Abstract
Ovarian fibrosis is a pathological condition associated with aging and is responsible for a variety of ovarian dysfunctions. Given the known contributions of tissue fibrosis to tumorigenesis, it is anticipated that ovarian fibrosis may contribute to ovarian cancer risk. We recently reported that diabetic postmenopausal women using metformin had ovarian collagen abundance and organization that were similar to premenopausal ovaries from nondiabetic women. In this study, we investigated the effects of aging and metformin on mouse ovarian fibrosis at a single-cell level. We discovered that metformin treatment prevented age-associated ovarian fibrosis by modulating the proportion of fibroblasts, myofibroblasts, and immune cells. Senescence-associated secretory phenotype (SASP)–producing fibroblasts increased in aged ovaries, and a unique metformin-responsive subpopulation of macrophages emerged in aged mice treated with metformin. The results demonstrate that metformin can modulate specific populations of immune cells and fibroblasts to prevent age-associated ovarian fibrosis and offers a new strategy to prevent ovarian fibrosis.
Metformin can prevent age-associated ovarian fibrosis by modulating the immune system and promoting tissue homeostasis.
INTRODUCTION
Aging is a multifactorial process characterized by a series of gradual changes such as increasing mitochondrial dysfunction, oxidative stress, DNA damage, and systemic chronic inflammation (1). Although the cellular mechanisms of aging are based on various hypotheses, it has become clear that the process of cellular senescence is a key biological process underlying aging (2). As a terminal state of growth arrest, senescent cells are unable to proliferate but remain metabolically active and secrete a variety of proinflammatory chemokines, cytokines, and interleukins, which are known as the “senescence-associated secretory phenotype” (SASP) (3). Although aging is also associated with a decline in immune response and efficacy, known as immunosenescence (4), it often leads to age-associated inflammation (inflamm-aging) (5). Chronic, low-grade, sterile, and systemic inflammation is one of the main mechanistic pillars of aging that is shared by many age-related diseases and tissue fibrosis (6).
Tissue fibrosis is attributed to excess deposition of extracellular matrix (ECM) components and typically results from chronic inflammation (7). Although ovarian inflammation is a normal process essential for oocyte quality, ovulation, and ovulatory wound repair (8, 9), the repeated states of inflammation, wound healing, and constant remodeling throughout a reproductive lifetime are thought to be persistent irritants that promote ovarian fibrosis (10). We recently reported the accumulation of anisotropic (linearized) collagen, characteristic of organ fibrosis, in postmenopausal ovaries in association with a tissue-resident shift in immune cells (11). Every organ ages at a different pace, and the ovary is the first organ to show signs of aging and lose its function following menopause (12). Moreover, several known pathologies are characterized by ovarian fibrosis such as the ovarian chocolate cyst, polycystic ovarian syndrome, and premature ovarian failure, all of which are more susceptible to infertility and early menopause (13). Nevertheless, given the known contributions of general tissue fibrosis to tumorigenesis, metastasis, and the “seed and soil” hypothesis (14, 15), it is strongly anticipated that ovarian fibrosis is a major risk factor for ovarian cancer (11, 16). As a matter of fact, not only is ovarian stromal fibrosis capable of creating a permissive “soil” for metastasis, but fibrosis within ovarian tumors promotes cancer progression (17), supporting the urgent need for an ovarian fibrosis management strategy in aging women and patients with ovarian cancer (10).
In our previous study, we reported normal ovarian collagen organization and reduced M2 macrophage polarization in type 2 diabetic (T2D) postmenopausal women using metformin, compared with the fibrotic ovaries of postmenopausal nondiabetic women (11). Metformin is a well-known drug prescribed for the treatment of T2D (18) to decrease glucose absorption, improve peripheral glucose uptake, and increase insulin sensitivity to reduce blood glucose levels (19). The polarity of metformin makes it dependent on its transporter OCT1, coded by Slc22a1, to enter hepatic cells (20), where it can inhibit gluconeogenesis (19). Numerous studies have demonstrated the potential of metformin as an anticancer and an antiaging agent among other health benefits (21).
Given the possible effects of metformin on age-associated ovarian fibrosis, we sought to investigate whether metformin can prevent and/or reverse ovarian fibrosis. Using single-cell RNA sequencing (scRNA-seq), we evaluated phenotypic changes associated with ovarian aging and assessed the impact of metformin on these dynamics. We show that metformin treatment modulates the immune cells toward the emergence of a unique subpopulation of macrophages and prevents the establishment of age-associated ovarian fibrosis, likely by promoting ECM remodeling and the clearance of SASP-producing fibroblasts to maintain tissue homeostasis.
RESULTS
Metformin prevents but does not reverse age-associated ovarian fibrosis
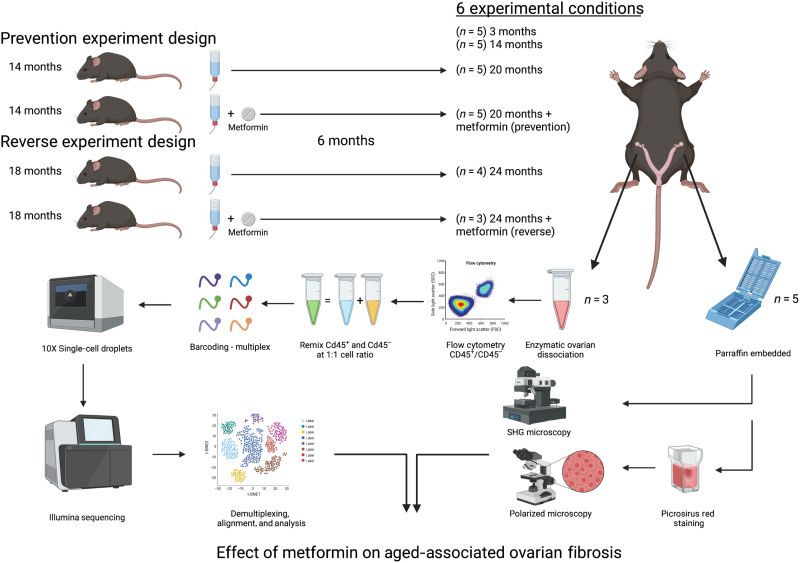
Mice were acquired at 14 and 18 months of age and aged in-house for 6 months with or without metformin in their drinking water as prevention and as a reversal treatment against ovarian fibrosis, respectively (Fig. 1). To assess the impact of age on ovarian fibrosis, we used ovarian tissue from sexually mature mice at 3 months of age (equivalent to 20 years old in humans), middle-aged mice from 12 to 17 months (ranging from 38 to 50 years old in humans, premenopausal stage), and aged mice between 20 and 24 months (ranging from 60 to 69 years old in humans, postmenopausal stage) (www.jax.org). Using Picrosirius red (PSR) staining, we found a prominent network of intense PSR-stained collagen fibers throughout the ovarian stroma and albuginea of aged mice under bright light microscopy (Fig. 2A). Mice treated with metformin as a prevention treatment had no to minimal staining of PSR. However, mice treated with metformin as a reverse treatment showed no difference in PSR staining compared with age-matched mice.
Fig. 1. Schematic of the ovarian fibrosis prevention and reversal experimental designs.
Fourteen- and 18-month-old mice were administrated metformin in drinking water (no metformin as control) for 6 months in prevention and reverse experimental settings, respectively. At the end point, 3- and 14-month-old control mice were also euthanized with the experimental mice to collect the ovaries for histology. The left ovary of the first three mice was used for single-cell analysis; single cells were flow sorted for viability and CD45 positivity and remixed at a ratio of 1:1 of immune (CD45+) and nonimmune (CD45−) cells for immune cell enrichment before multiplex barcoding and single-cell sequencing. Right ovaries were collected, formalin fixed, and paraffin embedded for second-harmonic generation (SHG) microscopy and with PSR staining assessed under polarized light microscopy. Together, histologic and transcriptomic data were used to analyze the effects of metformin on aged-associated ovarian fibrosis.
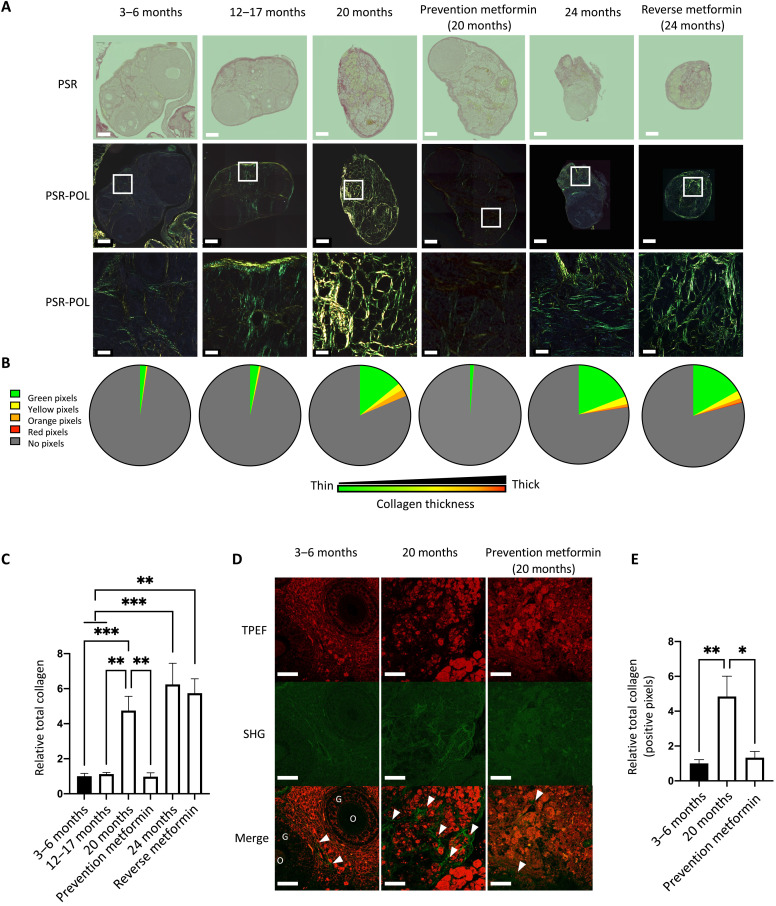
Fig. 2. Metformin prevents age-associated ovarian fibrosis as assessed by polarized light and SHG microscopy.
PSR-stained whole ovaries from young (3 to 6 months), middle-aged (12 to 17 months), and aged (20 and 24 months), with and without metformin treatment during aging (timelines shown in Fig. 1). (A) Ovaries were visualized under bright field (PSR) in the top and polarized light (PSR-POL) in the middle. Scale bars, 250 μm. The bottom panel is an enlargement of the white square in PSR-POL, for better visualization of the collagen fiber distribution and color. Scale bars, 20 μm (B) Quantification of collagen thickness by HUE is represented in a pie chart for each experimental condition for the green (thin fibers), yellow, orange, and red (thick fibers) pixels. (C) Histogram comparing the average total collagen in whole ovaries from all experimental conditions relative to young mice. (D) SHG microscopy of mouse ovaries in young (3 to 6 months), aged (20 months), and prevention groups. In the merged image, the white arrows indicate fibrotic ovarian regions, and the letters indicate ovarian structures such as the oocyte (O) or the granulosa cells (G). Scale bars, 20 μm. (E) Quantification of collagen deposition. Total positive pixels were normalized to total pixel count and averaged within each group. Data are means ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001).
PSR dye alone is not specific to collagen, but paired with polarized light, fibrillar collagen undergoes specific birefringence enhancement enabling fiber thickness visualization and quantification (22, 23). To examine the birefringence properties of PSR staining in ovarian tissue, we acquired multiple images under circularly polarized microscopy at 20× and stitched them together to obtain a figure showing the whole ovary, designated as POL-PSR (Fig. 2A). First, we observed intense birefringence in the ovarian regions that stained intensely red under bright-field light. Because the color of collagen fibers stained with PSR and viewed under polarized light depends on fiber thickness, we determined the ratio of color (HUE) in each pixel from the whole ovary. We reported the group average for each HUE (representative of thickness) in a pie chart for quick visualization between ages and treatments (Fig. 2B). There was no statistical difference between the ovarian size in pixels; however, older ovaries tended to be smaller (fig. S1A). Aged mice have increased collagen bundles associated with increased thickness. Similar to PSR under bright field, the prevention group had minimal bundles of collagen, while the reverse group had no change in collagen proportion and thickness compared with age-matched mice. Collagen fibers and their thickness increased with age in ovaries, and, most importantly, metformin treatment effectively prevented the formation of collagen bundles and reduced collagen thickness when provided during the period when age-associated ovarian fibrosis is normally established (Fig. 2C and fig. S1B). However, metformin failed to reverse preestablished age-associated ovarian fibrosis.
Using second-harmonic generation (SHG) microscopy and imaging, high-resolution quantifiable images of collagen fibers in our groups of interest were obtained. To further validate the PSR-POL observation, we acquired 20 images of three groups of three mice: young, aged, and aged plus metformin (prevention group; Fig. 2D). By quantifying the positive pixels of each SHG image above a constant threshold and calculating the average per group, we found a significant increase in the abundance of collagen fibers in aged mice and no difference in collagen fibers in aged mice treated with metformin in the prevention setting compared with young mice (Fig. 2E). The SHG analysis validated the birefringence analysis, demonstrating that PSR staining under polarized light is an efficient and cost-effective method to assess and quantify ovarian fibrosis. Together, multiple imaging approaches (PSR, POL-PSR, and SHG) have provided strong evidence that the abundance and thickness of collagen fibers are increased in the aged ovary and can be prevented by the use of metformin if administered before the establishment of ovarian fibrosis.
Establishment of ovarian fibrosis is accelerated in immunocompromised mice
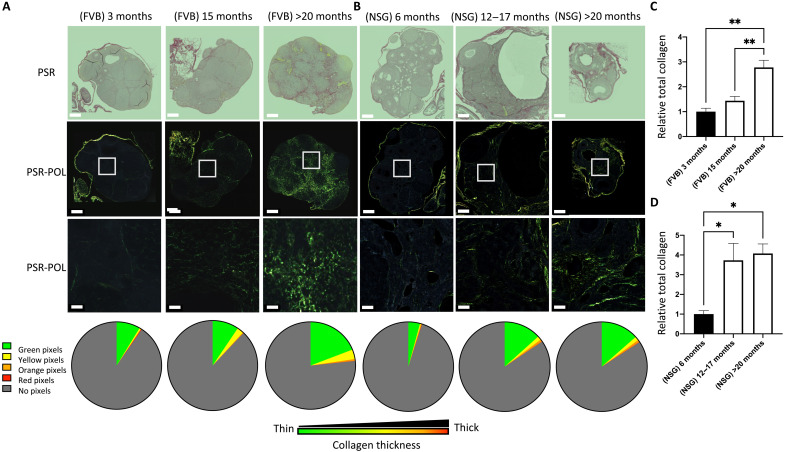
While the initial study was done with C57BL/6, the mouse strain used most often in research, we sought to investigate whether other immune-competent or immunocompromised mouse strains show the development of age-associated ovarian fibrosis. We examined the collagen composition of aging FVB/N and NSG mice using PSR staining under polarized light. First, we confirmed our observation in a different mouse strain and showed that aged FVB/N mice at 20 months of age had significantly higher relative total collagen compared with young mice at 3 months of age (Fig. 3, A and C). Immunocompromised mice developed ovarian fibrosis earlier than immune-competent strains, with a significantly higher collagen staining in the middle-aged group and as early as 12 months of age compared with young mice (Fig. 3, B and D). This observation suggests the immune system plays a major role in regulating the ovarian stroma during aging.
Fig. 3. Ovarian fibrosis significantly increases in advanced age in both FVB/N and NSG mice.
PSR-stained whole ovarian tissue from (A) immune-competent FVB/N mice and (B) immunodeficient NSG mice under bright-field (PSR) and polarized light (PSR-POL). Scale bars, 250 μm. The bottom is an enlargement of the white square in PSR-POL, for better visualization of the collagen fiber distribution and color. Scale bars, 20 μm. For each group, collagen thickness quantification is represented in a pie chart for each HUE pixel: green (thin fibers), and yellow, orange, and red (thick fibers). (C) Quantification of collagen deposition for immune-competent FVB/N mice and (D) immunodeficient NSG mice by quantifying the average of total collagen in whole ovaries from all experimental conditions relative to young mice. Data are means ± SEM (*P < 0.05 and **P < 0.01).
A comprehensive map of ovarian cell types after aging and metformin treatment
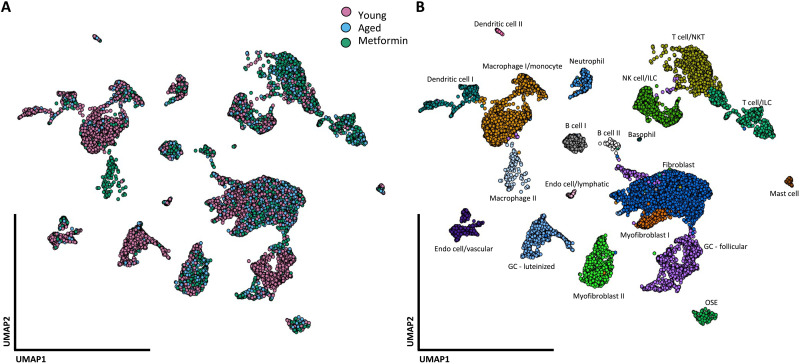
To examine the shifts in populations of cells that comprise the aging ovary and the effects of metformin treatment, we collected and dissociated whole ovaries from six groups of C57BL/6 mice. For each group, we flow sorted for CD45-positive cells and remixed the immune cells at a 1:1 ratio with nonimmune cells, which allowed us to have a better representation of the immune landscape within the ovary. The proportions of immune (CD45+) and nonimmune (CD45−) cells are shown in table S1 and indicate that aging is associated with an increase in ovarian immune cell infiltration, but metformin treatment at both ages seems to reduce the infiltration compared with their untreated counterpart. On the basis of the fibrosis prevention effect of metformin and the lack of reversal, we performed scRNA-seq on three groups (young mice, aged mice at 20 months, and aged mice at 20 months treated with metformin as a prevention treatment—hereafter referred to as the metformin group) to construct the ovarian cellular map. We profiled a total of 8082 CD45− (nonimmune) and 5726 CD45+ (immune) single cells summarized in Fig. 4A and identified a total of 20 distinct clusters that represent the cell types within the mouse ovary (Fig. 4B). We defined cell types using the ImmGen Project reference transcriptomes (24) and the automated annotation tool singleR.
Fig. 4. A single-cell map of mouse ovarian cells.
Single-cell RNA-seq data from immune and nonimmune compartments were analyzed and clustered by the Seurat packages, resulting in a two-dimensional projection of single cells onto a graph representation. (A) A total of 14,353 single cells from nine mice that were young (3 months), aged (20 months), and aged + metformin (20 months—Prevention study) were analyzed and marked by color for each condition. (B) Cells were associated with 20 cell types, annotated, and marked by color for each cluster.
Single-cell sequencing revealed shifts in fibroblast subpopulations in the aging ovary treated with metformin
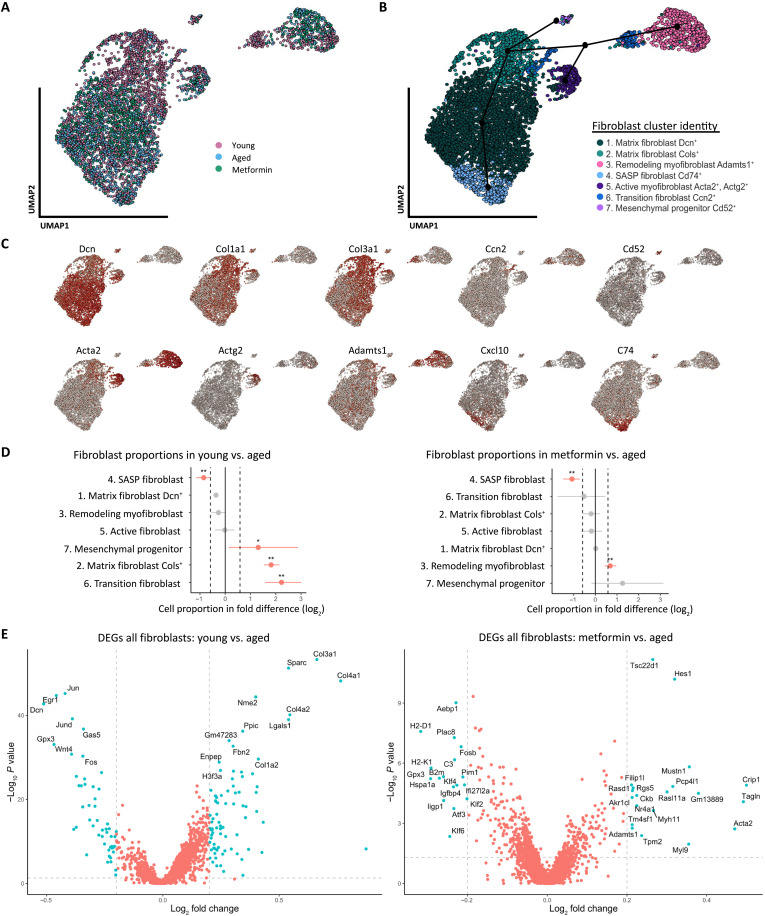
Because ECM components from tissue fibrosis are produced and maintained primarily by fibroblasts (25), we next sought to investigate the fibroblast subtypes and gene expression associated with aging and metformin treatment. From the original data, we subset three clusters identified as Fibroblast, Myofibroblast I, and Myofibroblast II, and remapped the single cells on a UMAP (Fig. 5A). The reclustering of fibroblast single cells identified a total of seven clusters (Fig. 5B). Differentially expressed gene (DEG) analysis, gene set enrichment analysis (GSEA), and module scoring were used to identify specific markers of each cluster for identification (table S2). We then assessed specific DEGs of each fibroblast subtype to confirm cluster identification (Fig. 5C). On the basis of the DEGs and pathway analysis, we labeled the clusters as follows: (1) matrix fibroblasts with high Decorin (Dcn) expression; (2) matrix fibroblasts with high collagen (Cols) expression; (3) remodeling myofibroblasts with high expression of alpha-actin 2 (Acta2) and A disintegrin and metalloproteinase with thrombospondin type 1 motif 1 (Adamts1); (4) SASP fibroblasts with high human leukocyte antigen (HLA) class II histocompatibility antigen gamma chain (also known as Cd74) expression and SASP marker expression such as C-X-C motif chemokine ligand 10 (Cxcl10); (5) myofibroblasts with high Acta2 and Actin gamma 2 (Actg2) expression; (6) transitioning fibroblasts with high expression of cellular communication network factor 2 (Ccn2); and (7) mesenchymal progenitor with high expression of Cd52, also known as CAMPATH-1 antigen.
Fig. 5. Dynamic changes in ovarian fibroblasts revealed distinct shifts in population proportions in aged and metformin-treated mice.
(A) Projection of the 5361 single cells belonging to fibroblast/myofibroblast I and II compartments of the original data, analyzed, and reclustered in a two-dimensional (2D) projection of single cells onto a graph representation by color for each condition. (B) Cells were associated with seven fibroblast states, annotated, and marked by color. The suggested pseudotrajectory from all fibroblast states is represented by black dots and lines on the 2D map. (C) Expression of key cell state–specific marker genes on top of the 2D map of ovarian fibroblasts. (D) Graph of the permutation test of each fibroblast cluster and the single-cell proportion differences between young versus aged and metformin versus aged. Dotted lines indicate the threshold for a fold change of 1.5 (log2 = 0.58), and red dots represent the significance of a change in cell proportion between groups compared [*FDR (false discovery rate) < 0.05, **FDR < 0.01]. (E) Differential gene expression of all fibroblasts derived from young versus aged and metformin versus aged ovaries. Vertical dotted lines indicate the threshold for a fold change of 1.25 (log2 = 0.3), and the horizontal dotted line indicates the P value threshold of 0.05 (−log10 = 1.3), Blue dots are significant DEGs that meet 1.25-fold change and P value <0.05. Red dots are not significant DEGs.
To determine how aging and metformin affect the composition of the fibroblast population, we used a Monte Carlo/permutation test that calculates the cell proportions of each cluster between aging (young versus aged) and metformin treatment (aged versus aged + metformin) using the scProportionTest package (26). We found a distinct shift in SASP fibroblasts with a higher proportion [false discovery rate (FDR) < 0.01] in aged mice compared with the young and metformin groups. We also found a significant increase in the proportion of the remodeling myofibroblast subtype in the metformin group (FDR < 0.01; Fig. 5D). Figure 5E shows the main significant DEGs between all fibroblasts for young versus aged and metformin versus aged. Notably, fibroblasts from young mice have higher gene expression for various collagen genes (Col1a2, Col3a1, and Col4a1), the aged group has higher expression of antigen presentation genes (H2-D1 and H2-K1), and the metformin group has higher expression of genes associated with activated remodeling by fibroblasts (Acta2 and Adamts1).
To uncover the global structure of fibroblast lineages within our data, we performed a trajectory inference analysis using the “slingshot” package (27) that characterizes the trajectory of fibroblast clusters into distinctive lineages in a semisupervised manner (Black dots and lines in Fig. 5B). The analysis revealed that cluster 4 is at the end of the matrix fibroblast lineage, and clusters 3 and 5 are at the end of a differentiated lineage of Act2+ activated fibroblasts, also called myofibroblasts and are separated by a transitioning fibroblast cell state, represented by cluster 6. The transitioning fibroblasts also had low to medium expression of Acta2 and high Ccn2 expression, which is known to promote fibroblast adhesion and is required for fibroblast activation (28). On the other end of the trajectory, the fibroblasts belonging to cluster 7 had high expression of Cd52, which is a known surface marker for the mesenchymal progenitors (29).
Fibroblast gene expression revealed specific and distinctive pathways among clusters
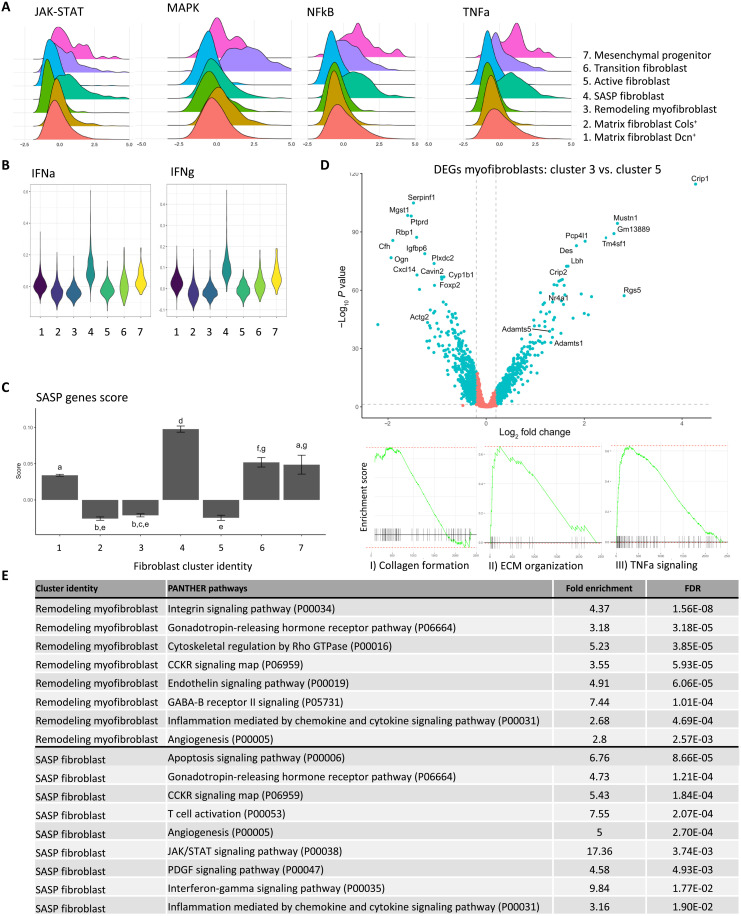
To investigate the cluster identity and functional pathways associated with each cluster, we identified the DEGs associated with each fibroblast cluster (table S2). We used the PROGENY R package to infer signaling pathway activity from 14 pathways based on consensus gene signature (30). Janus kinase (JAK)–signal transducers and activator of transcription (STAT), nuclear factor κB (NFκB), and tumor necrosis factor alpha (TNFa) activity were higher within the SASP fibroblasts, transition fibroblasts, and mesenchymal progenitors, while mitogen-activated protein kinase (MAPK) signaling pathway had a higher pathway score in the fibroblasts transitioning from matrix fibroblast to myofibroblast (Fig. 6A). Interferon (IFN) gamma and alpha activities were also increased in SASP fibroblasts using relevant modules from the MSigDB Hallmark gene sets (Fig. 6B) (31). A list of senescence marker genes was obtained from the literature (32), including senescence core genes, senescence-associated effector genes, and genes that encode SASP factors. We identified the fibroblasts in cluster 4 as SASP-associated fibroblasts as they scored highly for the panel of SASP factors (Fig. 6C). However, they show no higher expression of senescence core genes or senescence effector genes (fig. S2).
Fig. 6. Downstream analysis of ovarian fibroblasts revealed cluster identity and function.
(A) Ridge plot of the embedded value score from progeny pathway gene set of each fibroblast cluster for JAK-STAT, MAPK, NFκB, and TNFa signaling. (B) Violin plot of the module score value from the Hallmark gene set of each fibroblast cluster for IFN gamma and alpha (IFNg and IFNa). (C) Module score for SASP genes for each fibroblast cluster. Different letters above error bars (means ± SEM) indicate significant differences (P < 0.05) among groups (Tukey’s post hoc test). (D) Differential gene expression analysis of the two myofibroblast populations from all ages. Vertical dotted lines indicate the threshold for a fold change of 1.25 (log2 = 0.3), and the horizontal dotted line indicates the P value threshold of 0.05 (−log10 = 1.3). Blue dots are significant DEGs that meets 1.25-fold change and P value <0.05. Red dots are not significant DEGs. Bottom panels are plot enrichments of Kyoto Encyclopedia of Genes and Genomes pathways based on the DEGs associated to cluster 5 (active myofibroblasts): (i) collagen formation and (ii) ECM organization, and on the DEGs associated to cluster 3 (remodeling myofibroblasts): (iii) TNFa signaling. (E) Results from the statistical overrepresentation test of pathways in PANTHER based on the DEGs of the remodeling myofibroblasts or SASP Cd74+ fibroblasts, showing the fold enrichment of each pathway and the adjusted P value (FDR).
Two distinct populations of myofibroblasts (Acta2+) were also identified, and DEGs were determined (Fig. 6D). Myofibroblasts in cluster 5 enriched for genes associated with collagen formation and extracellular organization, suggesting an active myofibroblast population contributing to ECM formation (Fig. 6D, I and II). Myofibroblasts in cluster 3 enriched for the TNFa signaling pathway, which is associated with the remodeling of ECM by myofibroblasts in other tissues (Fig. 6D, III) (33) and had high expression of ADAM metallopeptidase 1 and 5 (Admts1 and 5), known to be required for the remodeling during ovarian folliculogenesis (Fig. 6D) (34).
To identify biological processes associated with each population, we used PANTHER to evaluate pathway enrichment for each cluster’s DEGs (table S3). The top pathways from the DEGS in remodeling fibroblasts and SASP fibroblasts are shown in Fig. 6E. Briefly, remodeling myofibroblasts from cluster 3 showed increased adhesion (integrin signaling pathway) and regulation of ECM production [cytoskeletal regulation by Rho GTPase (guanosine triphosphatase)]. In contrast, SASP fibroblasts from cluster 4 showed increased apoptosis, and JAK-STAT, IFN-gamma (IFNg), and PDGF signaling pathway activities.
Effects of metformin may be indirect in the mouse ovary
Metformin is known to act in the liver, where it inhibits gluconeogenesis by targeting complex 1 of the mitochondrial electron transport chain and by the activation of AMPK [reviewed in (19)]. Because of its structure, metformin is dependent on membrane transporters for cellular uptake and secretion, such as the solute carrier family 22 members 1, 2, 3, and 4 (Slc22a1, Slc22a2, Slc22a3, and Slc22a4), solute carrier family 47 members 1 and 2 (Slc47a1 and Slc47a2), and solute carrier family 29 member 4 (Slc29a4) (18). To determine whether metformin might be transported into the ovarian cells, we explored the gene expression of all known transporters of metformin in every ovarian cell population reported in our scRNA-seq analysis (fig. S3A) and discovered no expression of any known transporter, except for Slc22a1 in myofibroblast, which was reported in less than 6% of the cells from that cluster. On the basis of the human protein atlas, the SLC22A1 gene (also known as OCT1) is not expressed in human ovarian tissue [(35), humanproteinatlas.org], suggesting that metformin effects in the ovary are not mediated by direct action on ovarian fibroblasts.
Because metformin mainly targets the inhibition of mitochondrial complex I (NADH: ubiquinone oxidoreductase) and the oxidative stress response pathway (19, 36), we accessed public gene lists for these pathways to assess their expression in our fibroblasts (fig. S3B). We found no significant decrease in gene score from the complex I gene module or increase in oxidative responsive gene score in the metformin group, as we would expect if metformin had targeted fibroblasts directly. The slight increase in the score of the mitochondria gene module score in the metformin group compared with aged mice may be attributable to the shift in SASP-associated fibroblasts, which have lower mitochondrial activity (37, 38), to a higher proportion of activated fibroblasts seen in the metformin group. The lack of metformin transporter expression or change in cellular bioenergy suggests that metformin may have no direct effect on ovarian tissue.
A responsive population of macrophages is identified in the metformin treatment group
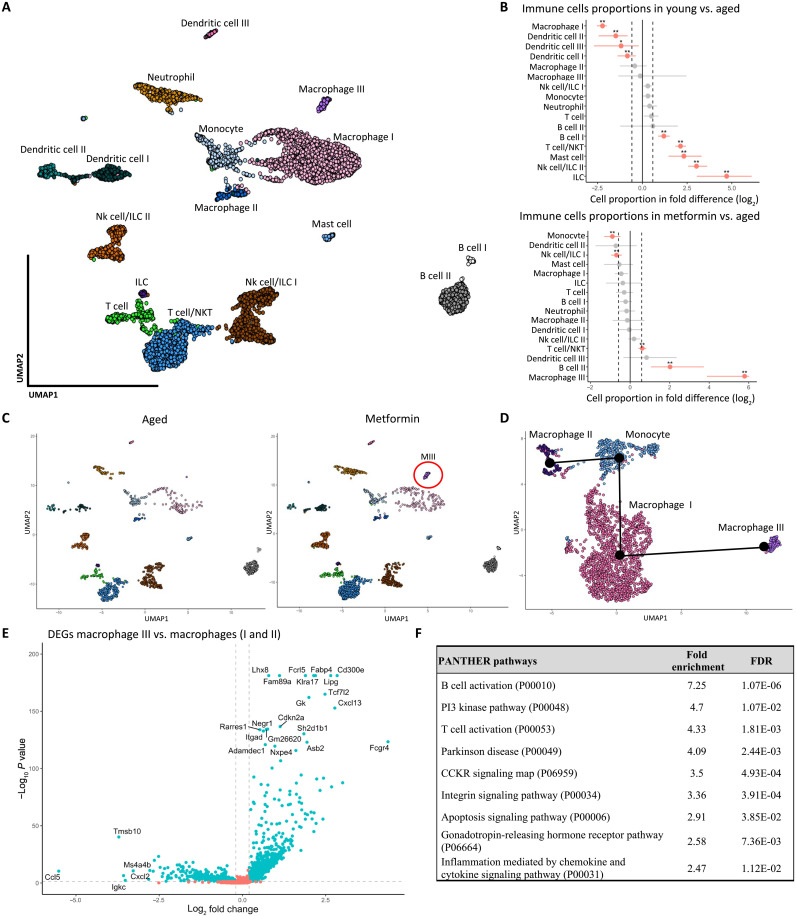
Because our observations in the NSG mice suggest that immune cells have an important role to play in the establishment of ovarian fibrosis, we sought to further investigate the immune cells in the single-cell RNA expression data and cell population proportions associated with aging and metformin treatment. From the original dataset (Fig. 4B), we subset 12 clusters identified as CD45+ and then reclustered these immune cells, which identified a total of 16 distinctive CD45+ clusters (Fig. 7A). As expected, aging altered the immune landscape of the ovary, which was associated with an increase in the proportions of B cells, T cells, natural killer T cells, innate lymphoid cells (ILCs), and mast cells and a decrease in macrophage and dendritic cells (Fig. 7B). Metformin prevention treatment in aged mice resulted in an increase in B cells and T cells and the recruitment of a metformin-responsive population of macrophages compared with the aged mice. Thus, we identified a population of macrophages specific to the metformin group, noted as “macrophage III” (Fig. 7C). From the original CD45+ data, we subset four clusters: macrophages I, II, and III, and monocytes and reclustered these populations as seen in the UMAP (Fig. 7D). We then performed a trajectory inference using the slingshot package to determine whether that population is from the same lineage of tissue-resident monocytes or alternatively recruited to the ovary (black dots and lines in Fig. 7D). One possible explanation for the separation between the macrophage I and macrophage III subpopulations and the lack of lineage association between the monocytes and the macrophage III cluster would be a possible differentiation of these macrophages in a different tissue before recruitment to and infiltration of the ovary. Further work is needed to determine the origin of that subpopulation of macrophages in the metformin-treated ovary.
Fig. 7. A single-cell map of ovarian immune cells revealed a metformin-responsive macrophage population.
(A) Projection of the 6171 single cells belonging to immune (CD45+) compartments. Cells were associated with 16 immune cell types, annotated, and marked by color. (B) Graph of the permutation test of each immune cluster and the single-cell proportion differences between young versus aged and metformin versus aged. Dotted lines indicate the threshold for a fold change of 1.5, and red dots represent a significant change in cell proportion (*FDR < 0.05, **FDR < 0.01). (C) Comparison of the single cells belonging to aged and metformin-treated groups in a 2D map. The red circle shows the emergence of a metformin-responsive population of macrophages (MIII), annotated macrophage III. (D) Projection of the 2201 single cells belonging to the macrophage and monocyte compartments. The suggested pseudotrajectory from all compartments is represented by black lines on the 2D map. (E) Differential gene expression analysis of the macrophage III against all macrophages. Dotted lines indicate the threshold for a fold change of 1.25 and the P value of 0.05. (F) Results from the statistical overrepresentation test of pathways in PANTHER based on the DEGs from macrophage III, showing the fold enrichment of each pathway and the adjusted P value (FDR).
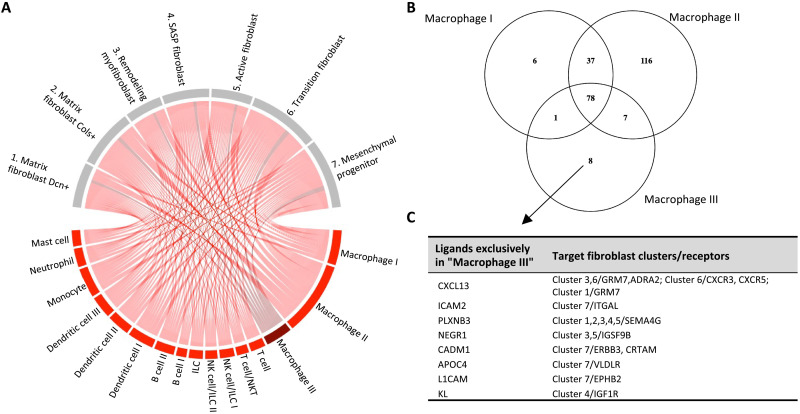
To determine the unique features of this metformin-induced macrophage population, we compared its gene expression profile relative to all other macrophages (Fig. 7E and table S4 for the full list). On the basis of the DEGs and a statistical enrichment analysis using the PANTHER pathway annotation, we found that the macrophage III cluster enriches for the activation of B cells and T cells, and phosphatidylinositol 3-kinase (PI3K) kinase activity, as well as integrin and apoptosis signaling pathway activity (Fig. 7F). Because we discovered that inflammation mediated by cytokines and chemokines, T cell activation, and IFNg signaling in the fibroblasts was associated with aging and ovarian fibrosis (Fig. 6E), we further investigated the cell communication network of that unique macrophage population by performing a ligand-receptor analysis using the LIANA R package on the single-cell data (table S5) (39). We limited the analysis to communications with immune cells as the ligand source and the seven fibroblast clusters as the receiver populations expressing cognate receptors to obtain a list of possible signaling ligands secreted by immune cells and acting on fibroblasts. It is worth noting that of all immune cells, macrophages appear to have the most interactions with fibroblasts (Fig. 8A). To identify the specific ligands secreted by the macrophage III cells, we further filtered the ligands exclusively expressed by all macrophages and compared that list of ligands within the three populations of macrophages (Fig. 8B). We found eight ligands specific to the macrophage III population and their target receptors in the fibroblast clusters (Fig. 8C). The genes for Cxcl13, Icam2, Plxnb3, Negr1, Cadm1, Apoc4, L1cam, and Kl are expressed by the macrophage III cells and have different receptors such as Igf1r, Cxcr3, Cxcr5, Grm7, Itgal, Sma4g, Igsf9b, Erbb3, Crtam, Vldlr, and Ephb2 in various fibroblast clusters of interest, including the SASP fibroblasts (cluster 4) and the remodeling myofibroblasts (cluster 3). The identification of specific DEGs and ligands expressed by the macrophage III subpopulation in the metformin group suggests unique signaling by the immune system on ovarian fibroblasts in response to metformin treatment.
Fig. 8. Ligand-receptor analysis of ovarian immune cells revealed a unique set of ligands to the metformin group.
(A) LIANA ligand-receptor analysis of ligand (red) produced from immune cells that have receptors (gray) in the fibroblast clusters. Dark red represents the ligand produced by the responsive population of macrophage III unique to the metformin group. (B) Venn Diagram comparing the expression of ligands in each macrophage compartment, revealing eight ligands that are exclusively expressed in the macrophage III cells. (C) List of the eight ligands from macrophage III cells and their respective fibroblast cluster target and receptor.
DISCUSSION
Age-associated ovarian fibrosis is most evident around the age of 20 months in female mice and is associated with increases in collagen fiber deposition and thickness (11, 40, 41). Using scRNA-seq, we found a decrease in macrophage and dendritic cell proportions in the aged ovary, which is characteristic of immunosenescence [reviewed in (42)]. We also found a higher proportion of ovarian fibroblasts in the aged fibrotic ovary that express various signaling factors that might be associated with a senescence phenotype and are enriched for pathways involved in the development and regulation of SASP, such as IFNg, JAK-STAT, NFκB, and TNFa signaling pathways (43, 44). The acquisition of the SASP by senescent cells is generally controlled by NFκB (45) and maintained by IFNg and TNFa, which induce and promote senescent cell survival (44). As a result of aging and immunosenescence (46), the immune system becomes less able to eradicate senescent cells, resulting in their persistence (4).
IFNg has been shown to induce cellular growth arrest and to stimulate major histocompatibility complex (MHC) class II surface expression by activating JAK-STAT in fibroblasts (47). It was recently shown that the expression of nonclassical MHC molecules on senescent cells allows them to evade immune clearance by inhibiting the immune responses (48). We found high expression for MHC class I and II genes (Cd74, H2-Aa, H2-Ab1, H2-Eb, H2-D1, and H2-K1) in SASP Cd74Hi fibroblasts (cluster 4) and aged fibroblasts. In support of this observation, aging in hepatocytes increased the expression of immune signature genes such as H2-Aa, H2-Ab1, H2-D1, H2-Eb1, and Cd74, which was described as an aging and cellular senescence gene signature (49). Our data suggest that aged SASP-associated Cd74Hi fibroblasts may be capable of evading the immune system clearance and persist in the aging ovary.
A recent review on the evasion of apoptosis by myofibroblasts suggests that those responsible for age-associated tissue fibrosis are evading cellular death and are persisting by becoming senescent, further contributing to the pathology of fibrosis (50). Moreover, we discovered an earlier establishment of ovarian fibrosis in immunocompromised NSG female mice, lacking functional immune cells such as macrophages. To our knowledge, ovarian fibrosis in immunocompromised mice has never been documented, and this is the first evidence of a link between the immune system and ovarian fibrosis. However, Fransolet et al. (51) noted that sheep ovarian tissue grafted into SCID and NOD-SCID female mice were more fibrotic after transplantation, suggesting that healthy ovarian tissue became fibrotic in an immune-compromised environment, which supports our finding that the immune cells play a central role in the establishment of ovarian fibrosis. In our study, metformin treatment prevented age-associated ovarian fibrosis, which was also associated with a decrease in the proportion of SASP Cd74Hi fibroblasts. Together, age-associated ovarian fibrosis appears to be the result of immunosenescence and an imbalance in fibroblast subpopulations, such that there is an increase and/or persistence of SASP-producing fibroblasts, potentially capable of evading immune cells and further stimulating ECM deposition as previously described (52).
Metformin has received a lot of attention for its capacity to regulate immune cells, especially macrophages [reviewed in (53)]. In this study, we discovered that metformin not only increases the populations of T cells and B cells but also promotes the establishment of a distinct macrophage subpopulation in the aged ovary. Metformin can modulate macrophage polarization directly (54) or indirectly by regulating glucose and insulin blood levels (55). As macrophages are a heterogeneous population specialized in sensing the microenvironment and modifying their properties accordingly (56), they can rewire their responses to different metabolic signals through activation of the PI3K pathway (57). This supports our finding that the macrophage population that appears in metformin-treated ovaries is a population that is responsive to treatment. The mechanism by which metformin modulates the immune cells remains unclear. We identified a distinct set of ligands expressed by the macrophage population specific to metformin treatment, which includes chemokine (C-X-C motif) ligand 13 (Cxcl13), Klotho (Kl), neural growth regulator 1 (Negr1), and plexin B3 (Plxnb3), which have known receptors in the SASP Cd74Hi fibroblasts and/or the remodeling myofibroblast subpopulations. CXC-chemokines and their receptors have consistently exhibited important regulatory roles in fibroblast and tissue fibrosis (7). Current evidence indicates that CXCL13 orchestrates cell-cell interactions with ECM components and regulates lymphocyte infiltration (58, 59). Klotho is known as an anti-aging protein that stimulates the production of anti-inflammatory factors and helps immune cells regain their immune functionality (60, 61). In various tissues, declines in Klotho have been reported to promote cellular senescence (62). Klotho can bind the insulin-like growth factor-1-receptor (IGF1R) to prevent activation of the signaling pathway (63, 64). In lung fibrosis, IGF1/IGF1R plays a major role in fibroblast survival and myofibroblast activation (65). By blocking the IGF1 signaling pathway, the resolution of bleomycin-induced lung fibrosis is accelerated (66). However, no role has been described yet for NEGR1 and PLXNB3 in tissue fibrosis. Our results suggest the emergence of a subpopulation of metformin-responsive macrophages that helps shape the fibroblast populations and immune landscape by secreting a unique subset of ligands to maintain optimal tissue homeostasis.
In other mouse models, metformin has been shown to prevent liver fibrosis and accelerate the reversal of lung fibrosis by modulating cellular proliferation/survival signaling pathways (67, 68). It is worth mentioning that these studies used relatively young mice with an optimal immune system. Without any intervention, both models of carbon tetrachloride–induced liver fibrosis and bleomycin-induced lung fibrosis will resolve on their own and are accompanied by an increase in macrophages (69, 70). Organ fibrosis can be resolved by removing and replacing the fibrotic tissue with healthy ECM in a controlled manner (71). It generally involves the immune cells to promote ECM degradation and collagen uptake by secreting matrix metalloproteinases (72), and the clearance of profibrotic myofibroblasts and senescent fibroblasts (50). Alternatively, fibrosis can be prevented by promoting collagen turnover to maintain healthy ECM production by fibroblasts (73). Although myofibroblasts are key players in the pathological conditions of fibrosis, they also play a pivotal role in tissue repair and remodeling by promoting ECM degradation, collagen uptake, and healthy tissue homeostasis (7). In the arterial adventitia, myofibroblasts can adopt one of two functions; positive remodeling, associated with a loss in collagen, or negative remodeling, associated with a gain in collagen fibers (74), which are under the control of the microenvironment (75). Our results suggest the presence of two similar myofibroblast types in the ovary: A population of negative remodeling myofibroblasts that enrich for extracellular structure organization and collagen formation and a population of positive remodeling myofibroblasts that express metalloproteinases. We discovered that metformin treatment increased the proportion of positive remodeling myofibroblasts compared with untreated aged ovaries, supporting the role of positive remodeling myofibroblasts in preventing ovarian fibrosis and maintaining healthy tissue homeostasis. Alternatively, metformin is known to inhibit SASP production by interfering with NFκB in cultured lung fibroblasts (76). Because SASP inhibitor can attenuate the deleterious consequences of SASP signaling and reduce senescence-induced inflammation (52), it is possible that metformin suppresses the formation of ovarian fibrosis by regulating the SASP fibroblasts.
To conclude, our results show that age-associated ovarian fibrosis is associated with immunosenescence and a higher proportion of fibroblasts with a SASP-like phenotype and expressing nonclassical MHC molecules. As a limitation of this study, it is not clear whether that higher proportion of SASP Cd74Hi fibroblasts is a result of ovarian fibrosis or is responsible for it. Nonetheless, we demonstrated that metformin prevents ovarian fibrosis by modulating the immune cells and that is associated with a decreased proportion of SASP Cd74Hi fibroblasts and increased proportion of positive remodeling myofibroblasts. Future work should focus on the involvement of the immune system during ovarian fibrosis and the mechanisms by which metformin modulates the actions of these cells toward the prevention of fibrosis in the ovary. Last, we know that young and healthy ovaries are continuously under extensive ECM remodeling during reproductive life (77). As a result, the ovarian stroma is constantly remodeled to accommodate folliculogenesis, ovulation, corpus luteum formation, and tissue repair, which is dependent on the estrus cycle (41). At menopause, ovaries stop cycling, which limits the stromal remodeling pressure of the estrus cycle and may promote permanent collagen deposition, resulting in ovarian fibrosis. As an additional contributing factor, immunosenescence prevents the clearance of SASP-producing fibroblasts, which further promotes a profibrotic microenvironment. Our results show that the intake of metformin can prevent the persistence of SASP-producing fibroblasts and maintain healthy tissue homeostasis by recruiting immune cells to the aging ovary.
MATERIALS AND METHODS
Animal models
This study was performed under a protocol approved by the Animal Care Committee of the University of Ottawa and conducted in accordance with the guidelines of the Canadian Council on Animal Care. Mice were housed under controlled environmental conditions with 12-hour alternating light/dark cycles, with free access to water and food. Young and aged female C57BL/6J mice were purchased from the Jackson Laboratory. Young FVB/N female mice were purchased from Charles Rivers and aged in-house. Ovaries from young and aged female NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice were generously donated by C. Addison (Ottawa Hospital Research Institute).
Experimental design
We previously found that metformin use by T2D postmenopausal women may abrogate ovarian fibrosis. In this study, we sought to determine whether metformin can prevent and/or reverse the formation of ovarian fibrosis. Because ovarian fibrosis is substantially increased between 15 and 18 months of age in mice, we designed our prevention study by treating adult female mice C57BL/6 beginning at the age 14 months (prefibrosis development; five per group) with 350 mg/kg per day of metformin (Sigma-Aldrich) in their drinking water for 6 months (no drug as control). We also determined whether existing fibrosis can be reversed by using naturally aged mice at 18 months of age, when collagen deposition is high. We treated the aged mice with the same metformin treatments in the drinking water for 6 months as a fibrosis reversal study. We selected this drug concentration in the drinking water as it achieves plasma concentrations similar to the doses taken by patients with T2D (i.e., 5 to 10 μM metformin) (78). At the end of the 6-month period, experimental mice were euthanized in static microisolators by CO2 asphyxiation delivered at a flow of 20 to 30% chamber volume per minute followed by cervical dislocation. We also euthanized two more experimental conditions based on age with no treatments, defined as young (n = 5; 3 months old) and middle-aged (n = 5; 14 months old). All mice were dissected to collect the ovaries. The left ovary of the first three mice of each group was used for scRNA-seq, while the right ovary of the same mice was collected for histologic analysis. The remaining two mice in each group had both ovaries collected for histologic analysis (Fig. 1).
Histologic preparation PSR staining and imaging
Histopathologic assessment of murine ovaries was performed using 5-μm sections of formalin-fixed paraffin-embedded tissue. Briefly, ovaries were incubated in 10% (w/v) neutered formaldehyde buffer (NFB) for 24 hours at room temperature and transferred to 70% ethanol to be paraffin embedded. Sections were prepared at a thickness of 5 μm and stained with PSR. Bright-field images were acquired using the ScanScope CS2 (Leica Biosystems, Concord, Canada), and polarized images were acquired using a Zeiss AxioObserver 7 (inverted) with a tunable polarizer and analyzer (Zeiss, Oberkochen, Germany). For collagen quantification, polarized images were analyzed using FIJI (ImageJ).
For each image, we performed the same subtraction operation to remove any background and noncollagen birefringence. We then determined the hue (color) of each pixel within the subtracted image. Because of the birefringence properties of collagen stained with PSR, the thickness of collagen fiber can be viewed under polarized light from green (thin) to yellow to orange to red (thick) (22). To determine the value of each hue pixel, the hue component was retained, and a histogram of hue frequency and saturation was obtained from the resolved 8-bit images (which contain 256 colors). We used the following hue definitions as previously described in (23): red, 2 to 9 and 230 to 256; orange, 10 to 38; yellow, 39 to 51; and green, 52 to 128. The hue between 129 and 229 consists of nonbirefringent tissue and is labeled as no pixel. The number of pixels within each hue range was determined and expressed as a percentage of the total ovarian area in pixels. The analysis of total collagen fiber content per area was the total sum of all hue range pixels for the total ovarian area expressed as a percentage. Last, groups were normalized by dividing each total collagen per area value by the average of the control group (young mice), which provided a relative total collagen per area.
SHG analysis
Unstained paraffinized 5-mm sections of mouse ovaries were imaged using SHG microscopy. Twenty regions of the ovarian area were annotated for imaging in each mouse (five per group). Images were acquired using a laser scanning microscope (Fluoview FVMPE-RS, Olympus) to scan the sample with a Ti:saphire femtosecond laser (Mai Tai HP, Spectra Physics). The pulse duration was 150 fs at an 80-MHz pulse frequency, and wavelength was set to 840 nm. The average laser power at the sample was 28 mW ± 1. SHG image analysis was performed using Fiji software. First, images with approximately >15% of nonovarian structure were removed from the analysis to prevent bias. Images were converted in 8-bit and based on the histogram of our control group (young mice); we selected a threshold with minimal pixel identification. All images were analyzed using the very same threshold and process. Imaging and analysis of SHG were performed blinded. The analysis of total collagen fiber content per area was the total positive pixels for the total image area relative to the young mice group.
scRNA-seq sample preparation
Ovaries were collected immediately after euthanasia, cleaned from any extraneous remaining tissue (fat, oviduct, etc.) and washed in phosphate-buffered saline (PBS) three times. All three ovaries from each group were pooled together in 3 ml of enzymatic dissociation solution [Hanks’ balanced salt solution with Pronase (1.25 mg/ml), Elastase (9.2 μg/ml), deoxyribonuclease I (100 μg/ml), Dispase (100 μg/ml), and Collagenase A (1.5 mg/ml)]. Ovaries were further chopped using surgical scissors in dissociation media and incubated for 30 min at 37°C in a rotatory shaker. Single-cell suspensions were filtered using a 40-μm cell strainer (which removed all oocytes as well), spun down, and resuspended in ACK lysis buffer for 30 s to remove red blood cells. Cells were washed in sterile PBS (PBS; Corning, no. 21-031-CV) before preparing for flow sorting.
Flow sorting samples for immune cell enrichment
Cells in suspension were incubated in Zombie Live Dead dye NIR solution (at 1/500 in PBS) for 20 min at room temperature and washed with the flow buffer [PBS–2% fetal bovine serum (FBS)]. Cells were then incubated in a CD45.2–fluorescein isothiocyanate solution (at 1/750 in flow buffer) for 15 min at room temperature. Cells were washed twice and resuspended in flow buffer before flow sorting. For each group, a gate was set for live cells (NIR+) and separated the live immune cells (CD45+) and live nonimmune cells (CD45−) in their respective 15-ml falcon tube. All flow cytometry was carried out on a Beckman Coulter MoFlo XDP (Beckman Coulter, Indianapolis, IN). Samples were kept on ice during the flow sorting process, which was performed within 15 min following the end of enzymatic dissociation. Cells were remixed together at a ratio of 1:1 immune and nonimmune before labeling.
Multiplex scRNA-seq preparation, sequencing, and processing
Following procedures previously described in (79), we performed multiplexing following the MULTI-seq protocol. After the final wash, cells were resuspended in PBS + 1% bovine serum albumin (BSA), pooled together, pelleted, and resuspended in PBS + 1% BSA. Viability and cell counts were then performed before preparation of the scRNA-seq libraries. Single-cell suspensions were processed using the 10× Genomics Single Cell 3′ RNA-seq Kit (v3). Gene expression libraries were prepared according to the manufacturer’s protocol. MULTI-seq barcode libraries were retrieved from the samples, and libraries were prepared independently according to the MULTI-seq library preparation protocol. Briefly, barcode libraries are separated from the cDNA libraries during the first round of size selection in the 10× Genomics library preparation protocol and polymerase chain reaction amplified before sequencing. Final libraries were sequenced on a NextSeq500 (Illumina). Expression libraries were sequenced so that time course libraries reached an approximate depth of 20,000 reads per cell (v3 scRNA-seq kit). We detected a median of 1718 genes and 4201 unique molecular identifiers (UMIs) per cell.
Raw sequencing reads from the gene expression libraries were processed using “CellRanger” v3.0.2. The GRCh38 build of the mouse genome was used. Except for explicitly setting --expect-cells = 25,000, default parameters were used for all samples. MULTI-seq barcode libraries were simply trimmed to 28 base pairs (v3 kit) using Trimmomatic48 (v0.36) before demultiplexing.
Demultiplexing was performed using the “demultiplex” R package (v1.0.2). The key concepts for demultiplexing are described in McGinnis et al. (80) Briefly, the tool takes the barcode sequencing reads and counts the number of times each barcode appears for each cell. Then, for each barcode, it assesses the distribution of counts in cells and determines an optimal quantile threshold to deem a cell positive for a given barcode. Cells positive for more than one barcode are classified as doublets and are removed. Only cells positive for a single barcode are retained for downstream analysis. As each barcode corresponds to a specific sample in the experiment, the sample annotations can then be added to all cells in the dataset.
Quality control was first performed independently on each 10× Genomics library, and all main processing steps were performed with Seurat v3.0.2. Expression matrices for each sample were loaded into R as Seurat objects, only retaining cells with >200 genes detected. Cells with a high percentage of mitochondrial gene expression were also removed. We then subset the data, making independent Seurat objects for immune and nonimmune cells (i.e., Fibroblast, CD45+, CD45−, etc.). Each condition was then processed independently with a standard workflow. We first removed genes detected in <1% of the cells for the given experiment. The expression values were then normalized with standard library size scaling and log transformation. The top 3000 variable genes were detected using the variance-stabilizing transformation (vst) selection method in Seurat. Expression values were scaled, and the following technical factors were regressed out: percentage of mitochondrial reads, number of RNA molecules detected, and cycle scores. UMAP dimensionality reduction and embeddings were mapped after applying “SCTransform” normalization and standardization algorithm.
Computational analyses
To determine the identity of the cell type in each cluster, we used the “celldex for cell-type identification” package (24) with the immunological Genome Project (ImmGen) as the reference with the automated annotation tool “singleR,” and we manually validated each cluster with specific markers (table S2). Differential expression analysis was performed using the R package “Seurat.” We assessed the number of DEGs in various specified contexts (e.g., Metformin versus Aged, Cluster#4) using the FindMarkers() function, and default parameters were used with idents and sub-idents specific to the analysis. GSEA was then performed using the R package “FGSEA.” Input genes were ranked by log2fold change and adjusted P value from the differential expression analysis. Reference gene sets were collected from the Molecular Signatures Database (MSigDB) v6.2 (31).
Gene set scoring was performed using the AddModuleScore() function provided by the Seurat package; default parameters were used. Gene lists from the public domain were acquired from Geneglobe Qiagen website (geneglobe.qiagen.com): Cellular Senescence (Core, Effective, and SASP gene set), Mitochondria Energy Metabolism [Complex I (NADH-Coenzyme Q Reductase) gene set], and Oxidative Stress (Oxidative Stress Responsive Genes gene set).
Pathway responsive genes for activity inference from gene expression analysis were performed using the R package “PROGENy” (30). Pathway activity score was computed on the scRNA-seq data from the fibroblasts subset Seurat object using the progeny() function with the mouse reference dataset. Alongside, we used the protein analysis through evolutionary relationships (PANTHER) classification system to perform an overrepresentation analysis (http://pantherdb.org). We imported positive DEGs associated with specific clusters (e.g., SASP fibroblast) and performed the analysis using the PANTHER pathway with the mouse whole genome as a reference.
Ligand-receptor analysis was performed by applying the “LIANA” package (39). Briefly, our Seurat object with relevant cellular population identities was used as the data source for the package. We converted all mouse genes to human nomenclature using a list of 1-to-1 human to mouse orthologs and ran LIANA to identify all possible ligand-receptor communications between all clusters. We chose the following implementations to calculate consensus-based receptor-ligand links: LogFC, NATMI, Connectome, SCA, SquidPy, CellChat, iTALK, and CytoTalk. After each implementation finished calculating consensus ranks, we aggregated the data into a single table with a meta P value that represents the strongest and most likely receptor-ligand interactions before focusing on relevant populations and their receptor-ligand interactome. Visualization of receptor-ligand interactions was done using the “Circlize” package.
Statistical analyses
All statistical analyses were performed using Prism 9 software (GraphPad Software Inc., La Jolla, CA, USA). Datasets were compared using analysis of variance (with Tukey’s or Dunnett’s multiple comparisons posttest) or otherwise when specified. All data are presented as means ± SEM.
Acknowledgments
We thank E. Macdonald and V. Maranda for the technical support, J. Coulombe and C. Addison for providing ovaries from NSG mice, C. van Oostende-Triplet for the PSR imaging technical support, Z. Gartner for providing the MULTI-seq protocol and reagents, and C. McCloskey for the careful review of this manuscript. D.A.L. was supported by postdoctoral fellowships from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Lalor Foundation.
Funding: We would like to acknowledge and honor the late M. Craig, whose generous support funded much of this study. This work was also funded by a grant from the Canadian Institutes of Health Research (E-436754).
Author contributions: D.A.L. and B.C.V. conceived the study. D.A.L. wrote the manuscript. D.A.L., E.Y., D.P.C., S.F., J.U., and B.C.V. reviewed the manuscript. D.A.L. performed mouse experiments, tissue collection, and PSR imaging. S.F. and J.U. performed the SHG imaging. D.A.L. performed all image analysis and statistical analysis. D.A.L., E.Y., and D.P.C. performed the scRNA-seq and computational analyses.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Raw sequencing files and processed UMI count matrices have been deposited in the NCBI Gene Expression Omnibus under the accession GSE199914. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S3
Table S1
Other Supplementary Material for this manuscript includes the following:
Tables S2 to S5
REFERENCES AND NOTES
- 1.Di Micco R., Krizhanovsky V., Baker D., d’Adda di Fagagna F., Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Almeida A. J. P. O., Ribeiro T. P., de Medeiros I. A., Aging: Molecular pathways and implications on the cardiovascular system. Oxid. Med. Cell. Longev. 2017, 7941563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coppé J.-P., Desprez P.-Y., Krtolica A., Campisi J., The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song P., An J., Zou M.-H., Immune clearance of senescent cells to combat ageing and chronic diseases. Cells 9, 671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fulop T., Larbi A., Dupuis G., Le Page A., Frost E. H., Cohen A. A., Witkowski J. M., Franceschi C., Immunosenescence and inflamm-aging as two sides of the same coin: Friends or foes? Front. Immunol. 8, 1960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A., Inflammaging: A new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14, 576–590 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Wynn T. A., Cellular and molecular mechanisms of fibrosis. J. Pathol. 214, 199–210 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landry D. A., Sirard M.-A., Follicle capacitation: A meta-analysis to investigate the transcriptome dynamics following follicle-stimulating hormone decline in bovine granulosa cells. Biol. Reprod. 99, 877–887 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Boots C. E., Jungheim E. S., Inflammation and human ovarian follicular dynamics. Semin. Reprod. Med. 33, 270–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landry D. A., Vaishnav H. T., Vanderhyden B. C., The significance of ovarian fibrosis. Oncotarget 11, 4366–4370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCloskey C. W., Cook D. P., Kelly B. S., Azzi F., Allen C. H., Forsyth A., Upham J., Rayner K. J., Gray D. A., Boyd R. W., Murugkar S., Lo B., Trudel D., Senterman M. K., Vanderhyden B. C., Metformin abrogates age-associated ovarian fibrosis. Clin. Cancer Res. 26, 632–642 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Broekmans F. J., Soules M. R., Fauser B. C., Ovarian aging: Mechanisms and clinical consequences. Endocr. Rev. 30, 465–493 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Zhou F., Shi L.-B., Zhang S.-Y., Ovarian fibrosis: A phenomenon of concern. Chin. Med. J. 130, 365–371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox T. R., Erler J. T., Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer. Dis. Model. Mech. 4, 165–178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox T. R., Erler J. T., Molecular pathways: Connecting fibrosis and solid tumor metastasis. Clin. Cancer Res. 20, 3637–3643 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Harper E. I., Sheedy E. F., Stack M. S., With great age comes great metastatic ability: Ovarian cancer and the appeal of the aging peritoneal microenvironment. Cancer 10, 230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeung T.-L., Leung C. S., Yip K.-P., Sheng J., Vien L., Bover L. C., Birrer M. J., Wong S. T. C., Mok S. C., Anticancer immunotherapy by MFAP5 blockade inhibits fibrosis and enhances chemosensitivity in ovarian and pancreatic cancer. Clin. Cancer Res. 25, 6417–6428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pernicova I., Korbonits M., Metformin—Mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 10, 143–156 (2014). [DOI] [PubMed] [Google Scholar]
- 19.LaMoia T. E., Shulman G. I., Cellular and molecular mechanisms of metformin action. Endocr. Rev. 42, 77–96 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D.-S., Jonker J. W., Kato Y., Kusuhara H., Schinkel A. H., Sugiyama Y., Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J. Pharmacol. Exp. Ther. 302, 510–515 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Wang Y.-W., He S.-J., Feng X., Cheng J., Luo Y.-T., Tian L., Huang Q., Metformin: A review of its potential indications. Drug Des. Devel. Ther. 11, 2421–2429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drifka C. R., Loeffler A. G., Mathewson K., Mehta G., Keikhosravi A., Liu Y., Lemancik S., Ricke W. A., Weber S. M., Kao W. J., Eliceiri K. W., Comparison of picrosirius red staining with second harmonic generation imaging for the quantification of clinically relevant collagen fiber features in histopathology samples. J. Histochem. Cytochem. 64, 519–529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rich L., Whittaker P., Collagen and Picrosirius red staining: A polarized light assessment of fibrillar hue and spatial distribution. J. Morphol. Sci. 22, 97–104 (2005). [Google Scholar]
- 24.Heng T. S. P., Painter M. W.; Immunological Genome Project Consortium , The immunological genome project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Kendall R. T., Feghali-Bostwick C. A., Fibroblasts in fibrosis: Novel roles and mediators. Front. Pharmacol. 5, 123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller S. A., Policastro R. A., Savant S. S., Sriramkumar S., Ding N., Lu X., Mohammad H. P., Cao S., Kalin J. H., Cole P. A., Zentner G. E., O’Hagan H. M., Lysine-specific demethylase 1 mediates akt activity and promotes epithelial-to-mesenchymal transition in PIK3CA-mutant colorectal cancer. Mol. Cancer Res. 18, 264–277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Street K., Risso D., Fletcher R. B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S., Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorn L. E., Petrosino J. M., Wright P., Accornero F., CTGF/CCN2 is an autocrine regulator of cardiac fibrosis. J. Mol. Cell. Cardiol. 121, 205–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie T., Wang Y., Deng N., Huang G., Taghavifar F., Geng Y., Liu N., Kulur V., Yao C., Chen P., Liu Z., Stripp B., Tang J., Liang J., Noble P. W., Jiang D., Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 22, 3625–3640 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert M., Klinger B., Klünemann M., Sieber A., Uhlitz F., Sauer S., Garnett M. J., Blüthgen N., Saez-Rodriguez J., Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 9, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P., The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiss T., Nyúl-Tóth Á., Balasubramanian P., Tarantini S., Ahire C., DelFavero J., Yabluchanskiy A., Csipo T., Farkas E., Wiley G., Garman L., Csiszar A., Ungvari Z., Single-cell RNA sequencing identifies senescent cerebromicrovascular endothelial cells in the aged mouse brain. Geroscience 42, 429–444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redente E., Riches D. W. H., Tumor necrosis factor-α–induced myofibroblast plasticity as a mechanism of repair after fibrotic lung injury. Ann. Am. Thorac. Soc. 12, S71–S72 (2015). [Google Scholar]
- 34.Brown H. M., Dunning K. R., Robker R. L., Pritchard M., Russell D. L., Requirement for ADAMTS-1 in extracellular matrix remodeling during ovarian folliculogenesis and lymphangiogenesis. Dev. Biol. 300, 699–709 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Karlsson M., Zhang C., Méar L., Zhong W., Digre A., Katona B., Sjöstedt E., Butler L., Odeberg J., Dusart P., Edfors F., Oksvold P., von Feilitzen K., Zwahlen M., Arif M., Altay O., Li X., Ozcan M., Mardinoglu A., Fagerberg L., Mulder J., Luo Y., Ponten F., Uhlén M., Lindskog C., A single-cell type transcriptomics map of human tissues. Sci. Adv. 7, eabh2169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulkarni A. S., Gubbi S., Barzilai N., Benefits of metformin in attenuating the hallmarks of aging. Cell Metab. 32, 15–30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang J. L., Shukla P., Pagidas K., Ahmed N. S., Karri S., Gunn D. D., Hurd W. W., Singh K. K., Mitochondria in ovarian aging and reproductive longevity. Ageing Res. Rev. 63, 101168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman J., Fielder E., Passos J. F., Mitochondrial dysfunction and cell senescence: Deciphering a complex relationship. FEBS Lett. 593, 1566–1579 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Dimitrov D., Türei D., Boys C., Nagai J. S., Ramirez Flores R. O., Kim H., Szalai B., Costa I. G., Dugourd A., Valdeolivas A., Saez-Rodriguez J., Comparison of ressources and methods to infer cell-cell communication from single-cell RNA data. bioRxiv , 2021.05.21.445160 (2021). [Google Scholar]
- 40.Briley S. M., Jasti S., McCracken J. M., Hornick J. E., Fegley B., Pritchard M. T., Duncan F. E., Reproductive age-associated fibrosis in the stroma of the mammalian ovary. Reproduction 152, 245–260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umehara T., Richards J. S., Shimada M., The stromal fibrosis in aging ovary. Aging 10, 9–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lian J., Yue Y., Yu W., Zhang Y., Immunosenescence: A key player in cancer development. J. Hematol. Oncol. 13, 151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumari R., Jat P., Mechanisms of cellular senescence: Cell cycle arrest and senescence associated secretory phenotype. Front. Cell Dev. Biol. 9, 645593 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim K. S., Kang K. W., Seu Y. B., Baek S.-H., Kim J.-R., Interferon-gamma induces cellular senescence through p53-dependent DNA damage signaling in human endothelial cells. Mech. Ageing Dev. 130, 179–188 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Chien Y., Scuoppo C., Wang X., Fang X., Balgley B., Bolden J. E., Premsrirut P., Luo W., Chicas A., Lee C. S., Kogan S. C., Lowe S. W., Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25, 2125–2136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aw D., Silva A. B., Palmer D. B., Immunosenescence: Emerging challenges for an ageing population. Immunology 120, 435–446 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ilangumaran S., Finan D., La Rose J., Raine J., Silverstein A., De Sepulveda P., Rottapel R., A positive regulatory role for suppressor of cytokine signaling 1 in IFN-gamma-induced MHC class II expression in fibroblasts. J. Immunol. 169, 5010–5020 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Pereira B. I., Devine O. P., Vukmanovic-Stejic M., Chambers E. S., Subramanian P., Patel N., Virasami A., Sebire N. J., Kinsler V., Valdovinos A., LeSaux C. J., Passos J. F., Antoniou A., Rustin M. H. A., Campisi J., Akbar A. N., Senescent cells evade immune clearance via HLA-E-mediated NK and CD8+ T cell inhibition. Nat. Commun. 10, 2387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabula Muris Consortium , A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hinz B., Lagares D., Evasion of apoptosis by myofibroblasts: A hallmark of fibrotic diseases. Nat. Rev. Rheumatol. 16, 11–31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fransolet M., Henry L., Labied S., Masereel M.-C., Blacher S., Noël A., Foidart J.-M., Nisolle M., Munaut C., Influence of mouse strain on ovarian tissue recovery after engraftment with angiogenic factor. J. Ovarian Res. 8, 14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehdizadeh M., Aguilar M., Thorin E., Ferbeyre G., Nattel S., The role of cellular senescence in cardiac disease: Basic biology and clinical relevance. Nat. Rev. Cardiol. 19, 250–264 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Ursini F., Russo E., Pellino G., D’Angelo S., Chiaravalloti A., De Sarro G., Manfredini R., De Giorgio R., Metformin and autoimmunity: A “new deal” of an old drug. Front. Immunol. 9, 1236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiang C.-F., Chao T.-T., Su Y.-F., Hsu C.-C., Chien C.-Y., Chiu K.-C., Shiah S.-G., Lee C.-H., Liu S.-Y., Shieh Y.-S., Metformin-treated cancer cells modulate macrophage polarization through AMPK-NF-κB signaling. Oncotarget 8, 20706–20718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ayala T. S., Tessaro F. H. G., Jannuzzi G. P., Bella L. M., Ferreira K. S., Martins J. O., High glucose environments interfere with bone marrow-derived macrophage inflammatory mediator release, the TLR4 pathway and glucose metabolism. Sci. Rep. 9, 11447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Viola A., Munari F., Sánchez-Rodríguez R., Scolaro T., Castegna A., The metabolic signature of macrophage responses. Front. Immunol. 10, 1462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharif O., Brunner J. S., Vogel A., Schabbauer G., Macrophage rewiring by nutrient associated pi3k dependent pathways. Front. Immunol. 10, 2002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kazanietz M. G., Durando M., Cooke M., CXCL13 and its receptor CXCR5 in cancer: Inflammation, immune response, and beyond. Front. Endocrinol. 10, 471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cosgrove J., Novkovic M., Albrecht S., Pikor N. B., Zhou Z., Onder L., Mörbe U., Cupovic J., Miller H., Alden K., Thuery A., O’Toole P., Pinter R., Jarrett S., Taylor E., Venetz D., Heller M., Uguccioni M., Legler D. F., Lacey C. J., Coatesworth A., Polak W. G., Cupedo T., Manoury B., Thelen M., Stein J. V., Wolf M., Leake M. C., Timmis J., Ludewig B., Coles M. C., B cell zone reticular cell microenvironments shape CXCL13 gradient formation. Nat. Commun. 11, 3677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garth J., Easter M., Skylar Harris E., Sailland J., Kuenzi L., Chung S., Dennis J. S., Baumlin N., Adewale A. T., Rowe S. M., King G., Faul C., Barnes J. W., Salathe M., Krick S., The effects of the anti-aging protein Klotho on mucociliary clearance. Front. Med. 6, 339 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Typiak M., Piwkowska A., Antiinflammatory actions of Klotho: Implications for therapy of diabetic nephropathy. Int. J. Mol. Sci. 22, 956 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuro-o M., Klotho as a regulator of oxidative stress and senescence. Biol. Chem. 389, 233–241 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Kurosu H., Yamamoto M., Clark J. D., Pastor J. V., Nandi A., Gurnani P., McGuinness O. P., Chikuda H., Yamaguchi M., Kawaguchi H., Shimomura I., Takayama Y., Herz J., Kahn C. R., Rosenblatt K. P., Kuro-o M., Suppression of aging in mice by the hormone Klotho. Science 309, 1829–1833 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delcroix V., Mauduit O., Tessier N., Montillaud A., Lesluyes T., Ducret T., Chibon F., Van Coppenolle F., Ducreux S., Vacher P., The role of the anti-aging protein Klotho in IGF-1 signaling and reticular calcium leak: Impact on the chemosensitivity of dedifferentiated liposarcomas. Cancers 10, 439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi Y., Yuan D., Rajkovic A., Germ cell-specific transcriptional regulator sohlh2 is essential for early mouse folliculogenesis and oocyte-specific gene expression. Biol. Reprod. 79, 1176–1182 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hung C. F., Rohani M. G., Lee S., Chen P., Schnapp L. M., Role of IGF-1 pathway in lung fibroblast activation. Respir. Res. 14, 102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shankaraiah R. C., Callegari E., Guerriero P., Rimessi A., Pinton P., Gramantieri L., Silini E. M., Sabbioni S., Negrini M., Metformin prevents liver tumourigenesis by attenuating fibrosis in a transgenic mouse model of hepatocellular carcinoma. Oncogene 38, 7035–7045 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Rangarajan S., Bone N. B., Zmijewska A. A., Jiang S., Park D. W., Bernard K., Locy M. L., Ravi S., Deshane J., Mannon R. B., Abraham E., Darley-Usmar V., Thannickal V. J., Zmijewski J. W., Metformin reverses established lung fibrosis in a bleomycin model. Nat. Med. 24, 1121–1127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fallowfield J. A., Mizuno M., Kendall T. J., Constandinou C. M., Benyon R. C., Duffield J. S., Iredale J. P., Scar-associated macrophages are a major source of hepatic matrix metalloproteinase-13 and facilitate the resolution of murine hepatic fibrosis. J. Immunol. 178, 5288–5295 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Cabrera S., Maciel M., Hernández-Barrientos D., Calyeca J., Gaxiola M., Selman M., Pardo A., Delayed resolution of bleomycin-induced pulmonary fibrosis in absence of MMP13 (collagenase 3). Am. J. Physiol. Lung Cell. Mol. Physiol. 316, L961–L976 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Jun J.-I., Lau L. F., Resolution of organ fibrosis. J. Clin. Invest. 128, 97–107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giannandrea M., Parks W. C., Diverse functions of matrix metalloproteinases during fibrosis. Dis. Model. Mech. 7, 193–203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Herrera J., Henke C. A., Bitterman P. B., Extracellular matrix as a driver of progressive fibrosis. J. Clin. Invest. 128, 45–53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coen M., Gabbiani G., Bochaton-Piallat M.-L., Chen Y. E., Myofibroblast-mediated adventitial remodeling. Arterioscler. Thromb. Vasc. Biol. 31, 2391–2396 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Desmoulière A., Darby I. A., Gabbiani G., Normal and pathologic soft tissue remodeling: Role of the myofibroblast, with special emphasis on liver and kidney fibrosis. Lab. Invest. 83, 1689–1707 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Moiseeva O., Deschênes-Simard X., St-Germain E., Igelmann S., Huot G., Cadar A. E., Bourdeau V., Pollak M. N., Ferbeyre G., Metformin inhibits the senescence-associated secretory phenotype by interfering with IKK/NF-κB activation. Aging Cell 12, 489–498 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Duffy D. M., Ko C., Jo M., Brannstrom M., Curry T. E. Jr., Ovulation: Parallels with inflammatory processes. Endocr. Rev. 40, 369–416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chandel N. S., Avizonis D., Reczek C. R., Weinberg S. E., Menz S., Neuhaus R., Christian S., Haegebarth A., Algire C., Pollak M., Are metformin doses used in murine cancer models clinically relevant? Cell Metab. 23, 569–570 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Cook D. P., Vanderhyden B. C., Context specificity of the EMT transcriptional response. Nat. Commun. 11, 2142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McGinnis C. S., Patterson D. M., Winkler J., Conrad D. N., Hein M. Y., Srivastava V., Hu J. L., Murrow L. M., Weissman J. S., Werb Z., Chow E. D., Gartner Z. J., MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods 16, 619–626 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3
Table S1
Tables S2 to S5