Abstract
Background.
Clinicians increasingly utilize polymyxins for treatment of serious infections caused by multidrug-resistant gram-negative bacteria. Emergence of plasmid-mediated, mobile colistin resistance genes creates potential for rapid spread of polymyxin resistance. We investigated the possible transmission of Klebsiella pneumoniae carrying mcr-1 via duodenoscope and report the first documented healthcare transmission of mcr-1–harboring bacteria in the United States.
Methods.
A field investigation, including screening targeted high-risk groups, evaluation of the duodenoscope, and genome sequencing of isolated organisms, was conducted. The study site included a tertiary care academic health center in Boston, Massachusetts, and extended to community locations in New England.
Results.
Two patients had highly related mcr-1–positive K. pneumoniae isolated from clinical cultures; a duodenoscope was the only identified epidemiological link. Screening tests for mcr-1 in 20 healthcare contacts and 2 household contacts were negative. Klebsiella pneumoniae and Escherichia coli were recovered from the duodenoscope; neither carried mcr-1. Evaluation of the duodenoscope identified intrusion of biomaterial under the sealed distal cap; devices were recalled to repair this defect.
Conclusions.
We identified transmission of mcr-1 in a United States acute care hospital that likely occurred via duodenoscope despite no identifiable breaches in reprocessing or infection control practices. Duodenoscope design flaws leading to transmission of multidrug-resistant organsisms persist despite recent initiatives to improve device safety. Reliable detection of colistin resistance is currently challenging for clinical laboratories, particularly given the absence of a US Food and Drug Administration–cleared test; improved clinical laboratory capacity for colistin susceptibility testing is needed to prevent the spread of mcr-carrying bacteria in healthcare settings.
Keywords: duodenoscope, infection control, medical device safety, mobile colistin resistance, Klebsiella pneumoniae
Contaminated endoscopes can transmit infectious microorganisms, with most transmission attributed to limitations in reprocessing or infection control [1, 2]. In 2014, however, an outbreak of New Delhi metallo-β-lactamase–producing carbapenem-resistant Escherichia coli was caused by persistently contaminated duodenoscopes, and no such deficiencies were found [3]. Following this report, similar outbreaks were identified [4–7]. Investigation by the US Food and Drug Administration (FDA) determined that the complex design of duodenoscopes, in particular their unique cantilevered elevator mechanism, impedes effective reprocessing [8]. Subsequently, all manufacturers released updated, validated reprocessing instructions and some duodenoscope models were recalled for repair [9]. To further reduce the risk of transmission of infectious agents, the FDA provided a set of supplementary duodenoscope reprocessing measures that facilities might consider, including microbiological culturing, ethylene oxide sterilization, use of a liquid chemical sterilant processing system, and repeat high-level disinfection (HLD) [10].
The plasmid-mediated, mobile colistin resistance gene mcr-1 was first reported in 2015 after being recognized in Enterobacteriaceae isolated in China [11]. Subsequently, mobile colistin resistance genes have been identified in Enterobacteriaceae worldwide [12–16], including in isolates from 2 pigs and >25 patients in the United States [17–19]. Most United States mcr-1–positive isolates have been extended-spectrum β-lactamase (ESBL)–producing E. coli or Salmonella enterica isolated from outpatients with recent international travel and limited prior healthcare exposure (US Centers for Disease Control and Prevention [CDC], unpublished data) [18]. The emergence of transmissible colistin resistance is important because clinicians are increasingly turning to polymyxins as a last resort for treatment of serious infections caused by multidrug-resistant gram-negative bacteria, especially carbapenem-resistant Enterobacteriaceae (CRE). The spread of mcr genes into CRE, including in the United States [20], has raised the specter of potentially untreatable infections. To slow the spread of mcr-1, the CDC recommends screening healthcare contacts of infected and colonized patients for asymptomatic carriage and conducting prospective surveillance at inpatient healthcare facilities where mcr-1–positive patients were admitted [21]; among 3 case reports describing such activities, none detected transmission [22–24].
We report the first known transmission of mcr-1–carrying bacteria in a US healthcare setting. This transmission of colistin-resistant Klebsiella pneumoniae was associated with a duodenoscope, and occurred despite adherence to the device manufacturer’s updated reprocessing instructions and implementation of enhanced measures recommended by the FDA; this represents the first documented duodenoscope-linked transmission of an infectious organism reported to the CDC since publication of updated reprocessing guidelines.
METHODS
The activities conducted as part of this investigation were considered routine infection control response at Massachusetts General Hospital (MGH) and routine public health response at the CDC and the Massachusetts Department of Public Health (MDPH); as such, submission to the Partners Human Subjects Committee and the institutional review boards was not required. Persons providing surveillance samples gave verbal consent.
Case Identification and Epidemiologic Investigation
Case Definition
We defined a case as isolation of mcr-1–positive K. pneumoniae from a patient undergoing evaluation at MGH after 16 April 2017, the date when the index patient was admitted. Prior to this investigation, mcr-1 had not been identified at MGH.
Identification of Index Case
The index patient (C1), a Rhode Island resident with recurrent bacterial cholangitis, was admitted to MGH with fever in April 2017 and underwent endoscopic retrograde cholangiopancreatography (ERCP) on the first full day of admission (timeline day 1) (Figure 1). A culture of bile obtained during the procedure grew K. pneumoniae of 2 colony morphotypes. After routine antimicrobial susceptibility testing (AST) results were reported, a clinician requested colistin AST in anticipation of future use given the patient’s medical complexity. Colistin minimum inhibitory concentrations (MICs) were >4 μg/mL, a value above the Clinical and Laboratory Standards Institute (CLSI) epidemiological cutoff value (ECV) for Enterobacteriaceae (wild type, ≤2 μg/mL) [25]. The laboratory alerted the MGH Infection Control Unit and the MDPH of possible mcr-1–mediated colistin resistance. Isolates submitted via the Massachusetts State Public Health Laboratory to the CDC were confirmed by real-time polymerase chain reaction (PCR) to harbor mcr-1.
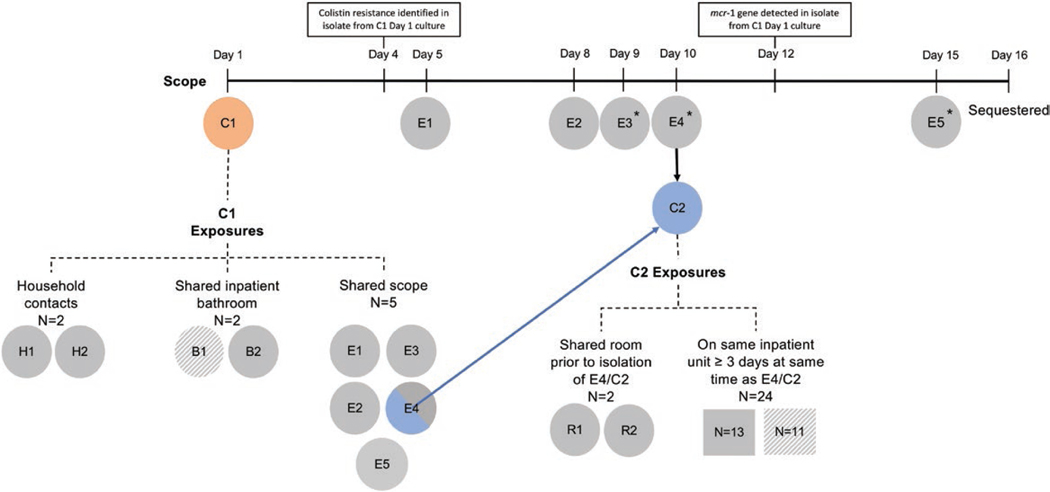
Figure 1.
Exposure investigation timeline and surveillance results. Patients exposed to the duodenoscope are shown below the timeline, from duodenoscope use during case 1’s (C1) procedure on day 1 through sequestration of the scope on day 16. The following designations were used: C for case-patients; E denoting endoscopic retrograde cholangiopancreatography (ERCP) exposure; R denoting roommate exposure; B indicating bathroom exposure, and N indicating patients identified as unit contacts. An asterisk indicates those patients who either had a stent placed during the ERCP (E3 and E4/C2) or who had an indwelling stent that was left in place following the ERCP (E5). Exposed patients who were identified as cases of mcr-1 are shown in color (orange for case 1 [C1], blue for case 2 [E4/C2]). Secondary exposures to C1 and E4/C2 were identified using a definition of shared environment informed by public health recommendations. Together, these individuals constituted the cohort for the exposure investigation. Exposed patients who were tested and had an mcr-1–negative result are shaded in solid gray. Exposed patients for whom no information was available are shown in hatched gray (eg, patient died or was discharged and lost to follow-up). Of the 7 individuals identified as healthcare-associated contacts of C1, all had been discharged from the hospital to home at the time the investigation was initiated, apart from E4/C2. Of the 26 contacts of E4/C2, 9 had been discharged to home, 13 had been transferred to a long-term acute care hospital or rehabilitation hospital, and 4 had died either in the hospital or since discharge; swabs could not be obtained from 11 of these 26 contacts.
Identification of Case 2
On day 21, while the C1 contact investigation was under way, a patient with walled-off pancreatic necrosis who had been admitted on day 7 and undergone ERCP on day 10 had 2 fluid specimens from abdominal flank drains cultured as part of an evaluation for suspected sepsis. This patient was identified as a contact of C1 due to exposure to the same duodenoscope and was designated E4/C2. Each culture grew 2 morphotypes of K. pneumoniae. One morphotype in each culture was found to be resistant to meropenem; reflex AST demonstrated colistin MICs of >4 μg/mL. Given this finding and the epidemiologic link to C1, colistin AST was also performed on the meropenem-susceptible K. pneumoniae isolates; colistin MICs were >4 μg/mL. Both morphotypes from both cultures were positive for mcr-1 by real-time PCR.
Epidemiologic and Contact Investigation
C1 was interviewed about healthcare and community mcr-1 risk factors (Supplementary Table 1); E4/C2 was unable to provide reliable history. The case-patients’ medical records, including clinical histories, contact with staff members and ancillary departments, and movement within the hospital, were systematically reviewed and abstracted to identify shared exposures and other patients at risk for mcr-1 acquisition. Patients considered at risk for mcr-1 acquisition were those who shared a room or bathroom with a case-patient, underwent endoscopy with the same duodenoscope as the case-patients, or overlapped on the same inpatient unit as a case-patient for ≥3 days before case-patient isolation. Persons living with a case-patient were also considered at risk for mcr-1 acquisition. Contacts were approached for rectal swab surveillance culturing to assess asymptomatic carriage.
Duodenoscope Investigation
Duodenoscope reprocessing at MGH was conducted using the manufacturer’s updated instructions for use [26] and also included: a second HLD step in the automatic endoscope reprocessor (Medivators, Inc, Minneapolis, Minnesota), testing for residual bioburden after manual cleaning and before first HLD (Channel Check, Healthmark, Fraser, Michigan), and periodic culturing and sequestering of duodenoscopes. The duodenoscope used for ERCP in C1 and E4/C2 was a closed-elevator wire channel duodenoscope in service since October 2014 and leased from the manufacturer. It was sequestered on day 16; surveillance samples to assess for bacterial contamination were collected by MGH staff following CDC guidance [27] and sent to a third-party laboratory for culture-based analysis (Aerobiology Laboratory Associates, Denver, Colorado). Additional sampling was performed at the CDC 86 days after last use of the duodenoscope [27, 28]. The duodenoscope was then returned to the manufacturer (PENTAX Medical, Montvale, New Jersey) for evaluation for mechanical defects.
Laboratory Analysis of Samples
Clinical, surveillance, and environmental samples were processed at the MGH Microbiology Laboratory and the CDC (Supplementary Appendix).
Genome Sequencing, Assembly, and Analysis
Genome sequencing, assembly, and analysis were conducted at the CDC using standard methods (Supplementary Appendix).
RESULTS
Case Identification and Contact Investigation
Interview of C1 revealed that he had traveled to multiple Caribbean islands 4 months before admission to MGH; Caribbean travel has also been reported among other individuals in the United States from whom bacteria harboring mcr-1 have been isolated [22, 24]. E4/C2 did not have documented international travel or recent hospitalizations. C1 and E4/C2 had no identified shared exposures (eg, providers, locations, procedures) except for ERCP with the same duodenoscope.
Figure 1 illustrates the nature of exposure for individuals identified through the contact investigation. Attempts were made to reach each contact to solicit consent for collection of a surveillance rectal swab (Supplementary Appendix). Over a period of 7 weeks, surveillance rectal swab samples were collected from both household contacts (H1 and H2) and 6 of 7 identified healthcare-associated contacts of C1 (B2, E1, E2, E3, E4/C2, and E5), and from 15 of 26 identified healthcare contacts of E4/C2 (R1, R2, and 13 of 24 individuals who had been admitted to the same patient care unit as E4/C2 for ≥3 days); except those collected from E4/C2, all were negative for mcr-1 or gram-negative bacilli with elevated colistin MICs. Long-term follow-up of cases to day 120 was conducted (Table 1 and Supplementary Appendix).
Table 1.
Characteristics of Specimens and Klebsiella pneumoniae Isolates Obtained From Case 1 (C1) and Case 2 (E4/C2)
| Isolate or Specimen Number | Timeline Day | Specimen Source | Klebsiella pneumoniae Colistin BMD MICs, μg/mLa,b | mcr-1 Gene Detected | WGS Performed |
|---|---|---|---|---|---|
| Case 1 | |||||
| C1-S1-01.1 | 1 | Bile | > 4; 8 | Yes | Yes |
| C1-S1-01.2 | 1 | Bile | > 4; 8 | Yes | Yes |
| C1-S2-01 | 30 | Rectal swab | ND | Yes | No |
| C1-S3-01 | 64 | Rectal swab | ND | Yes | No |
| C1-S4-01.1 | 85 | Rectal swab | ND | Yes | Yes |
| C1-S4-01.2 | 85 | Rectal swab | ND | Yes | Yes |
| C1-S4-01.3 | 85 | Rectal swab | ND | Yes | Yes |
| C1-S4-01.4 | 85 | Rectal swab | ND | Yes | Yes |
| Case 2 | |||||
| C2-S1-01.1 | 21 | Right abdominal flank drain | > 4; 8 | Yes | Yes |
| C2-S1-02.1 | 21 | Left abdominal flank drain | > 4; 8 | Yes | Yes |
| C2-S1-01.0 | 21 | Right abdominal flank drain | > 4 | Yes | No |
| C2-S1-02.0 | 21 | Left abdominal flank drain | > 4 | Yes | No |
| C2-S2-01.1 | 24 | Right lower quadrant abdominal collection | > 4 | Yes | Yes |
| C2-S2-01.2 | 24 | Right lower quadrant abdominal collection | > 4 | Yes | Yes |
| C2-S3-01 | 82 | Rectal swab | ND | Yes | No |
| C2-S3-02 | 82 | Rectal swab | 4 | Yes | No |
| C2-S3-02.1 | 82 | Abdominal fluid drain | ND | Yes | Yes |
| C2-S3-02.2 | 82 | Abdominal fluid drain | ND | Yes | Yes |
| C2-S3-02.3 | 82 | Abdominal fluid drain | ND | Yes | Yes |
| C2-S3-02.4 | 82 | Abdominal fluid drain | ND | Yes | Yes |
| C2-S4-01 | 120 | Rectal swab | NA | No | No |
Abbreviations: BMD, broth microdilution; MIC, minimum inhibitory concentration; NA, not applicable; ND, not done; WGS, whole-genome sequencing.
Where 2 colistin MICs are listed, the first is that determined at Massachusetts General Hospital (MGH) using a Sensititre BMD panel and the second is that determined at the US Centers for Disease Control and Prevention using reference broth microdilution. Where a single colistin MIC is listed, it is the MIC determined at MGH using a Sensititre BMD panel.
Once the phenotypic colistin susceptibility testing results and molecular mechanisms of colistin resistance of representative clinical isolates from both case-patients had been determined, additional testing to determine colistin MICs of the mcr-1-positive organisms isolated from the case-patients during surveillance for persistent colonization was not warranted for the purposes of the investigation.
Infection Control and Environment Assessment
MGH Infection Control Unit staff reviewed endoscope reprocessing, including automatic endoscope reprocessor logs and ChannelCheck verification test results, with no failures noted. The duodenoscope used for ERCP in C1 and E4/C2 had undergone complete reprocessing between each procedure in which it was used, and ChannelCheck assays were negative after each manual disinfection. Review of the duodenoscope preventive maintenance record demonstrated no significant findings; the scope had undergone preventive maintenance 3 months before C1’s ERCP. An on-site evaluation by MDPH on behalf of the Centers for Medicare and Medicaid Services did not identify breaches in relevant infection control or device reprocessing practices.
Laboratory Analysis of Case Isolates
Phenotypic and genotypic AST results for the clinical K. pneumoniae isolates from C1 and E4/C2, as well as the results of serial rectal surveillance swab tests for each case-patient, are shown in Table 1 and Supplementary Table 2. Isolates from both case-patients were multidrug-resistant and positive for ESBL production; a subset of isolates from E4/C2 was carbapenem-resistant.
Six K. pneumoniae isolates from C1 and 8 from E4/C2 underwent whole-genome sequencing (WGS); all were determined to be sequence type (ST) 15 carrying blaSHV-28 and blaOXA-1 β-lactamase genes and the catB4 chloramphenicol resistance gene. In all case-patient isolates, mcr-1 had a unique ATA start codon that has not been previously reported. In addition to the plasmid-mediated mcr-1 gene, isolates from both case-patients shared chromosomal mutations (an L26Q and an identical early truncation) of the phoP gene previously described to confer polymyxin resistance.
Pairwise comparisons revealed that case-patient isolates differed by 0–23 single-nucleotide variants (SNVs) across a core genome of 98% (Table 2). For comparison, 6 epidemiologically unlinked isolates with sequence read archive data available from the National Center for Biotechnology Information were 111–423 SNVs different over an 86% core genome (data not shown). The phylogenetic tree of the case-patient isolates shows 2 major clades, 1 for each case-patient, and a reference (an isolate from C1) that appears to be an outlier from the 2 groups (Figure 2). A greater number of SNVs was observed among the most distantly related isolates from C1 than between isolates from C1 and E4/C2.
Table 2.
Single-nucleotide Variant Matrix
| Sample | C1-S4-01.4 | C1-S1-01.1 (Reference) | C1-S4-01.3 | C1-S2-01 | C2-S1-01.1 | C1-S3-01 | C2-S1-02.1 | C2-S2-02.1 | C2-S2-01.1 | C2-S3-01.1 | C2-S3-02.3 | C2-S3-02.1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1-S4-01.4 | 0 | 23 | 7 | 4 | 16 | 4 | 15 | 14 | 13 | 13 | 13 | 13 |
| C1-S1-01.1 (Reference) | 23 | 0 | 22 | 19 | 19 | 19 | 18 | 17 | 16 | 16 | 16 | 16 |
| C1-S4-01.3 | 7 | 22 | 0 | 3 | 15 | 3 | 14 | 13 | 12 | 12 | 12 | 12 |
| C1-S2-01 | 4 | 19 | 3 | 0 | 12 | 0 | 11 | 10 | 9 | 9 | 9 | 9 |
| C2-S1-01.1 | 16 | 19 | 15 | 12 | 0 | 12 | 3 | 2 | 3 | 3 | 3 | 3 |
| C1-S3-01.1 | 4 | 19 | 3 | 0 | 12 | 0 | 11 | 10 | 9 | 9 | 9 | 9 |
| C2-S1-02.1 | 15 | 18 | 14 | 11 | 3 | 11 | 0 | 1 | 2 | 2 | 2 | 2 |
| C2-S2-02.1 | 14 | 17 | 13 | 10 | 2 | 10 | 1 | 0 | 1 | 1 | 1 | 1 |
| C2-S2-01.1 | 13 | 16 | 12 | 9 | 3 | 9 | 2 | 1 | 0 | 0 | 0 | 0 |
| C2-S3-01.1 | 13 | 16 | 12 | 9 | 3 | 9 | 2 | 1 | 0 | 0 | 0 | 0 |
| C2-S3-02.3 | 13 | 16 | 12 | 9 | 3 | 9 | 2 | 1 | 0 | 0 | 0 | 0 |
| C2-S3-02.1 | 13 | 16 | 12 | 9 | 3 | 9 | 2 | 1 | 0 | 0 | 0 | 0 |
Figure 2.
Phylogenetic tree. A maximum likelihood phylogenetic tree was constructed from an alignment of single-nucleotide variants extracted from the whole genomes of Klebsiella pneumoniae isolates from the 2 case-patients, C1 and E4/C2. Abbreviation: SNV, single-nucleotide variant.
Duodenoscope Investigation
Cultures of duodenoscope surveillance samples obtained at MGH were negative at the independent third-party laboratory. At the CDC, E. coli and K. pneumoniae were recovered from the biopsy channel flush/brush and tip flush/brush specimens. Scant low-concern organisms (Micrococcus luteus, Staphylococcus epidermidis, and Bacillus species) were recovered from the rinse simultaneous to elevator raising and lowering. WGS indicated that all K. pneumoniae isolated from duodenoscope samples, like the case-patient mcr-1–positive K. pneumoniae, were ST15. However, each duodenoscope K. pneumoniae isolate was negative for mcr-1, lacked the blaSHV-28, blaOXA-1, and catB4 genes seen in the case-patient isolates, and harbored antimicrobial resistance genes (aac(6)-IIc, floR, and blaOXA-10) not present in the case-patient isolates. Additionally, phylogenetic analyses (not shown) demonstrated that duodenoscope isolates formed a cluster distinct from the clinical isolates.
Manufacturer evaluation of the duodenoscope identified an area at the distal tip where adhesive had peeled off; after disassembly, foreign material was detected on the interior of the distal case and at the distal tip of the duodenoscope body. The substance had spectra similar to sheep blood and was concluded to be artificial test soil used in cleaning and HLD testing either at MGH or during manufacturer review.
DISCUSSION
We report the first healthcare-associated transmission of mcr-1 in the United States. The mcr-1–positive isolates from both case-patients were K. pneumoniae; no other Klebsiella has been reported among the 29 mcr-carrying isolates identified in the United States as of 31 January 2018 [18]. WGS data supported that the isolates from both case-patients, which shared antimicrobial resistance gene profiles and a unique mcr-1 gene start codon, were very closely related. Although a highly sensitive (core genome 98%) SNV analysis showed that genomic variation between isolates from C1 and E4/C2 was present, the isolates from C1’s index bile culture demonstrated both phenotypic and genotypic variability, suggesting preexisting intrapatient microbial diversity. Transmission was associated with shared exposure to a duodenoscope; no alternative transmission route was identified through a detailed epidemiological investigation, and no additional patients with mcr-1 were identified.
This investigation highlights 2 critical public health issues: ongoing duodenoscope safety concerns and limited capacity to detect mcr-1–carrying bacteria in US healthcare settings. Although the manufacturer’s updated reprocessing instructions and FDA-issued supplemental measures were used, the duodenoscope was persistently contaminated with enteric bacteria, most likely due to a distal cap defect that allowed material to penetrate a sealed area inaccessible to cleaning. The subsequent national recall to replace the forceps elevator mechanism, O-rings, and distal end covering with a new, FDA-cleared design intended to reduce risk of contamination [29] highlights that recent initiatives to improve the safety of duodenoscopes have not fully addressed design issues that can lead to patient harm.
The manufacturer and FDA reported issues with cracks in adhesive binding the distal cap of this duodenoscope model in January 2017, 11 months before the recall, and advised customers that risk of contamination could be minimized by inspection for damage, meticulous adherence to the manufacturer’s updated reprocessing instructions, and returning the scope for inspection of the distal tip during regularly scheduled maintenance [30]. Our investigation highlights limitations of these recommendations. First, facility and CDC experts were unable to identify damage to the adhesive, indicating that, in practice, it is difficult for even experienced users to identify subtle defects in these complex devices. Second, duodenoscope surveillance cultures using the standard CDC protocol were negative, possibly because damage was absent in the 2 areas sampled by the protocol: the biopsy channel and the elevator mechanism. Enterobacteriaceae, however, were recovered through sampling conducted at the CDC, several months after sequestration. Whether or not the extended period of sequestration resulted in desiccation of biomaterial that then allowed it to be accessible by flushing per the standard protocol is unknown. Finally, this duodenoscope was returned from routine maintenance 1 day before the January 2017 safety communication; therefore, it was not due for service in the interval between the safety communication and when transmission was identified. Taken together, these findings indicate that more aggressive measures might be needed to ensure patient safety when potential defects with a duodenoscope model are known. Changes in duodenoscope designs that better support effective reprocessing procedures are urgently needed; proposed changes include disposable components and heat-stable components to allow steam sterilization [31].
In this investigation, mcr-1–carrying bacteria were not identified through routine testing but through clinician and laboratorian vigilance, highlighting that the absence of routine colistin AST in clinical laboratories creates the opportunity for occult introduction and dissemination of pathogens carrying mcr-1. Currently, there is no FDA-approved colistin susceptibility test. Challenges to test development have included the lack of FDA or CLSI breakpoints for colistin and Enterobacteriaceae, the antimicrobial’s cationic properties and poor diffusion into agar, and heteroresistance in some bacterial species [12, 25, 32–34]. Because of performance problems, in 2016 CLSI and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) jointly recommended abandoning colistin disk diffusion and gradient diffusion [34]. The only colistin AST method currently recognized by CLSI and EUCAST is reference broth microdilution (BMD) [25, 35], which is too laborious and technically challenging to be practical for routine use in many clinical laboratories. The commercially available Research Use Only BMD test used to initially detect colistin resistance among the case-patient isolates in this investigation has been reported to produce results comparable to those generated by reference BMD [36, 37], but is more laborious and expensive than disk and gradient diffusion methods. More experience with the ability of this and other AST methods to reliably detect colistin resistance is needed, particularly as the colistin MICs observed in many MCR-1–producing isolates have been 2–4 μg/mL [36, 38, 39], abutting the CLSI ECV. Notably, the K. pneumoniae isolates identified in this investigation carried not only the plasmid-mediated mcr-1 gene but also common chromosomal mutations previously demonstrated to confer colistin resistance, potentially elevating colistin MICs and making resistance easier to detect. Accurate, inexpensive, easy-to-perform colistin AST methods practical to implement in clinical laboratories are urgently needed, not only for prediction of polymyxin effectiveness in the treatment of individual infections, but also for use in prospective laboratory surveillance, which is an important strategy in tracking the spread of antimicrobial resistance [21].
This investigation has several limitations. Although efforts were undertaken to notify all potentially exposed individuals, not all underwent screening and some were screened up to 2 months after possible exposure. In addition, because our rectal screening procedure included selection with ceftriaxone, we cannot exclude the possibility that a distinct, ceftriaxone-susceptible organism had acquired the mcr-1 gene in 1 or more screened individuals. The duodenoscope was reprocessed multiple times after E4/C2’s ERCP, and mcr-harboring bacteria may have been removed before scope culturing. Furthermore, the duodenoscope was not cultured after disassembly by the manufacturer; therefore, culture results of the damaged components are not available. Despite these limitations, recovery of Enterobacteriaceae after multiple reprocessing steps, and evidence of material penetrating the distal cap, provides evidence of persistent contamination of the duodenoscope and a mechanism by which mcr-1–carrying bacteria may have been transmitted. The WGS data represent a subset of the bacteria colonizing and infecting the case-patients; as demonstrated by the observed intrapatient diversity, it is likely that the complete genomic diversity of mcr-1–harboring bacteria is not represented.
CONCLUSIONS
This investigation highlights that new issues continue to be identified among some duodenoscopes, continuing their linkage to transmission of multidrug-resistant organisms. As with previously reported outbreaks, the distinct and uncommon shared phenotypic AST pattern of the K. pneumoniae isolates in this report prompted further investigation; duodenoscope-related transmission of organisms with common susceptibility phenotypes is likely under-recognized. Renewed focus on duodenoscope design, including an emphasis on disposable components, is needed. Facilities should maintain heightened awareness of the risk of transmission through duodenoscopes, with procedures and processes to ensure adherence to reprocessing guidelines, vigilance for clusters of infections that may represent transmission, and continued discussions between clinicians and patients evaluating the risk-benefit ratio between these often lifesaving procedures and the ongoing risk of duodenoscope-associated infections. Finally, the current limited capacity for colistin AST in clinical laboratories increases the risk that mcr-1 may spread silently in healthcare facilities; clinical laboratorian vigilance and collaboration between clinical and public health laboratories are currently essential for detection of mcr-1.
Supplementary Material
Acknowledgments.
The authors wish to honor the memory of Kathy Seiber and thank her for her skilled and dedicated coordination of CDC laboratory and epidemiology activities during this investigation; she is dearly missed. The authors thank Marwan Azar, MD, Lisa Desrosiers, MPH, MT (ASCP), Vedrana Eleta, BS, MT (ASCP), Mary Jane Ferraro, PhD, MPH, Lisa Gartland, BS, MT (ASCP), Victoria Giangreco, BS, MLS (ASCP), Rachelle P. Markham, BS, MLS (ASCP), Janice Stefanski, BS, MT (ASCP), and Heather Wilson, BS, MT (ASCP) of the MGH Microbiology Laboratory; Karen S. Manning, Joseline Guerrero, BSN, RN, Sarah Turbett, MD, and Cindy Alejunas of MGH Infectious Disease Associates; James Richter, MD, David Forcione, MD, Norman Nishioka, MD, Thomas Cerruto, CRCST, CFER, Margie Voltero, MSHI, RN-BC, Carol Shea, BSN, RN, and Gayle Fishman, DNP, MBA, RN, NEA-BC of the MGH Endoscopy Unit; Lauren R. West, MPH, MGH Infection Control Unit; Elizabeth Mort, MD, MGH Lawrence Center for Quality and Safety; John Belknap, MGH Compliance; Seth Peters, MPH, and Patricia McAuley, MSN, RN, CIC, Rhode Island Department of Health, Massachusetts State Public Health; Laboratory; Hannah Leeman, BA, New Hampshire Division of Public Health Services, Bureau of Infectious Disease Control, Healthcare-Associated Infections (HAI) Program; Maine Center for Disease Control and Prevention, Division of Infectious Disease, HAI/Antimicrobial Resistance Program.
Disclaimer.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Footnotes
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Seoane-Vazquez E, Rodriguez-Monguio R. Endoscopy-related infection: relic of the past? Curr Opin Infect Dis 2008; 21:362–6. [DOI] [PubMed] [Google Scholar]
- 2.Srinivasan A. Epidemiology and prevention of infections related to endoscopy. Curr Infect Dis Rep 2003; 5:467–72. [DOI] [PubMed] [Google Scholar]
- 3.Epstein L, Hunter JC, Arwady MA, et al. New Delhi metallo-β-lactamase-producing carbapenem-resistant Escherichia coli associated with exposure to duodenoscopes. JAMA 2014; 312:1447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wendorf KA, Kay M, Baliga C, et al. Endoscopic retrograde cholangiopancreatography-associated AmpC Escherichia coli outbreak. Infect Control Hosp Epidemiol 2015; 36:634–42. [DOI] [PubMed] [Google Scholar]
- 5.Yang S, Hemarajata P, Hindler J, et al. Evolution and transmission of carbapenem-resistant Klebsiella pneumoniae expressing the blaOXA-232 gene during an institutional outbreak associated with endoscopic retrograde cholangiopancreatography. Clin Infect Dis 2017; 64:894–901. [DOI] [PubMed] [Google Scholar]
- 6.Marsh JW, Krauland MG, Nelson JS, et al. Genomic epidemiology of an endoscope-associated outbreak of Klebsiella pneumoniae carbapenemase (KPC)–producing K. pneumoniae. PLoS One 2015; 10:e0144310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verfaillie CJ, Bruno MJ, Voor in ‘t Holt AF, et al. Withdrawal of a novel-design duodenoscope ends outbreak of a VIM-2-producing Pseudomonas aeruginosa. Endoscopy 2015; 47:502. [DOI] [PubMed] [Google Scholar]
- 8.US Food and Drug Administration (FDA). Design of endoscopic retrograde cholangiopancreatography (ERCP) duodenoscopes may impede effective cleaning: FDA safety communication. Available at: http://wayback.archive-it.org/7993/20170722213105/https://www.fda.gov/MedicalDevices/Safety/AlertsandNotices/ucm434871.htm. Accessed 13 February 2018.
- 9.US Food and Drug Administration (FDA). Infections associated with reprocessed duodenoscopes. Available at: https://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/ReprocessingofReusableMedicalDevices/ucm454630.htm. Accessed 13 February 2018.
- 10.US Food and Drug Administration (FDA). Duodenoscope reprocessing: FDA safety communication—supplemental measures to enhance reprocessing. Available at: https://www.fda.gov/Safety/MedWatch/SafetyInformation/SafetyAlertsforHumanMedicalProducts/ucm457132.htm. Accessed 13 February 2018.
- 11.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16:161–8. [DOI] [PubMed] [Google Scholar]
- 12.Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 2017; 30:557–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill 2016; 21:30155. [DOI] [PubMed] [Google Scholar]
- 14.Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 2016; 21. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 15.Yin W, Li H, Shen Y, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio 2017; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carattoli A, Villa L, Feudi C, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 2017; 22. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGann P, Snesrud E, Maybank R, et al. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 2016; 60:4420–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention (CDC). Tracking the mcr gene. Available at: https://www.cdc.gov/drugresistance/tracking-mcr1.html. Accessed 31 January 2018.
- 19.Meinersmann RJ, Ladely SR, Plumblee JR, Cook KL, Thacker E. Prevalence of mcr-1 in the cecal contents of food animals in the United States. Antimicrob Agents Chemother 2017; 61. doi: 10.1128/AAC.02244-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mediavilla JR, Patrawalla A, Chen L, et al. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio 2016; 7. doi: 10.1128/mBio.01191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention (CDC). Interim guidance for a health response to contain novel or targeted multidrug-resistant organisms (MDROs). Available at: https://www.cdc.gov/hai/outbreaks/mdro/index.html. Accessed 26 November 2017.
- 22.Vasquez AM, Montero N, Laughlin M, et al. Investigation of Escherichia coli harboring the mcr-1 resistance gene—Connecticut, 2016. MMWR Morb Mortal Wkly Rep 2016; 65:979–80. [DOI] [PubMed] [Google Scholar]
- 23.Kline KE, Shover J, Kallen AJ, Lonsway DR, Watkins S, Miller JR. Investigation of first identified mcr-1 gene in an isolate from a U.S. patient—Pennsylvania, 2016. MMWR Morb Mortal Wkly Rep 2016; 65:977–8. [DOI] [PubMed] [Google Scholar]
- 24.Macesic N, Green D, Wang Z, et al. Detection of mcr-1-carrying Escherichia coli causing bloodstream infection in a New York City Hospital: avian origins, human concerns? Open Forum Infect Dis 2017; 4:ofx115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 27th ed. CLSI supplement M100. Wayne, PA: CLSI, 2017. [Google Scholar]
- 26.PENTAX Medical Company. Instructions for use (reprocessing): PENTAX video duodenoscope ED-3490TK. Montvale, NJ: PENTAX of America, 2016. [Google Scholar]
- 27.Centers for Disease Control and Prevention (CDC). Interim duodenoscope sampling method. Available at: https://www.cdc.gov/hai/settings/lab/lab-duodenoscope-sampling.html. Accessed 26 November 2017.
- 28.Centers for Disease Control and Prevention (CDC). Interim duodenoscope culture method. Available at: https://www.cdc.gov/hai/settings/lab/lab-duodenoscope-culture-method.html. Accessed 26 November 2017.
- 29.US Food and Drug Administration (FDA). Pentax medical duodenoscope model ED-3490TK: FDA safety communication—updated design and labeling cleared. Available at: https://www.fda.gov/Safety/MedWatch/SafetyInformation/SafetyAlertsforHumanMedicalProducts/ucm595788.htm?utm_campaign=Pentax%20Medical%20Duodenoscope%20Model%20ED-3490TK%3A%20FDA%20Safety%20Communication&utm_medium=email&utm_source=Eloqua. Accessed 7 February 2018.
- 30.US Food and Drug Administration (FDA). ED-3490TK video duodenoscope by Pentax: FDA safety communication—UPDATE—follow Pentax validated reprocessing instructions. Available at: https://www.fda.gov/Safety/MedWatch/SafetyInformation/SafetyAlertsforHumanMedicalProducts/ucm537146.htm. Accessed 7 February 2018.
- 31.US Food and Drug Administration (FDA). FDA clears first duodenoscope with disposable distal cap. Available at: https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm576753.htm. Accessed 13 February 2018.
- 32.Humphries RM, Ferraro MJ, Hindler JA. Impact of 21st century cures act on breakpoints and commercial antimicrobial susceptibility test systems: progress and pitfalls. J Clin Microbiol 2018; 56. doi: 10.1128/JCM.00139-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphries RM, Hindler J, Jane Ferraro M, Mathers A. 21st Century Cures Act and antimicrobial susceptibility testing: clinical implications in the era of multidrug resistance [manuscript published online ahead of print 23 May 2018]. Clin Infect Dis 2018. doi: 10.1093/cid/ciy432. [DOI] [PubMed]
- 34.CLSI-EUCAST Polymyxin Breakpoints Working Group. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. Available at: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf. Accessed 4 December 2017.
- 35.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Antimicrobial susceptibility testing of colistin—problems detected with several commercially available products. Available at: http://www.eucast.org/ast_of_bacteria/warnings/#c13111.
- 36.Chew KL, La MV, Lin RTP, Teo JWP. Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-positive Enterobacteriaceae: comparison of Sensititre, MicroScan, Vitek 2, and Etest with broth microdilution. J Clin Microbiol 2017; 55:2609–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hindler JA, Humphries RM. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant gram-negative bacilli. J Clin Microbiol 2013; 51:1678–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Centers for Disease Control and Prevention (CDC). FDA-CDC antimicrobial resistance isolate bank. Available at: https://www.cdc.gov/drugresistance/resistance-bank/index.html. Accessed 3 December 2017.
- 39.Matuschek E, Ahman J, Webster C, Kahlmeter G. Antimicrobial susceptibility testing of colistin—evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect 2017. doi: 10.1016/j.cmi.2017.11.020. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.