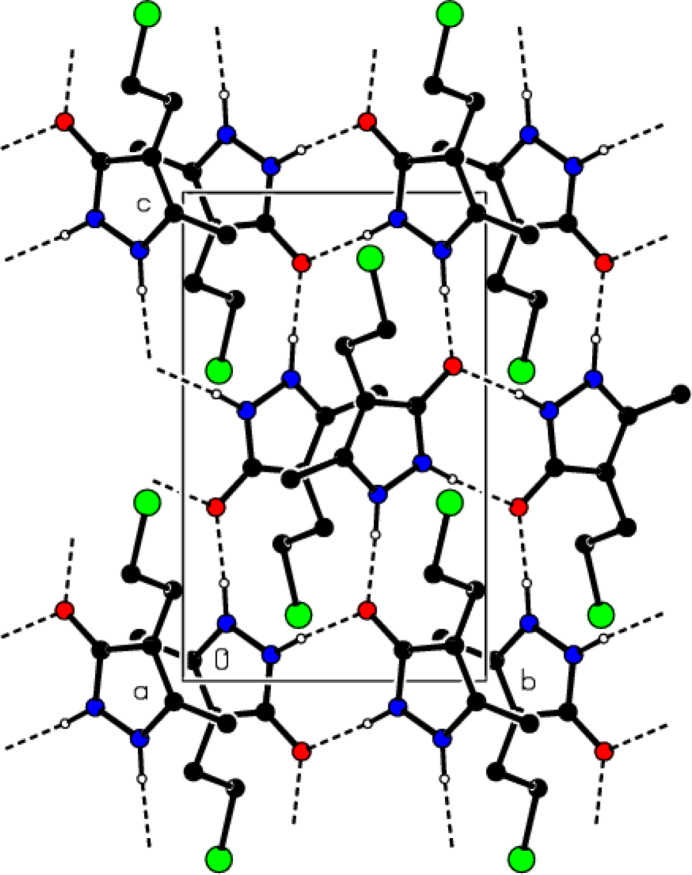
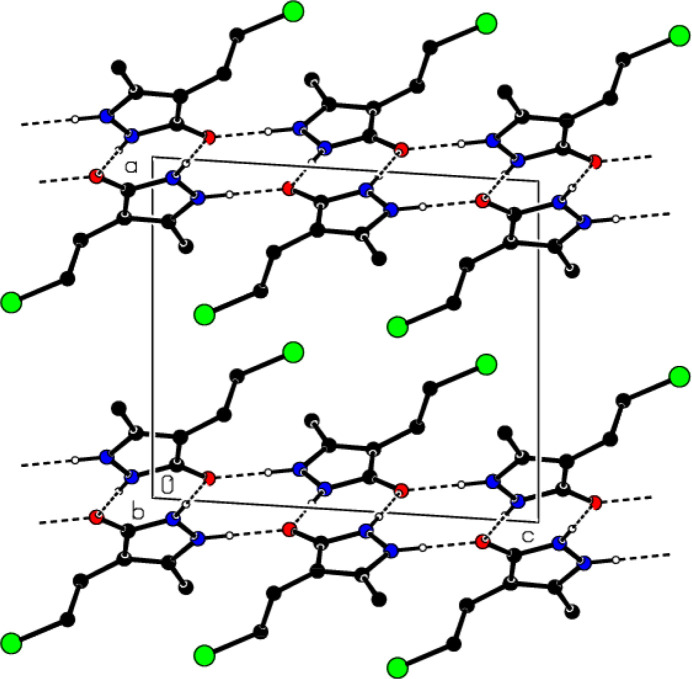
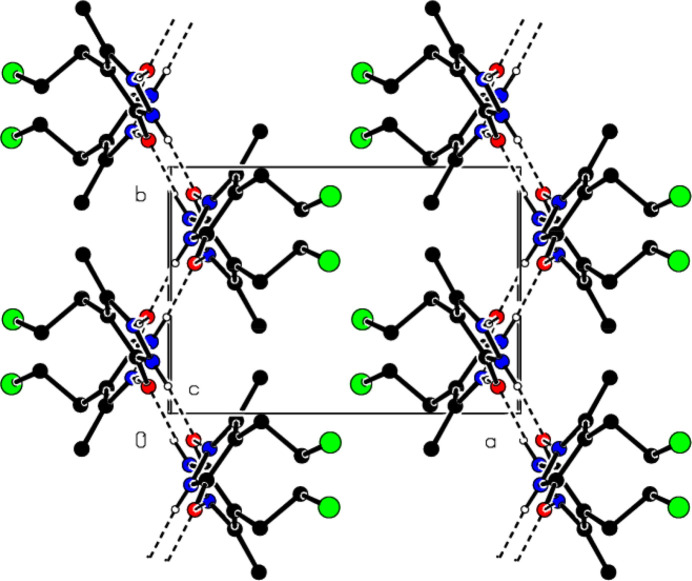
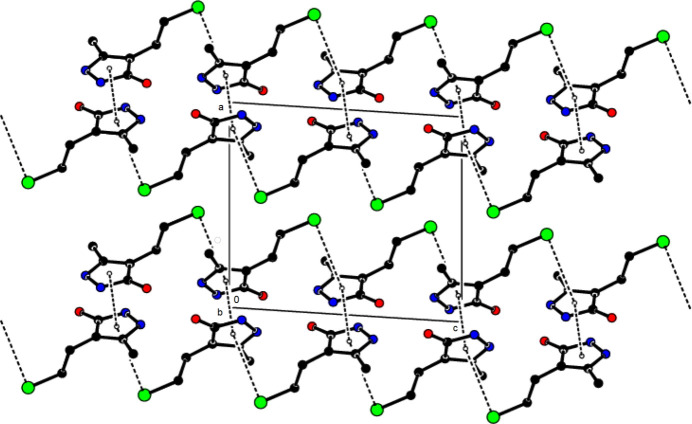
In the crystal, molecular pairs form dimers through N—H⋯O hydrogen bonds. These dimers are linked into ribbons parallel to the (100) plane by further N—H⋯O hydrogen bonds. In addition, π–π and C—Cl⋯π interactions between the ribbons form layers parallel to the (100) plane.
Keywords: crystal structure, hydrogen bonds, dimers, pyrazole ring, Hirshfeld surface analysis
Abstract
In the crystal of the title compound, C6H9ClN2O, molecular pairs form dimers with an R 2 2(8) motif through N—H⋯O hydrogen bonds. These dimers are connect into ribbons parallel to the (100) plane with R 4 4(10) motifs by N—H⋯O hydrogen bonds along the c-axis direction. In addition, π–π [centroid-to-centroid distance = 3.4635 (9) Å] and C—Cl⋯π interactions between the ribbons form layers parallel to the (100) plane. The three-dimensional consolidation of the crystal structure is also ensured by Cl⋯H and Cl⋯Cl interactions between these layers. According to a Hirshfeld surface study, H⋯H (43.3%), Cl⋯H/H⋯Cl (22.1%) and O⋯H/H⋯O (18.7%) interactions are the most significant contributors to the crystal packing.
1. Chemical context
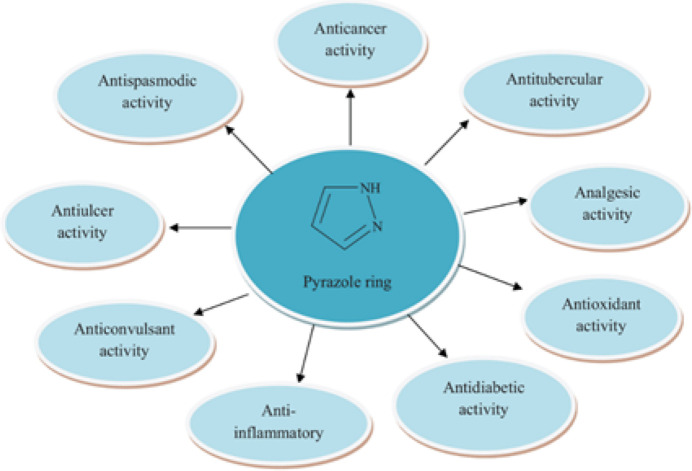
Nitrogen-based heterocyclic compounds are an important branch of organic chemistry. These systems have received increasing attention over the past two decades. Synthetic chemistry is growing extensively with recently developed heterocyclic systems for various research and commercial purposes (Maharramov et al., 2021 ▸, 2022 ▸; Erenler et al., 2022 ▸; Akkurt et al., 2023 ▸). These systems have found wide applications in diverse branches of chemistry, including the chemistry of coordination compounds (Gurbanov et al., 2021 ▸; Mahmoudi et al., 2021 ▸), drug development (Donmez & Turkyılmaz, 2022 ▸; Askerova, 2022 ▸) and material science (Velásquez et al., 2019 ▸; Afkhami et al., 2019 ▸). The pyrazole motif is the most widespread five-membered heteroaromatic ring system in nitrogen heterocycles. It is an essential structural motif present in many natural bioactive molecules such as l-α-amino-β-(pyrazolyl-N)-propanoic acid, withasomnine, pyrazofurin, pyrazofurin B, formycin, formycin B, oxoformycin B, nostocine A, fluviols (A, B, C, D and E), pyrazole-3(5)-carboxylic acid, 4-Methyl pyrazole-3(5)-carboxylic acid, 3-n-nonylpyrazole (Khalilov et al., 2022 ▸; Kumar et al., 2013 ▸; Sobhi & Faisal, 2023 ▸). The pyrazole ring incorporating derivatives with various biological activities (Singh et al., 2023 ▸), such as anticonvulsant, antidiabetic, anti-inflammatory, antioxidant, anticancer, antitubercular, antiulcer activities and other properties has been reviewed recently (Fig. 1 ▸).
Figure 1.
The biological activities of compounds incorporating the pyrazole motif.
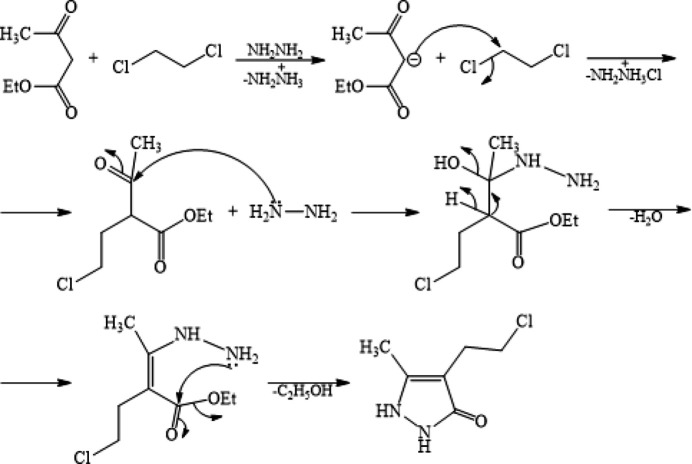
On the other hand, the incorporation of various pharmacophore groups in a pyrazole scaffold has led to the development of best-selling drugs such as ibrutinib, ruxolitinib, axitinib, niraparib and baricitinib (Atalay et al., 2022 ▸; Alam, 2023 ▸). Thus, in the framework of our studies in heterocyclic chemistry (Naghiyev et al., 2020 ▸, 2021 ▸, 2022 ▸), we herein report the crystal structure and Hirshfeld surface analysis of the title compound, 4-(2-chloroethyl)-5-methyl-1,2-dihydropyrazol-3-one, for which the proposed reaction mechanism is shown in Fig. 2 ▸.
Figure 2.
The proposed reaction mechanism for the formation of the title compound.
2. Structural commentary
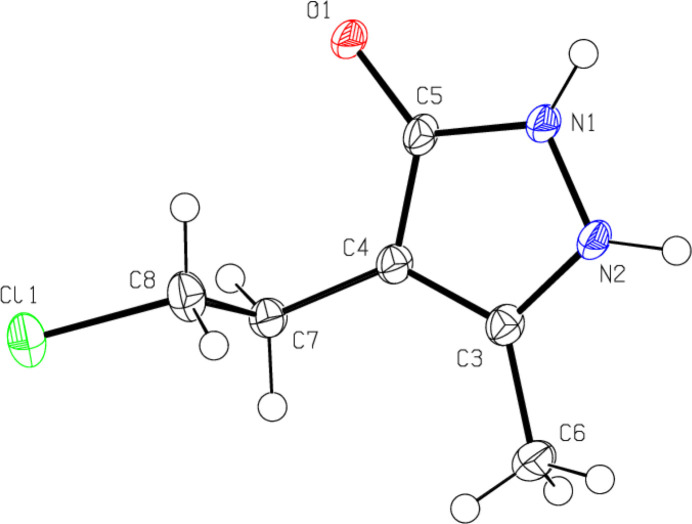
In the title compound (Fig. 3 ▸), the pyrazoline ring (N1/N2/C3–C5) has an essentially planar conformation [maximum deviation = 0.006 (1) Å for N1]. The C3—C4—C7—C8 and C4—C7—C8—Cl1 torsion angles are 105.67 (19) and 172.38 (11)°, respectively. The geometric parameters of the title compound are normal and comparable to those of related compounds given in the Database survey section.
Figure 3.
The molecular structure of the title compound, showing the atom labelling and displacement ellipsoids drawn at the 50% probability level.
3. Supramolecular features and Hirshfeld surface analysis
In the crystal, molecular pairs form dimers with an
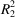 (8) motif (Bernstein et al., 1995 ▸) through N—H⋯O hydrogen bonds (Table 1 ▸ and Fig. 4 ▸). These dimers are also connected into ribbons parallel to the (100) plane by forming N—H⋯O hydrogen bonds with
(8) motif (Bernstein et al., 1995 ▸) through N—H⋯O hydrogen bonds (Table 1 ▸ and Fig. 4 ▸). These dimers are also connected into ribbons parallel to the (100) plane by forming N—H⋯O hydrogen bonds with
 (10) motifs along the c-axis direction (Figs. 5 ▸ and 6 ▸). In addition, π–π [Cg1⋯Cg1i = 3.4635 (9) Å, slippage = 0.511 Å; symmetry code: (i) − x, 1 − y, 1 − z; Cg1 is a centroid of the pyrazole ring (N1/N2/C3–C5)] and C—Cl⋯π [C8—Cl1⋯Cg1ii: C8—Cl1 = 1.8040 (18) Å, Cl1⋯Cg1ii = 3.8386 (8) Å, C8—Cl1⋯Cg1ii = 84.57 (6)°; symmetry code: (ii) x,
(10) motifs along the c-axis direction (Figs. 5 ▸ and 6 ▸). In addition, π–π [Cg1⋯Cg1i = 3.4635 (9) Å, slippage = 0.511 Å; symmetry code: (i) − x, 1 − y, 1 − z; Cg1 is a centroid of the pyrazole ring (N1/N2/C3–C5)] and C—Cl⋯π [C8—Cl1⋯Cg1ii: C8—Cl1 = 1.8040 (18) Å, Cl1⋯Cg1ii = 3.8386 (8) Å, C8—Cl1⋯Cg1ii = 84.57 (6)°; symmetry code: (ii) x,
 − y,
− y,
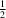 + z] interactions between the ribbons form layers parallel to the (100) plane. The three-dimensional consolidation of the crystal structure is also ensured by the Cl⋯H and Cl⋯Cl interactions [(C8)Cl1⋯H6B
iii = 3.12 (3) Å, C8—Cl1⋯H6B
iii = 135.3 (6)° and (C8)Cl1⋯Cl1iv = 3.5071 (7) Å, C8—Cl1⋯Cl1iv = 161.79 (7)°; symmetry codes: (iii) 1 − x,
+ z] interactions between the ribbons form layers parallel to the (100) plane. The three-dimensional consolidation of the crystal structure is also ensured by the Cl⋯H and Cl⋯Cl interactions [(C8)Cl1⋯H6B
iii = 3.12 (3) Å, C8—Cl1⋯H6B
iii = 135.3 (6)° and (C8)Cl1⋯Cl1iv = 3.5071 (7) Å, C8—Cl1⋯Cl1iv = 161.79 (7)°; symmetry codes: (iii) 1 − x,
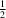 + y,
+ y,
 − z; (iv) 1 − x, 1 − y, 2 − z] between these layers (Table 2 ▸; Fig. 7 ▸).
− z; (iv) 1 − x, 1 − y, 2 − z] between these layers (Table 2 ▸; Fig. 7 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1i | 0.88 (3) | 1.81 (3) | 2.6861 (18) | 174 (2) |
| N2—H2⋯O1ii | 0.92 (3) | 1.75 (3) | 2.6772 (17) | 177 (2) |
Symmetry codes: (i)
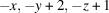 ; (ii)
; (ii)
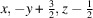 .
.
Figure 4.
View of the N—H⋯O hydrogen bonds of the title compound down the a-axis.
Figure 5.
View of the N—H⋯O hydrogen bonds of the title compound down the b-axis.
Figure 6.
View of the N—H⋯O hydrogen bonds of the title compound down the c-axis.
Table 2. Summary of short interatomic contacts (Å) in the title compound.
| Cl1⋯H6B | 3.12 | 1 − x,
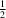 + y,
+ y,
 − z
− z
|
| Cl1⋯Cl1 | 3.51 | 1 − x, 1 − y, 2 − z |
| H1⋯O1 | 1.80 | −x, 2 − y, 1 − z |
| H6C⋯O1 | 2.89 | −x, 1 − y, 1 − z |
| O1⋯H2 | 1.76 |
x,
 − y,
− y,
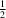 + z
+ z
|
| H6A⋯H7B | 2.60 |
x,
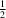 − y, −
− y, −
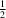 + z
+ z
|
Figure 7.
View of the π-π- and C—Cl⋯π interactions of the title compound down the b-axis.
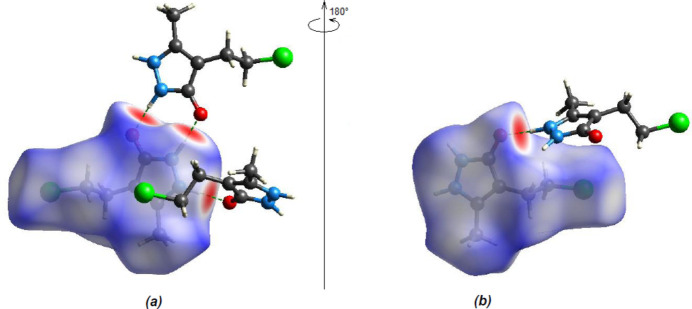
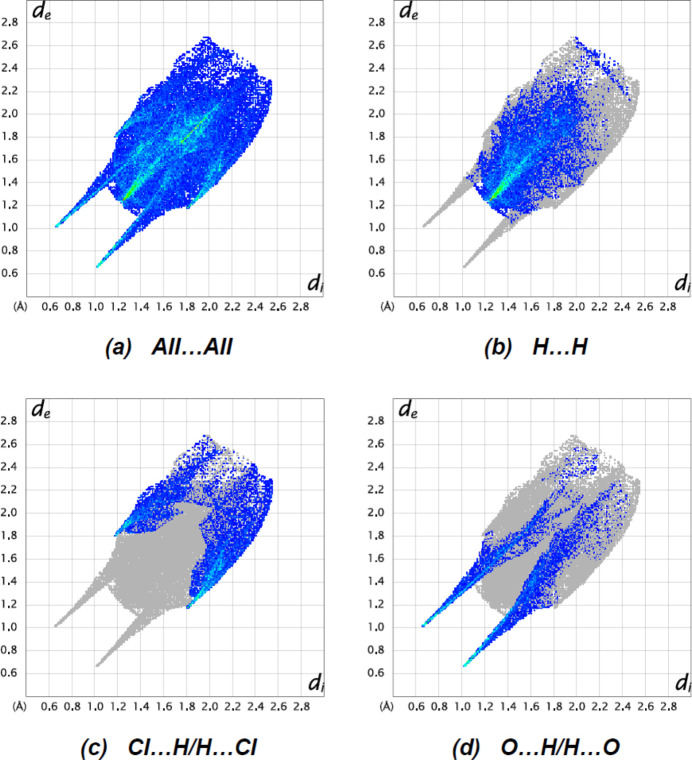
To quantify the intermolecular interactions in the crystal, two-dimensional fingerprint plots and Hirshfeld surfaces were produced using Crystal Explorer 17.5 (Spackman et al., 2021 ▸). Fig. 8 ▸ shows the mapping of the Hirshfeld surfaces over d norm in the range −0.7296 (red) to +1.3271 (blue) a.u. The interactions given in Tables 1 ▸ and 2 ▸ play a key role in the molecular packing of the title compound. H⋯H is the most significant interatomic contact because it contributes the most to the crystal packing (43.3%, Fig. 9 ▸ b). Other significant contributions are made by Cl⋯H/H⋯Cl (22.1%, Fig. 9 ▸ c) and O⋯H/H⋯O (18.7%, Fig. 9 ▸ d) interactions. The following interactions make minor contributions: Cl⋯C/C⋯Cl (2.4%), C⋯H/H⋯C (2.6%), N⋯H/H⋯N (4.3%), N⋯C/C⋯N (3.4%), Cl⋯N/N⋯Cl (0.7%), and C⋯C (0.7%).
Figure 8.
(a) Front and (b) back sides of the three-dimensional Hirshfeld surface of the title compound mapped over d norm, with a fixed colour scale of −0.7296 to +1.3271 a.u.
Figure 9.
The two-dimensional fingerprint plots of the title compound, showing (a) all interactions, and delineated into (b) H⋯H, (c) Cl⋯H/H⋯Cl and (d) O⋯H/H⋯O interactions. [d e and d i represent the distances from a point on the Hirshfeld surface to the nearest atoms outside (external) and inside (internal) the surface, respectively].
4. Database survey
A search of the Cambridge Structural Database (CSD, Version 5.43, last update November 2022; Groom et al., 2016 ▸) for the central five-membered ring 2,3-dihydro-1H-pyrazole yielded six compounds related to the title compound, viz. 3-methyl-5-(3-methylphenoxy)-1-phenyl-1H-pyrazole-4-carbaldehyde (CSD refcode TERZAV; Archana, et al., 2022 ▸), N-{3-cyano-1-[2,6-dichloro-4-(trifluoromethyl)phenyl]-4-(ethylsulfanyl)-1H-pyrazol-5-yl}-2,2,2-trifluoroacetamide (FERPOL; Priyanka et al., 2022 ▸), 4-[3-(4-hydroxyphenyl)-4,5-dihydro-1H-pyrazol-5-yl]-2-methoxyphenol monohydrate (KOXGAI; Duong Khanh et al., 2019 ▸), 5-chloro-N 1-(5-phenyl-1H-pyrazol-3-yl)benzene-1,2-diamine (CAXZUZ; Yartsev et al., 2017 ▸), 5-(butylamino)-3-methyl-1-(pyridin-2-yl)-1H-pyrazole-4-carbaldehyde (EYEHEX; Macías et al., 2016 ▸) and 5-amino-1-(2-chlorophenyl)-1H-pyrazole-4-carbonitrile (AFIJOP; Lin et al., 2007 ▸).
The molecular packing of TERZAV features aromatic π–π stacking and weak C—H⋯π interactions. In the crystal of FERPOL, strong N—H⋯O hydrogen bonds link the molecules into chains that extend parallel to the a-axis. In the crystal of KOXGAI, the molecules are connected into chains running in the b-axis direction by O—H⋯N hydrogen bonding. Parallel chains interact through N—H⋯O hydrogen bonds and π–π stacking of the trisubstituted phenyl rings. In the crystal of CAXZUZ, the A and B molecules are linked by two pairs of N—H⋯N hydrogen bonds, forming A–B dimers. These are further linked by a fifth N—H⋯N hydrogen bond, forming tetramer-like units that stack along the a-axis direction, forming columns, which are in turn linked by C—H⋯π interactions, forming layers parallel to the ac plane. The supramolecular structure of EYEHEX assembly has a three-dimensional arrangement controlled mainly by weak C—H⋯O and C—H⋯π interactions. The crystal structure of AFIJOP is consolidated by two N—H⋯N hydrogen bonds.
5. Synthesis and crystallization
Acetoacetic ether (7.7 mmol), dichloroethane (7.7 mmol) and hydrazine hydrate (15.4 mmol) were dissolved in 40 ml of ethanol and the reaction mixture was refluxed for 4 h. Then the reaction mixture was cooled to room temperature with the formation of white crystals. The crystals were separated by filtration and recrystallized from an ethanol–water mixture (m.p. 499–500 K, yield 78%).
1H NMR (300 MHz, DMSO-d 6, ppm.): 2.06 (s, 3H, CH3); 2.64 (t, 2H, CH2, H-H J 2 = 7.2); 3.49 (s, 2H, 2NH); 3.58 (t, 2H, ClCH2, H-H J 2 = 7.2). 13C NMR (75 MHz, DMSO-d 6, ppm.): 10.28 (CH3), 26.02 (CH2), 44.91 (CH2Cl), 97.63 (C tert. =), 160.12 (HN—C tert. =), 162.34 (N—C=O).
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The C-bound H atoms were placed in calculated positions (0.95–0.99 Å) and refined as riding with U iso(H) = 1.2 or 1.5U eq(C). The N-bound H atoms were located in a difference map and freely refined.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C6H9ClN2O |
| M r | 160.60 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 100 |
| a, b, c (Å) | 9.8420 (2), 6.9145 (2), 11.1807 (2) |
| β (°) | 93.618 (2) |
| V (Å3) | 759.36 (3) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 3.92 |
| Crystal size (mm) | 0.20 × 0.12 × 0.06 |
| Data collection | |
| Diffractometer | XtaLAB Synergy, Dualflex, HyPix |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2022 ▸) |
| T min, T max | 0.513, 0.750 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 6642, 1532, 1467 |
| R int | 0.027 |
| (sin θ/λ)max (Å−1) | 0.633 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.036, 0.097, 1.05 |
| No. of reflections | 1532 |
| No. of parameters | 127 |
| H-atom treatment | All H-atom parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.28, −0.41 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989024000835/nx2004sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989024000835/nx2004Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989024000835/nx2004Isup3.cml
CCDC reference: 2327646
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Authors contributions are as follows. Conceptualization, IGM, ANK and EAD; methodology, AB and MA; investigation, VNK and FNN; writing (original draft), MA, AB and ANK; writing (review and editing of the manuscript), MA and ANK; visualization, MA, IGM and FNN; funding acquisition, VNK, AB and FNN; resources, AB, VNK and MA; supervision, MA and ANK.
supplementary crystallographic information
Crystal data
| C6H9ClN2O | F(000) = 336 |
| Mr = 160.60 | Dx = 1.405 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54184 Å |
| a = 9.8420 (2) Å | Cell parameters from 4592 reflections |
| b = 6.9145 (2) Å | θ = 4.5–77.6° |
| c = 11.1807 (2) Å | µ = 3.92 mm−1 |
| β = 93.618 (2)° | T = 100 K |
| V = 759.36 (3) Å3 | Prism, colourless |
| Z = 4 | 0.20 × 0.12 × 0.06 mm |
Data collection
| XtaLAB Synergy, Dualflex, HyPix diffractometer | 1467 reflections with I > 2σ(I) |
| Radiation source: micro-focus sealed X-ray tube | Rint = 0.027 |
| ω scans | θmax = 77.5°, θmin = 4.5° |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2022) | h = −12→12 |
| Tmin = 0.513, Tmax = 0.750 | k = −8→7 |
| 6642 measured reflections | l = −14→8 |
| 1532 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: difference Fourier map |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.036 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.097 | All H-atom parameters refined |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0501P)2 + 0.606P] where P = (Fo2 + 2Fc2)/3 |
| 1532 reflections | (Δ/σ)max = 0.001 |
| 127 parameters | Δρmax = 0.28 e Å−3 |
| 0 restraints | Δρmin = −0.41 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.45324 (5) | 0.61848 (7) | 0.86496 (4) | 0.03447 (18) | |
| O1 | 0.06705 (13) | 0.89161 (17) | 0.64445 (10) | 0.0231 (3) | |
| N1 | 0.05777 (14) | 0.7905 (2) | 0.44644 (12) | 0.0197 (3) | |
| H1 | 0.013 (3) | 0.890 (4) | 0.413 (2) | 0.040 (7)* | |
| N2 | 0.11129 (14) | 0.6437 (2) | 0.38217 (12) | 0.0195 (3) | |
| H2 | 0.096 (2) | 0.636 (3) | 0.300 (2) | 0.038 (6)* | |
| C3 | 0.18687 (16) | 0.5302 (2) | 0.45765 (14) | 0.0189 (3) | |
| C4 | 0.18362 (16) | 0.6035 (2) | 0.57245 (14) | 0.0181 (3) | |
| C5 | 0.10154 (16) | 0.7717 (2) | 0.56311 (13) | 0.0188 (3) | |
| C6 | 0.2560 (2) | 0.3553 (3) | 0.41307 (16) | 0.0254 (4) | |
| H6A | 0.282 (3) | 0.372 (4) | 0.331 (3) | 0.058 (8)* | |
| H6B | 0.338 (3) | 0.328 (5) | 0.458 (3) | 0.065 (9)* | |
| H6C | 0.205 (3) | 0.250 (5) | 0.414 (3) | 0.066 (9)* | |
| C7 | 0.25721 (17) | 0.5342 (2) | 0.68559 (14) | 0.0205 (3) | |
| H7A | 0.194 (2) | 0.527 (3) | 0.7508 (18) | 0.021 (5)* | |
| H7B | 0.295 (2) | 0.406 (3) | 0.676 (2) | 0.029 (5)* | |
| C8 | 0.37260 (18) | 0.6724 (3) | 0.71941 (16) | 0.0254 (4) | |
| H8A | 0.340 (2) | 0.807 (4) | 0.724 (2) | 0.031 (6)* | |
| H8B | 0.443 (2) | 0.665 (4) | 0.664 (2) | 0.037 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0401 (3) | 0.0366 (3) | 0.0249 (3) | 0.00431 (18) | −0.01242 (19) | −0.00157 (17) |
| O1 | 0.0374 (7) | 0.0209 (6) | 0.0110 (5) | 0.0082 (5) | 0.0007 (5) | −0.0010 (4) |
| N1 | 0.0297 (7) | 0.0180 (7) | 0.0114 (6) | 0.0045 (5) | 0.0008 (5) | −0.0006 (5) |
| N2 | 0.0278 (7) | 0.0194 (7) | 0.0115 (7) | 0.0026 (5) | 0.0013 (5) | −0.0028 (5) |
| C3 | 0.0237 (7) | 0.0179 (7) | 0.0153 (7) | −0.0011 (6) | 0.0027 (6) | 0.0008 (6) |
| C4 | 0.0238 (7) | 0.0177 (7) | 0.0128 (7) | 0.0008 (6) | 0.0024 (6) | 0.0017 (6) |
| C5 | 0.0273 (8) | 0.0183 (7) | 0.0110 (7) | −0.0002 (6) | 0.0023 (6) | 0.0003 (6) |
| C6 | 0.0333 (9) | 0.0233 (8) | 0.0198 (9) | 0.0057 (7) | 0.0033 (7) | −0.0036 (7) |
| C7 | 0.0280 (8) | 0.0192 (8) | 0.0144 (8) | 0.0036 (6) | 0.0015 (6) | 0.0016 (6) |
| C8 | 0.0264 (8) | 0.0315 (9) | 0.0180 (8) | 0.0006 (7) | −0.0019 (6) | 0.0029 (7) |
Geometric parameters (Å, º)
| Cl1—C8 | 1.8040 (18) | C4—C7 | 1.496 (2) |
| O1—C5 | 1.2920 (19) | C6—H6A | 0.97 (3) |
| N1—C5 | 1.354 (2) | C6—H6B | 0.95 (3) |
| N1—N2 | 1.3685 (19) | C6—H6C | 0.88 (3) |
| N1—H1 | 0.88 (3) | C7—C8 | 1.514 (2) |
| N2—C3 | 1.342 (2) | C7—H7A | 0.99 (2) |
| N2—H2 | 0.92 (3) | C7—H7B | 0.97 (2) |
| C3—C4 | 1.382 (2) | C8—H8A | 0.99 (2) |
| C3—C6 | 1.489 (2) | C8—H8B | 0.95 (2) |
| C4—C5 | 1.416 (2) | ||
| C5—N1—N2 | 108.95 (13) | H6A—C6—H6B | 105 (2) |
| C5—N1—H1 | 126.8 (17) | C3—C6—H6C | 113 (2) |
| N2—N1—H1 | 123.6 (17) | H6A—C6—H6C | 107 (3) |
| C3—N2—N1 | 108.66 (13) | H6B—C6—H6C | 107 (3) |
| C3—N2—H2 | 129.9 (15) | C4—C7—C8 | 108.91 (14) |
| N1—N2—H2 | 121.4 (15) | C4—C7—H7A | 110.2 (12) |
| N2—C3—C4 | 108.96 (14) | C8—C7—H7A | 110.3 (12) |
| N2—C3—C6 | 120.74 (15) | C4—C7—H7B | 111.5 (13) |
| C4—C3—C6 | 130.29 (15) | C8—C7—H7B | 108.6 (13) |
| C3—C4—C5 | 106.22 (14) | H7A—C7—H7B | 107.3 (18) |
| C3—C4—C7 | 128.97 (15) | C7—C8—Cl1 | 112.04 (12) |
| C5—C4—C7 | 124.69 (14) | C7—C8—H8A | 111.5 (13) |
| O1—C5—N1 | 122.31 (15) | Cl1—C8—H8A | 105.7 (13) |
| O1—C5—C4 | 130.49 (14) | C7—C8—H8B | 111.5 (15) |
| N1—C5—C4 | 107.19 (13) | Cl1—C8—H8B | 106.1 (15) |
| C3—C6—H6A | 111.6 (17) | H8A—C8—H8B | 109.7 (19) |
| C3—C6—H6B | 111.9 (19) | ||
| C5—N1—N2—C3 | 0.98 (18) | N2—N1—C5—C4 | −1.24 (18) |
| N1—N2—C3—C4 | −0.30 (18) | C3—C4—C5—O1 | 179.99 (17) |
| N1—N2—C3—C6 | 178.79 (15) | C7—C4—C5—O1 | −3.7 (3) |
| N2—C3—C4—C5 | −0.45 (18) | C3—C4—C5—N1 | 1.04 (18) |
| C6—C3—C4—C5 | −179.43 (17) | C7—C4—C5—N1 | 177.36 (15) |
| N2—C3—C4—C7 | −176.56 (16) | C3—C4—C7—C8 | 105.67 (19) |
| C6—C3—C4—C7 | 4.5 (3) | C5—C4—C7—C8 | −69.8 (2) |
| N2—N1—C5—O1 | 179.70 (15) | C4—C7—C8—Cl1 | 172.38 (11) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1i | 0.88 (3) | 1.81 (3) | 2.6861 (18) | 174 (2) |
| N2—H2···O1ii | 0.92 (3) | 1.75 (3) | 2.6772 (17) | 177 (2) |
Symmetry codes: (i) −x, −y+2, −z+1; (ii) x, −y+3/2, z−1/2.
Funding Statement
This paper was supported by Baku State University and the RUDN University Strategic Academic Leadership Program.
References
- Afkhami, F. A., Mahmoudi, G., Khandar, A. A., Franconetti, A., Zangrando, E., Qureshi, N., Lipkowski, J., Gurbanov, A. V. & Frontera, A. (2019). Eur. J. Inorg. Chem. pp. 262–270.
- Akkurt, M., Maharramov, A. M., Shikhaliyev, N. G., Qajar, A. M., Atakishiyeva, G., Shikhaliyeva, I. M., Niyazova, A. A. & Bhattarai, A. (2023). UNEC J. Eng. Appl. Sci. 3, 33–39.
- Alam, M. A. (2023). Future Med. Chem. 15, 2011–2023. [DOI] [PMC free article] [PubMed]
- Archana, S. D., Nagma Banu, H. A., Kalluraya, B., Yathirajan, H. S., Balerao, R. & Butcher, R. J. (2022). IUCrData, 7, x220924. [DOI] [PMC free article] [PubMed]
- Askerova, U. F. (2022). UNEC J. Eng. Appl. Sci, 2, 58–64.
- Atalay, V. E., Atish, I. S., Shahin, K. F., Kashikchi, E. S. & Karahan, M. (2022). UNEC J. Eng. Appl. Sci. 2, 33–40.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Donmez, M. & Turkyılmaz, M. (2022). UNEC J. Eng. Appl. Sci, 2, 43–48.
- Duong Khanh, L., Hanh Trinh Thi, M., Quynh Bui Thi, T., Vu Quoc, T., Nguyen Thien, V. & Van Meervelt, L. (2019). Acta Cryst. E75, 1590–1594. [DOI] [PMC free article] [PubMed]
- Erenler, R., Dag, B. & Ozbek, B. B. (2022). UNEC J. Eng. Appl. Sci. 2, 26–32.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Gurbanov, A. V., Mertsalov, D. F., Zubkov, F. I., Nadirova, M. A., Nikitina, E. V., Truong, H. H., Grigoriev, M. S., Zaytsev, V. P., Mahmudov, K. T. & Pombeiro, A. J. L. (2021). Crystals, 11, 112.
- Khalilov, A. N., Khrustalev, V. N., Tereshina, T. A., Akkurt, M., Rzayev, R. M., Akobirshoeva, A. A. & Mamedov, İ. G. (2022). Acta Cryst. E78, 525–529. [DOI] [PMC free article] [PubMed]
- Kumar, V., Kaur, K., Gupta, G. K. & Sharma, A. K. (2013). Eur. J. Med. Chem. 69, 735–753. [DOI] [PubMed]
- Lin, Q.-L., Zhong, P. & Hu, M.-L. (2007). Acta Cryst. E63, o3813.
- Macías, M. A., Orrego-Hernández, J. & Portilla, J. (2016). Acta Cryst. E72, 1672–1674. [DOI] [PMC free article] [PubMed]
- Maharramov, A. M., Shikhaliyev, N. G., Zeynalli, N. R., Niyazova, A. A., Garazade, Kh. A. & Shikhaliyeva, I. M. (2021). UNEC J. Eng. Appl. Sci. 1, 5–11.
- Maharramov, A. M., Suleymanova, G. T., Qajar, A. M., Niyazova, A. A., Ahmadova, N. E., Shikhaliyeva, I. M., Garazade, Kh. A., Nenajdenko, V. G. & Shikaliyev, N. G. (2022). UNEC J. Eng. Appl. Sci. 2, 64–73.
- Mahmoudi, G., Zangrando, E., Miroslaw, B., Gurbanov, A. V., Babashkina, M. G., Frontera, A. & Safin, D. A. (2021). Inorg. Chim. Acta, 519, 120279.
- Naghiyev, F. N., Akkurt, M., Askerov, R. K., Mamedov, I. G., Rzayev, R. M., Chyrka, T. & Maharramov, A. M. (2020). Acta Cryst. E76, 720–723. [DOI] [PMC free article] [PubMed]
- Naghiyev, F. N., Khrustalev, V. N., Novikov, A. P., Akkurt, M., Rzayev, R. M., Akobirshoeva, A. A. & Mamedov, I. G. (2022). Acta Cryst. E78, 554–558. [DOI] [PMC free article] [PubMed]
- Naghiyev, F. N., Tereshina, T. A., Khrustalev, V. N., Akkurt, M., Rzayev, R. M., Akobirshoeva, A. A. & Mamedov, İ. G. (2021). Acta Cryst. E77, 516–521. [DOI] [PMC free article] [PubMed]
- Priyanka, P., Jayanna, B. K., Sunil Kumar, Y. C., Shreenivas, M. T., Srinivasa, G. R., Divakara, T. R., Yathirajan, H. S. & Parkin, S. (2022). Acta Cryst. E78, 1084–1088. [DOI] [PMC free article] [PubMed]
- Rigaku OD (2022). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Singh, S., Tehlan, S. & Kumar Verma, P. (2023). Mini Rev. Med. Chem. 23, 2142–2165. [DOI] [PubMed]
- Sobhi, R. M. & Faisal, R. M. (2023). UNEC J. Eng. Appl. Sci. 3, 21–32.
- Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D. & Spackman, M. A. (2021). J. Appl. Cryst. 54, 1006–1011. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Velásquez, J. D., Mahmoudi, G., Zangrando, E., Gurbanov, A. V., Zubkov, F. I., Zorlu, Y., Masoudiasl, A. & Echeverría, J. (2019). CrystEngComm, 21, 6018–6025.
- Yartsev, Y., Palchikov, V., Gaponov, A. & Shishkina, S. (2017). Acta Cryst. E73, 876–879. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989024000835/nx2004sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989024000835/nx2004Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989024000835/nx2004Isup3.cml
CCDC reference: 2327646
Additional supporting information: crystallographic information; 3D view; checkCIF report