The lung is a highly complex human organ, harboring over 40 different cell types with specialized functions to facilitate gas exchange and host defense (1). Single-cell omics technologies, such as single-cell RNA sequencing (scRNA-seq), have been leveraged to create multiple atlas-level single-cell datasets of the lung (2). The functions of individual cells have been characterized using image resolution technologies, such as immunofluorescence confocal microscopy, to elucidate the spatial distribution of cells, proteins, mRNA, and metabolites. Although omics data promise additional critical insights into the cellular biology of the lung, integrating omics data with existing single-cell atlases and image resolution technologies has posed a challenge for many mainstream biologists. Web servers and computational platforms have been designed for the general analysis of lung omics data, with minimal prior computational knowledge to enhance the accessibility of omics data analysis for mainstream biologists (3, 4). However, with the growing number of lung and airway datasets, there is an increasing need for an integrative, user-friendly online tool to revolutionize the identification of disease mechanisms. This will aid the design of novel diagnostic and personalized therapeutic regimens for complex lung diseases such as pulmonary fibrosis, bronchopulmonary dysplasia, chronic obstructive pulmonary disease, asthma, and others.
In this issue of the Journal, Gaddis and colleagues (pp. 129–139) propose LungMAP.net as a new gateway portal for lung research that integrates computational tools and multi-omics data resources (Figure 1) (5). This comprehensive website links eight independent lung research portals with omics and image data, adopts a core set of community schemas to interconnect metadata and file formats for each portal, and provides applications to map the user’s data to reference panels. Considering the complexity of LungMAP.net, the authors provide 10 tutorials, including over 3 hours of video tutorials that cater to users with different backgrounds and varying degrees of expertise. This effectively democratizes access to advanced lung biology research.
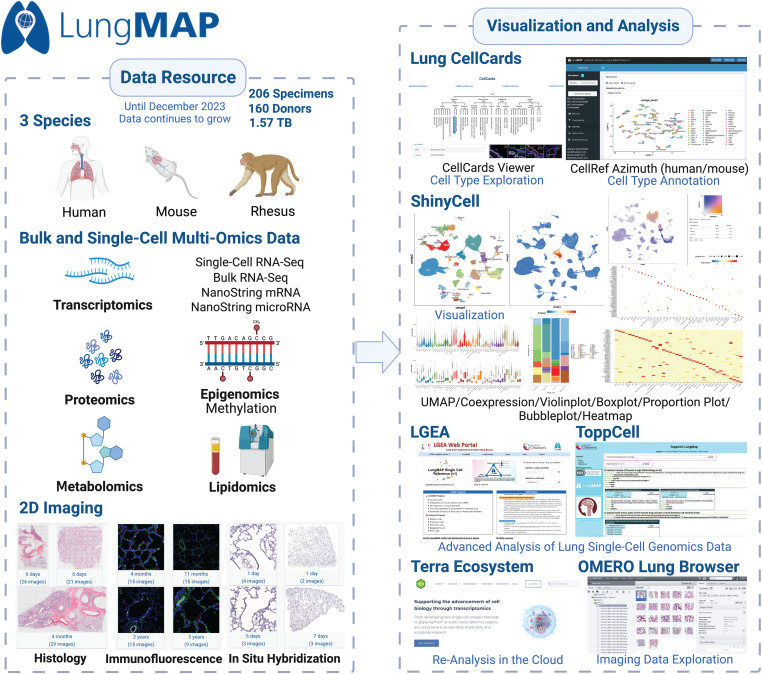
Figure 1.
Overview of LungMAP.net. The LungMAP.net portal mainly includes two sections: (Left) data resource and (Right) visualization and analysis. In the data resource section, LungMAP aggregates a comprehensive collection of bulk and single-cell multi-omic data, encompassing transcriptomics, proteomics, epigenomics, metabolomics, and lipidomics from human, mouse, and rhesus specimens together with extensive imaging datasets. In the Visualization and Analysis section, CellCards offers detailed biological insights into various curated lung cell types and a cell type annotation web tool for human and mouse through Azimuth. The portal also facilitates a wide range of data visualization methods through ShinyCell, including Uniform Manifest Approximation and Projection (or UMAP), boxplots, violin plots, bubble plots, and heatmaps. Additionally, it integrates with external portals such as Lung Gene Expression Analysis (or LGEA), ToppCell, and Terra, further enhancing its utility for advanced lung single-cell genomics research. The figure was created by BioRender.com. OMERO = open microscopy environment; RNA-seq = RNA sequencing; TB = terabyte.
The main advantages of LungMAP.net include the following:
-
1.
Interactive analysis. LungMAP.net provides interactive operations for lung data analysis and visualization. CellRef, a R Shiny application (Azimuth), allows users to map scRNA-seq data to references provided from CellCards (6). It integrates ShinyCell to visualize the clustering results and gene expression using boxplots, violin plots, heatmaps, and scatterplots (7). It also integrates ToppCell to curate gene signatures of cells on the basis of their lineages, cell classes, subclasses, clinical information, and other features (8). Users can reprocess the data from the LungMAP Data Browser through Terra using open-access single-cell analysis workflows in the cloud.
-
2.
Comprehensive integration. LungMAP.net collected: 1) omics array data from 206 specimens of different ages, including bulk and scRNA-seq, proteomics, lipidomics, DNA methylation, microRNA, and metabolomics measurements for diverse samples, including those matched for the same samples (multi-omics); 2) image data to understand the spatial and cellular localization of proteins, mRNA transcripts, and metabolites in the lung; 3) CellCards that integrate functional and emerging single-cell genomic datasets for accurate and evolvable annotation of important cell lineages within the respiratory system (6); and 4) Lung Gene Expression Analysis (LGEA) that explores multi-omics and imaging data through an interactive web interface to connect lung structure and histology to cell type–specific gene expression (9). In addition, LungMAP.net uses a standardized and iterative curation metadata approach, supporting new technologies and sample types.
This harmonized central resource supports hypothesis-driven research questions and broad exploratory investigations. The authors define how users can query specific genes, proteins, cell types, datasets, diseases, or developmental stages to discover affected molecular and cellular programs for subsequent validation in independent cohorts. LungMAP.net breaks down silos and prevents researchers from reinventing the wheel by enhancing and refining data and analysis sharing, thereby reducing unnecessary redundancy and increasing opportunity for novel exploration and discovery. Integrative resources such as LungMAP.net thus have the potential to accelerate scientific discoveries that will ultimately translate into improved care for patients with pulmonary disease.
Although LungMAP.net integrates comprehensive multi-omics data, the ecosystem has two potential directions for future improvement. First, LungMAP.net could collect spatial transcriptomics data for lung diseases, which encompasses a suite of innovative technologies that enable positional profiling of gene expression in the lung (10, 11). Further spatial transcriptomics data analysis tasks, such as spatial domain detection and spatial variable gene detection, can be facilitated through the Terra cloud environment (12–14). Second, LungMAP.net could develop an additional plug-in or webpage to integrate multi-omics data, such as bulk and scRNA-seq with newly developed analytic tools in the field (15).
LungMAP.net is a comprehensive lung ecosystem with multi-omics, multitechnology, and multispecies information. It also utilizes a cloud computing platform to make integrative analysis available to physicians and biologists who lack significant prior experience with these methodologies. Aggregating data from various research networks and promoting broader scientific community participation and collaboration, LungMAP.net paves the way for enhanced molecular biological education and research for the lung.
Footnotes
Originally Published in Press as DOI: 10.1165/rcmb.2023-0439ED on December 18, 2023
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Schiller HB, Montoro DT, Simon LM, Rawlins EL, Meyer KB, Strunz M, et al. The Human Lung Cell Atlas: a high-resolution reference map of the human lung in health and disease. Am J Respir Cell Mol Biol . 2019;61:31–41. doi: 10.1165/rcmb.2018-0416TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature . 2020;587:619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdulla S. Aevermann B. Assis P. Badajoz S. Bell SM. Bezzi E. CZI Single-Cell Biology Program; Cz CELL×GENE Discover: a single-cell data platform for scalable exploration, analysis and modeling of aggregated data. 2023. [DOI]
- 4. Franzén O, Gan L-M, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database . 2019;2019:baz046. doi: 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaddis N, Fortriede J, Guo M, Bardes EE, Kouril M, Tabar S, et al. LungMAP portal ecosystem: systems-level exploration of the lung. Am J Respir Cell Mol Biol . 2024;70:129–139. doi: 10.1165/rcmb.2022-0165OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun X, Perl A-K, Li R, Bell SM, Sajti E, Kalinichenko VV, et al. NHLBI LungMAP Consortium A census of the lung: CellCards from LungMAP. Dev Cell . 2022;57:112–145.e2. doi: 10.1016/j.devcel.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ouyang JF, Kamaraj US, Cao EY, Rackham OJL. ShinyCell: simple and sharable visualization of single-cell gene expression data. Bioinformatics . 2021;37:3374–3376. doi: 10.1093/bioinformatics/btab209. [DOI] [PubMed] [Google Scholar]
- 8. Jin K, Bardes EE, Mitelpunkt A, Wang JY, Bhatnagar S, Sengupta S, et al. An interactive single cell web portal identifies gene and cell networks in COVID-19 host responses. iScience . 2021;24:103115. doi: 10.1016/j.isci.2021.103115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Du Y, Ouyang W, Kitzmiller JA, Guo M, Zhao S, Whitsett JA, et al. Lung gene expression analysis web portal version 3: lung-at-a-glance. Am J Respir Cell Mol Biol . 2021;64:146–149. doi: 10.1165/rcmb.2020-0308LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moses L, Pachter L. Museum of spatial transcriptomics. Nat Methods . 2022;19:534–546. doi: 10.1038/s41592-022-01409-2. [DOI] [PubMed] [Google Scholar]
- 11. Zhu J, Fan Y, Xiong Y, Wang W, Chen J, Xia Y, et al. Delineating the dynamic evolution from preneoplasia to invasive lung adenocarcinoma by integrating single-cell RNA sequencing and spatial transcriptomics. Exp Mol Med . 2022;54:2060–2076. doi: 10.1038/s12276-022-00896-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun S, Zhu J, Zhou X. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat Methods . 2020;17:193–200. doi: 10.1038/s41592-019-0701-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shang L, Zhou X. Spatially aware dimension reduction for spatial transcriptomics. Nat Commun . 2022;13:7203. doi: 10.1038/s41467-022-34879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schatz MC, Philippakis AA, Afgan E, Banks E, Carey VJ, Carroll RJ, et al. Inverting the model of genomics data sharing with the NHGRI genomic data science analysis, visualization, and informatics lab-space. Cell Genom . 2022;2:100085. doi: 10.1016/j.xgen.2021.100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol . 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]