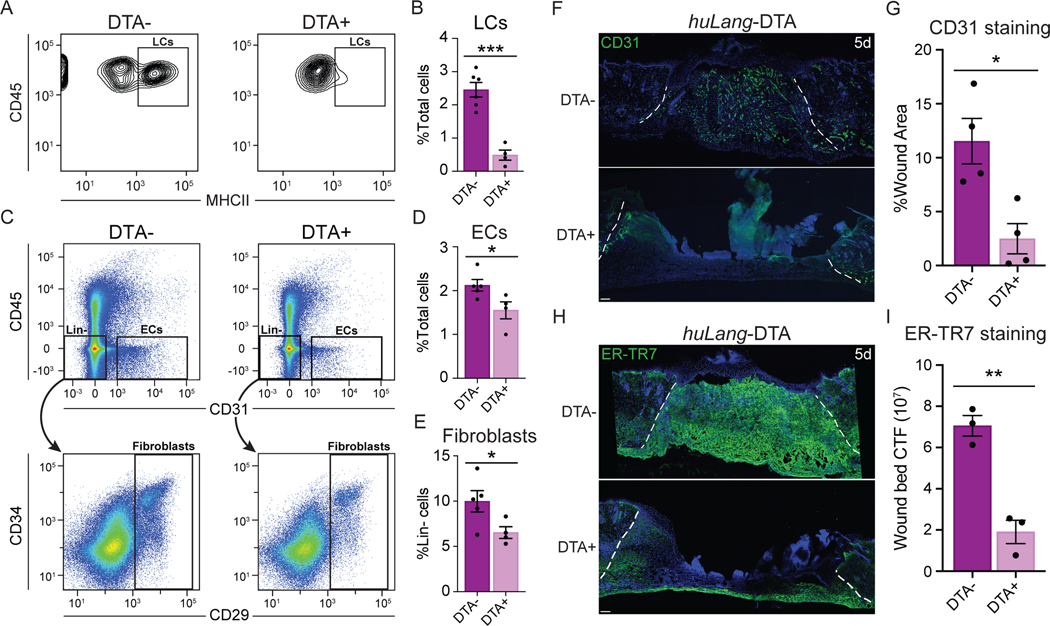
Since LCs are present in regions of skin wounds undergoing repair (Figs. 1–4), we sought to determine if LCs contribute to skin wound healing, and particularly to angiogenesis. To this end, we analyzed tissue repair in a mouse model that specifically lacks LCs using the huLangerin-DTA (huLang-DTA) mice, in which the promoter sequence for the human langerin gene drives expression of diphtheria toxin, resulting in constitutive and specific ablation of LCs without impacting langerin+ dDCs21 (Figs. 5A, S5A). Importantly, we did not detect any changes in skin structure or immune cells in uninjured adult huLang-DTA mice compared to control littermates (Fig. S5B). After injury, we noted that the epidermis bordering 1-day wounds in littermate control mice contained ~2.5% LCs, LCs were almost entirely depleted in huLang-DTA+ mice (Figs. 5A–B).
Figure 5: LCs are necessary for efficient wound healing.
(A-B) Flow cytometry plots (A) and quantification (B) of LCs (CD45+ MHCII+ epidermal cells) from wound-edge epidermal preps in huLang-DTA+ mice and DTA- littermate controls. Data are 3–5 mice. Error bars indicate mean +/− SEM. unpaired T-test, ***p < 0.0005.
(C-D) Flow cytometry plots (C) and quantification of endothelial cells (ECs; CD31+ CD45-) (D) and fibroblasts (Fb; lineage (Lin)- CD29+) (E) in 5-day wound beds from huLang-DTA+ mice and DTA- littermate controls. Data are 3–5 mice. Error bars indicate mean +/− SEM. unpaired T-test, ***p < 0.0005.
*p < 0.05.
(F-G) Images (F) and quantification (G) of CD31 immunostaining (green) and DAPI (blue) in cross-sections of 5-day wound beds from huLang-DTA and control mice. White dashed lines delineate wound edges. Scale bars, 100 μm. Data are 4 mice. Error bars indicate mean +/− SEM. unpaired T-test, *p < 0.05.
(H-I) Images (H) and quantification (I) of ER-TR7 immunostaining to label fibroblasts (green) in cross-sections of 5-day wound beds from huLang-DTA and control mice. White dashed lines delineate the wound edges. Scale bars, 100 μm. Data are 3 mice. unpaired T-test, **p < 0.005. See also Figures S5 and S6.
To examine skin wound repair in the absence of LCs, we analyzed skin wounds at various time points after injury in huLang-DTA mice. Interestingly, LC-deficient mice did not display defects in the inflammatory response 3 days after injury. LC-deficient mice possessed similar numbers of monocytes, macrophages, neutrophils, and T cells in 3-day wounds compared to DTA- littermate controls (Figs. S5C–G). Thus, despite the importance of the recruitment of immune cells during the inflammatory phase of healing 5 and the proposed role for LCs in this process 37, inflammatory cell recruitment after skin injury proceeded normally in the absence of LCs, as indicated by similar monocytes and macrophages numbers and polarity in huLang-DTA mice compared to control mice (Figs. S5D and S5E). Furthermore, we did detect a slightly more T cells as a percentage of immune cells (Fig. S5F), the absolute number of T cells at day 3 after injury is not significant (Fig. S5G) likely due to the slightly reduced numbers of CD45+ cells (Fig. S5D).
Examination of the proliferative phase of wound healing revealed that huLang-DTA+ mice exhibited profound regenerative defects at day 5 after injury compared to littermate controls. Histological analysis of hematoxylin and eosin-stained sections of skin wounds revealed that wounds of huLang-DTA+ mice were wider and thinner than control mice (Fig. S6A). To analyze which cell types were impaired in huLang-DTA+ mouse wounds, we analyzed the repair of keratinocytes, ECs, and fibroblasts. While keratinocyte re-epithelialization was variable in both control and huLang-DTA+ mice, no significant defect was detected in the percent of wound area covered by in huLang-DTA+ mouse wounds compared to control mice (Figs. S6A–B). However, FACS analysis of cells within skin wounds (Fig. S6C) revealed a significant reduction in the number of CD31+ ECs (Figs. 5C–D) and CD29+ fibroblasts (Figs. 5C and 5E) in the wound beds of huLang-DTA+ mouse wounds compared to control mice. Histological analysis of the localization of ER-TR7+ fibroblasts within 5 day wound beds revealed a ~70% reduction in fluorescence within skin wounds depleted for LCs (Figs. 5H–I). Similarly, huLang-DTA+ mice displayed a 4-fold decrease in fluorescence of CD31+ EC compared to wound beds of control mice (Figs. 5F–G). Since angiogenesis of CD31+ blood ECs precedes regeneration of Lyve1+ lymphatic ECs (Fig. S6D)38,39, we conclude that these defects are driven primarily by defects in blood ECs. Together, these data suggest that LCs directly promote proliferative repair processes, which are critical for efficient wound repair.
To investigate the angiogenic repair defects in LC depleted mice in more detail, we examined the vascular network in huLang-DTA mice. Naive skin of LC-depleted mice displayed normal vasculature organization (Fig. S7A) as indicated by whole mount imaging of cleared wound beds immunostained with antibodies against CD31. Control wounds displayed clear signs of blood vessel repair mechanisms, including higher coverage of the wound bed with CD31+ blood vessels and thick vessels concentrated distally to the wound bed (Fig. 6A). In contrast, LC-depleted wounds exhibited stunted vessel growth at the edge of the wound bed and lacked clusters of large vessels (Fig. 6A).
Figure 6: Blood vessel morphology and proliferation is defective in the absence of Langerhans cells.
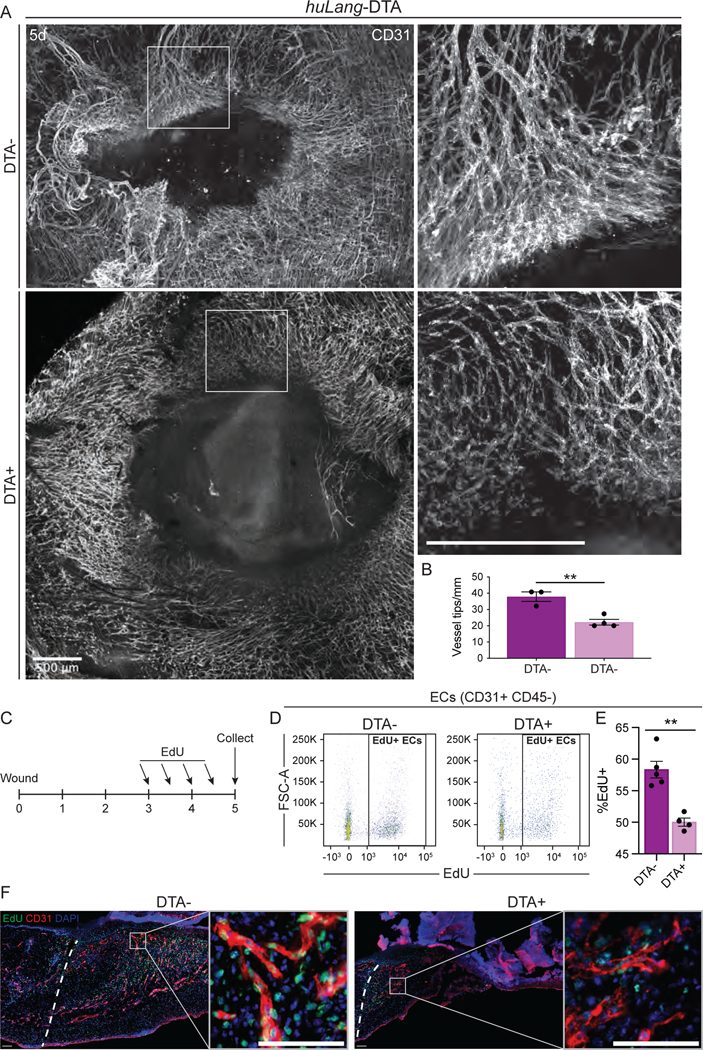
(A) Maximum intensity projection of confocal imaging of CD31+ blood vessels (white) in whole mounts of 5-day wound beds from huLang-DTA mice. Insets provide higher magnification view of branching vessels at wound edges. Scale bars, 500μm.
(B) Quantification of the number of blood vessel tips per mm wound edge in 5-day wounds from huLang-DTA mice. Data are 3–4 mice. Error bars indicate mean +/− SEM. unpaired T-test, **p < 0.005.
(C) Schematic depicting experimental design of EdU pulse-chase experiment in huLang-DTA mice and control mice.
(D-E) Representative flow cytometry plots (D) and quantification (E) of EdU+ labeling in endothelial cells (ECs; CD31+ CD45- cells) from 5-day huLang-DTA wounds. Data are 4–5 mice. Error bars indicate mean +/− SEM. unpaired T-test, **p < 0.005.
(F) Fluorescent imaging of EdU (green) and DAPI (blue) nuclei within CD31+ (red) blood vessels in 5-day wounds of huLang-DTA and control (DTA-) mice. See also Figure S7.
Examining the tip cells, which extend and proliferate to form new blood vessels at the leading edge of angiogenesis, we noted a high density of overlapping tip cells clustered at the leading edge of the sprouting angiogenic front in control mice (Figs. 6A–B), although the thickness of the leading edge was similar between control and DTA+ mice (Fig. S7B). However, in mice lacking LCs, tip cells were distant from neighboring tip cells at the leading edge of the blood vessel front, and the vessels appeared thinner and sparser than the wounds control mice (Figs. 6A–B).
To determine if LCs are required for proliferation of endothelial cells after injury, we pulsed huLang-DTA mice with EdU during the height of EC proliferation (3 and 4 days after injury) to label proliferating cells (Fig. 6C), and analyzed CD31+, EdU+ cells 5 days after injury using flow cytometry. Compared to control samples, wounds from LC-depleted mice contained significantly fewer total EdU+, CD31+ endothelial cells (Figs. 6D–E). Immunostaining of skin wound sections from mice pulsed with EdU confirmed that CD31+ cells in LC-depleted mice displayed reduced proliferation during this critical time for angiogenesis associated with tissue repair (Fig. 6F). Interestingly, significant changes in proliferation at day 5 in huLang-DTA mice compared to control mice in CD45+ immune cells and fibroblasts were not observed (Figs. S7C–D). Given that fibroblast repair is aberrant in mouse wounds lacking LCs, fibroblasts may receive migratory signals directly or indirectly from LCs, which will be explored in future studies. Taken together, our data provide evidence that LCs are necessary for angiogenesis and fibroblast repair in skin wounds.
Discussion
Here, by analyzing scRNA-sequencing data to identify cellular interactions during skin repair, we provide a map of the early angiogenic niche within skin wounds and implicate LCs as an essential regulator of angiogenesis during skin repair.
Our data indicate that Langerhans cells contribute to the early stages of angiogenesis, likely through multiple mechanisms. Several of the ligands expressed by LCs in skin wounds likely induce EC expression of cFos, a subunit of the Activator Protein-1 (AP-1) complex, which is known to activate VEGF expression in ECs40,41. LCs also upregulate several genes that impinge upon the VEGFA pathway, a classic angiogenesis regulator, including VEGFA, Neuropilin, and PGF. The expression of CD24a, which distinguishes LCs from dDCs, is predicted to regulate Slc30a1, a zinc transporter42. Interestingly, Zn2+ has been shown to regulate EC survival and growth through activation of GPR39, a Zn2+-sensing receptor43. LCs in skin wounds also upregulate many factors that promote EC migration including C-C Motif Chemokine Ligand (Ccl2), Cxcl16, (Matrix metalloproteinase) Mmp9, Metalloproteinase Inhibitor (TIMP)1. Our data resonate with studies that have implicated dendritic cells in promoting angiogenesis during inflammation in lymph nodes44 and during ovarian carcinoma tumorigenesis45. Our model is consistent with recent data demonstrating that a subset of LCs do not migrate to LNs and act locally in the skin46. Since we focused on early stages of skin repair, our data do not define whether additional mechanisms can compensate for the function of LCs during angiogenesis at later stages of repair, allowing angiogenesis to normalize at later timepoints. Overall, these data broaden the function of LCs in the skin beyond roles as specialized tissue-associated monocyte-derived cells involved in antigen presentation and T-cell immunity21.
Recent analysis of ECs from several tissues suggests that the surrounding tissue niche contributes to EC heterogeneity17. Interestingly, the wound healing phenotype in the LC-specific depletion model differed from the wound repair defects previously reported in which all langerin+ cells were depleted18, indicating that LCs and langerin+ dDCs play unique and opposing roles during wound healing. Our future studies will focus on defining the molecular basis for LCs function during wound repair.
Angiogenesis is a complex process that requires inputs from multiple cell types within tissue to promote EC migration, proliferation, and the formation of new vessels. Angiogenesis is impaired in wound repair of diabetic patients and mice, which likely contributes to chronic wound healing defects47. Interestingly, healing diabetic wounds contain more LCs than non-healing wounds from diabetic patients48, suggesting that LCs may be a convenient cellular therapy for these chronic wounds. While the bulk of our analysis focuses on early timepoints after injury, our data provides a basis for understanding EC behavior in pathological angiogenesis in cancer and chronic wounds, which are potential opportunities for controlling angiogenesis to prevent disease through a combinatorial approach that targets multiple aspects of the early angiogenic niche within tissues.
Limitations of Study
Our study identifies provides a map of the interactions between cells in the skin and endothelial cells, we only provide analysis of one timepoint in the dynamic process of wound repair and additional mechanisms might emerge by analyzing additional timepoints. Further, while LCs upregulate angiogenic factors and reside at the tip of repairing endothelial cells in mouse wounds, LCs may also drive angiogenesis indirectly through additional cell types including fibroblasts, which are aberrant in the absence of LCs. Future questions include the specific molecular mechanisms by which LCs are recruited to regenerating endothelial cells and drive angiogenesis and fibroblast repair in skin wounds.
STARS METHODS
Resource Availability:
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Valerie Horsley, valerie.horsley@yale.edu
Materials Availability:
This study did not generate new unique reagents.
Data and Code Availability:
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the Key resource Table.
Microscopy data reported in this paper will be shared by the lead contact upon request.
The code to reproduce analyses of single Cell RNA sequencing are deposited at the GitHub repository: https://github.com/khbridges/wasko-langerhans.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Wild-type C57BL6/J mice were purchased from Charles River. B6.FVB-Tg(CD207-Dta)312Dhka/J (huLang-DTA); Tg(CD207-cre/ERT2)1Dhka/J (huLang-CreER); and B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (mT/mG) mice were purchased from The Jackson Laboratories. Male mice were used to avoid the impact of variable hair cycling in female mice on wound healing. Mice were maintained through routine breeding in an Association for Assessment and Accreditation of Laboratory Animal Care (AALAC)-accredited animal facility at Yale University. Animals were maintained on a standard chow diet ad libitum (Harlan Laboratories, 2018S) in 12-hour light/dark cycling. Two or three injured mice were housed per cage. All experimental procedures were approved and in accordance with the Yale University Institutional Animal Care and Use Committee.
METHOD DETAILS:
Lineage tracing and EdU treatment
To label Langerhans cells, huLangerin-CreER; mT/mG mice received daily intraperitoneal (i.p.) injections of 50 μL of 30mg/mL tamoxifen (Sigma Aldrich) in sesame oil for 3 days.
For EdU experiments, 50mg/kg of EdU (Invitrogen) was injected intraperitoneally at indicated time points and detected per manufacturer protocols. Detection of EdU-incorporating cells was performed using Click-it EdU Imaging or Flow Cytometry Assay kits (Invitrogen).
Wound healing models
RNA sequencing data from GSE14247111 utilized a 6-mm punch biopsy to induce wounding and analyzed the proliferative phase of wound repair at day 4. The RNA sequencing data from wounds purified for CD45+ cells (GSE166950)22 and all other skin wound analyses utilized a 4-mm punch biopsy model. The proliferative phase of wound repair begins at day 4 and day 3 for the 6-mm and 4-mm wound models, respectively.
For all histological and FACS data, 7–9-week-old mice were wounded during the telogen phase of hair cycling. Mice were anesthetized using isoflurane and wounds were created on shaved back skin using a 4mm biopsy punch (Accuderm). Animals were sacrificed at noted intervals after injury and wound beds were processed for subsequent analysis.
Immunofluorescence and imaging
Skin sections:
Mouse skin and wound beds were embedded in O.C.T. and wound beds were sectioned through their entirety to identify the center. 14 μm cryosections were processed as previously described 49 and stained with antibodies listed in the Key resource Table. Composite images were acquired using the tiles module on a Zeiss AxioImager M1 (Zeiss) equipped with an Orca camera (Hamamatsu). Tiled and stitched images of wound sections were collected using a 20X objective, controlled by Zen software (Carl Zeiss). The percentage of the wound bed covered by DAPI staining (re-epithelialization), width of the wound bed, and ER-TR7 corrected total fluorescence were calculated from the 3 central most tissue sections using ImageJ software (National Institutes of Health, Bethesda, MD) as described previously50,51. Revascularization (CD31+) was calculated using Adobe Photoshop to measure the total pixels positive for antibody staining divided by the total number of pixels in wound beds. EdU labeling was performed using the Click-iT EdU™ Cell Proliferation Kit for Imaging per the manufacturer’s instructions (Invitrogen).
Skin whole mount:
Staining of whole mount adult mouse back skin was adapted from prior studies52. Briefly, mice were euthanized, and their back skin was chemically depilated (Nair, 5 minutes) and then cleansed with 70% ethanol. A 6mm-diameter biopsy punch was used to excise nonwounded back skin or wounds (captures 4mm wound with a 1mm border of surrounding nonwounded skin). Tissue was placed dermis-down on Whatman paper and fixed in 4% formaldehyde in PBS for 1 hour at room temperature, followed by extensive washing with PBS. Tissue was permeabilized with 0.3% Triton X-100 in PBS (PBS-T) overnight at 4°C, followed by incubation in blocking buffer (1% fish gelatin, 2.5% normal donkey serum, 2.5% normal goat serum, 1% BSA, 0.3% PBS-T) for 3–4 hours at room temperature. For immunolabeling, primary antibodies were incubated at room temperature overnight, followed by hourly washes with 0.3% PBS-T for 5 hours. Secondary antibodies conjugated to Alexa Fluor™ 488, RRX, or 647 (1:300, Invitrogen), were incubated at room temperature overnight, followed by hourly PBS-T washes for 5 hours, proceeded by tissue clearing.
Tissue clearing:
Tissue clearing was adapted from Gur-Cohen et al. (2019)52. Briefly, immunostained back skin tissues were dehydrated in increasing concentrations of ethanol (30%, 50%, and 70%, diluted in distilled water and adjusted to pH 9.0) for 45–60 minutes each at room temperature and with gentle agitation. Samples were then incubated in 2 rounds of 100% ethanol (no pH adjustment) for 60 minutes each, at room temperature with gentle agitation. Dehydrated samples were transferred into 500 μL ethyl cinnamate (Sigma) in polypropylene tubes for clearing overnight at room temperature. To acquire images, cleared skin was mounted dermis-down with ethyl cinnamate in a glass bottom microwell dish (35mm, MatTek) held in place with a coverslip (22 mm x 22 mm, Fisher Scientific).
Confocal microscopy:
Confocal images were acquired using a Zeiss LSM 880 confocal microscope. The LSM 980 confocal microscope is equipped with Zeiss Axio Observer Z1 inverted microscope with 405, 458, 488, 514, 561, and 633 laser lines, and Zen software (Zeiss). Stacks of 4–13 μm steps were collected (step sized determined by setting pinhole opening to 1 Airy Unit) with a 10x or 20x objective. Imaging data stitching, processing, and rendering was performed in ZEN (ZEISS) and FIJI (NIH). FIJI software was used to generate maximum intensity projections and 3D renderings of z-stacks.
Analysis of whole mount images:
To quantify endothelial vessel tips, Z-stack images were converted into maximum intensity projections, the length of leading endothelial edge was measured, and then the total number of vessel tips protruding into the wound was counted manually in FIJI. The number of vessel tips was then normalized to the length of the leading endothelial edge of the wound. To measure the thickness of the leading edge of the endothelial front, the FIJI was used to re-slice a Z-stack image of a whole mount wound bed into a series of cross-sectional images of the wound. The thickness of the tip of the endothelial front was then measured at 6 representative locations around the wound perimeter and averaged together for each mouse.
Flow cytometry and Cell Sorting
For all flow cytometry experiments, mouse back skin and wound beds were dissected and digested into a single cell suspension, resuspended in FACS staining buffer (1% BSA in PBS with 2mM EDTA), and then filtered with a 70 mm and a 40 mm cell strainer prior to centrifugation. Cell suspensions were stained with antibodies purchased from eBioscience, Biolegend, and BD Bioscience in the Key resource Table for 20–30 minutes on ice, washed, and then analyzed on the flow cytometer. Flow cytometry analysis was performed using FlowJo Software (FlowJo).
Immunophenotyping analysis:
For the quantification of myeloid cells and T cells, skin tissue was digested using Liberase TM (Roche). To exclude dead cells, Sytox Orange or Sytox Blue (Invitrogen, 1:1000) was added immediately before analysis. Flow cytometry was performed on a FACS Aria III with FACS DiVA software (BD Biosciences).
Dermal analysis:
For the analysis of dermal cell types (endothelial cells and fibroblasts), skin tissue was digested using Collagenase 1 (Worthington). Analysis of proliferation using EdU incorporation was performed using the Click-iT™ EdU Flow Cytometry Assay Kit per the manufacturer’s instructions (Invitrogen). Flow cytometry was performed on a BD FACS LSR Fortessa X20 with FACS DiVA software (BD Biosciences).
Epidermal cell analysis:
For the analysis of LCs in epidermal tissue, we adapted the protocol from 53. In short, naive skin or wound beds were dissected and the underlying facia and adipose tissue were scraped off. Skin pieces were floated dermis-down on 0.25% Trypsin-EDTA (Gibco) at 37˚C for 30–60 minutes, and then epidermal cells were gently scraped in the direction of hair growth into the solution. Cells were then washed, pelleted, and stained as described. To exclude dead cells, Sytox Orange or Sytox Blue (Invitrogen, 1:1000) was added immediately before analysis. Flow cytometry was performed on a FACS Aria III with FACS DiVA software (BD Biosciences).
Single-cell RNA-sequencing data analysis
Data for GSE166950:
Unwounded skin or wound beds with 0.25 mm perimeter of adjacent nonwounded skin were excised and digested in Liberase TL (Sigma) at 37°C for 2.5 hours. After placing samples on ice and adding EDTA and FBS, digested tissues were mechanically disrupted by syringe plunger and then filtered through a 70-micron filter to exclude tissue debris and obtain a single cell suspension for downstream analyses.
Single cell suspensions were stained with anti-CD16/32 before staining with surface fluorescent conjugated antibodies and/or oligo-tagged antibodies at predetermined concentrations in a 100 μL staining buffer (PBS containing 5% FBS and 1% HEPES) per 107 cells. Stained cells were re-suspended in 4’,6-diamidino-2-phenylindole (DAPI) in FACS buffer (Sigma-Aldrich) prior to analysis. Data were acquired on LSRII Analyzers (BD Biosciences) and then analyzed with FlowJo program. Fluorescence-activated cell sorting (FACS) was conducted using Aria Cell Sorters (BD Biosciences).
FACS purified live CD45.2+ CD90.2+ TCR Vγ3 − cells from unwounded skin, 3 and 5 days post wounding were prelabeled with surface epitope marking oligo-tagged antibodies and sample specific oligo-tagged Totalseq-A antibodies (Biolegend, see Key resource Table). Hashed samples were pooled at Ctrl,1: D3,1.5, D5:1 ratio prior to library preparation (Chromium Single Cell 3′ Library, 10x Genomics) and sequenced on an Illumina HiSeq 4000 as 150 bp paired-end reads. Sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. The Cell Ranger Single-Cell Software Suite was used to perform sample demultiplexing, barcode processing, and single-cell 3’ gene counting. The cDNA insert was aligned to the mm10/GRCm38 reference genome. Only confidently mapped, non-PCR duplicates with valid barcodes and UMIs were used to generate the gene-barcode matrix. Cell Ranger output was further analyzed in R using the Seurat package54. Surface epitope oligo sequences were merged with cell transcriptome data by matching the cell barcode IDs.
Further analysis including quality filtering, the identification of highly variable genes, dimensionality reduction, standard unsupervised clustering algorithms, and the discovery of differentially expressed genes was performed using the Seurat R package. Samples were demultiplexed to filter out multiplets (cells mapping to multiple hashtags) and negative cells (cells missing hashtags) with a positive quantile threshold of 0.99 between samples. Individual samples were further processed to remove cells with > 20% mitochondrial gene expression. To exclude low quality cells and remaining multiplets or cells that were extreme outliers, we calculated the distribution of total genes/ cells. Following that, we applied control parameters to filter cells with fewer than 200 detected genes and more than 3800 detected genes. After removing unwanted cells from the dataset, we normalized the data by the total expression, multiplied by a scale factor of 10,000, and log-transformed the result.
The downstream analysis was performed in R programming environment and primarily using the Seurat package55.The hashtag data was first demultiplexed using Seurat demultiplexing where the HTO data was normalized using centered log ratio transformation (CLR) followed by HTODemux function with positive quantile set to (0.99). The HTO demultiplexed data was further subset to only include the singlets. The RNA data was further filtered using standard QC steps where cells with number of genes less than 200 and greater 4500 were filtered out to remove any low-quality cells and any remaining doublets. Cells with overall mitochondrial gene expression greater 20% were also filtered to remove cells with poor survival rate.
Followed by quality control filtering, we performed standard data processing on RNA seq assay including normalization, scaling and PCA. First clustering results were generated using the first 7 dimensions and resolution set to 0.3 for the FindClusters function. UMAP plot (Fig. 2A) was generated using these results and the resulting 12 clusters were annotated using marker genes identified after using FindAllMarkers function and using predefined marker genes for each cell type (Fig. 2A, Fig. S2C). Feature expression plots were also generated for genes of interest using FeaturePlot function (Fig. 2B, Fig. S2C–D).
Data analysis of GSE142471:
Data from Haensel et al. (2020) was first preprocessed to remove low quality cells using Seurat tools adapted for Python (Scanpy)56, and then log-normalized to convert mRNA counts to gene expression. Dimensionality reduction and visualization were accomplished using the Scanpy implementation of Uniform Manifold Approximation and Projection (UMAP)56). To label the data by broad cell type, we adapted the annotation pipeline from 57 to use a simple feedforward neural network (NN). The NN was built with the TensorFlow module in Python (https://www.tensorflow.org/). Differentially expressed genes (DEGs) across wounding conditions were identified using the diffxpy package (log2FC > 1 and adjusted p-value < 0.05). Enrichment analysis on the top DEGs was performed using the Generally Applicable Gene-set Enrichment (GAGE) package in R58. Gene Ontology (GO) biological processes and the Kyoto Encyclopedia of Genes and Genomes (KEGG) were used as reference databases.
To characterize potential signaling to endothelial cells (ECs) from scRNA-seq, we took advantage of the NicheNet algorithm, which makes inferences about ligand binding from patterns in expression of target genes in receiving cells15. Target genes in the EC population were identified as genes significantly upregulated after wounding (log2FC > 0.25 and adjusted p-value < 0.05). NicheNet is available as an open-source software package in R.
QUANTIFICATION AND STATISTICAL ANALYSIS:
Histological quantification for each wound bed was conducted on the three central-most sections and the averages from two wounds were averaged for each animal. Image and FACS quantification were analyzed using unpaired t-tests to compare two groups and two-way ANOVA to compare 4 timepoints of LC cell numbers in wounds. Analyses were performed using GraphPad Prism.
For single-cell measurements, statistics were generally performed using two-sided Wilcoxon rank-sum tests and the Benjamini-Hochberg method of correction for pairwise multiple comparisons, or as specified in the figure legends. Values were considered significant at P < 0.05. Analyses were performed using custom Python and R scripts.
Supplementary Material
Highlights.
scRNA sequencing provides a map of the angiogenic niche during skin repair
Langerhans cells upregulate angiogenic mRNAs after injury
Langerhans cells localize to angiogenic blood vessel tips in wound beds
Langerhans cells are necessary for proper angiogenesis and repair after injury in mice
Acknowledgements
We would like to thank Dr. Themis Kyriakides and the members of the Horsley, Miller-Jensen, and Eichmann laboratory for their feedback and critical analysis of these data and manuscript. We would also like to thank the Yale Animal Resources Center (YARC) staff for animal husbandry and the Yale Science Building (YSB) Imaging core facility for confocal use. V.H. is funded by N.I.H- NIAMS R01s AR076938, AR0695505, AR075412, and AR079232. R.W. was funded by NIH T32 GM007499 and NIH F31 AR073094–01A1. K.M-J. is funded by NIH U01-CA238728, R01-CA238728, and R01-GM123011. S.N. is funded by R01-AI68462 and is a NYSCF Robertson Stem Cell Investigator.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Inclusion and Diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Herbert SP, and Stainier DY (2011). Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat Rev Mol Cell Biol 12, 551–564. 10.1038/nrm3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giacca M, and Zacchigna S. (2012). VEGF gene therapy: therapeutic angiogenesis in the clinic and beyond. Gene Ther 19, 622–629. 10.1038/gt.2012.17. [DOI] [PubMed] [Google Scholar]
- 3.Johnson KE, and Wilgus TA (2014). Vascular Endothelial Growth Factor and Angiogenesis in the Regulation of Cutaneous Wound Repair. Adv Wound Care (New Rochelle) 3, 647–661. 10.1089/wound.2013.0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams RH, and Alitalo K. (2007). Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol 8, 464–478. 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 5.Eming SA, Martin P, and Tomic-Canic M. (2014). Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med 6, 265sr266. 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stockmann C, Kirmse S, Helfrich I, Weidemann A, Takeda N, Doedens A, and Johnson RS (2011). A wound size-dependent effect of myeloid cell-derived vascular endothelial growth factor on wound healing. J Invest Dermatol 131, 797–801. 10.1038/jid.2010.345. [DOI] [PubMed] [Google Scholar]
- 7.Rossiter H., Barresi C., Pammer J., Rendl M., Haigh J., Wagner EF., and Tschachler E. (2004). Loss of vascular endothelial growth factor a activity in murine epidermal keratinocytes delays wound healing and inhibits tumor formation. Cancer Res 64, 3508–3516. 10.1158/0008-5472.CAN-03-2581. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J, Patel J, Kaur S, Sim SL, Wong HY, Styke C, Hogan I, Kahler S, Hamilton H, Wadlow R, et al. (2021). Sox9 and Rbpj differentially regulate endothelial to mesenchymal transition and wound scarring in murine endovascular progenitors. Nat Commun 12, 2564. 10.1038/s41467-021-22717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman PJ, Berndt MC, Gorski J, White GC, Lyman S, Paddock C, and Muller WA (1990). PECAM-1 (CD31) cloning and relation to adhesion molecules of the immunoglobulin gene superfamily. Science 247, 1219–1222. 10.1126/science.1690453. [DOI] [PubMed] [Google Scholar]
- 10.Eming SA, Brachvogel B, Odorisio T, and Koch M. (2007). Regulation of angiogenesis: wound healing as a model. Prog Histochem Cytochem 42, 115–170. 10.1016/j.proghi.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Haensel D, Jin S, Sun P, Cinco R, Dragan M, Nguyen Q, Cang Z, Gong Y, Vu R, MacLean AL, et al. (2020). Defining Epidermal Basal Cell States during Skin Homeostasis and Wound Healing Using Single-Cell Transcriptomics. Cell Rep 30, 3932–3947.e3936. 10.1016/j.celrep.2020.02.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang DH, Hsu CF, Lin CY, Guo JY, Yu WC, and Chang VH (2013). Krüppellike factor 10 upregulates the expression of cyclooxygenase 1 and further modulates angiogenesis in endothelial cell and platelet aggregation in gene-deficient mice. Int J Biochem Cell Biol 45, 419–428. 10.1016/j.biocel.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Murphy JF, Steele C, Belton O, and Fitzgerald DJ (2003). Induction of cyclooxygenase-1 and −2 modulates angiogenic responses to engagement of alphavbeta3. Br J Haematol 121, 157–164. 10.1046/j.1365-2141.2003.04247.x. [DOI] [PubMed] [Google Scholar]
- 14.Kumar VB, Viji RI, Kiran MS, and Sudhakaran PR (2012). Angiogenic response of endothelial cells to fibronectin. Adv Exp Med Biol 749, 131–151. 10.1007/978-1-4614-3381-1_10. [DOI] [PubMed] [Google Scholar]
- 15.Browaeys R, Saelens W, and Saeys Y. (2020). NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods 17, 159–162. 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 16.Koh TJ, Novak ML, and Mirza RE (2013). Assessing macrophage phenotype during tissue repair. Methods Mol Biol 1037, 507–518. 10.1007/978-1-62703-505-7_30. [DOI] [PubMed] [Google Scholar]
- 17.Kalucka J, de Rooij LPMH, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen LA, Veys K, et al. (2020). Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 180, 764–779.e720. 10.1016/j.cell.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Rajesh A, Stuart G, Real N, Ahn J, Tschirley A, Wise L, and Hibma M. (2020). Depletion of langerin. Immunology 160, 366–381. 10.1111/imm.13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vinish M., Cui W., Stafford E., Bae L., Hawkins H., Cox R., and Toliver-Kinsky T. (2016). Dendritic cells modulate burn wound healing by enhancing early proliferation. Wound Repair Regen 24, 6–13. 10.1111/wrr.12388. [DOI] [PubMed] [Google Scholar]
- 20.Joshi N, Pohlmeier L, Ben-Yehuda Greenwald M, Haertel E, Hiebert P, Kopf M, and Werner S. (2020). Comprehensive characterization of myeloid cells during wound healing in healthy and healing-impaired diabetic mice. Eur J Immunol 50, 1335–1349. 10.1002/eji.201948438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan DH, Jenison MC, Saeland S, Shlomchik WD, and Shlomchik MJ (2005). Epidermal langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity 23, 611–620. 10.1016/j.immuni.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Konieczny P, Xing Y, Sidhu I, Subudhi I, Mansfield KP, Hsieh B, Biancur DE, Larsen SB, Cammer M, Li D, et al. (2022). Interleukin-17 governs hypoxic adaptation of injured epithelium. Science 377, eabg9302. 10.1126/science.abg9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maglione D, Guerriero V, Viglietto G, Delli-Bovi P, and Persico MG (1991). Isolation of a human placenta cDNA coding for a protein related to the vascular permeability factor. Proc Natl Acad Sci U S A 88, 9267–9271. 10.1073/pnas.88.20.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuge X, Murayama T, Arai H, Yamauchi R, Tanaka M, Shimaoka T, Yonehara S, Kume N, Yokode M, and Kita T. (2005). CXCL16 is a novel angiogenic factor for human umbilical vein endothelial cells. Biochem Biophys Res Commun 331, 1295–1300. 10.1016/j.bbrc.2005.03.200. [DOI] [PubMed] [Google Scholar]
- 25.Yu X, Zhao R, Lin S, Bai X, Zhang L, Yuan S, and Sun L. (2016). CXCL16 induces angiogenesis in autocrine signaling pathway involving hypoxia-inducible factor 1α in human umbilical vein endothelial cells. Oncol Rep 35, 1557–1565. 10.3892/or.2015.4520. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Fan P, Chou H, Li J, Wang K, and Li H. (2019). Herbacetin suppressed MMP9 mediated angiogenesis of malignant melanoma through blocking EGFR-ERK/AKT signaling pathway. Biochimie 162, 198–207. 10.1016/j.biochi.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Soloviev DA, Hazen SL, Szpak D, Bledzka KM, Ballantyne CM, Plow EF, and Pluskota E. (2014). Dual role of the leukocyte integrin αMβ2 in angiogenesis. J Immunol 193, 4712–4721. 10.4049/jimmunol.1400202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu S, Liu Y, You T, and Zhu L. (2018). Semaphorin 7A Promotes VEGFA/VEGFR2Mediated Angiogenesis and Intraplaque Neovascularization in. Front Physiol 9, 1718. 10.3389/fphys.2018.01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan Y, Arif A, Gong Y, Jia J, Eswarappa SM, Willard B, Horowitz A, Graham LM, Penn MS, and Fox PL (2012). Stimulus-dependent phosphorylation of profiling-1 in angiogenesis. Nat Cell Biol 14, 1046–1056. 10.1038/ncb2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLisser HM, Christofidou-Solomidou M, Strieter RM, Burdick MD, Robinson CS, Wexler RS, Kerr JS, Garlanda C, Merwin JR, Madri JA, and Albelda SM (1997). Involvement of endothelial PECAM-1/CD31 in angiogenesis. Am J Pathol 151, 671–677. [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, Lin Z, Liu CH, Gong Y, Liegl R, Fredrick TW, Meng SS, Burnim SB, Wang Z, Akula JD, et al. (2017). Inflammatory signals from photoreceptor modulate pathological retinal angiogenesis via c-Fos. J Exp Med 214, 1753–1767. 10.1084/jem.20161645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoeffel G., Wang Y., Greter M., See P., Teo P., Malleret B., Leboeuf M., Low D., Oller G., Almeida F., et al. (2012). Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med 209, 1167–1181. 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merad M, Manz MG, Karsunky H, Wagers A, Peters W, Charo I, Weissman IL, Cyster JG, and Engleman EG (2002). Langerhans cells renew in the skin throughout life under steady-state conditions. Nat Immunol 3, 1135–1141. 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seré K, Baek JH, Ober-Blöbaum J, Müller-Newen G, Tacke F, Yokota Y, Zenke M, and Hieronymus T. (2012). Two distinct types of Langerhans cells populate the skin during steady state and inflammation. Immunity 37, 905–916. 10.1016/j.immuni.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 35.Bobr A, Igyarto BZ, Haley KM, Li MO, Flavell RA, and Kaplan DH (2012). Autocrine/paracrine TGF-β1 inhibits Langerhans cell migration. Proc Natl Acad Sci U S A 109, 10492–10497. 10.1073/pnas.1119178109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 37.Igyarto BZ, and Kaplan DH (2010). The evolving function of Langerhans cells in adaptive skin immunity. Immunol Cell Biol 88, 361–365. 10.1038/icb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong YK, Lange-Asschenfeldt B, Velasco P, Hirakawa S, Kunstfeld R, Brown LF, Bohlen P, Senger DR, and Detmar M. (2004). VEGF-A promotes tissue repair-associated lymphatic vessel formation via VEGFR-2 and the alpha1beta1 and alpha2beta1 integrins. FASEB J 18, 1111–1113. 10.1096/fj.03-1179fje. [DOI] [PubMed] [Google Scholar]
- 39.Shimamura K, Nakatani T, Ueda A, Sugama J, and Okuwa M. (2009). Relationship between lymphangiogenesis and exudates during the wound-healing process of mouse skin full-thickness wound. Wound Repair Regen 17, 598–605. 10.1111/j.1524-475X.2009.00512.x. [DOI] [PubMed] [Google Scholar]
- 40.Catar R, Moll G, Kamhieh-Milz J, Luecht C, Chen L, Zhao H, Ernst L, Willy K, Girndt M, Fiedler R, et al. (2021). Expanded Hemodialysis Therapy Ameliorates Uremia-Induced Systemic Microinflammation and Endothelial Dysfunction by Modulating VEGF, TNF-α and AP-1 Signaling. Front Immunol 12, 774052. 10.3389/fimmu.2021.774052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia J, Ye T, Cui P, Hua Q, Zeng H, and Zhao D. (2016). AP-1 transcription factor mediates VEGF-induced endothelial cell migration and proliferation. Microvasc Res 105, 103–108. 10.1016/j.mvr.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stutte S, Jux B, Esser C, and Förster I. (2008). CD24a expression levels discriminate Langerhans cells from dermal dendritic cells in murine skin and lymph nodes. J Invest Dermatol 128, 1470–1475. 10.1038/sj.jid.5701228. [DOI] [PubMed] [Google Scholar]
- 43.Sunuwar L, Asraf H, Donowitz M, Sekler I, and Hershfinkel M. (2017). The Zn. Biochim Biophys Acta Mol Basis Dis 1863, 947–960. 10.1016/j.bbadis.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosisio D, Ronca R, Salvi V, Presta M, and Sozzani S. (2018). Dendritic cells in inflammatory angiogenesis and lymphangiogenesis. Curr Opin Immunol 53, 180–186. 10.1016/j.coi.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 45.Curiel TJ, Cheng P, Mottram P, Alvarez X, Moons L, Evdemon-Hogan M, Wei S, Zou L, Kryczek I, Hoyle G, et al. (2004). Dendritic cell subsets differentially regulate angiogenesis in human ovarian cancer. Cancer Res 64, 5535–5538. 10.1158/0008-5472.CAN-04-1272. [DOI] [PubMed] [Google Scholar]
- 46.Sheng J, Chen Q, Wu X, Dong YW, Mayer J, Zhang J, Wang L, Bai X, Liang T, Sung YH, et al. (2021). Fate mapping analysis reveals a novel murine dermal migratory Langerhans-like cell population. Elife 10. 10.7554/eLife.65412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Falanga V. (2005). Wound healing and its impairment in the diabetic foot. Lancet 366, 1736–1743. 10.1016/S0140-6736(05)67700-8. [DOI] [PubMed] [Google Scholar]
- 48.Stojadinovic O, Yin N, Lehmann J, Pastar I, Kirsner RS, and Tomic-Canic M. (2013). Increased number of Langerhans cells in the epidermis of diabetic foot ulcers correlates with healing outcome. Immunol Res 57, 222–228. 10.1007/s12026-013-8474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shook B, Xiao E, Kumamoto Y, Iwasaki A, and Horsley V. (2016). CD301b+ Macrophages Are Essential for Effective Skin Wound Healing. J Invest Dermatol 136, 1885–1891. 10.1016/j.jid.2016.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shook BA., Wasko RR., Rivera-Gonzalez GC., Salazar-Gatzimas E., López-Giráldez F., Dash BC., Muñoz-Rojas AR., Aultman KD., Zwick RK., Lei V., et al. (2018). Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 362. 10.1126/science.aar2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt BA, and Horsley V. (2013). Intradermal adipocytes mediate fibroblast recruitment during skin wound healing. Development 140, 1517–1527. 10.1242/dev.087593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gur-Cohen S, Yang H, Baksh SC, Miao Y, Levorse J, Kataru RP, Liu X, de la Cruz-Racelis J, Mehrara BJ, and Fuchs E. (2019). Stem cell–driven lymphatic remodeling coordinates tissue regeneration. Science 366, 1218–1225. 10.1126/science.aay4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soteriou D, Kostic L, Sedov E, Yosefzon Y, Steller H, and Fuchs Y. (2016). Isolating Hair Follicle Stem Cells and Epidermal Keratinocytes from Dorsal Mouse Skin. J Vis Exp. 10.3791/53931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R. (2018). Integrating singlecell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal singlecell data. Cell 184, 3573–3587.e3529. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar MP, Du J, Lagoudas G, Jiao Y, Sawyer A, Drummond DC, Lauffenburger DA, and Raue A. (2018). Analysis of Single-Cell RNA-Seq Identifies Cell-Cell Communication Associated with Tumor Characteristics. Cell Rep 25, 1458–1468.e1454. 10.1016/j.celrep.2018.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo W, Friedman MS, Shedden K, Hankenson KD, and Woolf PJ (2009). GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161. 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the Key resource Table.
Microscopy data reported in this paper will be shared by the lead contact upon request.
The code to reproduce analyses of single Cell RNA sequencing are deposited at the GitHub repository: https://github.com/khbridges/wasko-langerhans.