Abstract
Radiotherapy (RT) combined with immunotherapy is promising; however, the immune response signature in the clinical setting after RT remains unclear. Here, by integrative spatial and single-cell analyses using multiplex immunostaining (CODEX), spatial transcriptome (VISIUM), and single-cell RNA sequencing, we substantiated the infiltration of immune cells into tumors with dynamic changes in immunostimulatory and immunosuppressive gene expression after RT. In addition, our comprehensive analysis uncovered time- and cell type–dependent alterations in the gene expression profile after RT. Furthermore, myeloid cells showed prominent up-regulation of immune response–associated genes after RT. Notably, a subset of infiltrating tumor-associated myeloid cells showing PD-L1 positivity exhibited significant up-regulation of immunostimulatory (HMGB1 and ISG15), immunosuppressive (SIRPA and IDO1), and protumor genes (CXCL8, CCL3, IL-6, and IL-1AB), which can be targets of immunotherapy in combination with PD-L1. These datasets will provide information on the RT-induced gene signature to seek an appropriate target for personalized immunotherapy combined with RT and guide the timing of combination therapy.
Single-cell analysis revealed radiotherapy-induced immune genes modulation that can be a promising target for immune-radiotherapy.
INTRODUCTION
Radiotherapy (RT) is an important component of cancer treatment and is given to approximately 50 to 60% of all patients with cancer. In the past century, the major antitumor effect of RT has been considered to be the induction of lethal DNA damage. However, accumulating evidence has recently revealed that immune responses and immunological cell death also contribute to the antitumor effect of RT, although the effect is limited by RT alone (1, 2). To support the notion of antitumor immune activation by RT, multiple recent studies have proposed that RT can stimulate both immunostimulatory and immunosuppressive effects in cancer tissue (3). For example, ionizing irradiation induces immune cell infiltration and major histocompatibility complex (MHC) class I expression as immune-stimulatory effects; in contrast, it up-regulates PD-L1 and exhibits cytotoxic effects on immune cells, which result in immunosuppressive effects (4–6). Although the immune responses in the tumor microenvironment (TME) after RT have been extensively investigated, it is unknown which cell types and which genes are responsive to RT in patients at the single-cell level. The characterization of the gene profile of specific cell types within the TME is critically important, which necessitates single-cell analysis, to determine a drug target for the improvement of RT-induced antitumor immunity (7–9).
Following the development of immune checkpoint inhibitors (ICIs), many clinical trials of RT combined with immunotherapy have been undertaken. Notably, the addition of ICIs to RT significantly improves the effect of RT on non–small cell lung cancer (NSCLC) (10, 11). Furthermore, phase 1 and 2 trials of combined RT and ICIs have shown promising responses in other types of cancer, such as head and neck, pancreatic, and colorectal cancer (12–14). Unfortunately, several trials have reported negative or inconclusive results with regard to which patients might benefit from combination therapy (15). This discrepancy suggests that a personalized strategy is required to maximize the antitumor immune impacts of combination therapy. There are two major parameters to consider for optimizing RT combined with immunotherapy: the timing of drug administration and the drug target, which could involve factors in PD-1/PD-L1–dependent and –independent pathways. At present, the only adjuvant anti–PD-L1 antibody applied after RT has proven to be effective in a phase 3 trial (10). In contrast, the efficacy of concurrent ICI administration with RT has not been fully investigated in the clinical setting, possibly due to the lack of evidence, although it can be a promising strategy. Therefore, an examination of the temporal changes in the gene expression profile after RT treatment is critical to optimize concurrent therapy.
As a partner of RT, combination therapy targeting the PD-1/PD-L1 pathway has been reported to improve outcomes in phase 3 trials (10); however, approximately half of the patients relapse even after anti–PD-L1 combination therapy. Therefore, the use of multiple ICIs targeting PD-1/PD-L1 as well as other targets is currently being explored. Phase 1 to 3 trials targeting more than 10 ICIs of other targets are ongoing (7, 15, 16). Notably, combinations of ICIs, such as the PD-1/CTLA-4 and PD-L1/TIGIT inhibitor combinations, have been shown to be more effective than ICI monotherapy (NCT03563716) (17). Other ICI targets, such as CD47/SIRPA and IDO1, are expected to be next-generation targets for immunotherapy, and phase 2 and 3 trials are ongoing (18–21). Meanwhile, the response rate of CD47/SIRPA and IDO1 target monotherapy is limited, suggesting that they appear to have benefits in combination therapy (22, 23). Furthermore, clinical trials independent of ICIs, e.g., immunotherapy targeting chemokines and interleukins (ILs), have been recently initiated (24, 25). As a cell-targeted therapy combined with RT, macrophage exclusion after RT has been shown to improve the tumor response to RT (NCT01977677) (26). Hence, multiple clinical trials of combined RT and ICIs or other immunotherapies are ongoing; however, the optimization of this approach necessitates information on the gene expression profile in each cell type after RT. It is therefore important to investigate the dynamic changes in RT-induced immune cell distribution and expression profile of immune genes in RT-treated patients.
To address this critical question and gain insight into the spatiotemporal dynamics of the gene profile in each cell type within the clinical setting of the TME after RT, we visualized the distribution of immune cells and characterized the gene expression profile in each immune cell type over time after RT. Methods for multiplex immunostaining (CODEX) and spatial transcriptome analysis (VISIUM) have been recently developed and are powerful tools to analyze the spatial distribution and gene expression in the TME at the single-cell level (27, 28). In addition, single-cell RNA sequencing (scRNA-seq) is an excellent tool to analyze the transcriptome and is able to determine the expression profile of ~2000 genes in each immune cell type, even with small tissue samples such as biopsy samples. In the present study, we explored the characteristics of gene expression patterns and the dynamic distribution of immune cells in esophageal squamous cell carcinoma (ESCC) patient tissues after RT because (i) ESCC at clinical stages I to IV is commonly treated with RT (29) and (ii) recent clinical trials revealed that combination with an anti–PD-1 or anti–PD-L1 antibody improves the efficacy of conventional RT in patients with ESCC (30, 31). Our comprehensive spatial and single-cell analysis uncovered dynamic tumor immune cell infiltration and identified a unique gene expression pattern during and after RT, particularly in myeloid cells. Furthermore, a subset of infiltrating tumor-associated myeloid cells, including macrophages, expressing PD-L1 exhibited significant up-regulation of both multiple immune-stimulatory and -suppressive genes, indicating these myeloid-expressing factors as promising targets for immunotherapy. In addition, our deposited datasets provide a wealth of information to facilitate the screening and study of cell types and therapeutic targets to optimize immune- and radiation-based cancer therapy.
RESULTS
Spatial identification of immune cell infiltration in the tumor microenvironment after radiotherapy
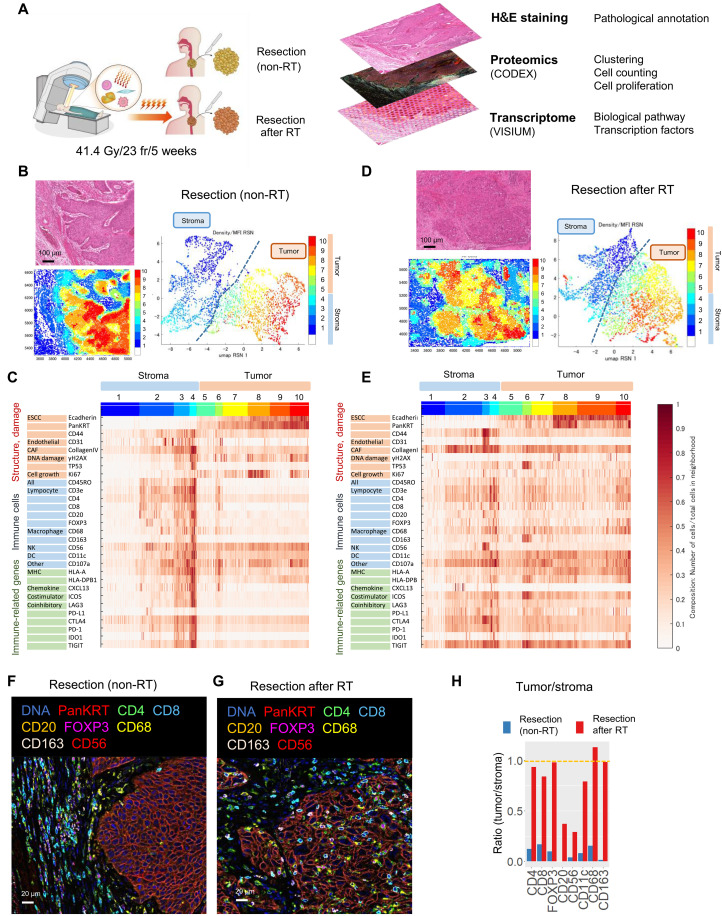
To comprehensively assess the spatial distribution of immune cells and their transcriptomic profiles within the TME after RT at the single-cell level, multi-immunostaining (CODEX) analysis was performed on surgically resected ESCC tissue. Resected tissue from a patient with ESCC who underwent surgery only (non-RT) and from a patient who received preoperative (RT) (41.4 grays (Gy)/23 fr) was applied for CODEX (Fig. 1A). Surgically resected tissue rather than biopsied tissue was used to analyze the spatial localization of cells for integrated analysis [the detailed scheme of the comprehensive analysis using hematoxylin and eosin (H&E) staining and CODEX analysis in combination with VISIUM analysis is shown in figs. S1 and S2]. First, to examine cell morphology, ESCC tissues were evaluated by H&E staining. As expected, meganuclei and tissue damage were only observed in RT-treated tissues (fig. S3A) (32, 33). To distinguish the tumor and stroma, the ESCC component was demarcated on the basis of morphological features, KRT5 expression by VISIUM and pan-keratin and E-cadherin by CODEX (fig. S3, C and D) (N.B.; the list of gene sets analyzed by CODEX in this study, predominantly focusing on immune-related proteins, is shown in figs. S1 and S2). To visualize the clustering pattern of gene expression after RT, the results of CODEX were classified into 10 clusters. The Uniform Manifold Approximation (UMAP) plots and heatmaps of non-RT and RT samples are shown (Fig. 1, B to E; the digitized heatmap is shown in fig. S4, A and B). The boundary of the clustering pattern was evident in non-RT tissue (Fig. 1, B and C); in contrast, the clustering pattern was markedly changed after RT (Fig. 1, D and E), suggesting that RT dynamically changes the proportion and distribution of cell types within the TME. Before RT, immune cells, mainly helper T (CD4+), cytotoxic T cells (CD8+), regulatory T cells (Tregs; FOXP3), CD20-positive B cells, natural killer (NK) cells (CD56+), dendritic cells (DCs) (CD11c+), and macrophages (CD68+ and CD163+), were present in the stroma (Fig. 1, B, C, and F). In contrast, CD4+, CD8+, CD68+, and CD163+ cells were enriched within the field of cluster numbers 5 to 10 (tumor) after RT, showing that immune cells infiltrated into the tumor after RT (Fig. 1, D, E, and G; representative images of immune cell infiltration are shown in Fig. 1, F and G). Large field images are shown in fig. S4 (C and D). These results substantiate that RT stimulates the infiltration of immune cells such as T cells, B cells, NK cells, DC cells, and macrophages in the TME. To further consolidate the heatmap results, the number of cells expressing each immune marker per area within the stroma and tumor was enumerated. The tumor/stroma ratio in immune cells after RT was nearly 1.0, suggesting that the level of immune cell distribution was equal between the stroma and tumor after RT (the tumor/stroma ratio is shown in Fig. 1H, and the number of cells per area is shown in fig. S4, E and F). Together, our spatial analysis shows the concept that RT stimulates immune cell infiltration into tumors.
Fig. 1. Spatial identification of immune cell infiltration in the tumor microenvironment after radiotherapy.
(A) Workflow of this research. See also figs. S1 to S3. For the specialty analyses (CODEX and VISIUM), the data were obtained from a single patient for each pre-radiotherapy (RT) and post-RT sample. (B) Hematoxylin and eosin (H&E) observation and Uniform Manifold Approximation (UMAP) visualization of esophageal squamous cell carcinoma (ESCC) cell, stromal cell, and immune cell clusters in resected ESCC tissue. (C) Expression patterns of signature genes in distinct ESCC clusters. Clusters 5 to 10 showed the ESCC part in resection. A numerical heatmap is shown in fig. S4A. (D) H&E observation and UMAP visualization of ESCC cell, stromal cell, and immune cell clusters in resected tissue after RT. (E) Expression patterns of signature genes in distinct ESCC clusters. Clusters 5 to 10 indicated the resected ESCC tumor after RT. A numerical heatmap is shown in fig. S4B. (F and G) RT increased immune cell infiltration into ESCC. Multicolor immunohistochemistry (IHC) staining in ESCC tissue after resection (F) and resection after RT (G). Scale bars, 20 μm. The low-power field is shown in fig. S4C. (H) Ratio of immune cells in the tumor/stroma. The absolute number of each cell type is shown in fig. S4 (E and F).
Characterization of gene expression profiles in the tumor microenvironment of ESCC after radiotherapy
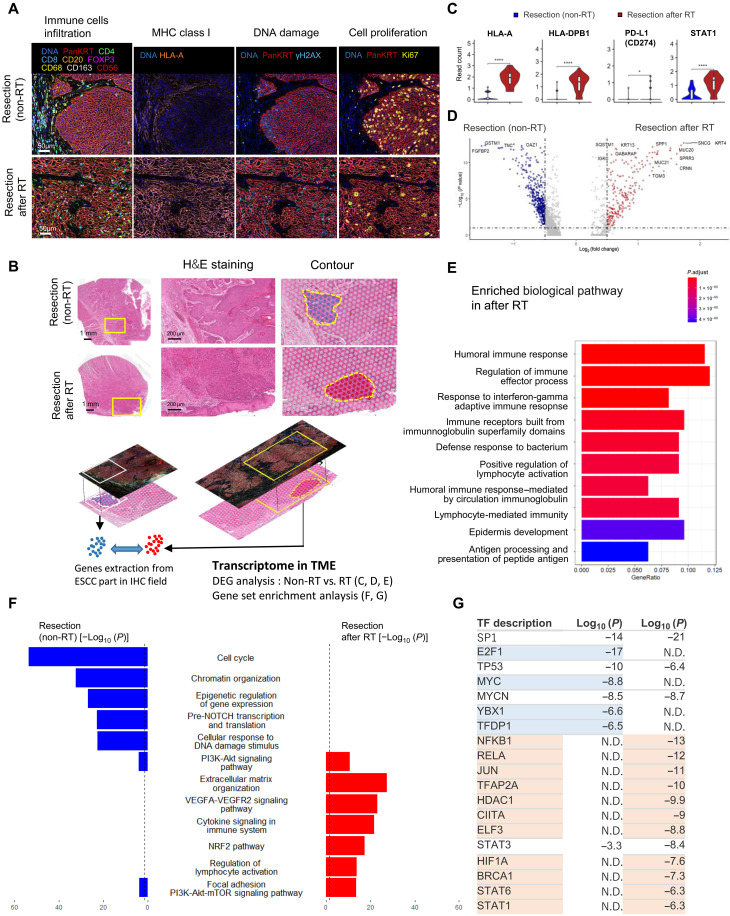
Next, to characterize gene signature with the spatial information within the TME after RT, spatial transcriptome analysis via VISIUM was performed on the same fields analyzed by CODEX. Before the CODEX-VISUM combination analysis, we confirmed the presence of immune cell infiltration (immune cell marker), MHC class I expression [human leukocyte antigen A (HLA-A)], DNA damage (γH2AX), and cell proliferation (Ki67) in the field of the combination analysis (Fig. 2A). The presence of DNA damage signals and proliferative cells in the sample after RT confirmed that cells within the TME remain alive and sustain signal transduction ability, which may be causally related to radioresistance even after curative-dose irradiation. For the integrated CODEX-VISIUM combination analysis, we examined the field that contained ~40 spots of VISIUM analysis and showed positivity of immune cell markers by CODEX. To validate the quality of our samples for VISIUM analysis, gene expression of KRT5 and the primary immune factors HLA-A, HLA-DPA1, and PD-L1 (CD274) were confirmed (fig. S5A). Following the contouring of the tumor area in the field, gene expression data were extracted from each sample (Fig. 2B and fig. S5A). Next, analysis of differentially expressed genes (DEGs) and gene signatures specific to non-RT and RT tissues were performed from the extracted gene expression data. Similar to the results of CODEX, the up-regulation of HLA-A, HLA-DPB1, PD-L1 (CD274), and STAT1 expression was observed after RT, confirming that RT up-regulates the expression of immune signaling genes (Fig. 2C). The volcano plots of the DEG analysis between non-RT and RT tissue are shown (Fig. 2D). In addition, gene set enrichment analysis (GSEA) revealed enrichment of pathways related to the immune response after RT (>1.5-fold) (Fig. 2E). Humoral immune response genes were enriched after RT (Fig. 2E; see also further scRNA-seq analysis results in the next section). In summary, VISIUM analysis substantiates the up-regulation of immune response genes within the tumor field of ESCC after RT. Furthermore, we identified several biological and molecular pathways activated in non-RT and RT tissues (Fig. 2F and fig. S5, B and C), showing a gene expression signature switch from cell cycle–related genes (including E2F1 and MYC) to stress response–related genes (including NRF2, VEGF, NFKB1, and HIF1A). Next, we examined transcription factors activated by RT (Fig. 2G). Notably, E2F1, MYC, YBX1, and TFDP1 were unique in non-RT tissue, whereas NFKB1, RELA, JUN, TFAP2A, HDAC1, CIITA, ELF3, HIF1A, BRCA1, STAT6, and STAT1 were specific to RT tissue. Thus, the CODEX-VISUM combination analysis validates the sample quality of our spatial integration analysis and substantiates the characteristics of the gene profile within the TME of ESCC cells after RT (7, 34, 35).
Fig. 2. Characterization of gene expression profiles in the tumor microenvironment (TME) of esophageal squamous cell carcinoma (ESCC) after radiotherapy.
(A) Identification of the microenvironment surrounding ESCCs after radiotherapy (RT). Multicolor immunohistochemistry (IHC) staining showed immune cell infiltration, major histocompatibility complex (MHC) class I expression [human leukocyte antigen A (HLA-A)], DNA damage (γH2AX), and cell proliferation (Ki67). (B) Workflow of integrated spatial analysis. The TME was identified by IHC, and TME transcriptome data were extracted from the IHC-merged field in VISIUM. H&E, hematoxylin and eosin. (C) Gene expression of MHC class I (HLA-A), MHC class II (HLA-DPB1), PD-L1 (CD274), and STAT1 according to 10x VISIUM (****P < 1 × 10−16, *P < 0.05, Bonferroni-adjusted Wilcoxon test; ns, no significance). Figure S5A shows gene expression in the total and IHC fields. (D) Differentially expressed gene (DEG) analysis between the Re and RT groups. Volcano plot showing gene expression in each contoured field in Fig. 2B. (E) Gene enrichment analysis showed up-regulated biological pathways in the viable ESCC field after RT. (F) Pathways enriched in non-RT and after RT tissue (adjusted P < 0.01). (G) Transcription factors that were significantly enriched in non-RT and post-RT tissues (P < 1 × 10−6).
Single-cell RNA sequencing uncovers the up-regulation of immune response–associated genes in myeloid cells by radiotherapy
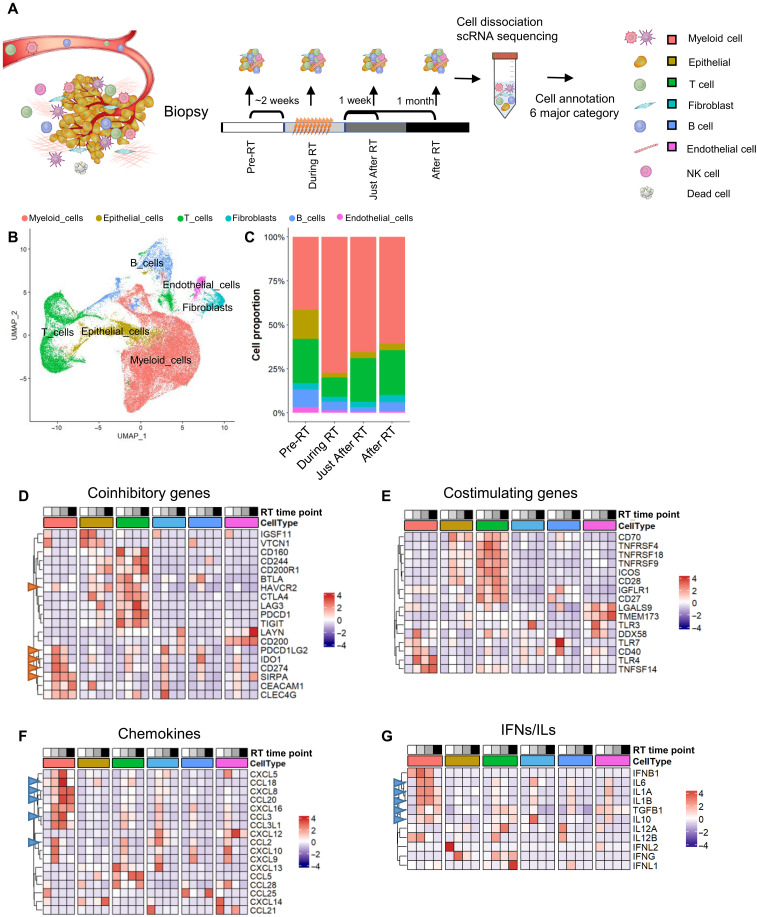
To understand the temporal changes in gene expression profile in the TME by RT at the single-cell level, scRNA-seq was performed in biopsy samples obtained “pre-RT,” “during RT,” “just after RT (7 days after RT),” and “after RT (~1 month after RT)” (the detailed definition of timing for these groups is shown in Fig. 3A and fig. S6A) (N.B.; 11 biopsy samples were obtained from five ESCC patients). Using SingleR package (36), cells were classified into six major classes: myeloid cells (DCs, macrophages, monocytes, and neutrophils), epithelial cells, T cells, fibroblasts, B cells, and endothelial cells (Fig. 3B and fig. S6B). The increase in the population of myeloid cells was most notably observed at the “during RT” time point, with the increase being sustained to “after RT (~1 month after RT)” albeit with some decrease. The initial increase is likely caused by a reduction in the population of T, B, and epithelial cells by RT (Fig. 3C) because these cells are more sensitive to ionizing radiation than myeloid cells (37). To identify cell type–specific alterations in the gene expression profile with time after RT, the dynamics of immune response gene expression were visualized on the basis of functional categorization, such as immuno-coinhibitory (Fig. 3D), costimulatory (Fig. 3E), chemokines (Fig. 3F), and interferons (IFNs)/ILs (Fig. 3G), as previously described (38) (the details of gene selection are described in Materials and Methods). Of the six cell types examined, the increase in immune response genes was most evident in myeloid cells. Although the magnitude of the increase in expression differs depending on the specific gene, the gene expression pattern in myeloid cells was markedly enhanced “during” and “just after” RT. Immune-inhibitory genes in the category of coinhibitory [PD-L1 (CD274) and SIRPA], chemokines (CCL3 and CXCL18), and IFNs/ILs (IFNG and IL-6) were significantly up-regulated in myeloid cells by RT (the representative immune-inhibitory genes of coinhibitory factors are indicated by red arrowheads in Fig. 3D). In addition, genes involved in protumor processes (e.g., CCL3, IL-6, and IL-1AB, which are involved in cancer growth and metastasis) (39, 40) were up-regulated, especially in myeloid cells (Fig. 3, F and G; these genes are indicated by blue arrowheads). To further comprehensively analyze the alteration of the gene expression profile in the context of TME after RT, gene enrichment analysis and heatmap analysis were performed. Activation of lymphocyte pathways was observed in almost all cell types (fig. S7A), confirming that RT promotes immune activation in the TME. Furthermore, the expression of genes related to metabolism, hypoxia, and cancer associated fibroblasts (CAFs) was also changed by RT (fig. S7, B to E). These results support the notion that, in addition to immune responses, RT converts the overall TME status by modulating the expression of multiple genes. Since the enrichment of gene expression in the humoral immune response family was observed by VISIUM analysis (Fig. 2E), the expression of related genes was examined by using an scRNA-seq dataset (fig. S7F). We found that S100A8/S100A9/HLA-A were up-regulated in myeloid cells after RT. In addition, SLP1/SPINK5/LCN2 expression in epithelial cells was increased by RT. Together, our scRNA-seq analysis substantiates that RT markedly changes the expression profile of immune response genes in a time- and cell type–dependent manner. Notably, we found significant up-regulation of immune-inhibitory genes, particularly in myeloid cells, including macrophages and DCs, within the TME after RT.
Fig. 3. Single-cell RNA sequencing (scRNA-seq) uncovers the up-regulation of immune response genes in myeloid cells by radiotherapy.
(A) Workflow of scRNA-seq analysis. (B) Uniform Manifold Approximation (UMAP) of major cell clusters based on scRNA-seq data after integration. (C) Proportions of the cell clusters annotated in (B). (D to G) Expression of representative markers of the indicated biological class as assessed by scRNA-seq. The grayscale bar indicates the time point during radiotherapy (RT): white, pre-RT; light gray, during RT; dark gray, immediately post-RT; black, post-RT. The red arrowhead shows immune-inhibitory genes. The blue arrowhead shows tumor progression–related genes. IFNs, interferons; ILs, interleukins.
Characterization of immune response gene expression profile in myeloid cells with time after radiotherapy
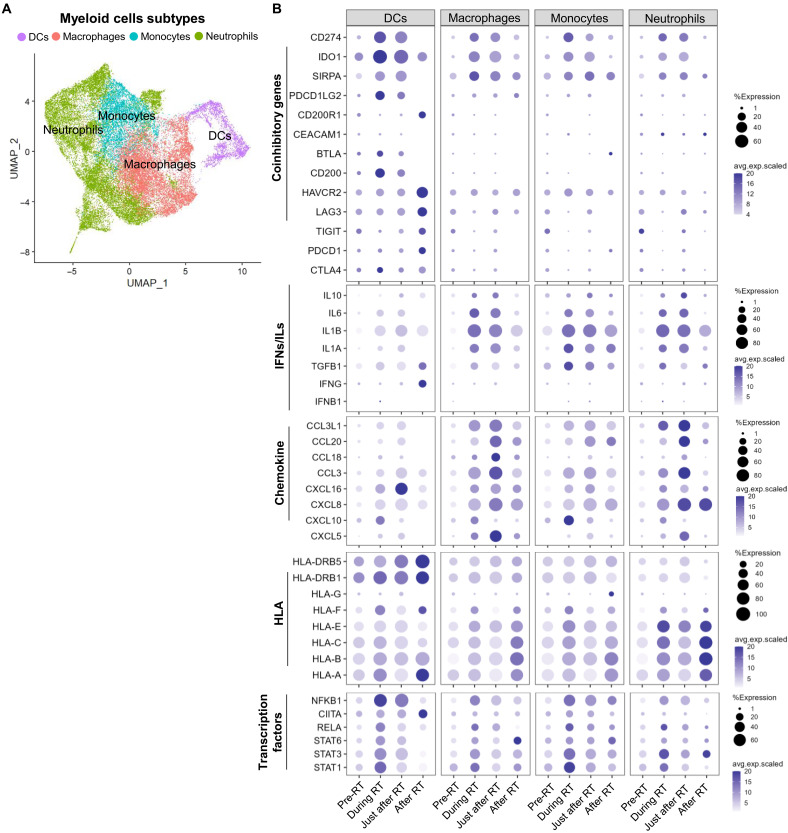
To investigate the expression profile of the immune response genes in infiltrated myeloid cells after RT, we further classified the myeloid dataset of scRNA-seq into subclasses, i.e., DCs, macrophages, monocytes, and neutrophils (Fig. 4A; the expression of key markers in the four myeloid subtypes is shown in fig. S8A). Among the coinhibitory genes targeted for ICI therapy, PD-L1 (CD274), IDO1, and SIRPA were significantly up-regulated by RT in all myeloid cell types, particularly during and just after RT (Fig. 4B). The expression profile of the individual patient is shown in fig. S8B. The timing of the peaks in immune response genes (coinhibitory, costimulatory, chemokines, and IFNs/ ILs) of each cell type differed over the course of RT.
Fig. 4. Characterization of the immune response gene expression profile in myeloid cells with time after radiotherapy.
(A) Uniform Manifold Approximation (UMAP) of subclusters in the myeloid cell cluster. (B) Bubble charts of the percentage of cells within the indicated myeloid cell subclusters that express immune coinhibitory, interleukin (IL) and interferon (IFN), chemokine, and human leukocyte antigen (HLA) genes and the corresponding average gene expression level. A larger dot indicates a higher percentage of cells expressing a particular gene; a darker color dot indicates a higher average gene expression level. DCs, dendritic cells.
Recently, macrophage-targeted therapy has been suggested to improve the efficacy of RT (26, 39). Therefore, we next sought to examine the time-dependent alteration of gene expression profile in macrophages after RT. In macrophages, coinhibitory and IFN/IL genes, e.g., IL-1, IL-1B, and IL-6, peaked during RT (i.e., acute phase), chemokine genes, e.g., CCL3 and CCL20, peaked just after RT (i.e., subacute phase), and HLA gene expression gradually increased and peaked after RT (i.e., the chronic phase). Hence, our scRNA-seq analysis identified the characteristics of the immunological shift from an acute to a chronic phase in macrophages following RT.
Identification of PD-L1–positive myeloid cells showing high expression of multiple immune-inhibitory genes after radiotherapy
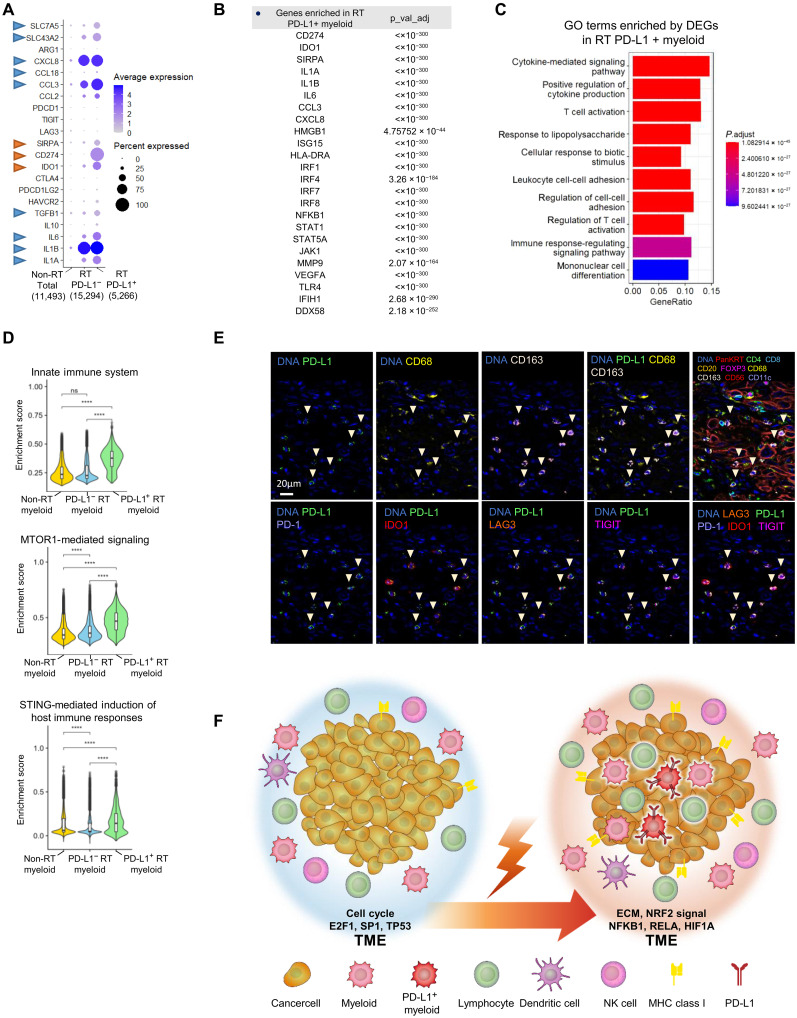
Recent studies have demonstrated that macrophage-specific PD-L1 expression can be a better target for the ICI response than cancer cell–specific PD-L1 expression in NSCLC (40). However, since PD-L1/PD-1 monotherapy alone is seldom effective, it is critical to investigate the expression of other immune-inhibitory genes in the PD-L1–positive population to optimize the benefit of multiple ICI therapy. We therefore characterized the gene expression profile of PD-L1 (CD274)–positive myeloid cells in ESCC after RT (N.B.; we examined myeloid cells rather than macrophages to obtain sufficient cell numbers for scRNA-seq analysis). The myeloid cells used for this scRNA-seq analysis were classified into three groups; those obtained from non-RT patients (non-RT total), PD-L1–negative myeloid cells obtained from post-RT patients (RT PD-L1−), and PD-L1–positive myeloid cells obtained from post-RT patients (RT PD-L1+) (Fig. 5A). Similar to the result in Fig. 4, we found the up-regulation of CXCL8, CCL3, IL-6, and IL-1A/B following RT. After RT, the PD-L1+ subgroup (designated RT PD-L1+) showed significantly greater expression of SIRPA, IDO1, CCL3, and IL-6 than the RT PD-L1− subgroup (Fig. 5A), indicating that a subset of RT PD-L1+ myeloid cells harbors high expression of the coinhibitory genes (red arrowhead) and protumor genes (blue arrowhead). Furthermore, the RT PD-L1+ subgroup showed high expression of CCL3 and IL-6 compared with the RT PD-L1− subgroup. To further characterize the gene expression signature in the RT PD-L1+ subgroup, GSEA was performed (Fig. 5, B and C). In addition to immune suppressive genes, immune activation surrogate genes such as HMGB1 and ISG15 and regulatory factors such as IRF1, -4, and -7 were significantly enriched in the RT PD-L1+ subgroup (Fig. 5B; the details are shown in table S1). The Gene Ontology (GO) enrichment analysis also revealed that multiple immune-stimulatory and antitumor pathways, such as the cytokine-mediated signaling pathway and T cell activation, were up-regulated in the RT PD-L1+ subgroup (Fig. 5C). To further clarify the differences in gene expression between the RT PD-L1− and RT PD-L1+ cells, the expression of genes, including genes encoding to coinhibitory molecules, IFNs/ILs, chemokines, HLAs, and transcription factors, were examined in PD-L1–positive and –negative populations after RT (fig. S9, A and B). In addition, a correlation heatmap in the three myeloid subsets was generated (fig. S9, C and D), showing the comprehensive relationships among the selected genes; of note, a stronger correlation was seen among the immune genes in the RT PD-L1+ myeloid cells. Furthermore, we identified that the innate immune system, MTORC1-mediated signaling, and STING pathways were significantly up-regulated in the RT PD-L1+ subgroup (Fig. 5D). Last, to confirm the expression of proteins encoded by the genes identified by scRNA-seq, the expression levels were assessed by CODEX. Consistent with the scRNA-seq results, PD-L1+ CD68+/CD168+ (PD-L1–expressing myeloid cells) exhibited high PD-1, IDO1, LAG3, and TIGIT expression [Fig. 5E; full images in the analysis are shown in figs. S10 and S11; N.B., the CODEX analysis showed high expression of TIGIT and PD-1 (PDCD1), although these genes were not detected by scRNA-seq, possibly due to the low gene detection sensitivity of scRNA-seq]. Together, our analyses uncovered the existence of PD-L1–positive myeloid cells showing high expression of other coinhibitory genes in the TME of ESCC after RT, suggesting that multiple ICIs targeting PD-L1 and the other targets such as SIRPA and IDO1 could be effective in combination with RT.
Fig. 5. Identification of PD-L1–positive myeloid cells showing high expression of multiple immune-inhibitory genes after radiotherapy.
(A) Gene expression signature of myeloid cells from non-radiotherapy (RT) and post-RT patients. Myeloid cells from patients after RT were classified as PD-L1–negative or PD-L1–positive. Bubble charts of total myeloid cells and non-RT (1,1493 cells), PD-L1–negative (15,294 cells), and PD-L1–positive (5,266 cells) myeloid cells after RT; PD-L1–positive myeloid cells showed high expression of immune checkpoint inhibitor (ICI) target genes (red arrowhead) and other protumor genes (blue arrowhead). (B) Differentially expressed genes (DEGs) between non-RT total myeloid cells and PD-L1–positive myeloid cells after RT. DEGs were defined by P < 0.05 (Bonferroni-adjusted Wilcoxon test) and fold change > 2. (C and D) GSEA of DEGs in PD-L1–positive myeloid cells after RT and total myeloid cells. (****P < 1 × 10−16, Bonferroni-adjusted Wilcoxon test; ns, no significance). (E) Identification of PD-L1–expressing cells after RT by multicolor immunohistochemistry (IHC) staining. In addition to PD-L1, the upper line shows CD68, CD163, PanKRT, CD4, CD8, CD20, FOXP3, CD56, and CD11c expression for cell orientation. The lower line shows the ICI target genes PD-1, IDO1, LAG3, and TIGIT. Figures S7 and S8 show the low-power field for each antibody staining experiment. (F) Graphical summary of tumor microenvironment (TME) alterations after RT in esophageal squamous cell carcinoma (ESCC) tissue.
DISCUSSION
In the present study, our integrated single-cell analysis using H&E staining, CODEX, and VISIUM revealed the infiltration of immune cells, including helper T cells, cytotoxic T cells, Tregs, B cells, NK cells, DC cells, and macrophages, into tumors following RT in a clinical setting. Although immune cell infiltration in the TME after RT has been previously reported by immunohistochemistry (IHC), our analysis specifically identified the cell types and their gene expression profiles at the single-cell level. Furthermore, among the cell types in the TME, myeloid cells were found to be strong responders exhibiting prominent up-regulation of immune response genes after RT. In addition, a subgroup of myeloid cells showing PD-L1 expression exhibited multiple immune-stimulatory and antitumor genes as well as activation of the innate immune system, MTORC1-mediated signaling, and the STING pathway. Hence, our comprehensive single-cell analysis revealed that RT elicits multiple immune response pathways in a cell type– and time-dependent manner. Our dataset provides invaluable single-cell–based information on RT-induced gene signatures, which could aid in optimizing targets for personalized immunotherapy combined with RT. A limitation of this study is the small sample size because it was conducted as a prospective observational study. In addition, case numbers were limited because endoscopy during and just after RT without medical justification was not permitted. Furthermore, since scRNA-seq requires fresh tissue, it was difficult to retroactively perform a comparative analysis between the during-RT and pre-RT time points of the same cases even for the rare patients who do undergo endoscopy for medical reasons. Thus far, barely any time-series analysis of standard clinical RT regimens in patient tissue has been undertaken. Nevertheless, despite the limited sample numbers, we believe that our results provide substantial information of relevance for radiation oncology and immunotherapy. Our control experiments verify the reliability of the single-cell analysis. For example, consistent with previous reports (34), cell cycle and chromatin abnormalities were enriched in non-RT ESCC tissue, while the VEGF and NRF2 pathways were significantly enriched in irradiated ESCC tissues after RT (35, 41–43). These results confirm that our single-cell analysis detects radiation-induced responses. We were concerned that the responses could be predominantly derived from dead or dying cells after the lethal dose of RT. However, we found cancer cells showing Ki67 positivity after RT (Fig. 2A), suggesting that these cancer cells remain viable despite the presence of severe genotoxic stress and the infiltration of immune cells into tumors. Hence, our dataset provides crucial information to understand resistance mechanisms and to develop a strategy to enhance cancer cell killing by targeting immune response genes after RT.
One of the most important aspects of the present study was the analysis of temporal changes in the gene expression sets over the course of the clinical RT schedule. A single dose of radiation is known to cause distinct immune responses depending on the time of analysis post-RT with time frames being classified as hours to days (acute), weeks (subacute), and months (chronic) after RT (3). Here, we defined the time periods as “pre-RT,” “during RT (acute),” “just after RT (subacute),” and “after RT (chronic).” For precise analysis, further investigation with additional time points is needed; nevertheless, our time-series analysis revealed distinct expression patterns in each phase after RT. Furthermore, we categorized genes associated with cancer immune responses as costimulatory genes, coinhibitory genes, IFNs, ILs, chemokines, or HLA genes. Notably, the expression of coinhibitory and IL genes peaked in the acute phase, the expression of chemokines peaked in the subacute phase, and the expression of HLAs peaked in the chronic phase, revealing a temporal modulation of immune responses by RT. The timing of combination therapy with RT and anticancer drugs is critical to optimize the efficacy of combination therapy; however, no comparative trials have been conducted thus far. Although trials of combinations of ICIs and RT have been ongoing in all settings (induction, concurrent, and adjuvant), these trials are designed to confirm efficacy (induction: NCT03217071, NCT03102242; concurrent: NCT0350912, NCT02343952; adjuvant: NCT03053856, NCT02125461). To date, no trials have been conducted to compare the impact of the time of drug administration. On the basis of our results, concurrent RT + ICI or ICI administration just after RT can be suitable for targeting a gene whose expression peaks during RT (acute) and just after RT (subacute). Adjuvant timing can also be suitable when targeting a gene whose expression peaks after RT (chronic). Consistent with this temporal analysis, the administration of anti–PD-L1 antibody within 2 weeks after RT significantly improved outcomes after RT, while administration later than 2 weeks after RT was without benefit (the PACIFIC trial, phase 3) (10). Thus, although further analysis is needed, our temporal gene expression data will help guide the timing of drug administration in clinical trials.
Our single-cell analysis also uncovered cell type– and time-dependent cancer immune responses within the TME after RT. Myeloid cells showed the most prominent alterations in gene expression profile in terms of both the levels and expression patterns of mRNA/proteins. Notably, PD-L1 in myeloid cells was significantly up-regulated by RT. Anti–PD-L1 therapy has been considered to be effective for PD-L1+ on tumor cells, with the therapeutic effect being predicted by the tumor proportion score (44). However, several recent reports have proposed that the therapeutic effect of anti–PD-L1 therapy should be assessed by scoring macrophages, lymphocytes, and tumor cells using the combined positive score (45, 46). Macrophages were identified as the prominent PD-L1–expressing cells in both the tumor and stromal compartments in mice and humans (47). High levels of PD-L1 expression in macrophages have been associated with a significantly longer overall survival in patients with NSCLC independent of PD-L1 expression in tumors (40). Our scRNA-seq analysis identified a subgroup of myeloid cells harboring high PD-L1 expression after RT. Since the PD-L1–positive myeloid subgroup strongly activates immune escape genes and antitumor pathways, the suppression of immune escape factors by targeting this subgroup of myeloid cells could be a promising treatment strategy. Together, our findings suggest that targeting myeloid cells, including macrophages, is a promising strategy, particularly in combination with RT.
Recent studies have suggested that coinhibitory factors other than PD-1/PD-L1, such as SIRPA and IDO1, could be useful in next-generation ICI therapy. Our analysis shows that RT up-regulates multiple genes involved in tumor growth and immune escape, including coinhibitory genes, chemokines, and ILs. Among the coinhibitory genes, SIRPA and IDO1 in myeloid cells showed kinetics similar to those of PD-L1. SIRPA directly suppresses the phagocytic ability of macrophages, and IDO1 impairs lymphocyte function via effects on tryptophan metabolism (25, 47, 48). Although the response rate of monotherapy for both SIRPA and IDO1 is approximately 10%, the efficacy is significantly improved when combined with PD-L1–targeted therapy (49, 50). Since our analysis identified the up-regulation of IDO1 and SIRPA after RT, these ICI combination therapies may be further improved by using RT. A very recent paper reported that RT combined with PD-1 (PDCD1) and SIRPA-targeted therapy for colorectal cancer elicits a highly effective cancer immune response, suggesting that such a combinational approach could be successful (51). As another strategy of immunotherapy, the combination of multiple modalities other than ICIs has been recently developed, e.g., targeting ILs or chemokines, cell therapy, and vaccines, to achieve synergistic effects (39, 52, 53). Our single-cell analysis identified the up-regulation of IL-1, IL-6, and CCL3 in myeloid cells and tumor cells after RT, suggesting the possibility of combining RT with such comprehensive immunotherapy for ESCC. Notably, many of the genes identified in this study are confirmed clinical drug targets, such as anti–PD-L1, anti-IL-1, and anti-IL-6 antibodies, which have Food and Drug Administration approval. Antibodies targeting IDO1 and SIRPA are also scheduled to be approved by 2023. Thus, our integrated single-cell analysis results provide further evidence to support the future application of RT combined with PD-L1–, SIRPA-, IDO1-, and IL-targeted therapy. In conclusion, our datasets obtained by comprehensive single-cell analysis provide an enormous resource containing information on RT-induced gene signatures that can be used to identify the appropriate targets of personalized immunotherapy combined with RT.
MATERIALS AND METHODS
Tissue material
ESCC samples were obtained from patients at the National Cancer Center Hospital East (NCCE). Written informed consent was obtained from all patients. The use of their tissues for this research was fully anonymized and approved by the NCCE IRB Administrative Panels (IRB2018-101). Surgically resected tissues were entered from past cases, and biopsy specimens were entered prospectively from another case. 5-Fluorouracil and cisplatin (CDDP) combined with 41.4 Gy/23 fr were used for preoperative RT. Endoscopic biopsies were performed before, during, just after, and following RT, and tissue samples were obtained from a minimum of two different time points.
Postradiation surgery is not commonly performed for surgically resectable ESCC in Japan based on phase 3 JCOG1109 (54). Therefore, it is difficult to collect surgical specimens after RT, resulting in a limited number of samples.
CODEX and QuPath analysis
Commercially available purified, carrier-free monoclonal and polyclonal anti-human antibodies [keratin 14, CD4, CD31, CD44, CD107a, CD20, E-cadherin, CD68, collagen IV, CD45RO CD223 (LAG3), HLA-A, CD8, CD11c, pan-cytokeratin, CD274 (PD-L1), IDO1, HLA-DPB1, Ki67, CXCL13, TP53, FOXP3, CTLA-4, CD163, CD56, CD3e, CD278 (ICOS), PDCD1 (PD-1), γH2AX, and TIGIT] were conjugated to maleimide-modified short DNA oligonucleotides (barcodes) according to the manufacturer’s protocol (CODEX User Manual, Akoya Biosciences) at a 2:1 w/w ratio of oligonucleotide to antibody, as previously described (28, 55). Thirty antibodies were selected, including those specific for ESCC markers (i.e., Ecadherin and PanKRT), lymphocyte markers (i.e., CD4, CD8, CD20, and FOXP3), NK cell markers (i.e., CD56), macrophage markers (i.e., CD68 and CD163), DC markers (i.e., CD11), and proteins targeted by ICIs (i.e., PD-L1, LAG3, IDO1, TIGIT, and ICOS) (table S2).
CODEX staining and imaging were performed according to the manufacturer’s protocol (CODEX User Manual) as previously described (28, 55). Briefly, the coverslip containing the tissue section was baked at 62°C for 1 hour, deparaffinized in xylene, rehydrated in ethanol, and washed in ddH2O. After antigen retrieval with citrate buffer (pH 6), the tissue section was stained with antibodies cocktail solution for 3 hours at RT. After fixation with 1.6% paraformaldehyde, 100% methanol, and a post-staining fixing solution, the coverslip was mounted onto a CODEX stage for the KEYENCE microscope. Imaging of the CODEX multicycle experiment was performed using an inverted fluorescence microscope (KEYENCE, Osaka, Japan; model BZ-X810) equipped with a CFI Plan Apo l 20×/0.75 objective (Nikon, Tokyo, Japan), a microfluidics instrument (Akoya Biosciences, Menlo Park, CA, USA), and CODEX Instrument Manager software (Akoya Biosciences, Menlo Park, CA, USA). Raw image files were processed using the CODEX Processor (version 1.8), as previously described (28, 55).
QPTIFF files were imported to QuPath (https://qupath.github.io/) version 0.3.2. We extracted a representative area of the image and analyzed the area. Cell segmentation was performed on the basis of nuclear stain using StarDist cell segmentation script. To classify cells for each marker, we used three 500-μm square regions outside the representative area. We made training data for cell classification by manually deciding the positive and negative for each 10 to 30 cells in the three regions based on fluorescence intensity. At the same time, we confirmed that the cell classification of all three regions displayed by machine learning was appropriate. This machine learning cell classification was applied to the representative area and spatial analysis was performed by detecting dimension centroid distances of two dimensions. The results, which include position, fluorescence intensity, and classification of each cell, were exported as a csv file. We analyzed the number of cells of each classification by using R version 4.2.2.
The csv file was imported to CytoMAP version 1.4.21 (56). We divide the representative area into 10 regions by 50-μm-radius raster scanned neighborhoods function, which uses the self-organizing map clustering methods, based on each fluorescence intensity. To examine the neighborhood of each region, we performed dimensionality reduction into a two-dimensional plot using UMAP and projection.
10x VISIUM spatial transcriptomic analysis
To analyze the TME after RT by using a formalin-fixed paraffin-embedded (FFPE) VISIUM spatial gene expression assay (10x Genomics), we selected a 6.5 × 6.5 mm2 optical region of the FFPE tumor block with DV200 ≥ 50% that contained cancer cells, stromal cells, and immune cells. VISIUM spatial gene expression slides and reagent kits were used according to the manufacturer’s instructions (10x Genomics). Each capture area contained approximately 5000 barcoded spots of 55 μm in diameter (100-μm center-to-center spacing between spots). FFPE tumor samples were prepared according to the recommended protocols (Tissue Preparation Guide, CG000408). H&E staining and imaging were performed according to the protocol (deparaffinization, H&E staining, imaging, and decrosslinking, CG000409). Illumina sequencing libraries were generated according to the protocol (Visium Spatial Gene Expression Reagent Kits for FFPE User Guide, CG000407) and sequenced on a NovaSeq 6000 system (Illumina). Sequencing was performed with the recommended protocol (read 1: 28 cycles; i7 index read: 10 cycles; i5 index read: 10 cycles; and read 2: 50 cycles), yielding between 150 million and 224 million sequenced reads. The Space Ranger pipeline was used to process the VISIUM spatial gene expression data, including the generation of FASTQ files, alignment to the hg38 human reference genome, tissue detection, fiducial detection, and barcode/UMI counting.
The VISIUM spatial expression object was further analyzed as follows to determine clusters and perform gene expression analysis. Only high-quality spots (≥180 genes per spot) were selected for subsequent analyses. The SCTransform function in Seurat was used for normalization, and principal components analysis (PCA) (RunPCA) was used for dimensionality reduction. On the basis of morphological characteristics and the expression of immunity-related marker genes, the spots were manually selected and annotated as epithelial and ESCC sites.
Integrated spatial analysis
H&E, CODEX, and VISIUM fields were manually merged into the total and partial fields. In integrated analysis, ESCC parts that matched IHC fields were morphologically confirmed on merged H&E, ESCC parts were contoured and transcriptome data in matched ESCC parts (resection, 43 spots; resection after RT, 46 spots) were extracted on the Loupe browser. The FindMarkers function in Seurat was used to identify DEGs between the resection and resection after RT using the criteria P ≤ 0.05 and fold change ≥ 2. The identified DEGs were used for GO enrichment analysis performed with the R package clusterProfiler (57).
Signature analysis was also performed on the extracted RNA data separately. The top 1000 genes expressed in resection and resection after RT were analyzed by Metascape, and those significantly enriched in terms of biological pathway, molecular function, and transcription factors were confirmed. Typical pathways and genes from each result are shown in Fig. 2 (F and G) and fig. S5 (B and C), and all are shown in table S3.
Single-cell RNA sequencing
scRNA-seq was performed as described previously (58). Briefly, tissue dissociation into single cells for scRNA-seq was performed with a Tumor Dissociation Kit (Miltenyi Biotec, 130-095-929) following the default protocol in the user guide of the Chromium Single Cell 5' Reagent Kit ver. 1 (10x Genomics). scRNA-seq libraries were prepared using a Chromium Single Cell 5' Reagent Kit ver. 1 (10x Genomics) and sequenced using the HiSeq 3000 platform (Illumina). Cell Ranger ver. 3.0.0 (10x Genomics) was used with default parameters to process the reference genome alignment and quantify cells and transcripts. The raw sequencing reads were mapped to the human genome assembly GRCh38.
Eleven scRNA-seq samples were collected, and the data were integrated using Seurat. The integrated object was then normalized and scaled. Next, PCA (RunPCA) was performed. The top 30 principal components were selected and submitted to FindNeighbors, FindClusters, and UMAP (RunUMAP) to obtain clusters. Cell type was annotated manually by referring to published papers and by SingleR (ver. 1.6.1) (34). The annotated cell clusters were verified using a known cell marker list. Myeloid cells, T cells, B cells, fibroblast, endothelial cells, and epithelial cells were selected for comparative analysis. Myeloid cells that simultaneously express coinhibitory genes PD-L1/CD274, IDO1, SIRPA, and LAG3 were categorized as “PD-L1+ myeloid,” and those that do not meet these criteria were categorized as other.
The FindMarkers function in Seurat was used to identify DEGs between the PD-L1+ myeloid and total myeloid cells using the criteria P ≤ 0.05 and fold change ≥ 2. The identified DEGs were used for GO enrichment analysis performed with the R package clusterProfiler (58). GSEA was performed on RT-induced tumor-associated macrophage (TAM) and other myeloid cells using the R package escape (57, 59).
The selection of gene sets to be used in the scRNA-seq analysis was based on the categorization proposed by Zhang et al. (38). Genes encoding ICI targets or candidates (VTCN1, HAVCR2, CTLA4, LAG3, PDCD1, TIGIT, CD200, PDCD1LG2, IDO1, CD274, and SIRPA) were also included. The selection of chemokine, protumor, and IL gene sets was performed as previously described (60). The information on gene selection is summarized in table S4. Genes showing low expression below the detection level were excluded from our list.
Summary of sample size in this study
The number of cells analyzed in CODEX and scRNA-seq is summarized in tables S5 and S6.
Figure 1 [sample size: single patient, 19,574 cells (non-RT), 22,958 cells (RT)] – (method: CODEX) – [analysis: clustering (B and D), gene expression signature (C and E), cell counting (H)].
Figure 2 [sample size: single patient, 3930 spots (non-RT), 4152 spots (RT)] – (method: VISIUM) – (analysis: gene set enrichment analysis).
Figure 3 (sample size: 5 patients, 58,074 cells) – (method: scRNA-seq) – (analysis: cell annotation, gene expression analysis).
Figure 4 (sample size: 5 patients, myeloid cells: 32,053 cells) – (method: scRNA-seq) − (analysis: cell annotation, gene expression analysis).
Figure 5, A to D [sample size: 5 patient, myeloid cells (total: 32,053 cells; non-RT: 11,493; RT PD-L1−: 15,294 cells; RT PD-L1+: 5266 cells) − (method: scRNA-seq) − (analysis: gene set enrichment analysis).
Figure 5E (single patient) − (method: CODEX) − (analysis: immunohistochemistry).
Acknowledgments
We thank the staff of National Cancer Center Hospital East for supporting the clinical sample collections and helpful advice and all patients and their families for contributing to this study. We thank P. A. Jeggo for the critical discussion about the manuscript.
Funding: This work was supported by Grants-in-Aid for Scientific Research KAKENHI 21 K07582, Grants-in-Aid for Scientific Research KAKENHI 16H06279 (PAGS), Grants-in-Aid for Scientific Research KAKENHI 22H04925 (PAGS), and AMED under grant numbers JP22zf0127008 and JP23zf0127008.
Author contributions: H.O., J.D., and S.-I.K. contributed to conceptualization, data curation, investigation, methodology, resources, validation, visualization, writing—original draft, writing—review and editing, and approval. R.Y. and S.A.S. contributed to gene expression analysis for VISIUM and scRNA-seq. J.Z., A.Su., and Y.S. contributed to the investigation, methodology, resources, validation, and visualization of scRNA-seq, CODEX, and VISIUM results. D.K., T.K., M.K., Y.N., and H.S. conducted endoscopic biopsies, determined clinical diagnoses, and administered treatment. Y.N. and M.K. contributed to the pathological diagnosis, sample preparation, and investigation for VISIUM and CODEX. M.O., M.N., H.Ho., and A.M. conducted radiotherapy and patient collection. H.Hi. and A.Sh. contributed to writing—original draft, writing—review and editing, and approval. S.-I.K. summarized all the data for the figures and tables with support from H.O., J.D., and S.A.S. S.-I.K. and A.Sh. wrote the manuscript with support from H.O., J.D., and Y.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The data of clinical specimens including scDNA-seq and VISIUM data generated in this study have been deposited in the DDBJ BioProject (BioProject accession: PRJDB15043). All the raw image of the CODEX analysis has been uploaded to the web server in the Human Genome Center, University of Tokyo (www.hgc.jp/~riuyamas/others/20230602_Oyoshi_et_al/CODEX.tar.gz). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Legends for tables S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
REFERENCES AND NOTES
- 1.L. Galluzzi, A. Buqué, O. Kepp, L. Zitvogel, G. Kroemer, Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 17, 97–111 (2017). [DOI] [PubMed] [Google Scholar]
- 2.E. A. Reits, J. W. Hodge, C. A. Herberts, T. A. Groothuis, M. Chakraborty, E. K.Wansley, K. Camphausen, R. M. Luiten, A. H. de Ru, J. Neijssen, A. Griekspoor, E. Mesman, F. A. Verreck, H. Spits, J. Schlom, P. van Veelen, J. J. Neefjes, Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. J. Exp. Med. 203, 1259–1271 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.U. M. Cytlak, D. P. Dyer, J. Honeychurch, K. J. Williams, M. A. Travis, T. M. Illidge, Immunomodulation by radiotherapy in tumour control and normal tissue toxicity. Nat. Rev. Immunol. 22, 124–138 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Y. Uchihara, T. B. M. Permata, H. Sato, R. Kawabata-Iwakawa, S. Katada, W. Gu, S. Kakoti, M. Yamauchi, R. Kato, S. Gondhowiardjo, N. Hosen, T. Yasuhara, A. Shibata, DNA damage promotes HLA class I presentation by stimulating a pioneer round of translation-associated antigen production. Mol. Cell 82, 2557–2570.e7 (2022). [DOI] [PubMed] [Google Scholar]
- 5.H. Sato, P. A. Jeggo, A. Shibata, Regulation of programmed death-ligand 1 expression in response to DNA damage in cancer cells: Implications for precision medicine. Cancer Sci. 110, 3415–3423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.H. Sato, A. Niimi, T. Yasuhara, T. B. M. Permata, Y. Hagiwara, M. Isono, E. Nuryadi, R. Sekine, T. Oike, S. Kakoti, Y. Yoshimoto, K. D. Held, Y. Suzuki, K. Kono, K. Miyagawa, T. Nakano, A. Shibata, DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 8, 1751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. McLaughlin, E. C. Patin, M. Pedersen, A. Wilkins, M. T. Dillon, A. A. Melcher, K. J. Harrington, Inflammatory microenvironment remodelling by tumour cells after radiotherapy. Nat. Rev. Cancer 20, 203–217 (2020). [DOI] [PubMed] [Google Scholar]
- 8.S. P. Pitroda, S. J. Chmura, R. R. Weichselbaum, Integration of radiotherapy and immunotherapy for treatment of oligometastases. Lancet Oncol. 20, e434–e442 (2019). [DOI] [PubMed] [Google Scholar]
- 9.E. C. Ko, D. Raben, S. C. Formenti, The integration of radiotherapy with immunotherapy for the treatment of non-small cell lung cancer. Clin. Cancer Res. 24, 5792–5806 (2018). [DOI] [PubMed] [Google Scholar]
- 10.S. J. Antonia, A. Villegas, D. Daniel, D. Vicente, S. Murakami, R. Hui, T. Kurata, A. Chiappori, K. H. Lee, M. de Wit, B. C. Cho, M. Bourhaba, X. Quantin, T. Tokito, T. Mekhail, D. Planchard, Y. C. Kim, C. S. Karapetis, S. Hiret, G. Ostoros, K. Kubota, J. E. Gray, L. Paz-Ares, J. de Castro Carpeño, C. Faivre-Finn, M. Reck, J. Vansteenkiste, D. R. Spigel, C. Wadsworth, G. Melillo, M. Taboada, P. A. Dennis, M. Özgüroğlu, Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N. Engl. J. Med. 379, 2342–2350 (2018). [DOI] [PubMed] [Google Scholar]
- 11.S. J. Antonia, A. Villegas, D. Daniel, D. Vicente, S. Murakami, R. Hui, T. Yokoi, A. Chiappori, K. H. Lee, M. de Wit, B. C. Cho, M. Bourhaba, X. Quantin, T. Tokito, T. Mekhail, D. Planchard, Y. C. Kim, C. S. Karapetis, S. Hiret, G. Ostoros, K. Kubota, J. E. Gray, L. Paz-Ares, J. de Castro Carpeño, C. Wadsworth, G. Melillo, H. Jiang, Y. Huang, P. A. Dennis, M. Özgüroğlu, Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N. Engl. J. Med. 377, 1919–1929 (2017). [DOI] [PubMed] [Google Scholar]
- 12.H. Bando, Y. Tsukada, M. Ito, T. Yoshino, Novel immunological approaches in the treatment of locally advanced rectal cancer. Clin. Colorectal Cancer 21, 3–9 (2022). [DOI] [PubMed] [Google Scholar]
- 13.X. Zhu, Y. Cao, W. Liu, X. Ju, X. Zhao, L. Jiang, Y. Ye, G. Jin, H. Zhang, Stereotactic body radiotherapy plus pembrolizumab and trametinib versus stereotactic body radiotherapy plus gemcitabine for locally recurrent pancreatic cancer after surgical resection: An open-label, randomised, controlled, phase 2 trial. Lancet Oncol. 22, 1093–1102 (2021). [DOI] [PubMed] [Google Scholar]
- 14.J. Weiss, S. Sheth, A. M. Deal, J. E. Grilley Olson, S. Patel, T. G. Hackman, J. M. Blumberg, T. J. Galloway, S. Patel, A. M. Zanation, C. J. Shen, D. N. Hayes, C. Hilliard, R. Mehra, K. P. McKinnon, H. H. Wang, M. C. Weissler, J. R. Bauman, B. S. Chera, B. G. Vincent, Concurrent definitive immunoradiotherapy for patients with stage III-IV head and neck cancer and cisplatin contraindication. Clin. Cancer Res. 26, 4260–4267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.K. B. Pointer, S. P. Pitroda, R. R. Weichselbaum, Radiotherapy and immunotherapy: Open questions and future strategies. Trends Cancer 8, 9–20 (2022). [DOI] [PubMed] [Google Scholar]
- 16.G. Petroni, L. C. Cantley, L. Santambrogio, S. C. Formenti, L. Galluzzi, Radiotherapy as a tool to elicit clinically actionable signalling pathways in cancer. Nat. Rev. Clin. Oncol. 19, 114–131 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J. Larkin, V. Chiarion-Sileni, R. Gonzalez, J. J. Grob, C. L. Cowey, C. D. Lao, D. Schadendorf, R. Dummer, M. Smylie, P. Rutkowski, P. F. Ferrucci, A. Hill, J. Wagstaff, M. S. Carlino, J. B. Haanen, M. Maio, I. Marquez-Rodas, G. A. McArthur, P. A. Ascierto, G. V. Long, M. K. Callahan, M. A. Postow, K. Grossmann, M. Sznol, B. Dreno, L. Bastholt, A. Yang, L. M. Rollin, C. Horak, F. S. Hodi, J. D. Wolchok, Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.K. Tang, Y. H. Wu, Y. Song, B. Yu, Indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors in clinical trials for cancer immunotherapy. J. Hematol. Oncol. 14, 68 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Z. Jiang, H. Sun, J. Yu, W. Tian, Y. Song, Targeting CD47 for cancer immunotherapy. J. Hematol. Oncol. 14, 180 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R. Advani, I. Flinn, L. Popplewell, A. Forero, N. L. Bartlett, N. Ghosh, J. Kline, M. Roschewski, A. LaCasce, G. P. Collins, T. Tran, J. Lynn, J. Y. Chen, J. P. Volkmer, B. Agoram, J. Huang, R. Majeti, I. L. Weissman, C. H. Takimoto, M. P. Chao, S. M. Smith, CD47 blockade by Hu5F9-G4 and rituximab in non-hodgkin's lymphoma. N. Engl. J. Med. 379, 1711–1721 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.M. T. Pallotta, C. Orabona, C. Volpi, C. Vacca, M. L. Belladonna, R. Bianchi, G. Servillo, C. Brunacci, M. Calvitti, S. Bicciato, E. M. C. Mazza, L. Boon, F. Grassi, M. C. Fioretti, F. Fallarino, P. Puccetti, U. Grohmann, Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat. Immunol. 12, 870–878 (2011). [DOI] [PubMed] [Google Scholar]
- 22.T. Ebata, T. Shimizu, Y. Fujiwara, K. Tamura, S. Kondo, S. Iwasa, K. Yonemori, A. Shimomura, S. Kitano, T. Koyama, N. Sato, K. Nakai, M. Inatani, N. Yamamoto, Phase I study of the indoleamine 2,3-dioxygenase 1 inhibitor navoximod (GDC-0919) as monotherapy and in combination with the PD-L1 inhibitor atezolizumab in Japanese patients with advanced solid tumours. Invest. New Drugs 38, 468–477 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.B. I. Sikic, N. Lakhani, A. Patnaik, S. A. Shah, S. R. Chandana, D. Rasco, A. D. Colevas, T. O’Rourke, S. Narayanan, K. Papadopoulos, G. A. Fisher, V. Villalobos, S. S. Prohaska, M. Howard, M. Beeram, M. P. Chao, B. Agoram, J. Y. Chen, J. Huang, M. Axt, J. Liu, J. P. Volkmer, R. Majeti, I. L. Weissman, C. H. Takimoto, D. Supan, H. A. Wakelee, R. Aoki, M. D. Pegram, S. K. Padda, First-in-human, first-in-class phase I trial of the anti-CD47 antibody Hu5F9-G4 in patients with advanced cancers. J. Clin. Oncol. 37, 946–953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Wu, Q. Huang, Y. Xie, X. Wu, H. Ma, Y. Zhang, Y. Xia, Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J. Hematol. Oncol. 15, 24 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. F. Sanmamed, P. Berraondo, M. E. Rodriguez-Ruiz, I. Melero, Charting roadmaps towards novel and safe synergistic immunotherapy combinations. Nat. Cancer 3, 665–680 (2022). [DOI] [PubMed] [Google Scholar]
- 26.R. P. Thomas, S. Nagpal, M. Iv, S. G. Soltys, S. Bertrand, J. S. Pelpola, R. Ball, J. Yang, V. Sundaram, S. B. Chernikova, J. Lavezo, D. Born, H. Vogel, J. M. Brown, L. D. Recht, Macrophage exclusion after radiation therapy (MERT): A first in human phase I/II trial using a CXCR4 inhibitor in glioblastoma. Clin. Cancer Res. 25, 6948–6957 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.L. Moses, L. Pachter, Museum of spatial transcriptomics. Nat. Methods 19, 534–546 (2022). [DOI] [PubMed] [Google Scholar]
- 28.S. Black, D. Phillips, J. W. Hickey, J. Kennedy-Darling, V. G. Venkataraaman, N. Samusik, Y. Goltsev, C. M. Schürch, G. P. Nolan, CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 16, 3802–3835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M. Schweigert, A. Dubecz, H. J. Stein, Oesophageal cancer—An overview. Nat. Rev. Gastroenterol. Hepatol. 10, 230–244 (2013). [DOI] [PubMed] [Google Scholar]
- 30.R. J. Kelly, J. A. Ajani, J. Kuzdzal, T. Zander, E. van Cutsem, G. Piessen, G. Mendez, J. Feliciano, S. Motoyama, A. Lièvre, H. Uronis, E. Elimova, C. Grootscholten, K. Geboes, S. Zafar, S. Snow, A. H. Ko, K. Feeney, M. Schenker, P. Kocon, J. Zhang, L. Zhu, M. Lei, P. Singh, K. Kondo, J. M. Cleary, M. Moehler, Adjuvant nivolumab in resected esophageal or gastroesophageal junction cancer. N. Engl. J. Med. 384, 1191–1203 (2021). [DOI] [PubMed] [Google Scholar]
- 31.H. Bando, D. Kotani, T. Tsushima, H. Hara, S. Kadowaki, K. Kato, K. Chin, K. Yamaguchi, S. I. Kageyama, H. Hojo, M. Nakamura, H. Tachibana, M. Wakabayashi, M. Fukutani, Y. Togashi, N. Fuse, H. Nishikawa, T. Kojima, TENERGY: Multicenter phase II study of atezolizumab monotherapy following definitive chemoradiotherapy with 5-FU plus cisplatin in patients with unresectable locally advanced esophageal squamous cell carcinoma. BMC Cancer 20, 336 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. F. Fajardo, The pathology of ionizing radiation as defined by morphologic patterns. Acta Oncol. 44, 13–22 (2005). [DOI] [PubMed] [Google Scholar]
- 33.P. Rubin, G. W. Casarett, Clinical radiation pathology as applied to curative radiotherapy. Cancer 22, 767–778 (1968). [DOI] [PubMed] [Google Scholar]
- 34.M. Li, Z. Zhang, Q. Wang, Y. Yi, B. Li, Integrated cohort of esophageal squamous cell cancer reveals genomic features underlying clinical characteristics. Nat. Commun. 13, 5268 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.H. E. Barker, J. T. Paget, A. A. Khan, K. J. Harrington, The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nat. Rev. Cancer 15, 409–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D. Aran, A. P. Looney, L. Liu, E. Wu, V. Fong, A. Hsu, S. Chak, R. P. Naikawadi, P. J. Wolters, A. R. Abate, A. J. Butte, M. Bhattacharya, Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D. Heylmann, V. Ponath, T. Kindler, B. Kaina, Comparison of DNA repair and radiosensitivity of different blood cell populations. Sci. Rep. 11, 2478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Y. Zhang, H. Chen, H. Mo, X. Hu, R. Gao, Y. Zhao, B. Liu, L. Niu, X. Sun, X. Yu, Y. Wang, Q. Chang, T. Gong, X. Guan, T. Hu, T. Qian, B. Xu, F. Ma, Z. Zhang, Z. Liu, Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell 39, 1578–1593.e8 (2021). [DOI] [PubMed] [Google Scholar]
- 39.A. Mantovani, P. Allavena, F. Marchesi, C. Garlanda, Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 21, 799–820 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Y. Liu, J. Zugazagoitia, F. S. Ahmed, B. S. Henick, S. N. Gettinger, R. S. Herbst, K. A. Schalper, D. L. Rimm, Immune Cell PD-L1 colocalizes with macrophages and is associated with outcome in PD-1 pathway blockade therapy. Clin. Cancer Res. 26, 970–977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Y. Matsuoka, R. Yoshida, K. Kawahara, J. Sakata, H. Arita, H. Nkashima, N. Takahashi, M. Hirayama, M. Nagata, A. Hirosue, Y. Kuwahara, M. Fukumoto, R. Toya, R. Murakami, H. Nakayama, The antioxidative stress regulator Nrf2 potentiates radioresistance of oral squamous cell carcinoma accompanied with metabolic modulation. Lab. Invest. 102, 896–907 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D. H. Gorski, M. A. Beckett, N. T. Jaskowiak, D. P. Calvin, H. J. Mauceri, R. M. Salloum, S. Seetharam, A. Koons, D. M. Hari, D. W. Kufe, R. R. Weichselbaum, Blockage of the vascular endothelial growth factor stress response increases the antitumor effects of ionizing radiation. Cancer Res. 59, 3374–3378 (1999). [PubMed] [Google Scholar]
- 43.H. J. Mauceri, N. N. Hanna, M. A. Beckett, D. H. Gorski, M. J. Staba, K. A. Stellato, K. Bigelow, R. Heimann, S. Gately, M. Dhanabal, G. A. Soff, V. P. Sukhatme, D. W. Kufe, R. R. Weichselbaum, Combined effects of angiostatin and ionizing radiation in antitumour therapy. Nature 394, 287–291 (1998). [DOI] [PubMed] [Google Scholar]
- 44.E. B. Garon, N. A. Rizvi, R. Hui, N. Leighl, A. S. Balmanoukian, J. P. Eder, A. Patnaik, C. Aggarwal, M. Gubens, L. Horn, E. Carcereny, M. J. Ahn, E. Felip, J. S. Lee, M. D. Hellmann, O. Hamid, J. W. Goldman, J. C. Soria, M. Dolled-Filhart, R. Z. Rutledge, J. Zhang, J. K. Lunceford, R. Rangwala, G. M. Lubiniecki, C. Roach, K. Emancipator, L. Gandhi, Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 372, 2018–2028 (2015). [DOI] [PubMed] [Google Scholar]
- 45.K. Yamashita, M. Iwatsuki, K. Harada, K. Eto, Y. Hiyoshi, T. Ishimoto, Y. Nagai, S. Iwagami, Y. Miyamoto, N. Yoshida, Y. Komohara, J. A. Ajani, H. Baba, Prognostic impacts of the combined positive score and the tumor proportion score for programmed death ligand-1 expression by double immunohistochemical staining in patients with advanced gastric cancer. Gastric Cancer 23, 95–104 (2020). [DOI] [PubMed] [Google Scholar]
- 46.D. B. Doroshow, S. Bhalla, M. B. Beasley, L. M. Sholl, K. M. Kerr, S. Gnjatic, I. I. Wistuba, D. L. Rimm, M. S. Tsao, F. R. Hirsch, PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 18, 345–362 (2021). [DOI] [PubMed] [Google Scholar]
- 47.S. Gholamin, S. S. Mitra, A. H. Feroze, J. Liu, S. A. Kahn, M. Zhang, R. Esparza, C. Richard, V. Ramaswamy, M. Remke, A. K. Volkmer, S. Willingham, A. Ponnuswami, A. McCarty, P. Lovelace, T. A. Storm, S. Schubert, G. Hutter, C. Narayanan, P. Chu, E. H. Raabe, G. Harsh IV, M. D. Taylor, M. Monje, Y. J. Cho, R. Majeti, J. P. Volkmer, P. G. Fisher, G. Grant, G. K. Steinberg, H. Vogel, M. Edwards, I. L. Weissman, S. H. Cheshier, Disrupting the CD47-SIRPα anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Sci. Transl. Med. 9, (2017). [DOI] [PubMed] [Google Scholar]
- 48.R. Majeti, M. P. Chao, A. A. Alizadeh, W. W. Pang, S. Jaiswal, K. D. Gibbs Jr., N. van Rooijen, I. L. Weissman, CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 138, 286–299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.H. Cai, M. Li, R. Deng, M. Wang, Y. Shi, Advances in molecular biomarkers research and clinical application progress for gastric cancer immunotherapy. Biomark. Res. 10, 67 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.J. W. Kjeldsen, C. L. Lorentzen, E. Martinenaite, E. Ellebaek, M. Donia, R. B. Holmstroem, T. W. Klausen, C. O. Madsen, S. M. Ahmed, S. E. Weis-Banke, M. O. Holmström, H. W. Hendel, E. Ehrnrooth, M. B. Zocca, A. W. Pedersen, M. H. Andersen, I. M. Svane, A phase 1/2 trial of an immune-modulatory vaccine against IDO/PD-L1 in combination with nivolumab in metastatic melanoma. Nat. Med. 27, 2212–2223 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R. C. Hsieh, S. Krishnan, R.-C. Wu, A. R. Boda, A. Liu, M. Winkler, W.-H. Hsu, S. H. Lin, M.-C. Hung, L.-C. Chan, K. R. Bhanu, A. Srinivasamani, R. A. De Azevedo, Y.-C. Chou, R. A. De Pinho, M. Gubin, E. Vilar, C. H. Chen, R. Slay, P. Jayaprakash, S. M. Hegde, G. Hartley, S. T. Lea, R. Prasad, B. Morrow, C. A. Couillault, M. Steiner, C.-C. Wang, B. P. Venkatesulu, C. Taniguchi, Y. S. B. Kim, J. Chen, N.-P. Rudqvist, M. A. Curran, ATR-mediated CD47 and PD-L1 up-regulation restricts radiotherapy-induced immune priming and abscopal responses in colorectal cancer. Sci. Immunol. 7, eabl9330 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D. Briukhovetska, J. Dörr, S. Endres, P. Libby, C. A. Dinarello, S. Kobold, Interleukins in cancer: From biology to therapy. Nat. Rev. Cancer 21, 481–499 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.V. Mollica Poeta, M. Massara, A. Capucetti, R. Bonecchi, Chemokines and chemokine receptors: New targets for cancer immunotherapy. Front. Immunol. 10, 379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.K. Kato, Y. Ito, H. Daiko, S. Ozawa, T. Ogata, H. Hara, T. Kojima, T. Abe, T. Bamba, M. Watanabe, H. Kawakubo, Y. Shibuya, Y. Tsubosa, N. Takegawa, T. Kajiwara, H. Baba, M. Ueno, R. Machida, K. Nakamura, Y. Kitagawa, A randomized controlled phase III trial comparing two chemotherapy regimen and chemoradiotherapy regimen as neoadjuvant treatment for locally advanced esophageal cancer, JCOG1109 NExT study. J. Clin. Oncol. 40(Suppl. 4), 238 (2022). [Google Scholar]
- 55.D. Phillips, C. M. Schürch, M. S. Khodadoust, Y. H. Kim, G. P. Nolan, S. Jiang, Highly multiplexed phenotyping of immunoregulatory proteins in the tumor microenvironment by CODEX tissue imaging. Front. Immunol. 12, 687673 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.C. R. Stoltzfus, J. Filipek, B. H. Gern, B. E. Olin, J. M. Leal, Y. Wu, M. R. Lyons-Cohen, J. Y. Huang, C. L. Paz-Stoltzfus, C. R. Plumlee, T. Pöschinger, K. B. Urdahl, M. Perro, M. Y. Gerner, CytoMAP: A spatial analysis toolbox reveals features of myeloid cell organization in lymphoid tissues. Cell Rep. 31, 107523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.G. Yu, L. G. Wang, Y. Han, Q. Y. He, clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Y. Kashima, D. Shibahara, A. Suzuki, K. Muto, I. S. Kobayashi, D. Plotnick, H. Udagawa, H. Izumi, Y. Shibata, K. Tanaka, M. Fujii, A. Ohashi, M. Seki, K. Goto, K. Tsuchihara, Y. Suzuki, S. S. Kobayashi, Single-cell analyses reveal diverse mechanisms of resistance to EGFR tyrosine kinase inhibitors in lung cancer. Cancer Res. 81, 4835–4848 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.N. Borcherding, A. Vishwakarma, A. P. Voigt, A. Bellizzi, J. Kaplan, K. Nepple, A. K. Salem, R. W. Jenkins, Y. Zakharia, W. Zhang, Mapping the immune environment in clear cell renal carcinoma by single-cell genomics. Commun. Biol. 4, 122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.N. Nagarsheth, M. S. Wicha, W. Zou, Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 17, 559–572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Legends for tables S1 to S6
Tables S1 to S6