Abstract
The master circadian clock generates 24-hour rhythms to orchestrate daily behavior, even running freely under constant conditions. Traditionally, the master clock is considered self-sufficient in sustaining free-running timekeeping via its cell-autonomous molecular clocks and interneuronal communications within the circadian neural network. Here, we find a set of bona fide ultradian oscillators in the Drosophila brain that support free-running timekeeping, despite being located outside the master clock circuit and lacking clock gene expression. These extra-clock electrical oscillators (xCEOs) generate cell-autonomous ultradian bursts, pacing widespread burst firing and promoting rhythmic resting membrane potentials in clock neurons via parallel monosynaptic connections. Silencing xCEOs disrupts daily electrical rhythms in clock neurons and impairs cycling of neuropeptide pigment dispersing factor, leading to the loss of free-running locomotor rhythms. Together, we conclude that the master clock is not self-sufficient to sustain free-running behavior rhythms but requires additional endogenous inputs to the clock from the extra-clock ultradian brain oscillators.
Bursting inputs from ultradian xCEOs in the brain are required for free-running circadian timekeeping.
INTRODUCTION
The master circadian clock in the brain comprises a circuit of interconnected neurons, each containing its own molecular clock. Molecular clocks generate approximately 24-hour cycles of activity via cell-autonomous transcription-translation feedback loops (TTFLs) (1, 2). Because these molecular clocks cycle in synchrony with each other (3–5), neurons within the central master clock can maintain unified timekeeping to persistently orchestrate circadian rhythms, even in constant darkness (DD).
Multiple pieces of evidence support the conventional view that the coherent circadian timekeeping relies on interneuronal connections within the central clock network (3–12). When electrically silenced or dissociated into single cells, central clock neurons in the mammalian suprachiasmatic nucleus (SCN) maintain molecular clock activity but lose synchronization (7, 13–18). Likewise, electrically silencing central clock neurons in Drosophila desynchronizes, and can even stop, their molecular clocks [(19, 20), but see (21)].
However, it remains unknown whether the master clock can maintain free-running timekeeping by relying solely on its cell-autonomous molecular clocks and interneuronal communications within the central circadian clock. To address this question, we performed multiple-electrode patch-clamp recordings of the Drosophila clock neurons to characterize the pattern and origin of basal electrical activity, which organizes circadian timekeeping in the absence of external time inputs. We found that most clock neuron subtypes exhibited a pattern of synchronous ultradian burst firing, which was driven by synaptic inputs from outside the master clock. Furthermore, we found a group of bona fide ultradian brain oscillators that mediate extra-clock bursting inputs to clock neurons. Moreover, the extra-clock oscillators and clock neurons form a hub circuit via parallel, monosynaptic connections at the accessory medulla (aMe). Genetic silencing of these extra-clock ultradian oscillators revealed that they are indispensable in sustaining free-running circadian timekeeping of locomotor rhythms by cooperating with the molecular clockworks to set circadian electrical rhythms, the key timing output from the central circadian clock.
RESULTS
Clock neurons display synchronous burst firing
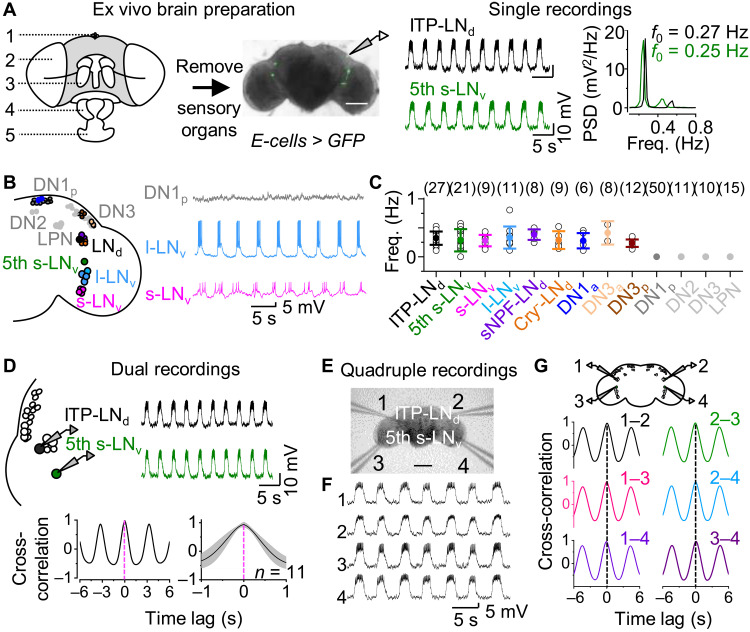
The central circadian neural network of Drosophila is composed of 150 clock neurons that distribute across the brain, including seven subtypes according to their anatomical positions and size (3, 9, 10). We conducted systematic patch-clamp recordings of all known clock neuron subtypes at the circadian time (CT) 0 to 8 in DD3 in the absence of external sensory inputs. Both the fifth small ventrolateral neuron (5th s-LNv) and ion transport peptide–expressing dorsolateral neuron (ITP-LNd)—two evening cells (E cells) that control peak activity at dusk (22, 23)—exhibited robust rhythmic burst firing in current-clamp recording mode (Fig. 1A). Bursts of up to 75 action potentials were superimposed on slow depolarizations (fig. S1, A and B), which occurred at frequencies of approximately 0.3 Hz (Fig. 1A). Similarly, the four other s-LNvs—morning cells (M cells) that control peak activity at dawn (22, 24)—and four large LNvs (l-LNvs)—the arousal clock neurons (25)—also exhibited rhythmic burst firing (Fig. 1B), consistent with prior reports (26–28). Moreover, dorsal neurons DN1a and anteriorly and posteriorly located DN3s (DN3a and DN3p) that have dendritic arborization in aMe and large cell bodies (29), together with dorsolateral neurons expressing short neuropeptide F (sNPF-LNds) and cryptochrome (CRY)–negative LNds, exhibited rhythmic burst firing (Fig. 1C). However, dorsal clock neurons DN1p, DN2, and DN3 and lateral posterior clock neurons (LPNs) did not display such bursting activity (Fig. 1, B and C).
Fig. 1. Clock neurons display synchronous burst firing.
(A) Left, ex vivo brain preparation: 1, ocellus; 2, compound eye; 3, antenna; 4, maxillary palp; and 5, labella; middle, representative recordings from ITP-LNd (top) and 5th s-LNv (bottom) neurons from two brain preparations at a controlled membrane potential of approximately −55 mV; right, power spectrum of subthreshold depolarization rhythms. Recording time: CT0-8 during DD3. Scale bar, 100 μm. (B) Left, schematic of clock neuron distribution; right, representative single recordings of DN1p (top), l-LNv (middle), and s-LNv (bottom). (C) Frequencies of slow rhythmic depolarization. Numbers represent recorded cells. (D) Top, schematic of dual recordings (left) and representative recordings of ITP-LNd and 5th s-LNv ipsilateral pair (right); bottom, cross-correlation analysis (left) and combined data (right). (E) Differential interference contrast (DIC) image of quadruple recordings. Scale bar, 50 μm. (F) Representative quadruple recordings. (G) Top, schematic of quadruple recordings; bottom, cross-correlation analysis of paired recordings in (F). Freq, frequency; PSD, power spectrum density. Combined data are presented as means ± SEM.
The similarity in frequency and pattern of this widespread rhythmic bursting activity suggested that the bursts may occur in synchrony. To test this, we performed dual patch-clamp recordings of ipsilateral pairs of E cells and observed synchronized bursts (Fig. 1D). Such synchrony was also evident between other ipsilateral pairs of bursting clock neurons (fig. S1, C and D). Furthermore, quadruple patch-clamp recordings of the four bilateral 5th s-LNvs and ITP-LNds revealed synchronized rhythmic bursts across the entire brain (Fig. 1, E and F). There was no time lag in synchrony among any of the four neurons (Fig. 1G and fig. S1E).
To examine whether clock neurons rhythmically burst in vivo, we conducted patch-clamp recordings in live flies (fig. S1F). The above bursting clock neurons showed similar rhythmic activity in vivo, but at a higher frequency than those in the ex vivo brain preparations (fig. S1, G and H), possibly due to additional inputs from sensory systems in vivo. Together, these data reveal the existence of widespread and synchronous bursting activity in Drosophila central clock neurons.
Extra-clock ultradian oscillators synchronize bursting
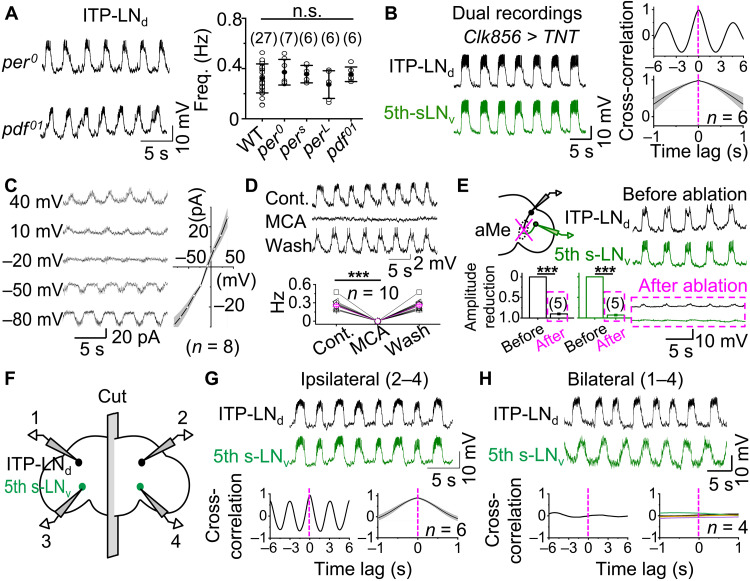
We next examined whether synchronized bursting in clock neurons depends on the molecular clockwork. In per0 flies, which lack the clock gene period and therefore functional circadian rhythms (30), rhythmic bursts were still observed in clock neurons in ex vivo brain preparations (Fig. 2A). Similarly, rhythmic bursts with the same frequency as those in wild-type (WT) flies were recorded in perS and perL flies (Fig. 2A), which exhibit short and long circadian periods (30), respectively. Therefore, burst firing in clock neurons is independent of the molecular clockwork.
Fig. 2. Extra-clock ultradian oscillators synchronize bursting.
(A) Left, representative rhythmic bursts in ITP-LNd from per0 (top) and pdf01 (bottom) flies at a controlled membrane potential of approximately −55 mV; right, combined data from WT, per0, perS, perL, and pdf01 flies, not statistically significant between WT and mutant flies by unpaired two-tailed Student’s t test. (B) Left, dual recordings of ipsilateral ITP-LNd (top) and 5th s-LNv (bottom) in Clk856-TNT flies; right, cross-correlation analysis (top) and combined data (bottom). (C) Current-voltage correlation relationship of rhythmic EPSCs in ITP-LNd. Left, representative voltage-clamp recordings at holding voltages indicated; right, plot of peak amplitudes of rhythmic EPSCs against voltages. (D) Top, 25 μM MCA blocked bursts in ITP-LNd; bottom, combined data. ***P < 0.001 by paired t test. (E) aMe ablation reduces clock neuron bursts. Left, schematic of aMe ablation and dual recordings (top) and combined data (bottom); right, representative dual recordings before (top) and after (bottom) aMe ablation. ***P < 0.001 by paired t test. (F) Schematic of dual recordings in split brain. (G) Representative recordings of ITP-LNd (top) and 5th s-LNv (middle) ipsilateral pair in split brains, cross-correlation analysis (bottom). (H) Representative recordings of ITP-LNd and 5th s-LNv bilateral pair in split brains (top), cross-correlation analysis (bottom). Combined data are presented as means ± SEM. n.s., not statistically significant, P > 0.05.
We subsequently investigated the role of intra-clock neuronal circuits within the central clock network. In pdf01 flies that display arrhythmic behavior due to a lack of neuropeptide pigment-dispersing factor (PDF) (31), normal burst rhythms were observed in E cells (Fig. 2A). Similarly, E and M cells were found to retain normal burst rhythms in flies in which synaptic transmission had been blocked by tetanus neurotoxin (TNT) expression using the pan-clock neuronal driver Clk856-Gal4 (Fig. 2B and fig. S2, A to D). Rhythmic excitatory postsynaptic potentials (EPSPs) also persisted when action potential firing was suppressed in clock neurons by expression of the inward rectifying opened potassium channel Kir2.1 (fig. S2, E to G), which hyperpolarizes clock neurons to −90 mV. Thus, neither the molecular clockwork nor intra-clock neuronal circuits are needed for synchronized bursting activity in clock neurons, implying an origin outside the central master clock network.
To investigate the ion channels that mediate extra-clock rhythmic inputs, we inhibited action potential firing in ITP-LNd neurons using the sodium channel blocker QX-314 (32) in the patch pipette. Rhythmic excitatory postsynaptic currents (EPSCs) reversed at approximately −5 mV (Fig. 2C), indicating that extra-clock rhythmic inputs are mediated by nonselective cation channels. Furthermore, bursts were reversibly abolished by the nicotinic acetylcholine receptor antagonist mecamylamine (MCA) (Fig. 2D), consistent with prior reports (27, 28). These results support the hypothesis that rhythmic bursting in clock neurons is driven by extra-clock synaptic inputs from a pacemaking ultradian oscillator.
We observed considerable overlap between clock neurons that display bursting activity and those that exhibit light-induced excitatory responses (29). A common feature of light-responsive clock neurons is their dendritic arborization in the aMe, a hub that relays light inputs from retinal photoreceptors to clock neurons (29). Localized ablation of the aMe with a two-photon laser beam eliminated rhythmic burst firing in the clock neurons (Fig. 2E) but left responses to current injection unaffected (fig. S2H), suggesting that synaptic inputs from an extra-clock ultradian oscillator are transmitted via the aMe.
Because each brain has two symmetrical aMe hubs and clock neurons are synchronized across both brain hemispheres, there are two possible origins for an extra-clock ultradian oscillator: a single oscillator assembly that simultaneously transmits rhythmic bursts to both aMe hubs or two coupled assemblies, each located in one hemisphere and connected to their ipsilateral aMe. To differentiate between these possibilities, we cut brain preparations along the midline and measured bursting using dual patch-clamp recordings (Fig. 2F). Recordings from ipsilateral pairs of E cells revealed synchronized bursts in both hemispheres (Fig. 2G). However, rhythmic bursting was desynchronized between bilateral pairs of E cells (Fig. 2H). These results suggest that an extra-clock ultradian oscillator exists in each hemisphere and that these oscillators likely couple through commissures to synchronize burst firing of central master clock neurons.
Identification of bona fide extra-clock ultradian electrical oscillators
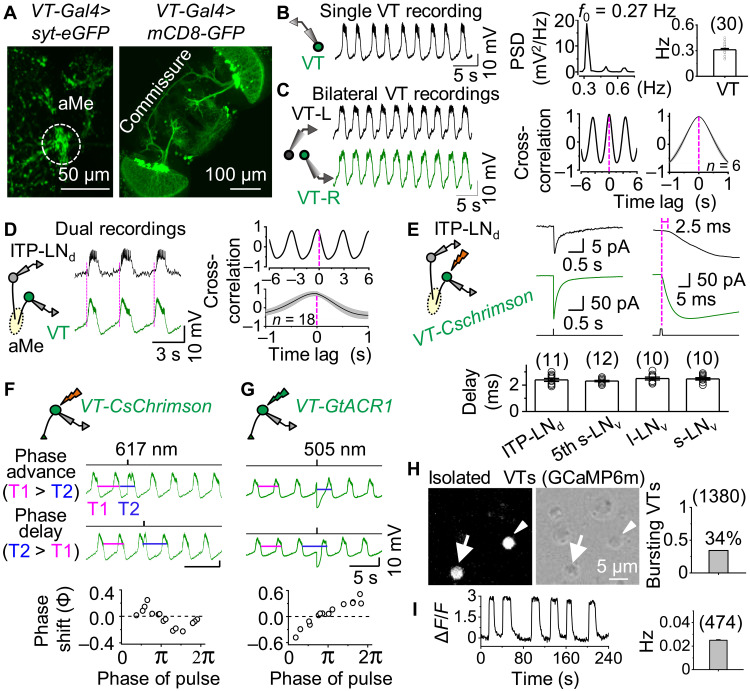
To determine the origin of the extra-clock burst inputs, we screened the Janelia GMR (33) and Vienna Tile (VT) Drosophila lines to search for neurons with axonal projections to the aMe and commissural connections. Of the screened lines, VT037867-Gal4 contained approximately a dozen pairs of neurons with both these features (Fig. 3A and fig. S3A). Most of these VT037867 neurons exhibited rhythmic bursts (Fig. 3B) with bilateral synchrony (Fig. 3C). In split brains, bilateral VT037867 pairs exhibited desynchronized burst firing, while ipsilateral pairs retained synchronous bursting activity (fig. S3, B and C).
Fig. 3. Identification of bona fide ultradian electrical oscillators.
(A) Expression pattern of VT037867-Gal4. Left, aMe projections revealed by syt.eGFP; right, commissure revealed by GFP. (B) Rhythmic bursts in VT037867 neurons. Representative recordings of VT037867 neurons (left), power spectral analysis (middle), and combined data (right). (C) Representative bilateral VT037867 pair recordings (left), cross-correlation analysis (middle), and combined data (right). (D) Schematic of dual recordings from ipsilateral VT037867 and ITP-LNd pair (left), representative recordings (middle), and cross-correlation analysis (right). The rising phase of each VT037867 burst is marked by the magenta dotted lines, appearing earlier than that of ITP-LNd bursts. (E) Schematic of optogenetic activation and dual recordings of ipsilateral VT037867 and ITP-LNd pair (top, left), representative recordings with time scales expanded (top, right), and combined data of synaptic delay (bottom). Optogenetic stimulation: 617 nm, 1 ms, and 600 μW. (F) Schematic of CsChrimson-induced depolarization of VT037867 neurons (top), phase advance and delay (middle), and phase response curve (PRC; bottom). Phase advance: T1 > T2; phase delay: T1 < T2. Optogenetic stimulation: 617 nm, 150 ms, and 30 μW. Similar results obtained in three other VT037867 neurons (fig. S3G). (G) Schematic of GtACR1-induced hyperpolarization in VT037867 neurons (top), phase advance and delay (middle), and PRC (bottom). Optogenetic stimulation: 505 nm, 300 ms, and 40 μW. Similar results obtained in five other VT037867 neurons (fig. S3H). (H) Fluorescence image of isolated GCaMP6m-expressing VT037867 neurons (left), DIC image of enzymatically dissociated VT037867 neurons (middle), and combined data (right). Arrow and arrowhead mark two GCaMP6m-expressing VT037867 neurons; arrowhead indicates VT037867 neuron exhibiting calcium oscillations (474 of 1380 exhibited oscillations in eight independent replicates). (I) Representative calcium oscillations in isolated VT037867 neurons (left) and combined frequencies of oscillating VT037867 neurons (right). ΔF/F, change of calcium fluorescent intensity. Combined data are presented as means ± SEM.
Dual patch-clamp recordings also revealed burst synchrony between the ipsilateral VT037867 neurons and ITP-LNd clock neuron but with a delay in ITP-LNd rhythmic depolarizations (Fig. 3D). This implies that either the ITP-LNd is downstream of VT037867 neurons or both neuronal types receive burst inputs from a common source with different delays. As chemogenetic activation of P2X2-expressing VT037867 neurons excited the ITP-LNd (fig. S3D), the former possibility seems most likely. Dual recordings revealed that optogenetic activation of VT037867 neurons expressing the light-sensitive cation channel CsChrimson resulted in excitation of the ipsilateral ITP-LNd with a delay of less than 3 ms (Fig. 3E), indicating a monosynaptic connection between VT037867 neurons and the ITP-LNd (34). A similar response delay was also observed between VT037867 neurons and other clock neurons, including l-LNvs, s-LNvs, and 5th s-LNv (Fig. 3E). Green fluorescent protein (GFP) reconstitution across synaptic partners (GRASP) (35) confirmed the monosynaptic contacts between the VT037867 neurons and clock neurons (fig. S3E). Moreover, the hemibrain connectome (36) showed that the VT037867-labeled aMe neurons form direct synaptic connections with clock neurons (fig. S3F). Thus, VT037867 neurons are positioned both anatomically and functionally to transmit extra-clock burst inputs to clock neurons.
A hallmark of bona fide neuronal oscillators is that perturbation of their membrane potentials causes phase shifts in their oscillatory activity (37). Consistent with this idea, brief depolarization of CsChrimson-expressing VT037867 neurons resulted in a phase shift to their rhythmic bursts, with an advance or a delay depending on the phase in which depolarization was applied (Fig. 3F and fig. S3G). Phase shifts were also observed following optogenetic hyperpolarization of VT037867 neurons by the light-sensitive chloride channel GtACR1, albeit with a distinct phase response curve (PRC; Fig. 3G and fig. S3H). These data suggest that VT037867 neurons are bona fide oscillators. We named these VT037867 neurons extra-clock electrical oscillators (xCEOs).
To test whether xCEOs can oscillate cell-autonomously, we enzymatically dissociated GCaMP6m-expressing VT037867 neurons into single cells (Fig. 3H). In total, 474 of 1380 VT037867-labeled neurons (34%) displayed spontaneous rhythmic oscillations of intracellular calcium in vitro (Fig. 3I and movie S1). Calcium oscillations occurred at lower frequencies and with less regularity than VT037867 bursting activity in intact brains (Fig. 3I), likely due to a lack of neuronal connections or enzymatic/mechanical damage. In contrast, spontaneous calcium oscillations were not observed in enzymatically dissociated clock neurons (fig. S3I), supporting the concept that bursting in clock neurons in situ is driven by extra-clock neuronal inputs.
We next sought to characterize the subpopulation of VT037867 neurons that spontaneously oscillate. Genetically crossing the VT037867-Gal4 line with enhancer-trap flippase lines of different neurotransmitters (38) revealed two subpopulations of VT037867 neurons, one glutamatergic and the other acetylcholinergic (fig. S3J). We expressed GCaMP6m in both subpopulations using their corresponding driver lines and found that 343 of 548 dissociated acetylcholinergic VT037867-labeled neurons (63%) displayed rhythmic calcium oscillations (fig. S3K). In contrast, none of the dissociated glutamatergic VT037867-labeled neurons showed rhythmic oscillations (fig. S3L). These results demonstrate that acetylcholinergic VT037867 neurons contain a population of bona fide neuronal oscillators that project axons to the aMe (fig. S3J), a structure required for relaying extra-clock burst inputs to clock neurons (Fig. 2E).
xCEOs drive bursting in clock neurons
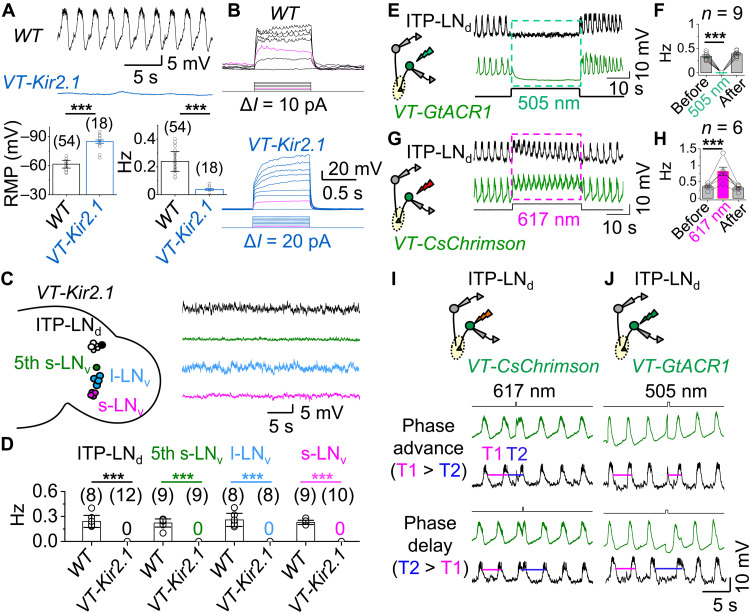
To investigate whether the xCEOs drive the bursting activity observed in clock neurons of intact brains, we expressed Kir2.1 in the xCEOs to hyperpolarize their resting membrane potentials (RMPs) to −90 mV (Fig. 4A) and block action potential firing (Fig. 4B). As a result, the xCEOs lost their rhythmic burst firing (Fig. 4A) but retained some small-amplitude, low-frequency rhythmic depolarizations (fig. S4A). In addition, clock neurons no longer exhibited rhythmic bursts (Fig. 4, C and D) but were able to fire action potentials in response to current injections (fig. S4B), indicating that burst firing in clock neurons originates from the xCEOs.
Fig. 4. xCEOs drive bursting in clock neurons.
(A) Representative recordings of xCEOs in WT (top) and VT037867-Kir2.1 flies (middle), and combined RMPs and burst frequencies (bottom). ***P < 0.001 by unpaired two-tailed Student’s t test. (B) Representative responses of xCEOs to current injection in WT (top) and VT037867-Kir2.1 flies (bottom). Magenta traces indicate injection of 20 pA, triggering spike firing in WT but not VT037867-Kir2.1 flies. (C) Schematic of clock neurons in VT037867-Kir2.1 flies (left) and representative recordings at approximately −55 mV (right). (D) Combined burst frequencies in WT and VT037867-Kir2.1 flies. ***P < 0.001 by unpaired two-tailed Student’s t test. (E) Schematic of xCEO inhibition by GtACR1 and dual recordings (left) and representative recordings of xCEO and ITP-LNd ipsilateral pair (right). Optogenetic stimulation (505 nm, 30 s, and 40 μW) indicated by the dashed green box. (F) Combined data from (E). ***P < 0.001 by paired t test. (G) Schematic of CsChrimson activation of xCEOs and dual recordings (left) and representative recordings of xCEOs and ITP-LNd ipsilateral pair (right). Optogenetic stimulation: 617 nm, 30 s, and 30 μW. (H) Combined data from (G). ***P < 0.001 by paired t test. (I) Schematic of xCEO depolarization by CsChrimson and dual recordings from ITP-LNd and xCEO (top), phase advance (middle), and phase delay (bottom) in ITP-LNd and xCEO. Optogenetic stimulation: 617 nm, 150 ms, and 30 μW. Similar results obtained in four independent replicates. (J) Schematic of xCEOs hyperpolarization by GtACR1 and dual recordings from ITP-LNd and xCEO (top), phase advance (middle), and phase delay (bottom) in ITP-LNd and xCEO. Optogenetic stimulation: 505 nm, 300 ms, and 40 μW. Similar results obtained in six independent replicates. RMP, resting membrane potential. Combined data are presented as means ± SEM.
Acute optogenetic manipulation of the xCEOs provided further evidence that these oscillators drive rhythmic bursting in clock neurons. Following the expression of GtACR1 in the xCEOs, 30-s light pulses induced hyperpolarization in xCEOs and reversibly abolished rhythmic bursts in both xCEOs and clock neurons (Fig. 4, E and F). On the other hand, 30-s light pulse activation of CsChrimson-expressing xCEOs induced depolarization and increased burst frequency in both xCEOs and clock neurons (Fig. 4, G and H). Moreover, ITP-LNd faithfully followed the phase advances or delays induced by brief optogenetic depolarization or hyperpolarization of the xCEOs (Fig. 4, I and J). In contrast, optogenetically induced depolarization and hyperpolarization of clock neurons had no effect on burst frequencies (fig. S4, C and D), consistent with our earlier result that membrane potential changes by current injection did not change the burst frequencies (fig. S1B). These results thus confirm that the xCEOs are responsible for the extra-clock bursting inputs to clock neurons.
xCEOs promote daily electrical rhythms in clock neurons
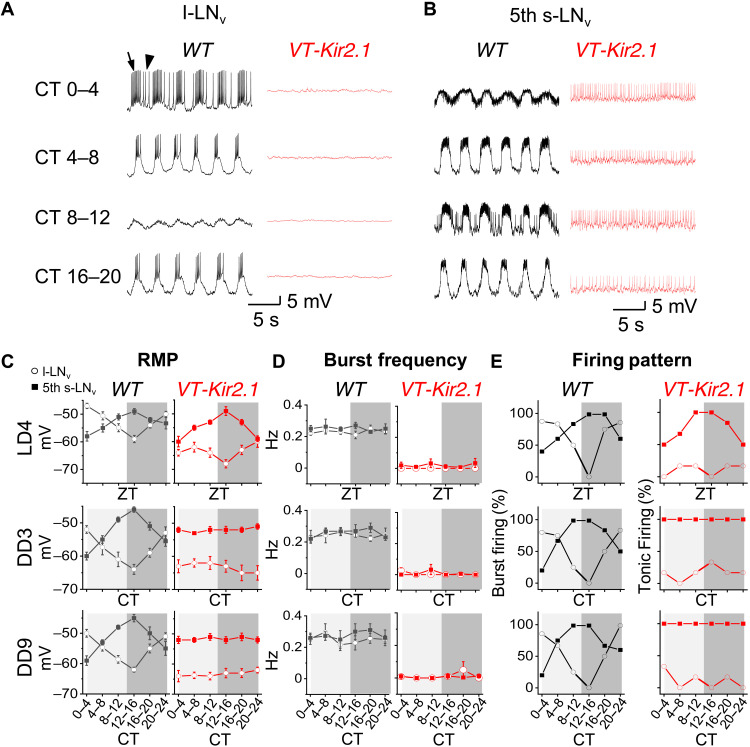
In addition to the 0.3-Hz rhythms described above, clock neurons exhibit daily rhythms in RMP and firing activity (39–44), which we observed in more than 1500 patch-clamp recordings. During DD, robust daily rhythms in RMP were observed in l-LNvs (arousal neurons), s-LNvs (M cells), and DN1ps: hyperpolarization from subjective dawn to dusk and depolarization from subjective dusk to dawn, whereas E cells (5th s-LNv and ITP-LNd) exhibited an RMP rhythm in antiphase to that of M cells (Fig. 5, A to C, and fig. S5, A to F). These opposing RMP rhythms were also evident during light/dark (LD) cycles (Fig. 5, C and fig. S5, A to F), consistent with the findings revealed by in vivo calcium imaging (45). Our recordings further revealed opposing daily rhythms in firing activity between M cells/l-LNvs and E cells, including both firing activity driven by the xCEOs and interburst tonic firing when RMPs were above threshold (Fig. 5, D and E, and fig. S5, A to F).
Fig. 5. xCEOs promote daily electrical rhythms in clock neurons.
(A) Representative recordings (zero holding current, or I = 0 pA) of l-LNv in WT (black) and VT037867-Kir2.1 (red) flies at different CT times during DD3. Arrow indicates burst firing; arrowhead indicates interburst tonic firing. (B) Representative recordings (zero holding current, or I = 0 pA) of 5th s-LNv in WT (black) and VT037867-Kir2.1 (red) flies at different CT times during DD3. (C) Combined daily RMP data. (D) Combined burst frequency data. (E) Combined firing pattern data. ZT, Zeitgeber time; CT, circadian time. Combined data are presented as means ± SEM. Each data point represents the average from 6 to 10 recorded cells.
The rhythmicity with which bursts occurred remained at the same frequency throughout the day (Fig. 5D and fig. S5, B and D), consistent with circadian-independent xCEO inputs (fig. S5, G to J). Nevertheless, we wondered whether inhibition of xCEO inputs by expressing Kir2.1 in the xCEOs could affect RMP and firing activity in clock neurons. Daily rhythms in RMP and firing activity were absent in clock neurons downstream of xCEOs in DD: M cells and l-LNvs remained silent at a steady hyperpolarized RMP, while E cells exhibited tonic firing superimposed on a steady depolarized RMP (Fig. 5 and fig. S5, A to D). In contrast, DN1ps, which lack xCEO inputs in WT flies, maintained daily rhythms of both RMP and firing activity (fig. S5, E and F). External light cycles during LD restored daily RMP rhythms in E cells of VT037867-Kir2.1 flies (Fig. 5C and fig. S5B) but not in M cells or l-LNvs (Fig. 5C and fig. S5D), possibly reflecting differential regulation of RMP by light.
In per0 flies, which lack functional molecular clocks but retain inputs from the xCEOs, the l-LNvs and M cells exhibited xCEO-driven rhythmic EPSPs superposed on a steady hyperpolarized RMP and E cells showed xCEO-driven burst firing superposed on a steady depolarized RMP (fig. S5, K and L). Collectively, these results indicate that xCEOs, together with the molecular clockwork, drive daily rhythms in RMP and enrich daily firing patterns in clock neurons.
xCEOs promote daily rhythms of PDF cycling and sustain free-running locomotor rhythms
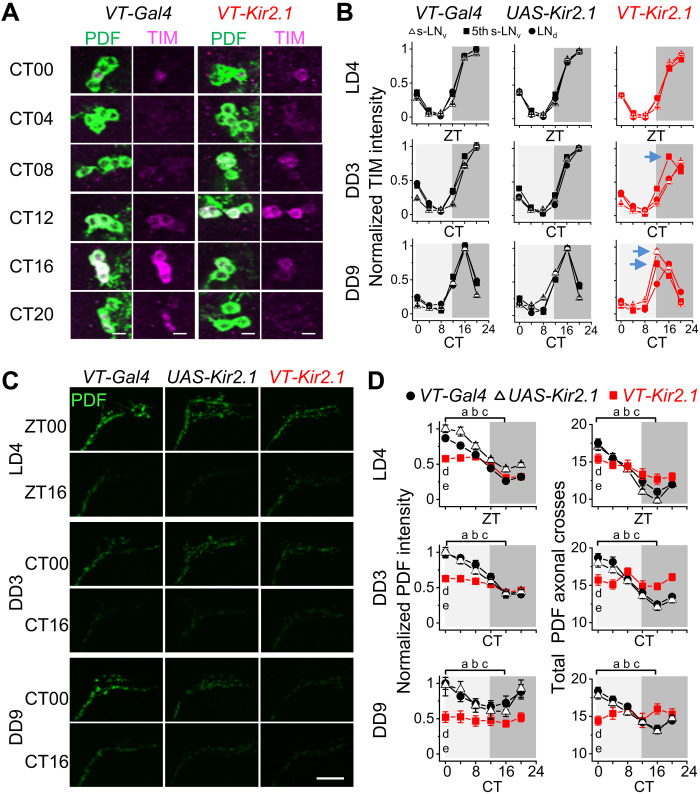
To investigate the physiological relevance of xCEO inputs on circadian timekeeping, we selectively suppressed these inputs using VT037867-Kir2.1 flies and examined the effect on the molecular clockwork and the daily PDF rhythms. By immunostaining the TIMELESS (TIM) protein, we observed slight dampened TIM cycling and slight phase desynchrony in M cells and E cells during DD but normal cycles during LD (Fig. 6, A and B, and fig. S6, A to C). In addition, daily cycling of TIM distribution in the nucleus and cytoplasm (fig. S6, D and E) was also slightly disrupted in the DD conditions. Furthermore, daily rhythms of PDF intensity and dorsal branches of s-LNvs were disrupted during DD, with significantly reduced intensity and axonal branches (Fig. 6, C and D).
Fig. 6. xCEOs promote daily rhythms of PDF cycling.
(A) TIM and PDF double immunostaining in s-LNvs during DD9. Green, anti-PDF; magenta, anti-TIM. Scale bars, 5 μm. (B) Normalized TIM intensity of s-LNv, 5th s-LNv, and LNd in VT037867-Gal4, UAS-Kir2.1, and VT037867-Kir2.1 flies. Slight phase desynchrony of TIM cycling among clock neuron subtypes in VT037867-Kir2.1 flies is indicated by arrows. (C) PDF immunostaining in dorsal branches of s-LNvs in VT037867-Gal4, UAS-Kir2.1, and VT037867-Kir2.1 flies. Scale bar, 20 μm. (D) Pooled PDF intensity (left) and axonal branches of s-LNvs (right) in VT037867-Gal4, UAS-Kir2.1, and VT037867-Kir2.1 flies. When ZT0 (or CT0) values are significantly larger than those at ZT16 (or CT16), PDF is considered to show daily cycling. a, b, and c represent the comparisons in VT037867-Gal4, UAS-Kir2.1, and VT037867-Kir2.1 flies, respectively. d represents comparisons of ZT0 (or CT0) values between VT037867-Kir2.1 and VT037867-Gal4 and e between VT037867-Kir2.1 and UAS-Kir2.1 flies. In LD4 and DD3, for intensity comparison, a, b, and c indicate P < 0.001 and d and e indicate P < 0.001; for axonal cross, a and b indicate P < 0.001, c indicates P < 0.01 in LD4 and n.s. in DD3, d and e indicate P < 0.05 (LD4) and P < 0.01 (DD3). In DD9, for intensity, a, b, and c indicate P < 0.05, P < 0.01, n.s., respectively, d indicates P < 0.001, e indicates P < 0.05; for axonal cross, a and b indicate P < 0.001, c indicates n.s, and d and e indicate P < 0.001. Two-tailed, unpaired Student’s t test. Combined data are presented as means ± SEM. Immunostaining was independently repeated four times, with at least a total of 25 brains examined at each time point.
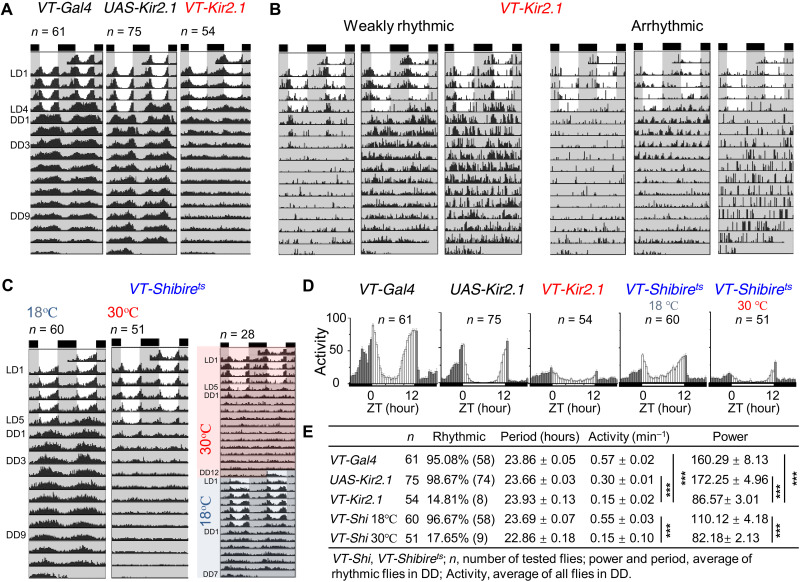
Having observed disrupted electrical circadian rhythms and PDF cycling during inhibition of xCEO inputs, we investigated the effects of VT037867-Kir2.1 flies on circadian behavior. Most xCEO-silenced flies showed weak rhythmic circadian behavior under LD, although some were arrhythmic (Fig. 7, A to D), indicating that xCEOs may contribute to circadian photoentrainment. However, after flies were released into DD, most became arrhythmic. Similar loss of rhythmicity was observed when xCEO inputs to clock neurons were conditionally suppressed in adult flies by expression of the temperature-sensitive shibirets allele (Fig. 7, C to E, and fig. S7), which inhibits endocytosis of synaptic vesicles at a restrictive temperature of 30°C. Locomotor rhythmicity recovered after restoration of xCEO inputs by switching to a permissive temperature of 18°C (Fig. 7C and fig. S7). These data reveal that xCEO inputs play a critical role in synchronizing free-running molecular clocks and promoting circadian locomotor rhythmicity in the absence of sensory inputs.
Fig. 7. xCEOs sustain free-running behavioral locomotor rhythms.
(A) Actograms of VT037867-Gal4 (left), UAS-Kir2.1 (middle), and VT037867-Kir2.1 flies (right). (B) Representative weak rhythmicity (left) and arrhythmicity (right) in VT037867-Kir2.1 flies. (C) VT037867-Shibirets flies maintained free-running rhythms at 18°C (left) but lost free-running rhythms at 30°C (middle) and the lost free-running rhythms at 30°C recovered at 18°C (right). (D) Activity histograms of VT037867-Gal4, UAS-Kir2.1, VT037867-Kir2.1, and VT037867-Shibirets flies in LD. (E) DD rhythmicity statistics (period, activity, and power) of VT037867-Gal4, UAS-Kir2.1, VT037867-Kir2.1, and VT037867-Shibirets flies. For activity, ***P(VT037867-Gal4/VT037867-Kir2.1) < 0.001, ***P(UAS-Kir2.1/VT037867-Kir2.1) < 0.001, and ***P(18°C/30°C) < 0.001 for VT037867-Shibirets flies; for power, ***P(VT037867-Gal4/VT037867-Kir2.1) < 0.001, ***P(UAS-Kir2.1/VT037867-Kir2.1) < 0.001, and ***P(18°C/30°C) < 0.001 for VT037867-Shibirets flies. These data are analyzed by Kruskal-Wallis ANOVA with Dunn’s test. Data are expressed as means ± SEM. Actograms and histograms are averaged from four independent replicates.
DISCUSSION
We have found a novel set of bona fide ultradian brain oscillators that support free-running circadian timekeeping of locomotor rhythms in Drosophila, despite their location outside the master clock network. The xCEOs generate cell-autonomous rhythmic burst firing independent of the molecular clockwork and drive widespread burst synchrony among clock neurons via parallel, monosynaptic connections in the aMe. Our data reveal that intraclock connectivity provides the neuronal infrastructure that allows communication and synchronization within the master clock but that xCEOs provide the endogenous extra-clock driving force. Thus, in contrast to current thinking, our work demonstrates that the master clock is not self-sufficient but requires endogenous burst inputs from xCEOs to sustain timekeeping of behavior rhythms in constant conditions.
Discovery of xCEOs
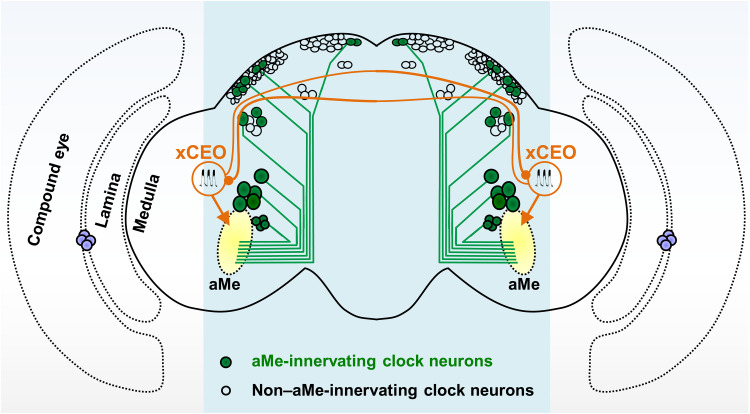
Our discovery of the xCEOs was facilitated by systematic patch-clamp recordings throughout the central clock in ex vivo Drosophila brain preparations. These recordings revealed burst firing in most clock neuron subtypes, including s-LNv, l-LNv, 5th s-LNv, ITP-LNd, sNPF-LNd, CRY-negative LNd, DN1a, DN3a, and DN3p. Bursting in these clock neurons was synchronous within and between ipsilateral and contralateral neurons, although synchrony between the latter was lost in split brain preparations. Local ablation of the aMe abolished burst firing in clock neurons, consistent with our previous finding that most of these neurons have dendritic arborizations to the aMe hub (29). Given these characteristic features (aMe projection and interhemispheric connection), we identified a Drosophila line, VT037867-Gal4, that labeled the xCEOs (Fig. 8).
Fig. 8. Widespread ultradian oscillation in clock neurons driven by xCEOs.
Among the 150 central clock neurons, many subtypes arborize their processes in the aMe and others do not. In each brain hemisphere, there is one set of xCEO neurons. The two sets of xCEO neurons mutually connect with each other via their dorsal commissure, producing synchronized ultradian bursting across the brain. By making monosynaptic connections with circadian clock neurons in the aMe, xCEOs pace synchronized ultradian bursting in most clock neuron subtypes to support free-running circadian locomotor rhythms.
Independent evidence that the xCEOs are bona fide ultradian electrical oscillators arose from five lines of enquiry. First, the xCEOs exhibit rhythmic burst firing that leads those of clock neurons. Second, silencing of the xCEOs abolished rhythmic burst firing in clock neurons. Third, steady depolarization or hyperpolarization of the xCEOs accelerated and slowed rhythmic bursts, respectively. Fourth, brief depolarization and hyperpolarization of the xCEOs induced phase shifts in the bursting behaviors of both the xCEOs and clock neurons. Last, single dissociated xCEO neurons exhibited spontaneous rhythmic calcium oscillations.
The frequency of rhythmic bursting in the xCEOs remained constant throughout the day, likely because xCEOs neither express clock genes such as TIM nor receive synaptic inputs from clock neurons. Thus, rhythmic inputs to clock neurons from xCEOs are endogenous and perpetual, regardless of circadian times.
xCEOs promote PDF cycling in M cells and electrical rhythms in clock neurons
Our patch-clamp recordings revealed that M and E cells exhibited opposing rhythms of RMP during both LD and DD. These rhythms were completely abolished during DD in VT037867-Kir2.1 flies. The similarity between RMP changes in VT037867-Kir2.1 and per0 flies suggested that the xCEOs may act on the molecular clockwork to regulate RMP. Consistent with this idea, we found that free-running molecular clock phases were desynchronized among clock neuron subtypes in VT037867-Kir2.1 flies. It is conceivable that the xCEOs regulate molecular clocks by modifying the electrical activity of clock neurons (23, 46). Alternatively, the xCEOs may modulate RMP rhythms in clock neurons via synaptic plasticity or homeostasis (47, 48). The second possibility is supported by our finding that RMP rhythms in M cells were abolished and PDF cycling in their terminals was also impaired, even though their molecular clock rhythms were maintained, in VT037867-Kir2.1 flies during LD conditions. A distinct mechanism of RMP regulation may also exist in DN1ps, which do not receive xCEO inputs, given that RMP rhythms persisted in these cells in VT037867-Kir2.1 flies (fig. S5, E and F).
Because the aMe hub receives inputs from the visual system and xCEOs, it is possible that both systems interact with the same set of clock neurons. Our finding that all light-excitable clock neurons exhibit rhythmic electrical bursting supports this hypothesis. aMe-mediated visual inputs have been implicated in resetting clock neurons so that endogenous rhythms are aligned to the same external time (29, 49–51). By regulating the electrical activity of the same set of clock neurons, the xCEOs likely synchronize the central clock in the absence of external time inputs, such as during DD. Therefore, the two aMe-mediated inputs act together to synchronize the central clock, regardless of light conditions.
xCEOs facilitate communication within the clock
The widespread xCEO-driven bursting in clock neurons has implications for communication within the master clock. Clock neurons in Drosophila express a variety of classical neurotransmitters and neuropeptides, including PDF (31, 52, 53). Paracrine secretion of neuropeptides relies on calcium influx, typically driven by bursts of action potentials (53, 54). Our results revealed that xCEO activity is required for normal rhythms of PDF intensity in dorsal branches of s-LNvs. In addition, other studies have shown that subtle changes in the burst frequency of s-LNvs affects PDF release (26) and phase-locked bursting enables more reliable synaptic transmission via classical neurotransmitters (55). The perpetual burst inputs from the xCEOs thus maintain efficient communication within the central clock, reinforcing its coherent timekeeping. Furthermore, bursting in clock neurons facilitates communication between clock neurons and their downstream circuits.
RMP gates daily burst firing in clock neurons
The discovery of the xCEOs raises the question of how the central clock integrates these oscillatory inputs with intrinsic RMP rhythms. When RMP was above threshold, clock neurons exhibited rhythmic burst firing accompanied by tonic firing between bursts. When RMP was below threshold, clock neurons generated rhythmic burst firing only when the xCEOs drove depolarization above threshold; otherwise, they generated only rhythmic subthreshold EPSPs. Thus, although xCEO inputs are constant, their postsynaptic effects on clock neurons is gated by the RMP of clock neurons, producing distinct firing patterns in different cells at different times. Our experimental results demonstrate that relying on the intrinsic RMP activities of clock neurons without the xCEOs would produce a limited binary output (either firing or no firing) and circadian locomotor rhythms would be lost in DD.
Conserved extra-clock ultradian oscillatory inputs to the central circadian clock
In cockroach, some neurons innervating the aMe also exhibit ultradian calcium oscillations when enzymatically isolated (56). In addition, ultradian calcium oscillations driven by extra-SCN inputs were also reported in mouse SCN clock neurons (57). Similarly, fast calcium oscillations were also recently reported in in vivo Drosophila clock neurons (58), likely driven by ultradian electrical bursting reported here. Our discovery that ultradian oscillatory xCEO inputs support circadian timekeeping reveals a previously unknown mechanism by which endogenous extra-clock brain inputs support free-running circadian timekeeping in Drosophila and, likely, other animals including mammals. Although not required for cell-autonomous molecular clocks, these rhythmic bursts promote the phase synchrony of molecular clocks and enrich firing patterns in clock outputs. Thus, both cell-autonomous TTFLs and endogenous extra-clock ultradian oscillatory inputs work together to produce proper circadian timekeeping (59).
Limitations of this study
This study raises several questions, some of which we highlight here. First, although we have demonstrated that xCEO inputs pace widespread synchronous bursts in clock neurons and support free-running circadian behavior rhythms, the xCEOs may also drive postsynaptic metabolic changes via G protein–coupled receptors in clock neurons. Second, our work reveals that the intact molecular clocks are not sufficient to maintain normal locomotor rhythms but require additional endogenous brain inputs from the xCEOs. How the xCEOs and molecular clocks work together to generate proper timekeeping of free-running circadian rhythms needs more future studies. Third, more in vivo electrophysiological studies are needed to further characterize the physiology of the novel xCEOs. Fourth, VT037867-Gal4 also label some non–aMe-innervating neurons in the brain and ventral nerve cord. Further genetic refinement in labeling VT037867 subgroups helps rule out contribution from those non-xCEO VT037867 neurons to perturbation-induced changes in circadian timekeeping. Last, the mechanisms underlying the rhythmogenesis of the ultradian xCEOs remain to be elucidated. Nonetheless, our genetic identification of the xCEOs will promote further experimental investigation in these various directions.
MATERIALS AND METHODS
Drosophila
Flies were reared on standard cornmeal agar medium under 12-hour/12-hour LD cycles (60% humidity and 25°C). For electrophysiological experiments, flies (one day after eclosion) were subjected to four LD cycles and then released into DD. Most patch-clamp recordings were performed at CT0-8 on the third day of DD (DD3), unless otherwise specified. Flies were backcrossed for seven generations to w1118 flies (BL5905).
DvPdf-Gal4, Clk4.1M-Gal4, and Mai179-Gal4 were provided by P. Emery; per0, perS, perL, and Clk4.1M-LexA were from A. Sehgal; UAS-mCD8-GFP (2nd), UAS-mCD8-GFP (3rd), and UAS-TNT (2nd) were from C. Potter; Clk856-Gal4, pdf01, w1118, and tim01 were from T. Yoshii; Pdf-LexA, Cha-Flp, vGlut-Flp, LexAop-mCD4::spGFP11, UAS-mCD4::spGFP1–10, UAS-Kir2.1-GFP (3rd), vGlut-p65.AD, LexAop2-GFP (2nd), UAS-Shibirets (2nd), and UAS-Shibirets (3rd) were from Y. Rao; UAS-FRT-Stop-FRT-GFP, UAS-FRT-Stop-FRT-GCaMP6m, and UAS-Kir2.1 (2nd) were from C. Zhou; R54D11-Gal4 (BDSC_41279), UAS-syt.eGFP (BDSC_6926), UAS-GCaMP6m (BDSC_42748), UAS-GCaMP6m (BDSC_4750), LexAop2-GCaMP6m (BDSC_44276), UAS-P2X2 (BDSC_91222), UAS-CsChrimson (BDSC_55135), VT037867-Gal4.DBD (BDSC_74594), R54D11-Gal4.DBD (BDSC_69620), LexAop2-GCaMP6f (BDSC_44277), and LexAop2-CsChrimson (BDSC_55138) were from the Bloomington Stock Center; VT037867-Gal4 was from the Vienna Drosophila Resource Center (VDRC_203797); DvPdf-LexA (2nd) and DvPdf-LexA (3rd) were from our own stock (29); and VT037867-LexA (2nd), LexAop2-GtACR1 (2nd), UAS-GtACR1 (2nd), and R18F07-p65.AD (2nd) were generated by us. Genotypes used in the experiments are listed in table S1.
Ex vivo patch-clamp recordings
For ex vivo recordings, both male and female flies, unless otherwise specified, were selected at different times (LD4, DD3, or DD9) and dissected as described previously (29). Briefly, under dim red light, the flies were anesthetized in a glass tube with ice water. The fly head was separated from the body and then transferred to a recording chamber filled with pre-oxygenized Drosophila dissection saline composed of the following: 124 mM NaCl, 3 mM KCl, 4 mM MgCl2, 1.5 mM CaCl2, 10 mM Hepes (pH 7.3), 1 mM NaH2PO4, 5 mM N-tri-(hydroxymethyl)-methyl-2-aminoethane-sulfonic acid (TES), 20 mM d-glucose, 17 mM sucrose, and 5 mM trehalose.
Under a dissection microscope (M125, Leica), the external sensory organs, including compound eyes, ocelli, proboscis, antenna, and maxillary palps, were removed. Next, the cuticle covering the brain was detached. The isolated brain was then stabilized in the recording chamber with blue histoacryl (B. Braun, Germany). To record the VT037867, LNv, LNd, DN1a, DN3a, and DN3 clock neurons, the brain was positioned anterior side up. To record DN1p, DN2, DN3p, and LPNs, the brain was positioned posterior side up.
The stabilized brain preparations were perfused with Drosophila perfusion saline bubbled with 95% O2/5% CO2, at a flow rate of 1 ml/min. The perfusion saline was composed of the following: 103 mM NaCl, 3 mM KCl, 4 mM MgCl2, 1.5 mM CaCl2, 26 mM NaHCO3 (pH 7.3), 1 mM NaH2PO4, 5 mM TES, 20 mM d-glucose, 17 mM sucrose, and 5 mM trehalose. The final osmolarity was ~280 mosM. Before recordings, the brain preparations were kept in complete darkness for ~5 min.
Target neurons were identified by GFP expression and visualized using an upright microscope (Pro 6000, Scientifica) with a water-immersion objective (Olympus, 60×). The image was displayed on a monitor (PVM-122CE, Sony) through infrared differential interference contrast (IR-DIC; DAGE-1000, MTI). To expose the cell body for recording pipette access, the brain sheath above the target neuron was physically removed with sharp forceps. A 3-μm suction pipette filled with protease XIV (0.67 mg/ml) was used to remove tissue debris and other neurons that covered the target cell. During exposure of the cell body, a minimal enzymatic and mechanical treatment was applied to reduce the circuitry damage. Cell-body exposure and recordings were performed under IR-DIC.
Patch-clamp recording electrodes (~10 megohms) were pulled from borosilicate glass (PG10165-4, WPI) with a puller (P-1000, Sutter Instrument). For whole-cell recordings, the recording pipette was filled with an internal solution consisting of the following: 140 mM K-gluconate, 6 mM NaCl, 2 mM MgCl2, 0.1 mM CaCl2, 4 mM Mg–adenosine triphosphate (ATP), 0.5 mM guanosine triphosphate-tris, 1 mM EGTA-Na4, and 10 mM Hepes (pH 7.3), with an osmolarity of 275 mosM. For perforated patch-clamp recordings, the pipette was filled with a solution consisting of the following: 140 mM K-gluconate, 6 mM NaCl, 2 mM MgCl2, 0.1 mM CaCl2, 1 mM EGTA-Na4, 10 mM Hepes (pH 7.3), and amphotericin B (100 μg/ml). All chemicals were from Sigma-Aldrich.
After the establishment of patch-clamp recordings, both access resistance and membrane capacitance were compensated. Signals were amplified with two MultiClamp 700B amplifiers (Molecular Devices), digitized via Digidata 1440A (Molecular Devices), recorded via Clampex 10.6 (Molecular Devices), and filtered and sampled at 2 and 5 kHz, respectively. Measured voltages were corrected for a liquid junction potential of 13 mV.
In vivo patch-clamp recordings
Live-fly preparations were prepared as described previously (60). Briefly, an ice-anesthetized fly was inserted into the recording platform hole, i.e., 10-μm-thick stainless steel plate with a small rectangular hole (800 μm by 1200 μm). With the dorsal side of its head facing up, the fly was stabilized in the recording platform with low-melting wax. The gap between the fly head and hole edge was sealed with wax to prevent leakage.
The recording platform was then filled with pre-oxygenated Drosophila dissection saline. Under a dissection microscope (M125, Leica), the head cuticle, fat, and air sacs/trachea were removed to expose the target neurons. Muscle 16 was removed to prevent brain movement due to its contraction.
Next, the live fly preparation was transferred to the patch-clamp recording stage and perfused with regular Drosophila perfusion saline used in ex vivo recordings. The target neuron was identified by its GFP fluorescence and anatomical position. Cell-body exposure and recordings were performed as per ex vivo recordings.
Dual and quadruple patch-clamp recordings
Dual and quadruple patch-clamp recordings were performed similarly as single patch-clamp recordings. Briefly, the target neurons were identified by GFP expression, and their cell bodies were exposed with minimal enzymatic and mechanical treatment of the covering tissues. Next, two or four patch electrodes were sequentially positioned ~5 μm above the individual target neurons. Last, the target neurons were sequentially patched one by one. The success rate of the dual recordings was much lower than that of the single recordings. The success rate of the quadruple recordings was even lower (7 of 42 preparations), mainly due to the loss of some recordings when patching the third or fourth neuron.
RMP measurements
RMP was measured immediately after the establishment of whole-cell patch-clamp recordings under current-clamp (I = 0 pA) configuration. For nonbursting neurons, RMP was directly acquired from the steady membrane potential. For rhythmic bursting neurons, the trough voltage was taken as the RMP. This method was validated by experimental observations showing that the trough voltage was unchanged in clock neurons when their rhythmic bursts were eliminated by acute VT037867 silencing with GtACR1 (Fig. 4E).
Drug delivery by fast-solution changes
As described in our previous work (61), a three-barrel tube (SF77B, Warner Instruments) was positioned ~200 μm from the recorded neurons, controlled by a stepper (SF77B, Warner Instruments). The middle barrel was connected to the Drosophila perfusion saline, and each side barrel was connected to an outport of an eight-to-one manifold connected to different solutions. Drug application was achieved by switching the lamina flow between the middle and side barrels, which could be achieved within milliseconds (61).
For direct intracellular delivery of QX-314 (TOCRIS), the drug was dissolved in whole-cell recording internal solution to a final concentration of 25 mM. After obtaining whole-cell recordings, QX-314 was diffused from the recording pipette into the recorded neuron. Diffusion occurred within 3 to 5 min, as judged by complete blockade of action potential firing.
Brain splitting
After removal of the sensory organs and head cuticle, the isolated brains were cut with a razor blade along the midline. The two separate brain hemispheres were then stabilized in the recording chamber with blue histoacryl (B. Braun, Germany). Dual patch-clamp recordings were performed on either ipsilateral or bilateral pairs of neurons.
Laser ablation of aMe
As described in our previous work (29), the aMe was ablated with a laser beam. Briefly, the aMe was identified by GFP expression with R54D11-Gal4. A Maitai DeepSee Ti:Sapphire ultrafast laser (Spectra-Physics) tuned to 800 nm was used to illuminate the region of interest (ROI) in the aMe for 3 to 5 s with a dwell time of 4.8 μs/pixel. Laser size, duration, and power (50 to 70 mW) were optimized for different preparations. Successful ablation was a visible cavitation bubble and physical destruction of the aMe.
Chemogenetic manipulation
The ATP-gated ion channel P2X2, UAS-P2X2, was expressed in target neurons by specific Gal4 drivers. ATP was delivered to the target neurons through the fast solution-change system with SF77B. Concentrations and duration of ATP application were optimized for different preparations.
Optogenetic manipulation
CsChrimson or GtACR1 was expressed in the target neurons with specific Gal4 or LexA drivers. The flies were fed with fly food containing 100 μM all trans-retinal (ATR), which was diluted from 40 mM ATR stock in alcohol. After 5 days of ATR food, the flies were dissected for recordings. High-power light-emitting diodes (LED4D206 for both CsChrimson and GtACR1 excitation) driven by DC4100 (Thorlabs) were used to deliver light stimulation through the epi-fluorescence port of an upright microscope (Pro 6000, Scientifica).
Phase response curve
Single patch-clamp VT037867 recordings were performed on flies expressing either CsChrimson or GtACR1 in VT037867 neurons. To generate a full PRC, membrane-potential perturbation is applied at different phases of ongoing VT037867 bursting. As VT037867 bursting is internally generated, it is difficult to predetermine the VT037867 phase at which membrane-potential perturbation is applied. To solve this issue, we first recorded the ongoing VT037867 bursting to determine its period. Next, we delivered a set of brief light pulses to optogenetically depolarize or hyperpolarize the VT037867 neurons at a fixed pulse interval, which differs from the ongoing bursting period. By repeating multiple rounds of optogenetic stimulations, we obtained a full PRC with membrane-potential perturbation at different phases of VT037867 bursting. For optimal membrane-potential perturbation, the intensity and duration of light flashes were adjusted. To examine phase changes in clock neurons, dual patch-clamp recordings were performed on pairs of VT037867 and clock neurons in flies expressing either CsChrimson or GtACR1 in VT037867 neurons.
Calcium imaging of dissociated single neurons
Flies (2 to 5 days after eclosion) were dissected under dim red light, with ~25 to 30 brains collected. The sensory organs and head cuticles were removed from the heads. Next, the fly heads were transferred to a tube containing 1.5 ml of protease XIV solution (0.67 mg/ml in Drosophila perfusion saline). After incubation for 20 min at 23°C, the enzymatic reaction was stopped, and the fly heads were rinsed with fresh Drosophila dissection saline. The brain tissues were then dispersed into single cells by mechanical trituration with a glass pipette. The dissociated cells were loaded onto a coverslip precoated with poly-l-lysine (0.5 mg/ml; Sigma-Aldrich) and laminin (10 μg/ml; Sigma-Aldrich). After 30 min for settling, the coverslip was transferred to a recording chamber and perfused with Drosophila perfusion saline bubbled with 95% O2/5% CO2. Calcium imaging was performed with a scientific complementary metal-oxide semiconductor camera (Zyla, Andor).
Generation of transgenic flies
To generate VT037867-LexA construct, a 2.1-kb enhancer was amplified from VT037867-Gal4 flies (VDRC_203797) using the following primers and digested with Not I and Asc I. The enhancer fragment was then inserted into DSCP-LexA [5′-CGAAGTTATGCTAGCGGAGCGGCCGCTGCGTAGCCGAACTTCAAGACAAAA-3′ (forward) and 5′-TCGATCCCCGGGCGAGCTCGGCGCGCCCATCCCGAGTCGGAAAAGTCCCAG-3′ (reverse)].
To generate LexAop-GtACR1 and UAS-GtACR1 constructs, the coding sequence of GtACR1-EYFP was amplified from UAS-GtACR1.d.EYFP flies (BDSC_92983) using the following primers and digested with Not I and Xba I and then inserted into LexAop2-IVS-p10 (constructed from Addgene, 36431) and UAS-IVS-p10 (constructed from Addgene, 36431), respectively [5′-TCTTATCCTTTACTTCAGGCGGCCGCCACCATGAGTAGCATTACCTGCGACCCA-3′ (forward) and 5′-GTTATTTTAAAAACGATTCATTCTAGATCACTTATACAGCTCGTCCATTCC-3′ (reverse)].
To generate R18F07-p65.AD construct, a 2.9-kb enhancer was amplified from R18F07-Gal4 flies (BDSC_47867) using the following primers and digested with Not I and Asc I. The enhancer fragment was then inserted into DSCP-p65AD [5′-TACGAAGTTATGCTAGCGGAGCGGCCGCCAGCCATCGTAGTTTTGAGCGAAGA-3′ (forward) and 5′-TCGATCCCCGGGCGAGCTCGGCGCGCCCACACCCATTTTGCTCAGTGCATCT-3′ (reverse)]. To generate transgenic flies, the above constructs were injected and integrated into the attp5 or attP40 landing site through phiC31-mediated gene integration.
Immunohistochemistry
Rabbit polyclonal anti-TIM (TIMELESS) was raised against a TIM fragment (amino acids 222 to 557) (62). The antigens were expressed and purified in an Escherichia coli system. The rabbits were immunized four times over a 2-week interval. After protein A/G affinity purification of serum, anti-TIM was obtained. Anti-TIM was validated by both Western blotting (fig. S6A) and immunostaining (fig. S6B).
Whole flies were fixed in 4% paraformaldehyde in 0.1% Triton X-100 in phosphate-buffered saline (PBST) (pH 7.4) for 2 hours and 30 min. The flies were rinsed with PBST and then dissected in PBST. Subsequently, the brains were blocked in 10% normal goat serum in PBST for 4 hours at room temperature. Primary antibodies (rabbit anti-TIM 1:250; mouse anti-PDF, 1:4000, Drosophila Studies Hybridoma Library, DSHB, PDF-C7) were applied for 16 hours at 4°C, and brains were subsequently rinsed three times with PBST (20 min each time). Secondary antibodies Alexa Fluor 488–conjugated anti-mouse (1:200, Invitrogen, catalog no. A11001) and Alexa Fluor 568–conjugated anti-rabbit (1:200, Invitrogen, catalog no. A11036) were applied for 7 hours at room temperature. Brains were rinsed three times with PBST (20 min each time), then mounted in FocusClear™ (CelExplorer) with secure-seal spacer (Invitrogen, Thermo Fisher Scientific). Images were acquired in 1.5-μm sections with a confocal microscope (A1R+, Nikon, Japan) and a water-immersion objective (25×).
Quantification of immunostaining
To quantify TIM immunostaining intensity, the boundaries of single clock neurons were manually delimited using the ROI tool in the imaging software (NIS-Elements, Nikon). Background intensity was measured from a nearby region in each section. After subtracting the background, the section with the brightest TIM intensity among the entire set of confocal sections was selected to represent the target neuron.
To quantify PDF immunostaining of the dorsal projections of s-LNvs, we assembled a maximum intensity projection of the entire set of confocal sections. We measured staining intensity using MATLAB, subtracting the average background from three pixels outside the ROI (26). The Sholl method was used to quantify the axonal arborization of s-LNvs (63, 64). Briefly, evenly spaced (10 μm) concentric rings were drawn, with the first dorsal ramification as the center point. To quantify the immunostaining measurements, at least 25 brains were used at each time point and the experiments were repeated four times.
TIM subcellular distribution
We quantified the TIM subcellular distribution according to a prior publication (65). Here, we mainly focused on s-LNv, LNd, and 5th s-LNv clock neuron subgroups. Individual confocal sections were stacked. Cytoplasmic distribution was defined if TIM showed a ring-shaped pattern, and nuclear distribution was defined when TIM encompassed exclusively the nucleus or distributed in both the nucleus and cytoplasm. TIM distribution was evaluated by three independent observers.
Synaptic labeling by GRASP
GFP reconstitution across synaptic partners (GRASP) (35) was used to examine whether the VT037867 and clock neurons could form monosynaptic connections. Nonfluorescent complementary fragments of GFP, LexAop-mCD4::GFP11 and UAS-mCD4::spGFP1–10, were expressed with VT037867-Gal4 and Pdf-LexA or VT037867-LexA and Pdf-Gal80;R54D11-Gal4. Brains from flies expressing the two complementary GFP fragments were dissected for GFP imaging directly without immunostaining.
Hemibrain connectome analysis and visualization
Synaptic connectivity between the presynaptic VT037867-labeled aMe neurons and postsynaptic clock neurons of interest was collected from the Hemibrain dataset v1.2.1 (36). The postsynaptic l-LNvs (body IDs: 1884625521, 2065745704, 5813001741, and 5813026773), 5th s-LNv (511051477), and s-LNvs (1664980698, 1975347348, 2007068523, and 2068801704) were tagged in the Hemibrain dataset, and the ITP-LNd (5813069648) was identified on the basis of its morphological similarity to the 5th s-LNv. We identified the body IDs of VT037867-labeled aMe neurons in Hemibrain through the NeuronBridge (https://neuronbridge.janelia.org) database. The connectivity was visualized with Sankey diagram by the plotly Python library (https://plotly.com/python).
Behavioral assays
Male flies (1 to 2 days after eclosion) were used for locomotor activity assays at 25°C. A single fly was placed in a glass tube with food (2% agar and 5% sucrose) and recorded using DAM2 monitors (TriKinetics). Flies were subjected to four 12-hour/12-hour LD cycles with a light intensity of 200 lux and then released into DD for 12 days.
The actograms were analyzed with ActogramJ (66). Double-plotted actograms represent average activity over 30-min intervals from groups of flies. Free-running periods under DD were determined using Chi-Square periodogram analysis. The period was considered statistically significant when the periodogram showed a peak above the 0.05 confidence level. In LD experiments, for VT037867-Gal4, UAS-Kir2.1, and VT037867-Kir2.1 flies, the activity histograms represent averaged activity levels in LD3-4; for VT037867-Shibirets flies, the activity histograms represent averaged activity levels in LD4-5. All behavioral experiments were repeated four times.
Adult-specific VT037867 silencing
Transgenic flies expressing shibirets in VT037867 neurons were raised at 18°C. Male flies (1 to 2 days after eclosion) were first subjected to five LD cycles at 30°C and then released into DD for 12 cycles at 30°C, followed by seven LD cycles at 18°C and then seven DD cycles at 18°C. Experiments were repeated four times.
Western blot analysis
Twenty fly heads of each genotype were collected at ZT20 during LD4, quickly frozen in liquid nitrogen and then homogenized in EB1 lysis buffer [1× cOmplete EDTA-free protease inhibitor (Roche), 20 mM Hepes (pH 7.5), 100 mM KCl, 25 mM NaF, 2.5 mM EDTA, 5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 0.1% Triton X-100, and 5% glycerol].
Fly head extracts were boiled and resolved on 8% tris-glycine gels. Gels were transferred to polyvinylidene fluoride membranes and probed with antibodies rabbit anti-TIM (1:2000) and mouse anti-β-tubulin (1:5000, CWBIO, CW00998M). Western blot assays were repeated four times with similar results.
Statistics
Sample sizes were determined on the basis of standards in the field. The Wilks-Shapiro test was used to determine normality of data. Normally distributed data were analyzed using two-tailed, unpaired Student’s t test, or paired t test. Nonparametrically distributed data were analyzed using Kruskal-Wallis analysis of variance (ANOVA) with Dunn’s test. Data were presented as means ± SEM. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Acknowledgments
We thank W. W. S. Yue, Y. H. Lin, H. Q. Zhao, R. C. Li, Z. Y. Chai, F. Liu, L. Carey, X. H. Zhang, E. Zhang, Y. C. Yu, Y. Zhong, J. L. Du, M. Luo, Y. Naya, K. W. Yau, and Y. Rao for discussion of the results and comments on the manuscript; M. Rosbash, M. Young, and L. Luo for discussion of the results; T. Yoshii for help with immunochemistry and locomotor rhythms; F. R. Chang for help with Western blotting; members of the Luo laboratory for discussion and comments; and H. Wu, C. Tang, and Q. Ouyang for discussion.
Funding: This work was supported by National Natural Science Foundation of China grants 31930043, 31871058, and 62088102; Collaborative Research Fund of Chinese Institute for Brain Research, Beijing (2021-NKX-XM01); grant 2021ZD0203300; State Key Laboratory of Membrane Biology, Peking University; and Peking-Tsinghua Joint Center for Life Sciences.
Author contributions: Conceptualization: L.-H.C. and D.-G.L. Methodology: M.T., L.-H.C., T.Y., N.X., B.-Y.J., S.-X.M., S.X., K.-R.L., D.Y., M.-T.L., and D.-G.L. Investigation: M.T., L.-H.C., T.Y., N.X., B.-Y.J., S.-X.M., S.X., K.-R.L., D.Y., and M.-T.L. Visualization: M.T. and L.-H.C. Supervision: L.-H.C. and D.-G.L. Writing—original draft: M.T., L.-H.C., and D.-G.L. Writing—review and editing: M.T., L.-H.C., and D.-G.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Sehgal A., Physiology flies with time. Cell 171, 1232–1235 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allada R., Chung B. Y., Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol. 72, 605–624 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hastings M. H., Maywood E. S., Brancaccio M., Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Patke A., Young M. W., Axelrod S., Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 21, 67–84 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Allen C. N., Nitabach M. N., Colwell C. S., Membrane currents, gene expression, and circadian clocks. Cold Spring Harb. Perspect. Biol. 9, a027714 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aton S. J., Herzog E. D., Come together, right...now: Synchronization of rhythms in a mammalian circadian clock. Neuron 48, 531–534 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang X., Holy T. E., Taghert P. H., A series of suppressive signals within the drosophila circadian neural circuit generates sequential daily outputs. Neuron 94, 1173–1189.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nitabach M. N., Taghert P. H., Organization of the Drosophila circadian control circuit. Curr. Biol. 18, R84–R93 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Top D., Young M. W., Coordination between differentially regulated circadian clocks generates rhythmic behavior. Cold Spring Harb. Perspect. Biol. 10, a033589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welsh D. K., Takahashi J. S., Kay S. A., Suprachiasmatic nucleus: Cell autonomy and network properties. Annu. Rev. Physiol. 72, 551–577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Z., Shafer O. T., The drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343, 1516–1520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herzog E. D., Takahashi J. S., Block G. D., Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat. Neurosci. 1, 708–713 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Liu A. C., Welsh D. K., Ko C. H., Tran H. G., Zhang E. E., Priest A. A., Buhr E. D., Singer O., Meeker K., Verma I. M., Doyle F. J., Takahashi J. S., Kay S. A., Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundkvist G. B., Kwak Y., Davis E. K., Tei H., Block G. D., A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J. Neurosci. 25, 7682–7686 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Neill J. S., Maywood E. S., Chesham J. E., Takahashi J. S., Hastings M. H., cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320, 949–953 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welsh D. K., Logothetis D. E., Meister M., Reppert S. M., Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi S., Isejima H., Matsuo T., Okura R., Yagita K., Kobayashi M., Okamura H., Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Nitabach M. N., Blau J., Holmes T. C., Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109, 485–495 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Schlichting M., Diaz M. M., Xin J., Rosbash M., Neuron-specific knockouts indicate the importance of network communication to Drosophila rhythmicity. eLife 8, e48301 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Depetris-Chauvin A., Berni J., Aranovich E. J., Muraro N. I., Beckwith E. J., Ceriani M. F., Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr. Biol. 21, 1783–1793 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grima B., Chélot E., Xia R., Rouyer F., Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Guo F., Cerullo I., Chen X., Rosbash M., PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. eLife 3, e02780 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoleru D., Peng Y., Agosto J., Rosbash M., Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Shang Y., Griffith L. C., Rosbash M., Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc. Natl. Acad. Sci. U.S.A. 105, 19587–19594 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez-Chiappe F., Frenkel L., Colque C. C., Ricciuti A., Hahm B., Cerredo K., Muraro N. I., Ceriani M. F., High-frequency neuronal bursting is essential for circadian and sleep behaviors in Drosophila. J. Neurosci. 41, 689–710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy E. V., Wu Y., deCarvalho T., Brandt C., Cao G., Nitabach M. N., Synchronized bilateral synaptic inputs to Drosophila melanogaster neuropeptidergic rest/arousal neurons. J. Neurosci. 31, 8181–8193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muraro N. I., Ceriani M. F., Acetylcholine from visual circuits modulates the activity of arousal neurons in Drosophila. J. Neurosci. 35, 16315–16327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M.-T., Cao L.-H., Xiao N., Tang M., Deng B. W., Yang T., Yoshii T., Luo D.-G., Hub-organized parallel circuits of central circadian pacemaker neurons for visual photoentrainment in Drosophila. Nat. Commun. 9, 4247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konopka R. J., Benzer S., Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renn S. C. P., Park J. H., Rosbash M., Hall J. C., Taghert P. H., A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Wilson C. J., Kawaguchi Y., The origins of two-state spontaneous membrane potential fluctuations of neostriatal spiny neurons. J. Neurosci. 16, 2397–2410 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenett A., Rubin G. M., Ngo T.-T. B., Shepherd D., Murphy C., Dionne H., Pfeiffer B. D., Cavallaro A., Hall D., Jeter J., Iyer N., Fetter D., Hausenfluck J. H., Peng H., Trautman E. T., Svirskas R. R., Myers E. W., Iwinski Z. R., Aso Y., DePasquale G. M., Enos A., Hulamm P., Lam S. C. B., Li H.-H., Laverty T. R., Long F., Qu L., Murphy S. D., Rokicki K., Safford T., Shaw K., Simpson J. H., Sowell A., Tae S., Yu Y., Zugates C. T., A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y., Cao L.-H., Sui X.-W., Guo X.-Q., Luo D.-G., Mechanosensory circuits coordinate two opposing motor actions in Drosophila feeding. Sci. Adv. 5, eaaw5141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feinberg E. H., VanHoven M. K., Bendesky A., Wang G., Fetter R. D., Shen K., Bargmannl C. I., GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Scheffer L. K., Xu C. S., Januszewski M., Lu Z., Takemura S.-Y., Hayworth K. J., Huang G. B., Shinomiya K., Maitlin-Shepard J., Berg S., Clements J., Hubbard P. M., Katz W. T., Umayam L., Zhao T., Ackerman D., Blakely T., Bogovic J., Dolafi T., Kainmueller D., Kawase T., Khairy K. A., Leavitt L., Li P. H., Lindsey L., Neubarth N., Olbris D. J., Otsuna H., Trautman E. T., Ito M., Bates A. S., Goldammer J., Wolff T., Svirskas R., Schlegel P., Neace E., Knecht C. J., Alvarado C. X., Bailey D. A., Ballinger S., Borycz J. A., Canino B. S., Cheatham N., Cook M., Dreher M., Duclos O., Eubanks B., Fairbanks K., Finley S., Forknall N., Francis A., Hopkins G. P., Joyce E. M., Kim S., Kirk N. A., Kovalyak J., Lauchie S. A., Lohff A., Maldonado C., Manley E. A., McLin S., Mooney C., Ndama M., Ogundeyi O., Okeoma N., Ordish C., Padilla N., Patrick C. M., Paterson T., Phillips E. E., Phillips E. M., Rampally N., Ribeiro C., Robertson M. K., Rymer J. T., Ryan S. M., Sammons M., Scott A. K., Scott A. L., Shinomiya A., Smith C., Smith K., Smith N. L., Sobeski M. A., Suleiman A., Swift J., Takemura S., Talebi I., Tarnogorska D., Tenshaw E., Tokhi T., Walsh J. J., Yang T., Horne J. A., Li F., Parekh R., Rivlin P. K., Jayaraman V., Costa M., Jefferis G. S., Ito K., Saalfeld S., George R., Meinertzhagen I. A., Rubin G. M., Hess H. F., Jain V., Plaza S. M., A connectome and analysis of the adult Drosophila central brain. Elife 9, e57443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinsker H. M., Aplysia bursting neurons as endogenous oscillators. II. synchronization and entrainment by pulsed inhibitory synaptic input. J. Neurophysiol. 40, 544–556 (1977). [DOI] [PubMed] [Google Scholar]
- 38.Deng B., Li Q., Liu X., Cao Y., Li B., Qian Y., Xu R., Mao R., Zhou E., Zhang W., Huang J., Rao Y., Chemoconnectomics: Mapping chemical transmission in Drosophila. Neuron 101, 876–893 e4 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Cao G., Nitabach M. N., Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J. Neurosci. 28, 6493–6501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flourakis M., Kula-Eversole E., Hutchison A. L., Han T. H., Aranda K., Moose D. L., White K. P., Dinner A. R., Lear B. C., Ren D. J., Diekman C. O., Raman I. M., Allada R., A conserved bicycle model for circadian clock control of membrane excitability. Cell 162, 836–848 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green D. J., Gillette R., Circadian-rhythm of firing rate recorded from single cells in the rat suprachiasmatic brain slice. Brain Res. 245, 198–200 (1982). [DOI] [PubMed] [Google Scholar]
- 42.Kuhlman S. J., McMahon D. G., Rhythmic regulation of membrane potential and potassium current persists in SCN neurons in the absence of environmental input. Eur. J. Neurosci. 20, 1113–1117 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Michel S., Geusz M. E., Zaritsky J. J., Block G. D., Circadian rhythm in membrane conductance expressed in isolated neurons. Science 259, 239–241 (1993). [DOI] [PubMed] [Google Scholar]
- 44.Sheeba V., Gu H., Sharma V. K., O’Dowd D. K., Holmes T. C., Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J. Neurophysiol. 99, 976–988 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang X., Holy T. E., Taghert P. H., Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science 351, 976–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eck S., Helfrich-Förster C., Rieger D., The timed depolarization of morning and evening oscillators phase shifts the circadian clock of Drosophila. J. Biol. Rhythms 31, 428–442 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Mizrak D., Ruben M., Myers G. N., Rhrissorrakrai K., Gunsalus K. C., Blau J., Electrical activity can impose time of day on the circadian transcriptome of pacemaker neurons. Curr. Biol. 22, 1871–1880 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yap E.-L., Greenberg M. E., Activity-regulated transcription: Bridging the gap between neural activity and behavior. Neuron 100, 330–348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helfrich-Förster C., Winter C., Hofbauer A., Hall J. C., Stanewsky R., The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30, 249–261 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Helfrich-Förster C., Light input pathways to the circadian clock of insects with an emphasis on the fruit fly Drosophila melanogaster. J. Comp. Physiol. A 206, 259–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogueta M., Hardie R. C., Stanewsky R., Non-canonical phototransduction mediates synchronization of the Drosophila melanogaster circadian clock and retinal light responses. Curr. Biol. 28, 1725–1735.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helfrich-Förster C., Täuber M., Park J. H., Mühlig-Versen M., Schneuwly S., Hofbauer A., Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J. Neurosci. 20, 3339–3353 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taghert P. H., Nitabach M. N., Peptide neuromodulation in invertebrate model systems. Neuron 76, 82–97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nassel D. R., Zandawala M., Hormonal axes in Drosophila: Regulation of hormone release and multiplicity of actions. Cell Tissue Res. 382, 233–266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buzsáki G., Draguhn A., Neuronal oscillations in cortical networks. Science 304, 1926–1929 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Baz E.-S., Wei H., Grosshans J., Stengl M., Calcium responses of circadian pacemaker neurons of the cockroach Rhyparobia maderae to acetylcholine and histamine. J. Comp. Physiol. A 199, 365–374 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Wu Y.-E., Enoki R., Oda Y., Huang Z.-L., Honma K.-I., Honma S., Ultradian calcium rhythms in the paraventricular nucleus and subparaventricular zone in the hypothalamus. Proc. Natl. Acad. Sci. U.S.A. 115, E9469–E9478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang X., Holy T. E., Taghert P. H., Circadian pacemaker neurons display cophasic rhythms in basal calcium level and in fast calcium fluctuations. Proc. Natl. Acad. Sci. U.S.A. 119, e2109969119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nitabach M. N., Holmes T. C., Blau J., Membranes, ions, and clocks: Testing the Njus-Sulzman-Hastings model of the circadian oscillator. Methods Enzymol. 393, 682–693 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Murthy M., Turner G., Whole-cell in vivo patch-clamp recordings in the Drosophila brain. Cold Spring Harb. Protoc. 2013, 140–148 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Cao L.-H., Jing B.-Y., Yang D., Zeng X., Shen Y., Tu Y., Luo D.-G., Distinct signaling of Drosophila chemoreceptors in olfactory sensory neurons. Proc. Natl. Acad. Sci. U.S.A. 113, E902–E911 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grima B., Lamouroux A., Chélot E., Papin C., Limbourg-Bouchon B., Rouyer F., The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420, 178–182 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Sholl D. A., Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 87, 387–406 (1953). [PMC free article] [PubMed] [Google Scholar]
- 64.Fernández M. P., Berni J., Ceriani M. F., Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol. 6, e69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin Y., Stormo G. D., Taghert P. H., The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J. Neurosci. 24, 7951–7957 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmid B., Helfrich-Förster C., Yoshii T., A new ImageJ plug-in "ActogramJ" for chronobiological analyses. J. Biol. Rhythms 26, 464–467 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1
Movie S1