ABSTRACT
The design of inhibitors against metallo-β-lactamases (MBLs), the largest family of carbapenemases, has been a strategic goal in designing novel antimicrobial therapies. In this regard, the development of bicyclic boronates, such as taniborbactam (TAN) and xeruborbactam, is a major achievement that may help in overcoming the threat of MBL-producing and carbapenem-resistant Gram-negative pathogens. Of concern, a recent report has shown that New Delhi MBL-9 (NDM-9) escapes the inhibitory action of TAN by a single amino acid substitution with respect to New Delhi MBL-1 (NDM-1), the most widely disseminated MBL. Here, we report a docking and computational analysis that identifies that “escape variants” against TAN can arise by disruption of the electrostatic interaction of negative charges in the active site loops of MBLs with the N-(2-aminoethyl)cyclohexylamine side chain of TAN. These changes result in non-productive binding modes of TAN that preclude reaction with the MBLs, a phenomenon that is not restricted to NDM-9. This analysis demonstrates that single amino acid substitutions in non-essential residues in MBL loops can unexpectedly elicit resistance to TAN.
KEYWORDS: taniborbactam resistance, metallo-beta-lactamases, NDM-9
INTRODUCTION
The emergence and spread of antibiotic resistance is an issue of major concern in public health (1). In particular, the rapid evolution and dissemination of carbapenem-resistant Enterobacterales is threatening the use of these life-saving drugs (2, 3). Metallo-β-lactamases (MBLs) are zinc-dependent enzymes that confer resistance to most β-lactam antibiotics, including carbapenems. Moreover, all known MBLs are carbapenemases, representing one of the main targets to inhibit in order to prolong the lifetime of these antibiotics. Among them, the New Delhi MBL (NDM), Verona integron-encoded MBL (VIM), and Imipenemase (IMP) families, with 61, 86, and 100 variants, respectively, represent the main clinical challenge (2, 4–6). The administration of β-lactam inhibitors together with a β-lactam antibiotic has proven to be a successful strategy to address resistance (7). However, MBL inhibition has proven to be challenging due to the large diversity of these enzymes. At the moment, MBL inhibitors are not yet clinically available, but this scenario may change soon.
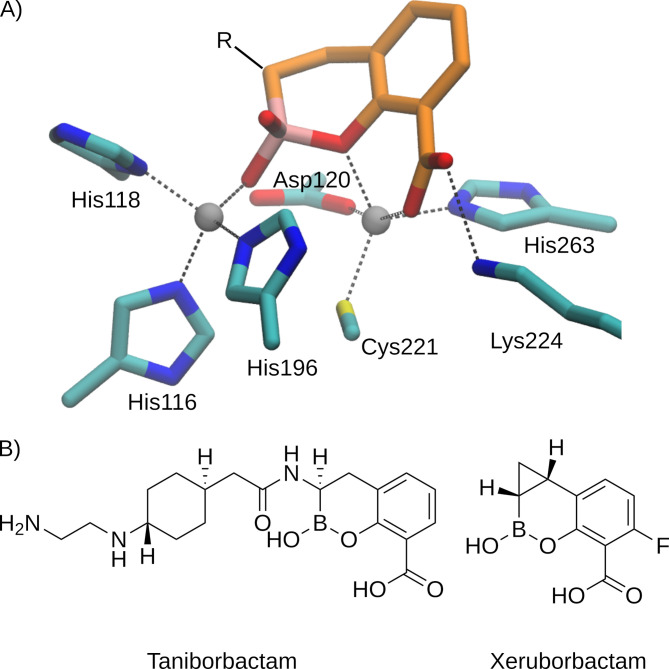
Recent progress in this field came from the development of boronic acid transition state inhibitors (BATSIs) that represent a new family of MBL inhibitors (8–12). These compounds are susceptible to nucleophilic attack by the zinc-bound hydroxide, which is the nucleophile responsible for β-lactam hydrolysis by MBLs, mimicking the reaction catalyzed by these enzymes in bacteria. This reaction gives rise to a stable covalent boron-oxygen bond in such a way that these compounds remain bound to the enzyme active site (Fig. 1A).
Fig 1.
(A) Adduct formed by a bicyclic boronate at the active site of a B1 MBL [based on Protein Data Bank structure 6SP7 (8)]. For clarity, hydrogen atoms are omitted and only the bicyclic boronate scaffold of the ligand is shown. (B) Chemical structures of taniborbactam and xeruborbactam.
Vaborbactam is an already approved cyclic boronate paired with meropenem for the treatment of complicated urinary tract infections (13, 14). However, vaborbactam is not able to inhibit MBLs. Instead, bicyclic boronates have been developed as potent MBL inhibitors. Taniborbactam (TAN) and xeruborbactam (XER) are bicyclic boronates currently under clinical trials that can target some of the most clinically relevant MBLs (Fig. 1B). TAN has demonstrated potent inhibitory activity against clinically relevant MBLs, with the notable exception of IMP-like enzymes (8). The TAN/cefepime combination has completed a phase III clinical trial and is currently under FDA review for approval, with promising efficacy against a wide range of multidrug-resistant Gram-negative bacteria, including carbapenem-resistant isolates. However, a recent report has revealed that NDM-9, a clinical variant of NDM, is refractory to inhibition by TAN (15, 16). This variant differs from NDM-1 by a single amino acid substitution, Glu149Lys (class B β-lactamases numbering), that predates the development of this inhibitor since it was initially reported in 2014 (17, 18). These results suggest that the inhibitory action of TAN can be bypassed by a single escape mutation. More recently, some of us reported that NDM-30 (a single Asp236Tyr substitution compared to NDM-1) and VIM-83 (a single Glu149Lys substitution compared to VIM-1) can also escape the inhibitory effect of TAN by single mutations (C. Le Terrier, C. Viguier, P. Nordmann, A. J. Vila, and L. Poirel, submitted for publication). TAN resistance is a rare but emerging threat that needs to be monitored and addressed to preserve the efficacy of this promising drug combination. With this in mind, we aimed to understand the molecular basis by which NDM-9 and other MBLs are not inhibited by this compound using docking simulations and electrostatic surface calculations.
RESULTS
Structural analysis of docking simulations
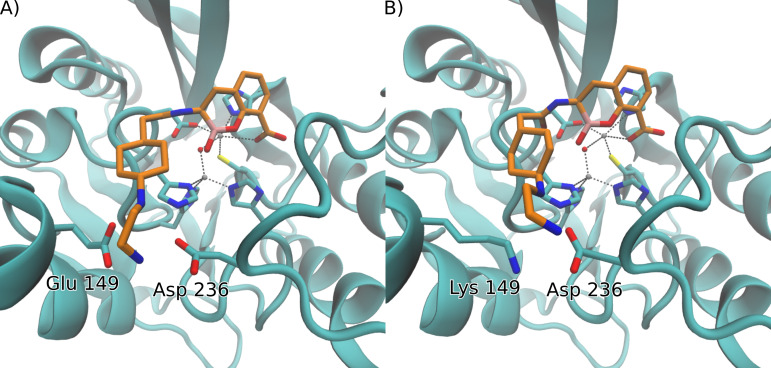
The Protein Data Bank (PDB) crystal structures of NDM-1 [PDB 6RMF (19)] and VIM-2 [PDB 6SP7 (8)] upon reaction with TAN reveal that inhibition of MBLs by this compound depends on two important interactions at the active site: (i) the formation of a covalent bond between the boron atom and the oxygen from the hydroxide bound to Zn1, leading to a tetrahedral boron geometry that mimics the transition state of the reaction mechanism; and (ii) the coordination of the carboxylate group of TAN with the Zn2 ion that mimics the interaction of the carboxylate moieties of β-lactam antibiotics with MBLs. In addition to these interactions subtended by the bicyclic core of TAN with the metal ions of the inhibited enzymes, the N-(2-aminoethyl)cyclohexylamine side chain interacts with the carboxylate group of Glu149 present in loop L2 and Asp236 located in loop L10 (Fig. 2A).
Fig 2.
Comparison of docking simulation results for TAN with (A) NDM-1 and (B) NDM-9. For visual clarity, only the side chains of residues of the active site and those that interact with the N-(2-aminoethyl)cyclohexylamine moiety of TAN are shown, and hydrogen atoms are omitted.
NDM-9 is not inhibited by TAN (15), and this can be attributed to the single amino acid substitution present in this variant, in which the Glu residue is replaced by a positively charged Lys. We hypothesized that this change could have an impact on binding, even though this residue is located outside the active site of the enzyme. More recent evidence has revealed that other mutations can also elicit resistance to TAN in other B1 MBLs (experimental data from the literature are summarized in Table S1). We therefore decided to explore this hypothesis by running a series of docking simulations of TAN to NDM-1, VIM-2, and variants on the glutamate residue in these MBLs (NDM-9 and VIM-2 Glu149Lys).
When analyzing the docking simulation results, we evaluated not only the binding energy of each obtained conformation, but also its population, which represents the percentage of individual docking results that were clustered as the same ligand pose. Given that the nucleophilic attack that results in the inhibited complex requires a specific positioning of the boron atom with respect to the OH- ion, the presence of several non-reactive conformations suggests a less efficient inhibition. We are aware that these values should not be extrapolated to retrieve thermodynamic parameters, but we regard them as a bona fide predictor of the experimentally observed trends.
NDM variants
Docking simulations of TAN into the crystal structure of unbound NDM-1 [PDB 3SPU (20)] were first performed. The best docking pose of TAN into the active site, shown in Fig. 2, is similar to the one reported in the crystal structure: the root mean square deviation (RMSD) of this pose with respect to the ligand atoms in crystal 6RMF is 1.11 Å. This conformation represents 95% of the population of all docking simulations, suggesting that no other significant conformations are predicted in the active site cavity. The B-OH- distance is 2.83 Å, which positions the boron atom adequately for a nucleophilic attack by the hydroxyl ion, in agreement with the crystallographic structure of the adduct after the reaction (PDB 6RMF) featuring a B-O bond. We conclude that this docked structure provides a bona fide description of the Michaelis complex which can elicit inhibition of NDM-1 by TAN. In contrast, docking simulations of TAN into the structure of NDM-9 [PDB 6OGO (21)] reveal that the bound structure that better resembles the binding to NDM-1 is represented by a population of only 38%, and its binding energy is reduced by 4 kcal/mol compared to NDM-1, revealing a disruptive role of the Lys residue in the binding of TAN to NDM-9. Other docking conformations represent non-productive poses of the boronate moiety of TAN. Indeed, the second most populated pose has a population of 10%, and the repulsive interaction between Lys149 and the amine groups of TAN leads to a conformation that places the ligand with a B-OH- distance of 3.65 Å that makes the reaction unfeasible, disfavoring, or preventing inhibition (Fig. S1). Given that surface lysine residues are flexible, which could potentially influence docking experiments, we further analyzed the position of Lys149 by performing 1 µs-long molecular dynamics simulations. Throughout the simulated time, the conformation of the lysine residue does not vary significantly from the one found in the crystal structure (Fig. S2), showing that the docking results should not be affected by the flexibility of this residue.
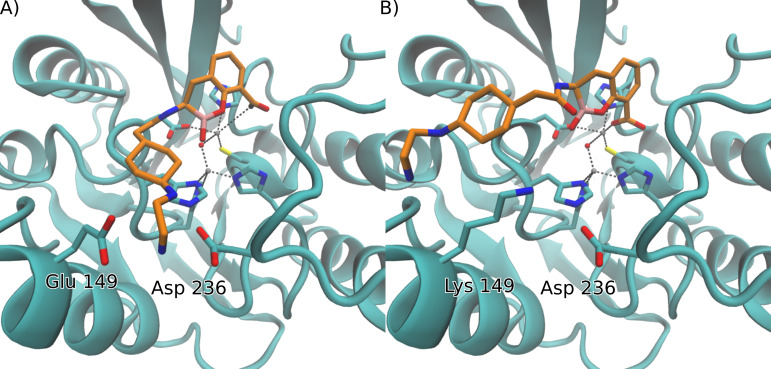
VIM variants
These insights are not restricted to the NDM family. A similar point mutation engineered into VIM-2 (Glu149Lys) also renders this enzyme refractory to inhibition by TAN (Table S1). The same behavior is displayed in VIM-83, which is a variant of VIM-1 that also replaces a Glu residue for Lys in position 149. We also explored the impact of the introduction of a Lys residue in this position in the structure of VIM-2 that led to similar results to those obtained for NDM (Fig. 3A). Binding of TAN to VIM-2 can be reproduced by docking simulations, which predict a highly populated productive pose (82%) that resembles the crystal structure (RMSD 1.42 Å with respect to PDB 6SP7). Introduction of a Lys residue in the Glu149Lys mutant also gives rise to a repulsive interaction with the amine moieties in TAN that significantly decrease the population of the productive pose (Fig. 3B), resulting in a higher amount of docking conformations with decreased binding energy (5 kcal/mol lower) and population (4%), none of which adopt a productive pose that could result in the nucleophilic attack from the hydroxyl group. VIM-83 displays the same behavior as the VIM-2 Glu149Lys mutant, in line with the decrease of the inhibitory potency of TAN against these two variants (Table S1).
Fig 3.
Comparison of docking simulation results for TAN with (A) VIM-2 and (B) VIM-2 Glu149Lys. For visual clarity, only the side chains of residues of the active site and those that interact with the N-(2-aminoethyl)cyclohexylamine moiety of TAN are shown, and hydrogen atoms are omitted.
IMP variants
The previously described docking studies in NDM and VIM variants show how the introduction of a positively charged residue in position 149 thwarts the inhibitory potency of TAN by eliciting non-productive binding to the active site of the MBLs. However, this does not explain why IMP-1, one of the most clinically relevant MBLs, is not inhibited by TAN, since IMP-1 contains an Asp residue in position 149 and in a similar orientation compared to NDM-1 and VIM-2 with respect to the active site. Mutation of this residue to a Glu did not enable inhibition by TAN, revealing that the presence of an Asp residue in this position is not an impediment for TAN to bind the enzyme (15). Docking of TAN to IMP-1 resulted in a productive binding pose with a 91% population. However, the calculated binding energy for this conformation is 2–3 kcal/mol smaller than for VIM-2 or NDM-1, suggesting that binding of TAN is less favorable for this enzyme. With these results, we decided to explore the presence of other structural features that could impede inhibition by TAN by calculating the electrostatic potential in the protein surface for all these variants.
Electrostatic features at the active sites of different MBLs
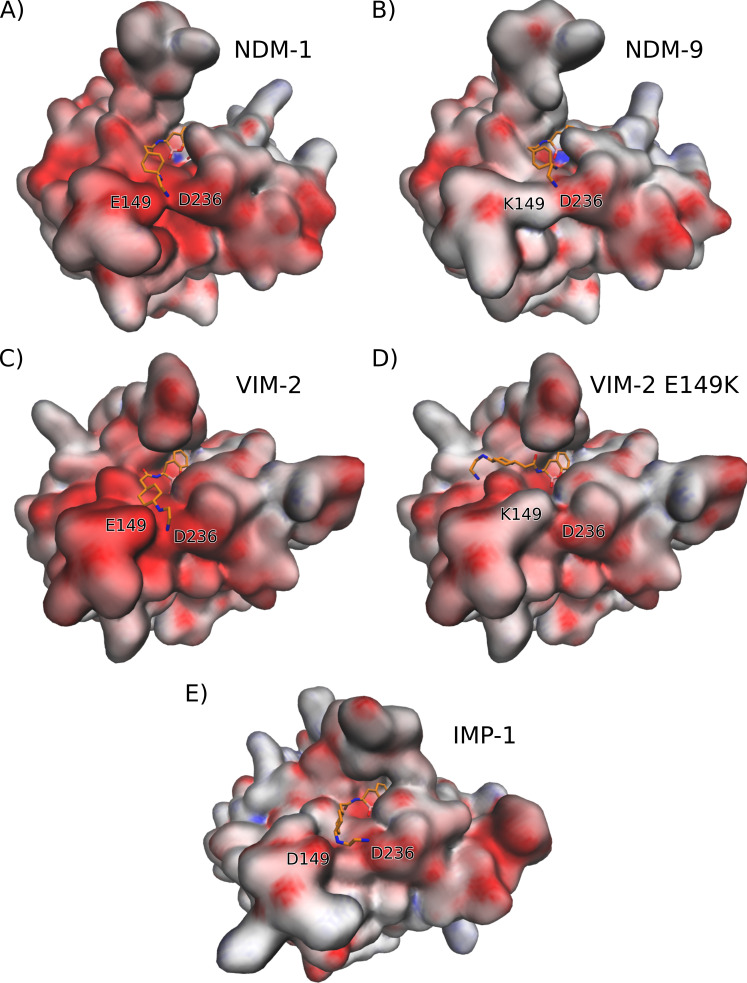
A systematic examination of other residues located near the binding pocket that could affect inhibition by TAN in IMP-1 revealed two positively charged Lys residues (Lys148 and Lys167) that surround the negatively charged Glu149 which could hinder its interaction with the amine groups present in the sidechain of the ligand. This compelled us to explore how changes in the electrostatics of the binding pocket can alter the interaction of each MBL with TAN. We therefore calculated the electrostatic potential surface of each variant, as shown in Fig. 4, where negative, neutral, and positive potentials are colored as red, white, and blue, respectively.
Fig 4.
Electrostatic potential surfaces of proteins (A) NDM-1, (B) NDM-9, (C) VIM-2, (D) VIM-2 Glu149Lys, and (E) IMP-1. The productive pose of each docking experiment of TAN to each protein are also shown (C, atoms of TAN are depicted in orange). Negative, neutral, and positive potentials are colored as red, white, and blue, respectively, in a scale from −10 to 10 kBT/ec.
Firstly, the electrostatic potentials in the surfaces of NDM-1 (Fig. 4A) and VIM-2 (Fig. 4C) reveal a negatively charged patch around residues Glu149 and Asp236 in the regions that provide a favorable interaction with the N-(2-aminoethyl)cyclohexylamine moiety of TAN. On the other hand, the substitution of Glu149 for a Lys residue in NDM-9 (Fig. 4B) and VIM-2 Glu149Lys (Fig. 4D) significantly decreases the negative charge of the region that interacts with the amine groups of TAN, which results in a less favorable interaction energy. The same effect is observed with the Lys substitution in VIM-83 (Fig. S3A). Additionally, the highly negative potential in the surface of VIM-2 compared to NDM-1 may account for the higher affinity of the former enzyme toward the inhibitor [0.06 vs 0.1 µM, according to half-maximal inhibitory concentration (IC50) values, Table S1].
Substitutions of the Asp236 residue of the loop L10 can also alter TAN binding. In this regard, changing this residue into a neutral or positively charged one will affect the negatively charged environment that can interact with the N-(2-aminoethyl)cyclohexylamine side chain. Indeed, NDM-30, which harbors the Asp236Tyr mutation, gives rise to a less negative surface (Fig. S3B), and is predicted to impair the binding energy (3 kcal/mol lower than in NDM-1). These results account for the poor inhibition of this enzyme by TAN (IC50 = 17 µM, Table S1).
The recent observation that TAN also lacks significant inhibitory activity against the B1 enzyme Seoul Imipenemase MBL-1 (SIM-1) is notable (Table S1). We therefore calculated the electrostatic surface in this enzyme (Fig. S3C). These calculations also show a decreased negative charge in the pocket that interacts with the amine groups of TAN, in agreement with the low interacting energy obtained from docking experiments (4 kcal/mol lower binding energy than NDM-1) and with the lack of inhibitory activity experimentally observed (Table S1).
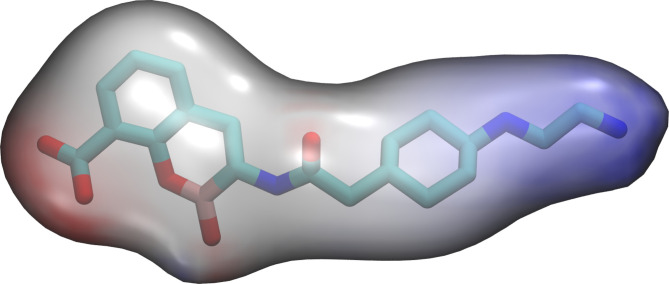
Finally, we explored why TAN is unable to inhibit IMP-1, the most widespread representative of enzymes from the IMP family. A closer inspection of the crystal structure of IMP-1 [PDB 7XHW (22)] revealed that Asp149 is flanked by two Lys residues pointing in the same direction, Lys148 and Lys167. Analysis of the electrostatic potential at the protein surface reveals that, despite the presence of this carboxylate group, the positive charge of the surrounding Lys residues counteracts its presence by providing a less negative binding surface that hinders the interaction with TAN. Therefore, although the lack of inhibition of IMP-1 cannot be attributed to a single residue, it can be described by the same type of interaction as found for NDM-9, NDM-30, and VIM-83, i.e., a repulsive interaction with the side chain of TAN. The amine groups in this side chain are protonated at physiological pH, concentrating a large positive partial charge that can favor these interactions, as revealed by calculation of the electrostatic potential of TAN (Fig. 5).
Fig 5.
Electrostatic potential surface of taniborbactam. Negative, neutral, and positive potentials are colored as red, white, and blue, respectively, in a scale from −10 to 10 kBT/ec.
DISCUSSION
The inhibitory potency of bicyclic boronates depends on the orientation of the boron atom toward the attacking nucleophile that is steered by the catalytic residues and the active site topology in each enzyme (8, 10, 19). Under this premise, understanding why successful BATSIs such as TAN are challenged by single amino acid substitutions can guide medicinal chemists in the improvement of novel boronate scaffolds by careful consideration of structure-activity relationship. The Glu149Lys mutation present in NDM-9 renders this MBL refractory to inhibition by TAN (IC50 = 53 µM). The N-(2-aminoethyl)cyclohexylamine moiety of TAN was critical in the development of this drug by enhancing membrane permeability and by interacting with the conserved anionic residues Glu149 and Asp236 in NDM-1 (Fig. 4A) and VIM-2 (Fig. 4C) (8, 19). By modeling the interactions of TAN with NDM-9, we identified that the presence of Lys152 disrupts this interaction because of its positive partial charge, diminishing its binding energy and, in turn, thwarting the inhibitory potency of TAN (Fig. 4B and D). Similar repulsive electrostatic interactions between the N-(2-aminoethyl)cyclohexylamine group and a patch of lysine residues in IMP-1, accounting for the failure of TAN in inhibiting this enzyme, were also found.
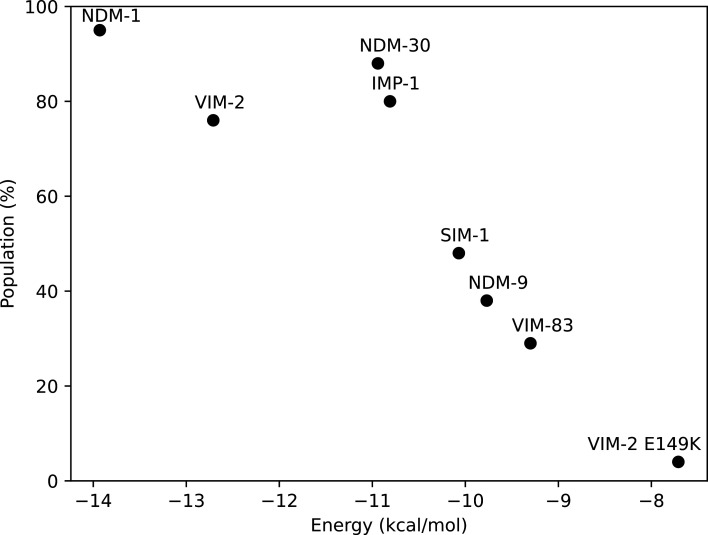
The docking experiments reveal that these substitutions can impair inhibition by decreasing the binding energy and/or by disfavoring the formation of a productive Michaelis complex. Figure 6 summarizes the results of the docking experiments depicting the interaction of TAN with the different MBLs, where the enzymes are located based on the population of the best docking pose and its binding energy. The enzymes showing potent inhibition by TAN are located in the upper left area, which is characterized by a high binding energy and a significant population of the productive binding mode. Instead, enzymes located outside this region are those that show an impaired inhibition by TAN.
Fig 6.
Docking experiment results plotted by the population and energy of the productive conformation.
In conclusion, the potency of TAN against MBLs can be overcome by variants accumulating substitutions in loops L2 and L10. These changes alter the conformation and electrostatics of the active site, reducing the affinity and stability of TAN binding through interaction with the N-(2-aminoethyl)cyclohexylamine group. In this regard, we do not expect that these substitutions could affect inhibition by XER, which has a more minimalistic boronate core that does not subtend these electrostatic interactions with the active site loops. These results are in agreement with a very recent paper from Lomovskaya et al. (23) that shows that XER is able to inhibit NDM-9 and proposes a similar explanation of the role of acidic-to-basic substitutions in TAN resistance based on the available crystallography data. Our analyses reveal the molecular mechanisms of resistance to TAN and provide insights for the design of novel boronate inhibitors that can overcome these challenges. Most importantly, this work highlights that the introduction of functional chemical moieties displaying electrostatic interactions with non-essential residues in active site loops of MBLs can rapidly elicit resistance to novel inhibitors. Finally, these results can guide therapy since the cefepime/TAN treatment may be successful or not depending on the identification of the MBL variant.
MATERIALS AND METHODS
Structural modeling
Available experimentally obtained structures for the MBLs were obtained from the Protein Data Bank: NDM-1 [PDB 3SPU (20)], NDM-9 [PDB 6OGO (21)], VIM-2 (PDB 5YD7), VIM-1 [PDB 5N5G (24)], and IMP-1 [PDB 7XHW (22)]. The structure for SIM-1 was obtained from the AlphaFold Protein Structure Database (25) (UniProt code A0A3Q9U5J7).
For variants with single amino acid substitutions that did not have an available structure, in silico mutations were performed using the Leap software from Amber22 (26): NDM-30 (Asp236Tyr mutation from NDM-1), VIM-2 Glu149Lys, VIM-83 (Glu149Lys substitution from VIM-1). In each structure created with Leap, the conformation of the altered residue was optimized using Sander software in the Amber22 package, using implicit solvent and restraining all atoms except the sidechain of that residue.
The structure of TAN was obtained by converting the canonical SMILES notation from PubChem (27) into a pdb file using Open Babel (28). A geometry optimization at B3LYP/6–31G* level was performed using Gaussian09 (29).
Docking simulations
Docking experiments were carried out using Autodock 4.2.6 (30). The grid maps were set to 70 × 70 × 70 points with a grid spacing of 0.375 Å centered on the oxygen atom of the hydroxyl ion in the catalytic site. Charges for the Zn2+ and hydroxyl ions present in the catalytic center of the MBLs were obtained from literature (31). To obtain boron Autodock parameters, the van der Waals radius for the desolvated atom and the associated van der Waals well-depth value were adapted from literature, and the default Autodock atomic solvation value for carbon was used (32, 33). For each docking calculation, 100 different docking runs were performed, which were clustered using an RMSD cut-off of 2 Å. To analyze the inhibitory potency of TAN to each MBL, we used the binding free energy score (ΔG) calculated by Autodock and the population, which is the number of individual docking results that were clustered as the same ligand pose. The poses selected as correct were those with the highest population and the most negative binding free energy. For RMSD calculations, only the atoms belonging to the bicyclic scaffold of TAN were used.
Molecular dynamics simulations
Molecular dynamics simulations performed on NDM-9 were done using the Amber22 package (26). Crystal structure (PDB 6OGO) was prepared using Tleap, by immersing the protein in a truncated octahedral box of TIP3P water (34). ff14SB parameters (35) were used for the protein, and the parameters for the active site were taken from literature (31). The initial structure was first minimized, then taken from a temperature of 0 K to 300 K at constant volume for 100 ps and equilibrated at a constant pressure of one bar for 200 ps, before the final production time of 1 µs at constant pressure and temperature. Langevin thermostat (36) and Berendsen barostat (37) were used for temperature and pressure control, respectively.
Electrostatic surface calculations
Electrostatic surface calculations were made with APBS software. The necessary PQR files were obtained using the PDB2PQR software included in the Amber22 package. Surface visualization was carried out using VMD software (38), using the quicksurf drawing method, with a radius scale of 0.8, a density isovalue of 0.7, and a color scale data range from −10 to 10 KBT/ec.
ACKNOWLEDGMENTS
This research was supported by grants from the National Institutes of Health (R01AI100560 to R.A.B. and A.J.V.) and Agencia I+D+I (grant PICT-2020-03769 to D.M.M. and PICT-2020-00031 to A.J.V.). A.J.V. and D.M.M are staff members from CONICET. S.I.D. is recipient of fellowship from CONICET, Argentina.
Contributor Information
Alejandro J. Vila, Email: vila@ibr-conicet.gov.ar.
Diego M. Moreno, Email: moreno@iquir-conicet.gov.ar.
Alessandra Carattoli, Universita degli studi di roma La Sapienza, Rome, Italy.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.01168-23.
Supplemental Table S1 and Figures S1-S2-S3.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, et al. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bahr G, González LJ, Vila AJ. 2021. Metallo-β-lactamases in the age of multidrug resistance: from structure and mechanism to evolution, dissemination, and inhibitor design. Chem Rev 121:7957–8094. doi: 10.1021/acs.chemrev.1c00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mojica MF, Rossi MA, Vila AJ, Bonomo RA. 2022. The urgent need for metallo-β-lactamase inhibitors: an unattended global threat. Lancet Infect Dis 22:e28–e34. doi: 10.1016/S1473-3099(20)30868-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. López C, Delmonti J, Bonomo RA, Vila AJ. 2022. Deciphering the evolution of metallo-β-lactamases: a journey from the test tube to the bacterial periplasm. J Biol Chem 298:101665. doi: 10.1016/j.jbc.2022.101665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ju LC, Cheng Z, Fast W, Bonomo RA, Crowder MW. 2018. The continuing challenge of metallo-β-lactamase inhibition: mechanism matters. Trends Pharmacol Sci 39:635–647. doi: 10.1016/j.tips.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mojica MF, Bonomo RA, Fast W. 2016. B1-metallo-β-lactamases: where do we stand? Curr Drug Targets 17:1029–1050. doi: 10.2174/1389450116666151001105622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bush K, Bradford PA. 2019. Interplay between β-lactamases and new β-lactamase inhibitors. Nat Rev Microbiol 17:459–460. doi: 10.1038/s41579-019-0221-6 [DOI] [PubMed] [Google Scholar]
- 8. Liu B, Trout REL, Chu G-H, McGarry D, Jackson RW, Hamrick JC, Daigle DM, Cusick SM, Pozzi C, De Luca F, Benvenuti M, Mangani S, Docquier J-D, Weiss WJ, Pevear DC, Xerri L, Burns CJ. 2020. Discovery of taniborbactam (VNRX-5133): a broad-spectrum serine- and metallo-β-lactamase inhibitor for carbapenem-resistant bacterial infections. J Med Chem 63:2789–2801. doi: 10.1021/acs.jmedchem.9b01518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hamrick JC, Docquier J-D, Uehara T, Myers CL, Six DA, Chatwin CL, John KJ, Vernacchio SF, Cusick SM, Trout REL, Pozzi C, De Luca F, Benvenuti M, Mangani S, Liu B, Jackson RW, Moeck G, Xerri L, Burns CJ, Pevear DC, Daigle DM. 2020. VNRX-5133 (Taniborbactam), a broad-spectrum inhibitor of Serine- and metallo-β- lactamases, restores activity of cefepime in enterobacterales and Pseudomonas aeruginosa. Antimicrob Agents Chemother 64:e01963-19. doi: 10.1128/AAC.01963-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brem J, Cain R, Cahill S, McDonough MA, Clifton IJ, Jiménez-Castellanos J-C, Avison MB, Spencer J, Fishwick C, Schofield CJ. 2016. Structural basis of metallo-β-lactamase, serine-β-lactamase and penicillin-binding protein inhibition by cyclic boronates. Nat Commun 7:12406. doi: 10.1038/ncomms12406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krajnc A, Lang PA, Panduwawala TD, Brem J, Schofield CJ. 2019. Will morphing boron-based inhibitors beat the β-lactamases? Curr Opin Chem Biol 50:101–110. doi: 10.1016/j.cbpa.2019.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hecker SJ, Reddy KR, Lomovskaya O, Griffith DC, Rubio-Aparicio D, Nelson K, Tsivkovski R, Sun D, Sabet M, Tarazi Z, Parkinson J, Totrov M, Boyer SH, Glinka TW, Pemberton OA, Chen Y, Dudley MN. 2020. Discovery of cyclic boronic acid QPX7728, an ultrabroad-spectrum inhibitor of serine and metallo-β-lactamases. J Med Chem 63:7491–7507. doi: 10.1021/acs.jmedchem.9b01976 [DOI] [PubMed] [Google Scholar]
- 13. Livermore DM, Mushtaq S. 2013. Activity of biapenem (RPX2003) combined with the boronate β-lactamase inhibitor RPX7009 against carbapenem-resistant enterobacteriaceae. J Antimicrob Chemother 68:1825–1831. doi: 10.1093/jac/dkt118 [DOI] [PubMed] [Google Scholar]
- 14. Drawz SM, Papp-Wallace KM, Bonomo RA. 2014. New Β-Lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob Agents Chemother 58:1835–1846. doi: 10.1128/AAC.00826-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le Terrier C, Gruenig V, Fournier C, Nordmann P, Poirel L. 2023. NDM-9 resistance to taniborbactam. Lancet Infect Dis 23:401–402. doi: 10.1016/S1473-3099(23)00069-5 [DOI] [PubMed] [Google Scholar]
- 16. Le Terrier C, Nordmann P, Buchs C, Di DYW, Rossolini GM, Stephan R, Castanheira M, Poirel L. 2023. Wide dissemination of gram-negative bacteria producing the taniborbactam-resistant NDM-9 variant: a one health concern. J Antimicrob Chemother 78:2382–2384. doi: 10.1093/jac/dkad210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X, Li H, Zhao C, Chen H, Liu J, Wang Z, Wang Q, Zhang Y, He W, Zhang F, Wang H. 2014. Novel NDM-9 metallo-β-lactamase identified from a ST107 Klebsiella pneumoniae strain isolated in China. Int J Antimicrob Agents 44:90–91. doi: 10.1016/j.ijantimicag.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 18. Yao X, Doi Y, Zeng L, Lv L, Liu JH. 2016. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis 16:288–289. doi: 10.1016/S1473-3099(16)00057-8 [DOI] [PubMed] [Google Scholar]
- 19. Krajnc A, Brem J, Hinchliffe P, Calvopiña K, Panduwawala TD, Lang PA, Kamps J, Tyrrell JM, Widlake E, Saward BG, Walsh TR, Spencer J, Schofield CJ. 2019. Bicyclic Boronate VNRX-5133 inhibits Metallo- and Serine-Β-Lactamases. J Med Chem 62:8544–8556. doi: 10.1021/acs.jmedchem.9b00911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. King D, Strynadka N. 2011. Crystal structure of new Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Sci 20:1484–1491. doi: 10.1002/pro.697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raczynska JE, Imiolczyk B, Komorowska M, Sliwiak J, Czyrko-Horczak J, Brzezinski K, Jaskolski M. 2020. Flexible loops of new Delhi metallo-β-lactamase modulate its activity towards different substrates. Int J Biol Macromol 158:104–115. doi: 10.1016/j.ijbiomac.2020.04.219 [DOI] [PubMed] [Google Scholar]
- 22. Kim H, Park J, Kim S, Shin DH. 2018. Crystal structure of D-glycero-α-d-manno-heptose-1-phosphate guanylyltransferase from Yersinia pseudotuberculosis. Biochim Biophys Acta Proteins Prot 1866:482–487. doi: 10.1016/j.bbapap.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 23. Lomovskaya O, Tsivkovski R, Totrov M, Dressel D, Castanheira M, Dudley M, Poirel L. 2023. New boronate drugs and evolving NDM-mediated beta-lactam resistance. Antimicrob Agents Chemother 67:e0057923. doi: 10.1128/aac.00579-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salimraj R, Hinchliffe P, Kosmopoulou M, Tyrrell JM, Brem J, van Berkel SS, Verma A, Owens RJ, McDonough MA, Walsh TR, Schofield CJ, Spencer J. 2019. Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-β-lactamases. FEBS J 286:169–183. doi: 10.1111/febs.14695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, et al. 2022. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50:D439–D444. doi: 10.1093/nar/gkab1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Case DA, Aktulga HM, Belfon K, Ben-Shalom IY, Berryman JT, Brozell SR, Cerutti DS, Cheatham TE, Cisneros GA, Cruzeiro VWD, et al. 2022. Amber 2022. University of California, San Francisco. [Google Scholar]
- 27. Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE. 2023. Pubchem 2023 update. Nucleic Acids Res 51:D1373–D1380. doi: 10.1093/nar/gkac956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. 2011. Open babel: an open chemical toolbox. J Cheminform 3:1–14. doi: 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, et al. 2009. Gaussian 09. Gaussian, Inc, Wallingford CT, USA. [Google Scholar]
- 30. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suárez D, Brothers EN, Merz KM. 2002. Insights into the structure and dynamics of the dinuclear zinc β-lactamase site from Bacteroides fragilis. Biochemistry 41:6615–6630. doi: 10.1021/bi0121860 [DOI] [PubMed] [Google Scholar]
- 32. Tiwari R, Mahasenan K, Pavlovicz R, Li C, Tjarks W. 2009. Carborane clusters in computational drug design: a comparative docking evaluation using autodock, flexx, glide, and surflex. J Chem Inf Model 49:1581–1589. doi: 10.1021/ci900031y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otkidach DS, Pletnev IV. 2001. Conformational analysis of boron-containing compounds using Gillespie–kepert version of molecular mechanics. J of Mol Struct: THEOCHEM 536:65–72. doi: 10.1016/S0166-1280(00)00602-3 [DOI] [Google Scholar]
- 34. Price DJ, Brooks CL III. 2004. A modified TIP3P water potential for simulation with ewald summation. J Chem Phys 121:10096–10103. doi: 10.1063/1.1808117 [DOI] [PubMed] [Google Scholar]
- 35. Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. 2015. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 11:3696–3713. doi: 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xiang T, Liu F, Grant DM. 1991. Generalized langevin equations for molecular dynamics in solution. J Chem Phys 94:4463–4471. doi: 10.1063/1.460602 [DOI] [Google Scholar]
- 37. Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. 1984. Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. doi: 10.1063/1.448118 [DOI] [Google Scholar]
- 38. Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. doi: 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1 and Figures S1-S2-S3.