Summary
Precise synaptic connectivity is a prerequisite for the function of neural circuits, yet individual neurons, taken out of their developmental context, readily form unspecific synapses. How does the genome encode brain wiring in light of this apparent contradiction? Synaptic specificity is the outcome of a long series of developmental processes and mechanisms before, during and after synapse formation. How much promiscuity is permissible or necessary at the moment of synaptic partner choice depends on the extent to which prior development restricts available partners or subsequent development corrects initially made synapses. Synaptic promiscuity at the moment of choice can thereby play important roles in the development of precise connectivity, but also facilitate developmental flexibility and robustness. In this review, we assess the experimental evidence for the prevalence and roles of promiscuous synapse formation during brain development. Many well-established experimental approaches are based on developmental genetic perturbation and an assessment of synaptic connectivity only in the adult; this can make it difficult to pinpoint when a given defect or mechanism occurred. In many cases, such studies reveal mechanisms that restrict partner availability already prior to synapse formation. Subsequently, at the moment of choice, factors including synaptic competency, interaction dynamics and molecular recognition further restrict synaptic partners. The discussion of the development of synaptic specificity through the lens of synaptic promiscuity suggests an algorithmic process based on neurons capable of promiscuous synapse formation that are continuously prevented from making the wrong choices, with no single mechanism or developmental time point sufficient to explain the outcome.
Wolterhoff and Hiesinger review roles and mechanisms of the prevalent, context-dependent ability of nerve cells to form synapses with varying partners for the development of precision and robustness of neural connectivity.
Main text
Introduction
The genome encodes developmental programs that produce remarkably precise synaptic connectivity of intricate neural circuits. Yet, taken out of the context of these developmental programs, many neurons have been shown to form synapses with alternative partners, including with themselves1,2,3,4,5,6. Similarly, during normal development, some degree of synaptic promiscuity is apparent at distinct developmental stages; for example, in the case of activity-dependent (or other competitive) pruning processes, initially exuberant synapse formation is a developmental requirement for the correct outcome7,8,9,10,11,12,13. The notion of promiscuous synapse formation is not at odds with precise outcomes. Instead, it offers the opportunity to explain precision in the context of developmental plasticity and robustness to perturbation14.
Synaptic specificity describes a developmental outcome: the precision of connectivity in the adult nervous system. The developmental program that leads to this outcome can be subdivided into, first, developmental growth processes prior to synapse formation, second, the actual moment of initial synaptic partner choice and, third, developmental growth processes that follow synapse formation (Figure 1A). Along this developmental timeline, synaptic promiscuity can only be defined for the moment of choice, i.e. when more or less promiscuous synapse formation actually occurs. Developmental processes prior to synapse formation ensure that only certain pre- and postsynaptic partners get to see each other at the moment of choice, a process we refer to as pre-specification; development after the moment of choice can either stabilize and strengthen or weaken and remove synapses in a process we refer to as post-specification (Figure 1A). Feedback and cross-activation between the three steps can blur their separation and create non-linearities, e.g. when post-specification mechanisms activate earlier developmental programs. In addition, there are neuron-specific variations of how the developmental steps transition and interact to lead to synaptic specificity in the outcome, as recently highlighted in a comparison of the development of different neural circuits in vertebrates15.
Figure 1.
Synaptic specificity is the outcome of development before, during and after the moment of synaptic partner choice.
(A) The developmental timeline for synaptic specificity in the outcome includes pre-specification, the moment of synaptic partner choice, and post-specification. As described in the text, processes can overlap and cross-activate each other. (B) At the moment of choice, more or less promiscuous synapse formation as the limiting cases of total promiscuity (any two neurons can form a synapse if given the opportunity) and no promiscuity (some molecular or other specification mechanism ensures that exclusively the correct two partners can form a synapse). Both limiting cases are very unlikely based on experimental evidence, indicating that the level of promiscuity is a quantitative property at the moment of synapse formation.
Neuronal development before and after the developmental time window of synapse formation greatly influences the specificity of the outcome. In fact, most studies on the development of brain wiring fall into these two categories, while the actual moment of choice is more difficult to study. Synaptic partner choice is often depicted as a particularly exquisite developmental period, when only highly specific partners are accepted out of a pool of all available partners at the time and place the choice is made. For most developing neurons, the pool of available partners at the moment of choice, and the degree to which the choice is promiscuous, remain unknown. The limiting case of ‘total promiscuity’, i.e. the ability of a given neuron to form synapses with any other neuron, is unlikely given examples of known molecular interactions that specify or bias synapse formation. At the other end of the spectrum, precise molecular key-and-lock mechanisms for all synapses represent the antithesis to promiscuous synapse formation: if the key does not fit the lock, a synapse should not form. This is also unlikely, given the known ability, and often developmental necessity, to form synapses with variable partners. Hence, synaptic partner choice is a developmental process that must settle somewhere between these two limiting cases (Figure 1B).
Each of the three developmental periods, pre-specification, the moment of choice and post-specification, contribute quantitatively, and neither alone suffices to explain specificity in the outcome. From the perspective of the outcome, only a highly selective subset out of all possible synaptic connections are realized. Each developmental process and mechanism can be viewed as a quantitative restriction, a stepwise sieving and selecting, that sculpts the final connectome (Figure 2). In this review, we therefore focus on assessing experimental evidence for when a given process or mechanism occurs in time. The majority of experimental evidence is based on perturbation experiments (e.g. gene manipulation), for which this assessment requires one to account for the experimental timing of two aspects: first, when a given perturbation occurs; second, when changes are measured, i.e. the readout. Perturbation at early developmental stages likely has a cascading effect that secondarily affects subsequent development, including the moment of choice and subsequent post-specification (Figure 3). Hence, pinpointing the precise moment of a perturbation is necessary to distinguish primary from secondary effects. Secondly, the later the readout, the more difficult it is to pinpoint when and how during development changes occurred (Figure 3). For example, assessing adult outcomes after developmental perturbation experiments is common practice, but makes it difficult to know whether a given process or mechanism actually occurred before, during or after the moment of choice. This assessment, in turn, affects to what extent synapse formation may have occurred more or less promiscuously.
Figure 2.
Stepwise sieving out of incorrect partners as a conceptual model for the successive restriction of synaptic partnerships during development.
(A) Pre-specification: development leading up to the moment of choice creates a local neighbourhood (encircled region) and selective adjacencies between neurons in this neighbourhood. Within this neighbourhood, all possible connections between five types of pre-synapses and five types of post-synapses (white, black, red, green, blue) are depicted. (B) Restriction at the moment of synaptic partner choice: at least three sieving mechanisms collaborate to restrict the choice: synaptic competency (represented by sieving out all connections between red/blue/green pre-synapses and black/white post-synapses), molecular recognition (represented by sieving out all connections between black/white pre-synapses and red/blue/green post-synapses) and interaction dynamics (represented by sieving out all connections where pre- and post-synapses have the same color). (C) Post-specification includes all developmental mechanisms following initial synapse formation that further restrict specificity, most prominently pruning (fine tuning; represented by sieving out multiples of any specific connection type). The output state of post-specification represents synaptic specificity in the outcome, i.e. all possible synaptic partnerships that were not actively prevented (sieved out) during preceding development.
Figure 3.

Developmental consequences of genetically induced defects at different times in the developmental decision tree.
(A) Genetic perturbation of a mechanism specific to the moment of choice (marked by semi-transparent red rectangle) may affect subsequent developmental processes minimally (blue line) or have a cascading effect to change all subsequent developmental processes (red lines). Assessment of the synaptic specificity phenotype in the outcome may not easily distinguish between these possibilities. (B) Genetic perturbation of a mechanism prior to synapse formation is likely to have a cascading effect on both the moment of choice as well as subsequent development. Assessment of the synaptic specificity defect in the outcome may reveal little about the primary defect at the beginning of the cascading developmental changes. (C) Perturbation of a single gene often causes multiple hits throughout the developmental decision tree, leading to compound cascading developmental changes that alter synaptic specificity in the outcome.
Prior to the moment of synaptic partner choice: creating neighborhoods and adjacencies
The more restrictive the neighborhood at the moment of choice, such that only a few axonal and dendritic processes are actually adjacent to each other, the more promiscuous synapse formation is possible without losing specificity (Figure 2A). If development prior to synapse formation has already achieved exclusive adjacencies between presumptive partners, then even an inherently promiscuous synapse formation process at the moment of choice will still only lead to synapses between exclusively adjacent partners. This idea of adjacency as a predictor for synaptic partnerships is sometimes referred to as Peters' rule based on a study by Peters and Feldman in 197616,17.
More recently, the idea has been explored with the aid of developmental electron microscopy reconstructions of several developmental stages of the Caenorhabditis elegans nervous system18,19,20. In effect, the reconstructions reveal not only connectomes, i.e. information about synaptic connectivity, but also contactomes, i.e. information about adjacencies between different neurons as a result of prior development. A recent analysis of contactomes at different developmental stages indeed indicated a high predictability of synaptic connectivity based on adjacency21, an idea that was originally proposed based on the very first C. elegans connectome in 198522,23. For a meaningful analysis, connectivity needs to be related to the developmental contactome at the moment of choice, while adult reconstructions are unlikely to represent developmental adjacencies from past synapse formation and therefore often reveal no obvious relationship between adjacency and connectivity24,25,26,27,28,29. Similar to the recent C. elegans work, stronger correlations and support for Peters' rule were previously found when analyzing axo-dendritic overlap during synapse formation in the cortex of juvenile mice (P11–17)30,31,32,33. Yet, the quantitative role of adjacency in bringing together synaptic partners likely differs amongst neurons. The recent C. elegans connectome analysis included different neuron types and supported such neuron-specific quantitative contributions to adjacency21,34. Of note, C. elegans synapses are of the ‘en passant’ type, i.e. they form along adjacent neuronal fibres, as opposed to the terminal synapses most commonly depicted in text books and schematics (e.g. Figure 2). Tracts of neuronal fibres create bundles with large adjacent surfaces and thus neighborhoods that increase the probability of synaptic partnerships when synapses form en passant22,35. Far from being specific to the worm, en passant synapses can be found across phyla36,37,38. The ability to form synapses along the entire length of axo-dendritic fibers implies that synapse formation is not restricted to a specific subcellular region, whereas terminal synapses represent a limiting case of locally restricted synaptic competency. The subcellular restriction of competency to form a synaptic contact may be understood as one of the ‘sieving’ mechanisms at the moment of choice, as discussed below (Figure 2B).
Molecular mechanisms that ensure adjacency prior to synapse formation
Mechanisms that generate adjacency during development prior to synapse formation arguably include most of early neuronal development, including differentiation, migration, and axonal and dendritic morphogenesis — all of which have been reviewed in depth elsewhere7,39,40,41,42,43. While thus a wide range of cell biological and molecular mechanisms ultimately contributes to the development of adjacencies, a particular focus has long been the molecular and cellular recognition of axonal and dendritic processes, i.e. the prospective synaptic partners. Disruption of such recognition mechanisms frequently affects synaptic specificity in the outcome, yet the developmental roles can occur long before the moment of choice (Figure 3B). For example, Drosophila Down Syndrome Cell Adhesion Molecule (DSCAM) has the remarkable property that a single gene locus can generate thousands of selectively, homophilically interacting proteins, suggestive of a solution to a choice amongst many potential partners44. However, an enormous body of work revealed developmental roles prior to synapse formation, in particular the mechanism of dendrite spreading through self-avoidance; an individual neuron's unique ‘self’ can be defined by a probabilistic expression of DSCAM isoforms that are unique to that neuron45,46. Hence, DSCAM exemplifies an instance of molecular recognition that does not constitute intercellular recognition, but instead an intracellular morphogenesis mechanism. This and other roles of DSCAM (and Protocadherins in vertebrates) occur long before synapse formation but, like a plethora of other axonal and dendritic patterning processes, clearly influence the neighborhood and adjacencies a neuron finds itself in at the moment of choice (Figure 3B).
In contrast to the indirect influences of morphogenesis on neighborhoods and adjacencies, intercellular recognition can directly promote adjacencies of prospective synaptic partners; however, many such mechanisms do so without being required for the process of synapse formation itself. For example, the interacting cell adhesion molecules Dpr11 and DIP-γ in the Drosophila visual system are in fact expressed on pre- and postsynaptic partner neurons, respectively. Yet, their mechanism of action requires axo-dendritic interactions to provide a survival signal long before synapse formation47,48,49. By providing a selective survival signal for the correct future synaptic partner, this molecular mechanism can plausibly be described as ‘matching’ two partners; however, it does so by creating adjacency prior to synapse formation. Yet, wild-type Dpr11-positive presynaptic neurons are capable of forming synapses with non-matched, neighboring ‘incorrect’ partners as shown in careful EM reconstructions49. Such rare, non-canonical synapses are common based on recent connectomes in diverse species19,50,51,52. Importantly, loss of either Dpr11 or DIP-γ does not alter synapse numbers53,54, but instead leads to the recruitment of other, ‘non-canonical’ synaptic partners54. Hence, restricted availability ensures a level of pre-specification that excludes most incorrect partners. The more availability is restricted, the more synaptic promiscuity may be permissible7.
Combinatorial molecular codes that affect developmental processes prior to the moment of choice have been suggested for several classes of cell surface proteins13,55. In the vertebrate retina and motor system, several Cadherins (a greater than 100-member superfamily) have been thoroughly characterized for combinatorial expression and function. For example, cell type-specific expression of Cadherins 8 and 9 in different bipolar cells of the retina function in restricting axonal arborizations in specific layers56. Similarly, retinal ganglion cell dendrites innervate specific layers in a manner dependent on specific Cadherins57. While these studies demonstrate the effects of axonal and dendritic developmental defects on adult synaptic specificity and circuit function, these effects likely occur prior to the moment of choice. Similar roles of a Cadherin code in motor neuron differentiation and the development of dendritic orientation have been characterized58. Finally, Cadherins are also known as trans-synaptically interacting proteins with roles in synaptic function and maintenance59. The many roles of Cadherins, from cell migration to synaptic maintenance, have highlighted their highly diverse functions and critical importance for synaptic specificity in the outcome but also have made it particularly difficult to pinpoint precise moments of action (Figure 3C)55,60.
Synaptic promiscuity revealed by ablation experiments
The selected examples discussed above highlight only a few of the mechanisms that create adjacency prior to the moment of choice, while their loss or manipulation can create adjacencies that would not normally occur during development. The extent to which neurons are able to form synapses in a new context reveals to what extent a neuron is not prevented from making synapses with non-canonical partners and is thus a measure of synaptic promiscuity. Ablation experiments are a classic means to test this idea. For example, in the mouse retina the amacrine cell A17 is specific to the rod pathway; it consequently forms highly specific synaptic connections with rod bipolar cells, while only very rarely making synapses with cone bipolar cells. However, ablation of the preferred synaptic partner changes the developmental context, i.e. adjacencies, and A17 readily forms numerous synapses with cone bipolar cells61. This observation mirrors earlier findings where rod bipolar cells form ectopic synapses with cones in the absence of rods62 and cone bipolar cells form ectopic synapses with rods in the absence of cones63. Note that in the rod bipolar cell ablation experiment the A17 neurons do not form synapses with potentially available amacrine or ganglion cell processes61. The ‘secondary’ preference for cone bipolar cells may be a direct consequence of the newly arranged neighborhood, but may also reflect subsequent restrictive mechanisms including interaction kinetics and time windows of synaptic competency, as discussed below. In either case, these experiments reveal a remarkable resilience of the synapse formation process per se. By contrast, disruption of the function of the trans-synaptic adhesion protein LRRTM4 leads to defects in the formation of A17 synapses with rod bipolar axon terminals. While this function is essential for the morphological and molecular organization of synapses, it is not clear whether it also serves a role in distinguishing between possible synaptic partners based on selective adhesion64. Hence, the molecular mechanisms for synapse induction and formation may be distinct from the process that creates the adjacencies.
In zebrafish, lateralis afferent axons form synaptic connections exclusively with hair cells of identical polarity. Ablation of most lateralis neurons leads to less competition between the remaining cells and the axons innervate and form functional synapses with hair cells of both polarities65. Ablation in the larval stage, after connections with the correct hair cells of identical polarity have already been established, leads to a rewiring of the remaining axons with the now vacant hair cells independent of polarity. Remarkably, these non-canonical synapses are pruned again if new lateralis axons are regrown 72 hours after they were ablated and as they seemingly competitively regrow into the same space65. Since the neurons are not principally prevented from forming ectopic synapses with non-canonical partners, the competitive growth likely creates the adjacencies that facilitate synapse formation between canonical partners.
In Drosophila, the genetic ablation of amacrine-like Dm8 cells leads their main presynaptic partners, R7 photoreceptor neurons, to form many synapses with non-canonical but clearly available partners54. This includes cell types including Mi1, Mi4, C2, C3, Mi8 and Tm1, none of which are canonical synaptic partners of R7 in the available connectome24. As in the cases of lateralis afferent axons in zebrafish or A17 in the mouse retina, this type of promiscuity likely results from new adjacencies created in the absence of the ablated cells, i.e. when other neurons become available in the spaces normally occupied by the ablated cells.
Synaptic promiscuity revealed by altered morphogenesis in vivo and in vitro
Experimental changes of developmental adjacencies can also be induced by altering morphological availability or rerouting axons into ectopic brain regions. For example, increasing or decreasing dendritic ‘claws’ of Drosophila Kenyon cells leads to more or less types of projection neurons forming synapses with the Kenyon cells, respectively66. In zebrafish, the optogenetically controlled rerouting of axons beyond molecular and cellular boundaries does not prevent synapse formation in the ectopic target regions3. Drosophila photoreceptor axon terminals readily form synapses in incorrect neuropil layers after genetic manipulations67,68, and axons rerouted to ectopic visual5 or olfactory4 brain regions surprisingly form morphologically normal tetrad synapses, i.e. single synapses with four distinct, albeit necessarily incorrect, partners. In fact, mutant analyses on some of the best candidates for trans-synaptic contact specification at the moment of choice in Drosophila do not cause loss of synapses, but instead ectopic synapse formation elsewhere53,69,70,71, as discussed in detail in the following section on the moment of choice.
Arguably the most dramatic changes to developmental adjacencies occur to neurons outside of their developmental context in culture. For example, co-cultures of neurons that would not normally interact with each other in vivo readily form synapses in culture (e.g. pontine nucleus neurons and Purkinje cells)72. Finally, primary cultured neurons taken out of their developmental context readily form synapses with themselves, so-called autapses1,2. Such autapses only form rarely in the developing brain, suggesting that normal development prevents a neuron from meeting itself in the first place. Conversely, no active molecular mechanism is needed to prevent synapse formation between two neurons that never get to interact during normal brain development. These studies further suggest that the intrinsic drive of many neurons to form synapses is such that, if the right partner is not available, they will do it with a wrong partner, and, if a wrong partner can't be found, they do it with themselves13,73.
To what extent the generation of adjacencies through preceding development is sufficient to ensure the specificity of subsequent synapse formation is unclear in most cases. An assessment of just how much pre-sorting can allow promiscuous synapse formation requires a detailed characterization of the developmental system just prior to synapse formation. For example, during Drosophila visual map formation the axonal sorting mechanisms ensure local neighborhoods of such precision that mostly, and maybe only, ‘correct’ synaptic partners are adjacent during subsequent synapse formation74,75,76. However, to what extent synaptic partner choice in this system is truly promiscuous or biased by differential interaction dynamics, time windows of synaptic competence, or molecular recognition still remains unknown. In most cases, further restrictions at the moment of choice contribute to the specificity of synaptic partnerships observed in the outcome.
The moment of synaptic partner choice
The preceding section suggests that most neurons in the developing brain will have a restricted choice, but a choice nonetheless, in their local neighborhoods at the time of synapse formation. Based on experimental evidence discussed in this section, this choice is influenced by at least three factors: molecular recognition, kinetic and geometric restriction (i.e. influences on how much two neurons get to interact in time and space), and synaptic competence (the ability to form a synapse at all, e.g. based on the presence of molecular machinery). Of these three mechanisms to restrict synapse formation at the moment of choice, molecular recognition has been studied extensively, while synaptic competency and interaction kinetics remain unknown for most neurons during brain development. In this context, the term ‘molecular recognition’ is meant to describe specific intercellular interactions through cell surface proteins on the membranes of two different cells55,77. This powerful idea dates back to the influential chemoaffinity hypothesis, as envisioned by Roger Sperry73,78,79. By contrast, kinetic and geometric restriction and synaptic competence are less intuitively linked to a single class of molecules and likely require a collaboration of more than one molecular mechanism12,80. For example, kinetic restriction may require the temporary stabilization or slowdown of continuously extending and retracting filopodia through an interplay of cytoskeletal and membrane regulators in order to allow synapse formation. Similarly, synaptic competence may be influenced by the translation, transport and retention of pre- or postsynaptic building materials of various kinds. All three factors may quantitatively contribute to sieving out incorrect partners at the moment of choice (Figure 2B), and may share molecular mechanisms, as discussed below.
Lessons from intercellular molecular recognition
The recent discovery of several families of heterophilically interacting cell adhesion molecules that are specifically expressed in synaptic partner neurons in Drosophila offers the promise to assess the contribution of molecular recognition between neurons. These families of cell surface proteins include the Dpr/DIP proteins48,81,82 and the Sidestep (Side)/Beaten Path (Beat) proteins81,83. The families of 21 Dpr and 11 DIP proteins form interacting pairs with some degeneracy (e.g. seven different Dpr proteins bind to DIP-β) and interacting pairs localize to many pre- and postsynaptic partner neurons with remarkable specificity48,82. Functional analyses of individual members of these proteins have so far uncovered a variety of phenotypes, from defects in cell survival to branching to synapse numbers48,53,71. The families of 14 Beat and 8 Side proteins form a similar heterophilic extracellular interaction network with remarkably complementary expression patterns on synaptic partner neurons81,83,84.
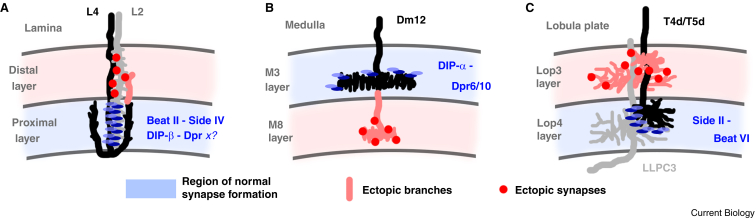
The most prominent and intuitive hypothesis for a role of Dpr/DIP and Side/Beat interactions at the moment of choice posits the specific induction of synapses at the time and place where the intercellular interaction takes place. However, loss of specific Dprs or DIPs has been shown to affect developmental processes prior to the moment of choice, e.g. cell survival in the case of Dpr11/DIP-γ as discussed above, or DIP-α and its interaction partners Dpr6 and Dpr1053. Yet, recent studies have begun to tease out putative roles at the moment of choice supporting a nuanced version of the first hypothesis. While loss of synapses at the place these molecules could induce synapse formation is not typically observed, ectopic synapses typically form nearby, often in a directly adjacent region (Figure 4). For example, loss of DIP-β in visual interneuron L4 does not lead to a loss of synapses at the place of normal DIP-β localization, but instead to ectopic synapse formation in an adjacent region where DIP-β is not normally localized (Figure 4A)71. Similarly, loss of Side-IV in L2, which forms reciprocal synapses with L4, does not lead to a loss of synapses where Side-IV could induce synapse formation, but instead to ectopic synapse formation in the same distal region where both Side-IV and DIP-β are not normally localized in L4 (Figure 4A)69. Likewise, loss of the DIP-α–Dpr6/10 interaction between Dm12 and its synaptic partner neurons in the medulla does not prevent synapse formation in medulla layer 3 where branches normally occur but leads to ectopic branching and synapse formation in layer 8 where neuronal branches are not normally localized (Figure 4B)53. Finally, Side-II is expressed in T4d/T5d neurons of the motion vision network and the interacting Beat-VI in the synaptic partner LLPC3 neurons (Figure 4C). Continuing the theme, loss of Side-II or Beat-VI does not prevent synapse formation between T4d/T5d and LLPC3, but instead leads to ectopic synapse formation in the adjacent region where these neurons normally do not form synapses (lobula plate layer 3). In summary, loss of Dpr/DIP or Side/Beat interactions does not typically lead to a reduction of synapses at the place where they are normally localized but commonly causes ectopic synapse formation in an adjacent region.
Figure 4.
Outcomes of perturbation experiments of interacting cell adhesion molecules in synaptically connected neurons in the Drosophila visual system.
In all examples, the molecular pairs are expressed specifically in neurons that form synapses in a distinct layer with subcellular specificity, yet loss of the inter-cellular interaction does not lead to a loss of synapses where they normally occur (blue regions) but instead to ectopic synapses in a nearby region (red regions; ectopic branches and ectopic synapses also marked in red). See text for possible mechanistic explanations. (A) Genetic perturbation of the Beat II–Side IV interaction or DIP-β causes synaptic specificity defects between the interneurons L2 and L4 in the Drosophila lamina. In both cases, synapses in the proximal layer of the lamina are still present, while ectopic synapses form in the distal lamina. (B) Genetic perturbation of the DIP-α–Dpr6/10 interaction does not lead to a loss of synapses in medulla layer M3 but to ectopic branching and synapse formation in M8 where Dm12 neurons do not normally branch or form synapses. (C) Genetic perturbation of the Side II–Beat VI interaction does not lead to a loss of synapses between T4d/T5d and LLPC3 neurons in the lobula plate layer 4 but to ectopic branching and synapse formation in the neighboring lobula plate layer 3.
In yet other examples, loss of members of the Dpr/DIP and Side/Beat families does not cause any obvious synapse formation defect in or nearby the regions where they are specifically expressed; for example, the same Side-IV/Beat-II interactions that are implicated in ectopic synapse formation in L4 neurons as discussed above mark another pair of synaptically connected neurons, T4c/T5c and LLPC2 in the lobula plate, where loss of either gene appears to not cause any obvious defect70. The hypothesis of locally specific synapse induction triggered by an intercellular interaction is not straight-forwardly consistent with these observations. The lack of reduced synapse formation is often interpreted as molecular redundancy, based on the multiple interacting cell adhesion molecules found in these interacting pairs by transcriptomics analyses84,85. However, why adjacent regions exhibit ectopic synapse formation while the synapse formation at the place of the lost intercellular molecular interaction is largely unaffected still requires an explanation. Here, the preferred interpretation in several studies is a ‘biasing’ of synapse formation at the place where the intercellular interaction normally occurs. According to this idea, following the loss of an intercellular molecular interaction, a ‘second best’ interaction at the ectopic location may take over70,71. However, just like the primary choice, a ‘second best‘ choice is likely influenced by a composite of molecular recognition, interaction kinetics and synaptic competency; hence, to account for the phenotypes described above may require a conceptual and mechanistic understanding that includes these different contributors at the moment of choice.
Integration of molecular recognition, synaptic competency and interaction kinetics
A hypothesis integrating molecular recognition with synaptic competency has been proposed in a recent preprint on the role of Side-IV in L4 neurons discussed above: intracellularly, Side-IV may recruit the synaptic seeding factor Syd-1 and thereby ensures synapse formation at a specific place. Loss of Side-IV may thus release Syd-1 and possibly other synapse building machinery to other axonal regions where synapse formation can now occur ectopically71. This is also consistent with the observation that ectopic L4 synapses in the DIP-β mutant are along the axon, not the dendrites, even though the dendrites are morphologically altered in the mutant (Figure 4A)71. The idea of an intracellular competition for synaptic seeding factors was first proposed for R7 photoreceptor axon terminals in Drosophila86. For these neurons, live observation at the moment of choice suggests the formation of one synapse at a time based on competition for synaptic seeding factors as a limiting resource. A side effect of this mechanism is a ‘counting’ of the number of synapses over time, ensuring a certain number of synapses are made in the developmental time available86. Loss of R7's main postsynaptic partner Dm8 (in the dpr11 or DIP-γ mutant as discussed above) does not affect this mechanism of serial synapse formation and hence leads to the same number of synapses, albeit with non-canonical postsynaptic partners54. Interestingly, a similar synapse constancy has been observed for several of the molecular recognition mutants described above; e.g. the total number of synapses formed by T4d/T5d (in the side-II mutant) or Dm12 (in the DIP-α mutant) remain relatively constant despite the addition of synapses in an ectopic layer (Figure 4B,C)53,70.
Both the ectopic synapse formation in an adjacent region and the stability of overall synapse numbers could be explained by a mechanism where the surface receptor recruits intracellular (presynaptic) building material that is only available as a limiting resource. In this model, the cell surface receptors increase the specificity of where synapses form by recruiting synaptic building material to a region where adjacency with a favoured synaptic partner has been created through its intercellular interaction. For this mechanism to work, the two functions — creating adjacency through intercellular adhesion and intracellular recruitment of synaptic building material — could be separated in time, as the creation of adjacency can occur both prior to as well as at the moment of choice.
Recruitment of intracellular synaptic building material represents a local regulation of synaptic competency with subcellular specificity; if synaptic building material is not available, e.g. because the neuron has not yet reached this developmental stage, then adjacency, molecular recognition and favourable kinetics do not lead to synapse formation. While a local increase of synaptic competency can explain ectopic synapse formation and synapse constancy in loss of function mutants, the missing link in this picture is the actual induction of synapse formation. In the case of Side-IV in L2 neurons, such a link has been proposed to be another cell surface receptor: loss of the immunoglobulin superfamily protein Kirre, in contrast to Side-IV or DIP-β, actually leads to a loss of reciprocal L4–L2 synapses87 and the Kirre protein directly interacts with Side-IV as co-receptor in L269. Kirre is a member of a smaller family of cell surface proteins that have been suggested to act both before and at the moment of choice88. Hence, a cell surface receptor that both creates intercellular adjacency with a synaptic partner and recruits intracellular building material may partner with a co-receptor that is required for actual synapse induction. Recruitment of a co-receptor might also explain why overexpression of several of the DIPs and Sides in particular have been shown to be sufficient for ectopic synapse formation53,69,71. Irrespective of whether the details of this mechanism are correct, these data suggest a molecular mechanistic separation of three functions prior to and at the moment of choice: creating intercellular adjacency, increasing local synaptic competency by recruiting intracellular synaptic building material, and induction of synapse formation. In particular, the mechanisms creating adjacency need not be executed by the same molecule or at the same time as the mechanism of synapse induction.
The idea of a requirement of two presynaptic receptors to control synapse numbers has recently been discussed as a form of coincidence detection in the formation of synapses by mouse hippocampal CA1-region pyramidal neurons89. Here, two presynaptic ligands (teneurins and fibronectin leucine-rich repeat transmembrane proteins [FLRTs]) have been shown to simultaneously bind to different postsynaptic latrophilins in different dendritic regions. The developmental and cell biological mechanism ensuring this exquisite subcellular localization does not seem to be an intrinsic property of the pyramidal neuron since in culture latrophilin-2 and latrophilin-3 localization are anatomically mixed; i.e. the subcellular separation requires the in vivo developmental context. One possibility is that the subcellularly restricted latrophilin localization domains develop in interaction with arriving axons that will subsequently provide synaptic input in vivo.
Critically, and in contrast to the Dpr/DIP and Side/Beat interactions in flies discussed above, loss of latrophilin-290 or latrophilin-389 leads to a significant loss of synapses in the regions where they are localized. Yet, earlier roles of teneurin–latrophilin interactions may create adjacencies prior to synapse formation91,92,93,94,95,96,97,98,99. For example, heterophilic teneurin-3/latrophilin-2-mediated repulsion and teneurin-3 homophilic attraction affect axon targeting in the hippocampus preceding synapse formation. Teneurin-3-expressing proximal CA1 neurons project to the distal subiculum where teneruin-3 expression is high, while avoiding the proximal subiculum where the teneurin-3 is low and latrophilin-2 expression is high93,94,98; conversely, latrophilin-2-expressing distal CA1 neurons project to the proximal subiculum where latrophilin-2 expression is high93,94. In zebrafish, teneurin-3 is required for axonal targeting of retinal ganglion cells, potentially via stabilizing branches that contact neighbouring teneurin-3-expressing cells96,97. Similarly, in the Drosophila antennal lobe, homophilic teneurin interactions have been suggested to regulate targeting of projection neurons to specific glomeruli. Subsequent synapse formation of projection neurons in an ectopic target region still occurred, suggesting a mechanistic separation of the development of adjacency and promiscuous synapse formation, as discussed above for multiple examples95. The axonal versus target (i.e. pre- versus postsynaptic) localization of these receptors has recently received some scrutiny, with evidence that teneurin-3 for example may function exclusively as a presynaptic receptor in the mouse cortex100, challenging some of the interpretation based on homophilic intercellular interactions discussed above.
Heterophilic teneurin interactions had previously been proposed to function at the moment of choice for synapse formation in the Drosophila central and peripheral nervous system101,102. In the Drosophila antennal lobe, loss of teneurin-a leds to reduced synapse numbers102. At the larval Drosophila neuromuscular junction, loss of heterophilic teneurin interactions led to defects of muscle 3 innervation, yet synapses did form in the nearby muscle 4101. In both systems pre-synaptic fine-structure was aberrant, suggesting that at least in theses contexts teneurins may contribute to synaptic competency. Since analyses in these studies were based on outcomes, it is further possible that synapses are initially formed but subsequently failed to be maintained.
In the mouse retina, many outcome-based studies similarly support roles of intercellular interactions that may shape synaptic specificity based on roles before, during or after synapse formation. A particularly striking example is the preferential pairing of highly specific pre- and postsynaptic neurons (the VG3 amacrine cell and W3B retinal ganglion cell subtypes) based on the function of the homophilic attractive cell adhesion molecule sidekick2103. A 10-fold increase of specific synapses between these two cell types (amongst many other nearby neuron types) is lost in the adult in the absence of sidekick2. Since the phenotype has only been analyzed in the adult outcome, however, it remains possible that Sidekick2's function involves the establishment of adjacencies prior to synapse formation or selective maintenance thereafter. Moreover, at the moment of choice, different modes of action are conceivable; for example, interaction dynamics like the extension and retraction speed of synaptogenic filopodia can restrict synapse formation, as discussed below for the example of fly photoreceptor neurons86,104. Indeed, the single Drosophila sidekick homolog has been shown to directly regulate extension and retraction dynamics in another tissue morphogenesis context based on cytoskeletal regulation105. Live observation in the developing mouse retina at the moment of choice would be required to test these possibilities.
In contrast to sidekick2's proposed homophilic attractive role in the mouse retina, the recent characterization of FLRT2 interaction with the UNC5 receptor revealed the intriguing mechanism of dendrite elimination upon intercellular interaction; loss of this mechanism leads to persistent arbors, which promptly leads to synapse formation with non-canonical partner neurons — again supporting the notion of a molecular separation between the mechanisms that create adjacencies and synapse formation itself106. Numerous other molecular intercellular interactions have been characterized based on synaptic specificity in the outcome of retina development. For example, Cadherin-6 is expressed by distinct retinal ganglion cells and their partner neurons during the period of target innervation; loss of Cadherin-6 leads to ectopic projections107. It remains unclear to what extent the stop signal for axonal growth is directly related to synapse formation. These studies highlight key roles of numerous cell surface proteins for synaptic specificity in the outcome that may well include a role at the moment of choice, yet do not exclude roles before or after (Figure 3). A more comprehensive review of cell adhesion-based mechanisms in retina circuitry is available elsewhere108.
Molecular mechanisms of synapse induction do not straight-forwardly predict synaptic partnerships
A synaptogenic interaction between two potential partner neurons at the moment of choice is a necessarily local occurrence that initiates synapse formation, i.e. synapse induction, between two membranous protrusions or surfaces7. The anatomical separation of latrophilin-2 and latrophilin-3 domains within the same neuron's dendrites showcased how subcellular specificity on the dendrites of the same cell can aid synapse formation with specific partners in restricted domains. In Drosophila mechanosensory axons, the subcellular restriction of an intracellular signalling factor has been shown to be necessary and sufficient to mark a subcellular region for synapse formation109. Here, subcellularly restricted localization of the membrane-anchored phosphatase Prl1 distinguishes parts of axonal branches where it is proposed as part of a synaptogenic signalling cascade. How Prl1 accumulates at the right time and place is not clear, and possible mechanisms include targeted protein trafficking, protein retention, or local translation109.
Amongst the best characterized synaptogenic cell surface proteins are the Neurexins and LAR-type receptor phosphatases. Members of the Neurexin family of cell surface proteins have prominently been discussed as a molecular code for the formation of specific types of synapses110,111. The ability of Neurexins to create many different splice variants has been intricately linked to the idea of the specificity of synapse development, but here the term ‘synaptic specificity’ does not refer to ‘partner choice’ but rather to the specificity of synaptic properties, e.g. the transmitter system. Correspondingly, Neurexins have most clearly been shown to play key roles in the instructive specification of synaptic properties, whereas a role in bringing together specific partner neurons remains less clear111,112,113. LAR receptors are similarly well characterized and implicated in both axonal targeting and synapse formation across species114,115,116,117,118. Remarkably, Neurexins and LAR receptors bind a large number of postsynaptic adhesion molecules to form diverse intercellular complexes and have been argued to account for the majority of trans-synaptic interactions in mice118.
Many possible interaction partners are less suggestive of a specific neuronal partnership code, but would be well-suited for robust synapse induction in light of a possible decoupling of the creation of adjacencies and synapse formation. Indeed, both LARs and Neurexins are strongly synaptogenic119,120. Surprisingly, however, neither LARs nor Neurexins appear to be generally required for synapse formation, supporting the notion of molecular redundancies121,122. Not even loss of three neurexins or all three LARs in mice appears to reduce synapse numbers, whereas only a conditional sixtuple-knockout of these LARs and Neurexins leads to an at least 50% reduction of synapse numbers between Purkinje cells and deep cerebellar nuclei118. Clearly, synapse formation per se is exquisitely robust to perturbation, even if the specification of properties or partners is less so. An integrative view of the diverse functions executed by intercellular molecular recognition posits that many parallel trans-synaptic signals together mediate different functions, including synapse induction and properties119.
In Drosophila, LAR functions as a pre-synaptic receptor on the tips of synaptogenic filopodia at the moment of synaptic partner choice of R7 photoreceptor axons86. This unequivocal mapping of the role of LAR to the very moment of choice is based on a live imaging analysis of the dynamics of synaptogenic axon terminal filopodia inside the intact developing Drosophila brain86,123. Loss of lar leads to a significant reduction of the formation of synaptogenic filopodia, and a quantitatively corresponding reduction in adult synapse numbers. Curiously, the most obvious defects in the adult outcome are R7 layer-specific targeting defects, which initially suggested a role of LAR earlier in development114,117. The live observations at the moment of choice indicate that the failure to stabilize is likely due to the primary defect in synapse formation86. The intercellular interaction partner of LAR on presynaptic filopodia has not been found, but a candidate interacting cell surface protein has recently been characterized124. On the presynaptic side, LAR directly recruits the synaptic seeding factor Liprin-α, as originally characterized in C. elegans125. Live observations of synaptogenic R7 filopodia in intact Drosophila brains support Liprin-α's role as a synaptic seeding factor downstream of the LAR receptor: in contrast to the lar mutant, synaptogenic filopodia do form in liprin-α or syd-1 mutant axon terminals, but the filopodia in both mutants fail to stabilize as presynaptic structures86. These findings in the R7 photoreceptor again highlight the links between three contributors to the moment of choice (Figure 2B): molecular recognition, interaction kinetics and synaptic competency. Phenotypes associated with the loss of lar are consistent with the loss of a molecular recognition event at the beginning of the synapse formation process, while those associated with loss of both liprin-α or syd-1 are consistent with a loss of synaptic competency.
Finally, interaction kinetics of R7 in Drosophila can be manipulated with developmental temperature54 or cell biologically through up- or downregulation of autophagy104. Autophagy is a ubiquitous membrane degradation mechanism that was previously shown to be required for normal synapse development in C. elegans126 and for the removal of excess synaptic spines in mice127. In Drosophila R7 axon terminals, loss of autophagy stabilizes synaptogenic filopodia, leading to excess synapse formation with non-canonical partners104. A comprehensive review of the roles of filopodia before and during the moment of choice is available elsewhere128. We interpret these findings as a mechanism of combinatorial, collaborative restriction when and where synapse induction can occur: just like developmental adjacencies restrict which neurons get to interact, so can interaction kinetics, synaptic competency and molecular recognition further restrict productive synaptogenetic events at the moment of choice (Figure 2B).
After the (initial) moment of synaptic partner choice
Throughout brain development, synapse formation occurs over extended periods in cell type-specific time windows. Initial synapse formation may be followed by more synapse formation, stabilization, or pruning before synaptic specificity in the outcome is achieved. Hence, after the first moment of choice, the separation into mechanisms that occur ‘before, during or after’ blurs. For example, following initial synapse formation, further axonal or dendritic growth may depend on successive and iterative steps of renewed rounds of synapse formation and branching, as observed in synaptotropic growth129,130. Here, new synapses lead to further branch growth to create new adjacencies in an iterative process. Conversely, destabilization of initially formed synapses and branches can be critical to achieve specificity in the outcome.
Synaptic pruning is one of the most studied processes to increase synaptic specificity after initial synapse formation8,9,131. In particular, activity-dependent 'fine tuning' of synaptic connectivity through coincidence-detection of spontaneous activity is often considered as a distinct developmental period following molecular specification mechanisms10,132,133. However, spontaneous activity that precedes sensory input is principally as much a part of genetically encoded development as other probabilistic, competitive developmental patterning processes13,73.
Neurotransmission at newly formed synapses can play critical roles for subsequent circuit development134,135. For example, disruption of GABA release from amacrine cells in the mammalian retina affects GABA receptor clustering on the cell bodies, but not the dendrites, of postsynaptic ganglion cells134. Similarly, GABAergic neurotransmission has been shown to affect cortical development through balancing excitation and inhibition in developing networks136.
Patterned spontaneous activity in developing neurons often occurs in waves following initial synapse formation and throughout the time synapses continue to form in the brains of diverse organisms10,137,138,139,140. In the mouse visual system, a major period of synapse formation occurs after birth but before eye opening, i.e. prior to sensory input. Remarkably, the spatial propagation of spontaneous retinal waves resembles the optic flow perceived by the mature animal during forward self-motion141. Earlier spontaneous activity already occurs in the mouse embryo, where it aids the assembly of thalamic and cortical sensory networks142. At birth, somatosensory and visual circuits that are initially intermingled in the superior colliculus segregate in a process that requires retinal activity143. Reduced cholinergic retinal waves in the first postnatal week lead to defects in direction-selective maps such that selectivity to horizontal motion is absent while selectivity to vertical motion remains144. In all these cases, correlated activity is considered a signal for selective synapse (and often branch) stabilization, while synapses that do not receive such a survival signal may be pruned. Conceptually, the activity-dependent process thereby represents another level of restriction to curb the internal drive of neurons to form synapses more promiscuously than the specificity observed in the outcome (Figure 2C). However, spontaneous activity may also serve other, non-Hebbian roles, as has been suggested for the remarkable patterned activity of neurons in the developing fly brain137,145. Here, brain-wide, yet cell type-specific patterns of spontaneous activity seem to be triggered by a small population of neurons, but the precise roles that these patterns may play for individual neuronal development remain an exciting, largely unanswered open question146.
Neuronal activity-dependent developmental mechanisms may directly feed back to molecular mechanisms that typically occur prior to or at the moment of choice. Odorant receptor neurons in mice express specific sets of cell surface proteins in a process that is regulated by neuronal activity; patterns of spontaneous activity are not spatially organized, but reflect odorant receptor cell types and have been proposed to directly alter the localization of cell surface proteins implicated in axon targeting choices147. In the mouse cortex, even earlier cell type specification may rely on neuronal activity; recent work revealed that, remarkably, late sensory input-dependent activity aids in the specification of some glutamatergic cell types in upper cortical layers in mice, i.e. a developmental process long before synapse formation148. Such mechanisms highlight how each subsequent developmental step before, during and after the initial moments of synaptic partner choice can feed back and cross-activate developmental programs. These programs and their underlying mechanisms thus function in transient, unique contexts in order to produce the synaptic specificity observed in the outcome.
Concluding remarks
Synaptic specificity, not synaptic promiscuity, is the focus of the study of brain development. The former is an assessment of an outcome, while the latter is a developmental process that occurs at the moment of synaptic partner choice and only makes sense in the context of prior and subsequent development. Arguably, selection of mutations at all developmental levels facilitates (re-)programming of brain wiring14,73. We speculate that promiscuous synapse formation may well reflect an ancestral state. Of course, molecular interactions between cells are critical for all developing multicellular structures, but the positioning and ‘connectivity’ of cells necessarily build on an evolutionary and developmental history that encompasses, but is not restricted to, intercellular recognition. Already the earliest neural nets developed in local neighbourhoods and with cell-specific properties of interaction dynamics and competency149. From there, evolutionary selection of any mutation in any gene that affected the restriction of promiscuous synapse formation at any level from pre- to post-specification may have allowed for ever more complicated network architectures.
Our review of recent and classic experimental evidence highlights how promiscuity at the moment of choice is not only permissible, but often required during algorithmic growth in order to produce synaptic specificity in the outcome. In particular, synaptic promiscuity allows for developmental plasticity, increases robustness of brain wiring to perturbation and facilitates evolutionary adaptation. In reviewing experimental evidence in this light, some observations seem to hold with considerable generality.
Firstly, synaptic promiscuity, together with other molecular or cellular mechanisms contributing to specificity in the outcome, only make sense in the unique developmental contexts in which they occur. At the moment of choice, the creation of adjacency, ensuring synaptic competency and synapse induction are typically executed by separate molecular mechanisms that are closely interlinked and provide context for each other. No single molecular mechanism at the moment of synaptic partner choice can explain synaptic specificity in the outcome without an understanding of the quantitative contributions summarily described as ‘context’.
Secondly, adjacency is opportunity. Given their intrinsic drive to form synapses, bringing together the right neurons at the right time and place is part and parcel of the development of brain wiring. But like other molecular and cellular mechanisms, adjacency, too, must be considered a quantitative contributor in the developmental time line of brain development.
Thirdly, neurons have a remarkably robust intrinsic drive to form synapses if not actively prevented from doing so. In the context of development, restriction of wrong choices may thus often be more important than attractively making the right choices. This especially makes sense from a perspective of evolutionary and developmental robustness, as fail-safe behaviors need not be separately encoded.
Maybe the development of brain wiring is best understood as a continuous process of trying to prevent neurons from making the wrong choices. Each consecutive step during development restricts the choices, up- and downwards from the moment two partners form a synapse, especially if the choice of the moment is rather promiscuous.
Acknowledgements
We would like to thank Drs Filipe Pinto Teixeira Sousa, Maria Ahmed and Joachim Fuchs for comments on the manuscript and all members of the Hiesinger lab for discussion. Work underlying this review in the Hiesinger lab is funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 101019191 ‘Synaptic Promiscuity in Brain Development’) and DFG Research Unit 5289 ‘RobustCircuit’. The authors declare that the text and figures of this article were prepared without the aid of any artificial intelligence.
Declaration of interests
The authors declare no competing interests.
References
- 1.Bekkers J.M., Stevens C.F. Excitatory and inhibitory autaptic currents in isolated hippocampal neurons maintained in cell culture. Proc. Natl. Acad. Sci. USA. 1991;88:7834–7838. doi: 10.1073/pnas.88.17.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van der Loos H., Glaser E.M. Autapses in neocortex cerebri: synapses between a pyramidal cell's axon and its own dendrites. Brain Res. 1972;48:355–360. doi: 10.1016/0006-8993(72)90189-8. [DOI] [PubMed] [Google Scholar]
- 3.Harris J.M., Wang A.Y., Boulanger-Weill J., Santoriello C., Foianini S., Lichtman J.W., Zon L.I., Arlotta P. Long-range optogenetic control of axon guidance overcomes developmental boundaries and defects. Dev. Cell. 2020;53:577–588.e7. doi: 10.1016/j.devcel.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clements J., Lu Z., Gehring W.J., Meinertzhagen I.A., Callaerts P. Central projections of photoreceptor axons originating from ectopic eyes in Drosophila. Proc. Natl. Acad. Sci. USA. 2008;105:8968–8973. doi: 10.1073/pnas.0803254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards T.N., Meinertzhagen I.A. Photoreceptor neurons find new synaptic targets when misdirected by overexpressing runt in Drosophila. J. Neurosci. 2009;29:828–841. doi: 10.1523/JNEUROSCI.1022-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiesinger P.R., Zhai R.G., Zhou Y., Koh T.W., Mehta S.Q., Schulze K.L., Cao Y., Verstreken P., Clandinin T.R., Fischbach K.F., et al. Activity-independent prespecification of synaptic partners in the visual map of Drosophila. Curr. Biol. 2006;16:1835–1843. doi: 10.1016/j.cub.2006.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agi E., Kulkarni A., Hiesinger P.R. Neuronal strategies for meeting the right partner during brain wiring. Curr. Opin. Neurobiol. 2020;63:1–8. doi: 10.1016/j.conb.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilton D.K., Dissing-Olesen L., Stevens B. Neuron-glia signaling in synapse elimination. Annu. Rev. Neurosci. 2019;42:107–127. doi: 10.1146/annurev-neuro-070918-050306. [DOI] [PubMed] [Google Scholar]
- 9.Lieberman O.J., McGuirt A.F., Tang G., Sulzer D. Roles for neuronal and glial autophagy in synaptic pruning during development. Neurobiol. Dis. 2019;122:49–63. doi: 10.1016/j.nbd.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shatz C.J. Emergence of order in visual system development. Proc. Natl. Acad. Sci. USA. 1996;93:602–608. doi: 10.1073/pnas.93.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrovic M., Schmucker D. Axonal wiring in neural development: Target-independent mechanisms help to establish precision and complexity. Bioessays. 2015;37:996–1004. doi: 10.1002/bies.201400222. [DOI] [PubMed] [Google Scholar]
- 12.Balaskas N., Abbott L.F., Jessell T.M., Ng D. Positional strategies for connection specificity and synaptic organization in spinal sensory-motor circuits. Neuron. 2019;102:1143–1156.e4. doi: 10.1016/j.neuron.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan B.A., Hiesinger P.R. Beyond molecular codes: Simple rules to wire complex brains. Cell. 2015;163:285–291. doi: 10.1016/j.cell.2015.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiesinger P.R., Hassan B.A. The evolution of variability and robustness in neural development. Trends Neurosci. 2018;41:577–586. doi: 10.1016/j.tins.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Sitko A.A., Goodrich L.V. Making sense of neural development by comparing wiring strategies for seeing and hearing. Science. 2021;371:eaaz6317. doi: 10.1126/science.aaz6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters A., Feldman M.L. The projection of the lateral geniculate nucleus to area 17 of the rat cerebral cortex. I. General description. J. Neurocytol. 1976;5:63–84. doi: 10.1007/BF01176183. [DOI] [PubMed] [Google Scholar]
- 17.Braitenberg V., Schüz A., editors. Cortex: Statistics and Geometry of Neuronal Connectivity. Springer; Berlin and Heidelberg: 1998. Peters’ rule and White's exceptions; pp. 99–101. [DOI] [Google Scholar]
- 18.Brittin C.A., Cook S.J., Hall D.H., Emmons S.W., Cohen N. A multi-scale brain map derived from whole-brain volumetric reconstructions. Nature. 2021;591:105–110. doi: 10.1038/s41586-021-03284-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witvliet D., Mulcahy B., Mitchell J.K., Meirovitch Y., Berger D.R., Wu Y., Liu Y., Koh W.X., Parvathala R., Holmyard D., et al. Connectomes across development reveal principles of brain maturation. Nature. 2021;596:257–261. doi: 10.1038/s41586-021-03778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook S.J., Jarrell T.A., Brittin C.A., Wang Y., Bloniarz A.E., Yakovlev M.A., Nguyen K.C.Q., Tang L.T., Bayer E.A., Duerr J.S., et al. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature. 2019;571:63–71. doi: 10.1038/s41586-019-1352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook S.J., Kalinski C.A., Hobert O. Neuronal contact predicts connectivity in the C. elegans brain. Curr. Biol. 2023;33:2315–2320.e2. doi: 10.1016/j.cub.2023.04.071. [DOI] [PubMed] [Google Scholar]
- 22.White J.G. Neuronal connectivity in Caenorhabditis elegans. Trends Neurosci. 1985;8:277–283. [Google Scholar]
- 23.White J.G., Southgate E., Thomson J.N., Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 24.Takemura S.Y., Xu C.S., Lu Z., Rivlin P.K., Parag T., Olbris D.J., Plaza S., Zhao T., Katz W.T., Umayam L., et al. Synaptic circuits and their variations within different columns in the visual system of Drosophila. Proc. Natl. Acad. Sci. USA. 2015;112:13711–13716. doi: 10.1073/pnas.1509820112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helmstaedter M., Briggman K.L., Turaga S.C., Jain V., Seung H.S., Denk W. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature. 2013;500:168–174. doi: 10.1038/nature12346. [DOI] [PubMed] [Google Scholar]
- 26.Briggman K.L., Helmstaedter M., Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- 27.Mishchenko Y., Hu T., Spacek J., Mendenhall J., Harris K.M., Chklovskii D.B. Ultrastructural analysis of hippocampal neuropil from the connectomics perspective. Neuron. 2010;67:1009–1020. doi: 10.1016/j.neuron.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee W.C., Bonin V., Reed M., Graham B.J., Hood G., Glattfelder K., Reid R.C. Anatomy and function of an excitatory network in the visual cortex. Nature. 2016;532:370–374. doi: 10.1038/nature17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasthuri N., Hayworth K.J., Berger D.R., Schalek R.L., Conchello J.A., Knowles-Barley S., Lee D., Vazquez-Reina A., Kaynig V., Jones T.R., et al. Saturated reconstruction of a volume of neocortex. Cell. 2015;162:648–661. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- 30.Fino E., Yuste R. Dense inhibitory connectivity in neocortex. Neuron. 2011;69:1188–1203. doi: 10.1016/j.neuron.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Packer A.M., Yuste R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J Neurosci. 2011;31:13260–13271. doi: 10.1523/JNEUROSCI.3131-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Packer A.M., McConnell D.J., Fino E., Yuste R. Axo-dendritic overlap and laminar projection can explain interneuron connectivity to pyramidal cells. Cereb. Cortex. 2013;23:2790–2802. doi: 10.1093/cercor/bhs210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W.C., Cooke T., Sautois B., Soffe S.R., Borisyuk R., Roberts A. Axon and dendrite geography predict the specificity of synaptic connections in a functioning spinal cord network. Neural Dev. 2007;2:17. doi: 10.1186/1749-8104-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuchs J., Hiesinger P.R. Brain wiring: Love the one you're with. Curr. Biol. 2023;33:R727–R729. doi: 10.1016/j.cub.2023.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Moyle M.W., Barnes K.M., Kuchroo M., Gonopolskiy A., Duncan L.H., Sengupta T., Shao L., Guo M., Santella A., Christensen R., et al. Structural and developmental principles of neuropil assembly in C. elegans. Nature. 2021;591:99–104. doi: 10.1038/s41586-020-03169-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takemura S.Y., Aso Y., Hige T., Wong A., Lu Z., Xu C.S., Rivlin P.K., Hess H., Zhao T., Parag T., et al. A connectome of a learning and memory center in the adult Drosophila brain. eLife. 2017;6:e26975. doi: 10.7554/eLife.26975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchizono K. Synaptic organization of the Purkinje cells in the cerebellum of the cat. Exp. Brain Res. 1967;4:97–113. doi: 10.1007/BF00240355. [DOI] [PubMed] [Google Scholar]
- 38.Stettler D.D., Yamahachi H., Li W., Denk W., Gilbert C.D. Axons and synaptic boutons are highly dynamic in adult visual cortex. Neuron. 2006;49:877–887. doi: 10.1016/j.neuron.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Hatten M.E. New directions in neuronal migration. Science. 2002;297:1660–1663. doi: 10.1126/science.1074572. [DOI] [PubMed] [Google Scholar]
- 40.Suter T., Jaworski A. Cell migration and axon guidance at the border between central and peripheral nervous system. Science. 2019;365:eaaw8231. doi: 10.1126/science.aaw8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holguera I., Desplan C. Neuronal specification in space and time. Science. 2018;362:176–180. doi: 10.1126/science.aas9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Danaf R.N., Rajesh R., Desplan C. Temporal regulation of neural diversity in Drosophila and vertebrates. Semin. Cell Dev. Biol. 2023;142:13–22. doi: 10.1016/j.semcdb.2022.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoersting A.K., Schmucker D. Axonal branch patterning and neuronal shape diversity: roles in developmental circuit assembly: Axonal branch patterning and neuronal shape diversity in developmental circuit assembly. Curr. Opin. Neurobiol. 2021;66:158–165. doi: 10.1016/j.conb.2020.10.019. [DOI] [PubMed] [Google Scholar]
- 44.Schmucker D., Flanagan J.G. Generation of recognition diversity in the nervous system. Neuron. 2004;44:219–222. doi: 10.1016/j.neuron.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 45.Kise Y., Schmucker D. Role of self-avoidance in neuronal wiring. Curr. Opin. Neurobiol. 2013;23:983–989. doi: 10.1016/j.conb.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 46.Zipursky S.L., Sanes J.R. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell. 2010;143:343–353. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Courgeon M., Desplan C. Coordination between stochastic and deterministic specification in the Drosophila visual system. Science. 2019;366:eaay6727. doi: 10.1126/science.aay6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carrillo R.A., Ozkan E., Menon K.P., Nagarkar-Jaiswal S., Lee P.T., Jeon M., Birnbaum M.E., Bellen H.J., Garcia K.C., Zinn K. Control of synaptic connectivity by a network of Drosophila IgSF cell surface proteins. Cell. 2015;163:1770–1782. doi: 10.1016/j.cell.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menon K.P., Kulkarni V., Takemura S.Y., Anaya M., Zinn K. Interactions between Dpr11 and DIP-gamma control selection of amacrine neurons in Drosophila color vision circuits. eLife. 2019;8:e48935. doi: 10.7554/eLife.48935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorkenwald S., Matsliah A., Sterling A.R., Schlegel P., Yu S.-c., McKellar C.E., Lin A., Costa M., Eichler K., Yin Y., et al. Neuronal wiring diagram of an adult brain. bioRxiv. 2023 doi: 10.1101/2023.06.27.546656. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winding M., Pedigo B.D., Barnes C.L., Patsolic H.G., Park Y., Kazimiers T., Fushiki A., Andrade I.V., Khandelwal A., Valdes-Aleman J., et al. The connectome of an insect brain. Science. 2023;379 doi: 10.1126/science.add9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loomba S., Straehle J., Gangadharan V., Heike N., Khalifa A., Motta A., Ju N., Sievers M., Gempt J., Meyer H.S., Helmstaedter M. Connectomic comparison of mouse and human cortex. Science. 2022;377 doi: 10.1126/science.abo0924. [DOI] [PubMed] [Google Scholar]
- 53.Xu S., Xiao Q., Cosmanescu F., Sergeeva A.P., Yoo J., Lin Y., Katsamba P.S., Ahlsen G., Kaufman J., Linaval N.T., et al. Interactions between the Ig-superfamily proteins DIP-alpha and Dpr6/10 regulate assembly of neural circuits. Neuron. 2018;100:1369–1384.e6. doi: 10.1016/j.neuron.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiral F.R., Dutta S.B., Linneweber G.A., Hilgert S., Poppa C., Duch C., von Kleist M., Hassan B.A., Hiesinger P.R. Brain connectivity inversely scales with developmental temperature in Drosophila. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.110145. [DOI] [PubMed] [Google Scholar]
- 55.Sanes J.R., Zipursky S.L. Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell. 2020;181:536–556. doi: 10.1016/j.cell.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 56.Duan X., Krishnaswamy A., De la Huerta I., Sanes J.R. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell. 2014;158:793–807. doi: 10.1016/j.cell.2014.06.047. [DOI] [PubMed] [Google Scholar]
- 57.Duan X., Krishnaswamy A., Laboulaye M.A., Liu J., Peng Y.R., Yamagata M., Toma K., Sanes J.R. Cadherin combinations recruit dendrites of distinct retinal neurons to a shared interneuronal scaffold. Neuron. 2018;99:1145–1154.e6. doi: 10.1016/j.neuron.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vagnozzi A.N., Moore M.T., Lin M., Brozost E.M., Kc R., Agarwal A., Schwarz L.A., Duan X., Zampieri N., Landmesser L.T., Philippidou P. Coordinated cadherin functions sculpt respiratory motor circuit connectivity. eLife. 2022;11:e82116. doi: 10.7554/eLife.82116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedman L.G., Benson D.L., Huntley G.W. Cadherin-based transsynaptic networks in establishing and modifying neural connectivity. Curr. Top Dev. Biol. 2015;112:415–465. doi: 10.1016/bs.ctdb.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gartner A., Fornasiero E.F., Dotti C.G. Cadherins as regulators of neuronal polarity. Cell Adh. Migr. 2015;9:175–182. doi: 10.4161/19336918.2014.983808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang C., Hellevik A., Takeuchi S., Wong R.O. Hierarchical partner selection shapes rod-cone pathway specificity in the inner retina. iScience. 2022;25 doi: 10.1016/j.isci.2022.105032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng Y.W., Hao Y., Petters R.M., Wong F. Ectopic synaptogenesis in the mammalian retina caused by rod photoreceptor-specific mutations. Nat. Neurosci. 2000;3:1121–1127. doi: 10.1038/80639. [DOI] [PubMed] [Google Scholar]
- 63.Haverkamp S., Michalakis S., Claes E., Seeliger M.W., Humphries P., Biel M., Feigenspan A. Synaptic plasticity in CNGA3(-/-) mice: cone bipolar cells react on the missing cone input and form ectopic synapses with rods. J. Neurosci. 2006;26:5248–5255. doi: 10.1523/JNEUROSCI.4483-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sinha R., Siddiqui T.J., Padmanabhan N., Wallin J., Zhang C., Karimi B., Rieke F., Craig A.M., Wong R.O., Hoon M. LRRTM4: A novel regulator of presynaptic inhibition and ribbon synapse arrangements of retinal bipolar cells. Neuron. 2020;105:1007–1017.e5. doi: 10.1016/j.neuron.2019.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pujol-Marti J., Faucherre A., Aziz-Bose R., Asgharsharghi A., Colombelli J., Trapani J.G., Lopez-Schier H. Converging axons collectively initiate and maintain synaptic selectivity in a constantly remodeling sensory organ. Curr. Biol. 2014;24:2968–2974. doi: 10.1016/j.cub.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 66.Ahmed M., Rajagopalan A.E., Pan Y., Li Y., Williams D.L., Pedersen E.A., Thakral M., Previero A., Close K.C., Christoforou C.P., et al. Input density tunes Kenyon cell sensory responses in the Drosophila mushroom body. Curr Biol. 2023;33:2742–2760.e12. doi: 10.1016/j.cub.2023.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berger-Muller S., Sugie A., Takahashi F., Tavosanis G., Hakeda-Suzuki S., Suzuki T. Assessing the role of cell-surface molecules in central synaptogenesis in the Drosophila visual system. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kulkarni A., Ertekin D., Lee C.H., Hummel T. Birth order dependent growth cone segregation determines synaptic layer identity in the Drosophila visual system. eLife. 2016;5 doi: 10.7554/eLife.13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Osaka J., Ishii A., Wang X., Iwanaga R., Kawamura H., Sugie A., Hakeda-Suzuki S., Suzuki T. Complex formation of immunoglobulin superfamily molecules Side-IV and Beat-IIb regulates synaptic specificity in the Drosophila visual system. bioRxiv. 2023 doi: 10.1101/2023.03.27.534487. Preprint at. [DOI] [PubMed] [Google Scholar]
- 70.Yoo J., Dombrovski M., Mirshahidi P., Nern A., LoCascio S.A., Zipursky S.L., Kurmangaliyev Y.Z. Brain wiring determinants uncovered by integrating connectomes and transcriptomes. Curr. Biol. 2023;33:3998–4005.e6. doi: 10.1016/j.cub.2023.08.020. [DOI] [PubMed] [Google Scholar]
- 71.Xu C., Theisen E., Maloney R., Peng J., Santiago I., Yapp C., Werkhoven Z., Rumbaut E., Shum B., Tarnogorska D., et al. Control of synaptic specificity by establishing a relative preference for synaptic partners. Neuron. 2019;103:865–877.e7. doi: 10.1016/j.neuron.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuwako K.I., Nishimoto Y., Kawase S., Okano H.J., Okano H. Cadherin-7 regulates mossy fiber connectivity in the cerebellum. Cell Rep. 2014;9:311–323. doi: 10.1016/j.celrep.2014.08.063. [DOI] [PubMed] [Google Scholar]
- 73.Hiesinger P.R. Princeton University Press; Princeton: 2021. The Self-Assembling Brain. How Neural Networks Grow Smarter. [Google Scholar]
- 74.Langen M., Agi E., Altschuler D.J., Wu L.F., Altschuler S.J., Hiesinger P.R. The developmental rules of neural superposition in Drosophila. Cell. 2015;162:120–133. doi: 10.1016/j.cell.2015.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agi E., Langen M., Altschuler S.J., Wu L.F., Zimmermann T., Hiesinger P.R. The evolution and development of neural superposition. J. Neurogenet. 2014;28:216–232. doi: 10.3109/01677063.2014.922557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwabe T., Neuert H., Clandinin T.R. A network of cadherin-mediated interactions polarizes growth cones to determine targeting specificity. Cell. 2013;154:351–364. doi: 10.1016/j.cell.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie Q., Li J., Li H., Udeshi N.D., Svinkina T., Orlin D., Kohani S., Guajardo R., Mani D.R., Xu C., et al. Transcription factor Acj6 controls dendrite targeting via a combinatorial cell-surface code. Neuron. 2022;110:2299–2314.e8. doi: 10.1016/j.neuron.2022.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sperry R.W. Regulative factors in the orderly growth of neural circuits. Growth. 1951;(Suppl. 10):63–87. [PubMed] [Google Scholar]
- 79.Sperry R.W. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc. Natl. Acad. Sci. USA. 1963;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hiesinger P.R. Brain wiring with composite instructions. Bioessays. 2021;43 doi: 10.1002/bies.202000166. [DOI] [PubMed] [Google Scholar]
- 81.Ozkan E., Carrillo R.A., Eastman C.L., Weiszmann R., Waghray D., Johnson K.G., Zinn K., Celniker S.E., Garcia K.C. An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell. 2013;154:228–239. doi: 10.1016/j.cell.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan L., Zhang K.X., Pecot M.Y., Nagarkar-Jaiswal S., Lee P.T., Takemura S.Y., McEwen J.M., Nern A., Xu S., Tadros W., et al. Ig superfamily ligand and receptor pairs expressed in synaptic partners in Drosophila. Cell. 2015;163:1756–1769. doi: 10.1016/j.cell.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H., Watson A., Olechwier A., Anaya M., Sorooshyari S.K., Harnett D.P., Lee H.P., Vielmetter J., Fares M.A., Garcia K.C., et al. Deconstruction of the beaten Path-Sidestep interaction network provides insights into neuromuscular system development. eLife. 2017;6:e28111. doi: 10.7554/eLife.28111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kurmangaliyev Y.Z., Yoo J., Valdes-Aleman J., Sanfilippo P., Zipursky S.L. Transcriptional programs of circuit assembly in the Drosophila visual system. Neuron. 2020;108:1045–1057.e6. doi: 10.1016/j.neuron.2020.10.006. [DOI] [PubMed] [Google Scholar]
- 85.Ozel M.N., Simon F., Jafari S., Holguera I., Chen Y.C., Benhra N., El-Danaf R.N., Kapuralin K., Malin J.A., Konstantinides N., Desplan C. Neuronal diversity and convergence in a visual system developmental atlas. Nature. 2021;589:88–95. doi: 10.1038/s41586-020-2879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ozel M.N., Kulkarni A., Hasan A., Brummer J., Moldenhauer M., Daumann I.M., Wolfenberg H., Dercksen V.J., Kiral F.R., Weiser M., et al. Serial synapse formation through filopodial competition for synaptic seeding factors. Dev. Cell. 2019;50:447–461.e8. doi: 10.1016/j.devcel.2019.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luthy K., Ahrens B., Rawal S., Lu Z., Tarnogorska D., Meinertzhagen I.A., Fischbach K.F. The irre cell recognition module (IRM) protein Kirre is required to form the reciprocal synaptic network of L4 neurons in the Drosophila lamina. J. Neurogenet. 2014;28:291–301. doi: 10.3109/01677063.2014.883390. [DOI] [PubMed] [Google Scholar]
- 88.Fischbach K.F., Linneweber G.A., Andlauer T.F., Hertenstein A., Bonengel B., Chaudhary K. The irre cell recognition module (IRM) proteins. J. Neurogenet. 2009;23:48–67. doi: 10.1080/01677060802471668. [DOI] [PubMed] [Google Scholar]
- 89.Sando R., Jiang X., Sudhof T.C. Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science. 2019;363:eaav7969. doi: 10.1126/science.aav7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anderson G.R., Maxeiner S., Sando R., Tsetsenis T., Malenka R.C., Sudhof T.C. Postsynaptic adhesion GPCR latrophilin-2 mediates target recognition in entorhinal-hippocampal synapse assembly. J. Cell Biol. 2017;216:3831–3846. doi: 10.1083/jcb.201703042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carr O.P., Glendining K.A., Leamey C.A., Marotte L.R. Overexpression of Ten-m3 in the retina alters ipsilateral retinocollicular projections in the wallaby (Macropus eugenii) Int. J. Dev. Neurosci. 2013;31:496–504. doi: 10.1016/j.ijdevneu.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 92.Dharmaratne N., Glendining K.A., Young T.R., Tran H., Sawatari A., Leamey C.A. Ten-m3 is required for the development of topography in the ipsilateral retinocollicular pathway. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pederick D.T., Lui J.H., Gingrich E.C., Xu C., Wagner M.J., Liu Y., He Z., Quake S.R., Luo L. Reciprocal repulsions instruct the precise assembly of parallel hippocampal networks. Science. 2021;372:1068–1073. doi: 10.1126/science.abg1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pederick D.T., Perry-Hauser N.A., Meng H., He Z., Javitch J.A., Luo L. Context-dependent requirement of G protein coupling for Latrophilin-2 in target selection of hippocampal axons. eLife. 2023;12:e83529. doi: 10.7554/eLife.83529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hong W., Mosca T.J., Luo L. Teneurins instruct synaptic partner matching in an olfactory map. Nature. 2012;484:201–207. doi: 10.1038/nature10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Antinucci P., Nikolaou N., Meyer M.P., Hindges R. Teneurin-3 specifies morphological and functional connectivity of retinal ganglion cells in the vertebrate visual system. Cell Rep. 2013;5:582–592. doi: 10.1016/j.celrep.2013.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Antinucci P., Suleyman O., Monfries C., Hindges R. Neural mechanisms generating orientation selectivity in the retina. Curr. Biol. 2016;26:1802–1815. doi: 10.1016/j.cub.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berns D.S., DeNardo L.A., Pederick D.T., Luo L. Teneurin-3 controls topographic circuit assembly in the hippocampus. Nature. 2018;554:328–333. doi: 10.1038/nature25463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Merlin S., Horng S., Marotte L.R., Sur M., Sawatari A., Leamey C.A. Deletion of Ten-m3 induces the formation of eye dominance domains in mouse visual cortex. Cereb. Cortex. 2013;23:763–774. doi: 10.1093/cercor/bhs030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang X., Lin P.Y., Liakath-Ali K., Sudhof T.C. Teneurins assemble into presynaptic nanoclusters that promote synapse formation via postsynaptic non-teneurin ligands. Nat. Commun. 2022;13:2297. doi: 10.1038/s41467-022-29751-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mosca T.J., Hong W., Dani V.S., Favaloro V., Luo L. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature. 2012;484:237–241. doi: 10.1038/nature10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mosca T.J., Luo L. Synaptic organization of the Drosophila antennal lobe and its regulation by the Teneurins. eLife. 2014;3 doi: 10.7554/eLife.03726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krishnaswamy A., Yamagata M., Duan X., Hong Y.K., Sanes J.R. Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature. 2015;524:466–470. doi: 10.1038/nature14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kiral F.R., Linneweber G.A., Mathejczyk T., Georgiev S.V., Wernet M.F., Hassan B.A., von Kleist M., Hiesinger P.R. Autophagy-dependent filopodial kinetics restrict synaptic partner choice during Drosophila brain wiring. Nat. Commun. 2020;11:1325. doi: 10.1038/s41467-020-14781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Malin J., Rosa Birriel C., Astigarraga S., Treisman J.E., Hatini V. Sidekick dynamically rebalances contractile and protrusive forces to control tissue morphogenesis. J. Cell Biol. 2022;221:e202107035. doi: 10.1083/jcb.202107035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prigge C.L., Dembla M., Sharma A., El-Quessny M., Kozlowski C., Paisley C.E., Miltner A.M., Johnson T.M., Della Santina L., Feller M.B., Kay J.N. Rejection of inappropriate synaptic partners in mouse retina mediated by transcellular FLRT2-UNC5 signaling. Dev. Cell. 2023;58:2080–2096.e7. doi: 10.1016/j.devcel.2023.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Osterhout J.A., Josten N., Yamada J., Pan F., Wu S.W., Nguyen P.L., Panagiotakos G., Inoue Y.U., Egusa S.F., Volgyi B., et al. Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron. 2011;71:632–639. doi: 10.1016/j.neuron.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Graham H.K., Duan X. Molecular mechanisms regulating synaptic specificity and retinal circuit formation. Wiley Interdiscip. Rev. Dev. Biol. 2021;10:e379. doi: 10.1002/wdev.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Urwyler O., Izadifar A., Vandenbogaerde S., Sachse S., Misbaer A., Schmucker D. Branch-restricted localization of phosphatase Prl-1 specifies axonal synaptogenesis domains. Science. 2019;364:eaau9952. doi: 10.1126/science.aau9952. [DOI] [PubMed] [Google Scholar]
- 110.Gomez A.M., Traunmuller L., Scheiffele P. Neurexins: molecular codes for shaping neuronal synapses. Nat. Rev. Neurosci. 2021;22:137–151. doi: 10.1038/s41583-020-00415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sudhof T.C. Synaptic neurexin complexes: A molecular code for the logic of neural circuits. Cell. 2017;171:745–769. doi: 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Traunmuller L., Gomez A.M., Nguyen T.M., Scheiffele P. Control of neuronal synapse specification by a highly dedicated alternative splicing program. Science. 2016;352:982–986. doi: 10.1126/science.aaf2397. [DOI] [PubMed] [Google Scholar]
- 113.Hart M.P., Hobert O. Neurexin controls plasticity of a mature, sexually dimorphic neuron. Nature. 2018;553:165–170. doi: 10.1038/nature25192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Clandinin T.R., Lee C.H., Herman T., Lee R.C., Yang A.Y., Ovasapyan S., Zipursky S.L. Drosophila LAR regulates R1-R6 and R7 target specificity in the visual system. Neuron. 2001;32:237–248. doi: 10.1016/s0896-6273(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 115.Krueger N.X., VanVactor D., Wan H.I., Gelbart W.M., Goodman C.S., Saito H. The transmembrane tyrosine phosphatase DLAR controls motor axon guidance in Drosophila. Cell. 1996;84:611–622. doi: 10.1016/S0092-8674(00)81036-3. [DOI] [PubMed] [Google Scholar]
- 116.Uhl G.R., Martinez M.J. PTPRD: neurobiology, genetics, and initial pharmacology of a pleiotropic contributor to brain phenotypes. Ann. NY Acad. Sci. 2019;1451:112–129. doi: 10.1111/nyas.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maurel-Zaffran C., Suzuki T., Gahmon G., Treisman J.E., Dickson B.J. Cell-autonomous and -nonautonomous functions of LAR in R7 photoreceptor axon targeting. Neuron. 2001;32:225–235. doi: 10.1016/s0896-6273(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 118.Sclip A., Sudhof T.C. Combinatorial expression of neurexins and LAR-type phosphotyrosine phosphatase receptors instructs assembly of a cerebellar circuit. Nat. Commun. 2023;14:4976. doi: 10.1038/s41467-023-40526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sudhof T.C. Towards an understanding of synapse formation. Neuron. 2018;100:276–293. doi: 10.1016/j.neuron.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Takahashi H., Craig A.M. Protein tyrosine phosphatases PTPdelta, PTPsigma, and LAR: presynaptic hubs for synapse organization. Trends Neurosci. 2013;36:522–534. doi: 10.1016/j.tins.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen L.Y., Jiang M., Zhang B., Gokce O., Sudhof T.C. Conditional deletion of all neurexins defines diversity of essential synaptic organizer functions for neurexins. Neuron. 2017;94:611–625.e4. doi: 10.1016/j.neuron.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 122.Emperador-Melero J., de Nola G., Kaeser P.S. Intact synapse structure and function after combined knockout of PTPdelta, PTPsigma, and LAR. eLife. 2021;10:e66638. doi: 10.7554/eLife.66638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ozel M.N., Langen M., Hassan B.A., Hiesinger P.R. Filopodial dynamics and growth cone stabilization in Drosophila visual circuit development. eLife. 2015;4:e10721. doi: 10.7554/eLife.10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bali N., Lee H.P., Zinn K. Sticks and Stones, a conserved cell surface ligand for the Type IIa RPTP Lar, regulates neural circuit wiring in Drosophila. eLife. 2022;11:e71469. doi: 10.7554/eLife.71469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhen M., Jin Y. The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature. 1999;401:371–375. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]
- 126.Stavoe A.K., Hill S.E., Hall D.H., Colon-Ramos D.A. KIF1A/UNC-104 transports ATG-9 to regulate neurodevelopment and autophagy at synapses. Dev. Cell. 2016;38:171–185. doi: 10.1016/j.devcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tang G., Gudsnuk K., Kuo S.H., Cotrina M.L., Rosoklija G., Sosunov A., Sonders M.S., Kanter E., Castagna C., Yamamoto A., et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wit C.B., Hiesinger P.R. Neuronal filopodia: From stochastic dynamics to robustness of brain morphogenesis. Semin. Cell Dev. Biol. 2023;133:10–19. doi: 10.1016/j.semcdb.2022.03.038. [DOI] [PubMed] [Google Scholar]
- 129.Niell C.M., Meyer M.P., Smith S.J. In vivo imaging of synapse formation on a growing dendritic arbor. Nat. Neurosci. 2004;7:254–260. doi: 10.1038/nn1191. [DOI] [PubMed] [Google Scholar]
- 130.Vaughn J.E. Fine structure of synaptogenesis in the vertebrate central nervous system. Synapse. 1989;3:255–285. doi: 10.1002/syn.890030312. [DOI] [PubMed] [Google Scholar]
- 131.Faust T.E., Gunner G., Schafer D.P. Mechanisms governing activity-dependent synaptic pruning in the developing mammalian CNS. Nat. Rev. Neurosci. 2021;22:657–673. doi: 10.1038/s41583-021-00507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tiriac A., Feller M.B. Roles of visually evoked and spontaneous activity in the development of retinal direction selectivity maps. Trends Neurosci. 2022;45:529–538. doi: 10.1016/j.tins.2022.04.002. [DOI] [PubMed] [Google Scholar]
- 133.Thompson A., Gribizis A., Chen C., Crair M.C. Activity-dependent development of visual receptive fields. Curr. Opin. Neurobiol. 2017;42:136–143. doi: 10.1016/j.conb.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bleckert A., Zhang C., Turner M.H., Koren D., Berson D.M., Park S.J.H., Demb J.B., Rieke F., Wei W., Wong R.O. GABA release selectively regulates synapse development at distinct inputs on direction-selective retinal ganglion cells. Proc. Natl. Acad. Sci. USA. 2018;115:E12083–E12090. doi: 10.1073/pnas.1803490115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bleckert A., Wong R.O. Identifying roles for neurotransmission in circuit assembly: insights gained from multiple model systems and experimental approaches. Bioessays. 2011;33:61–72. doi: 10.1002/bies.201000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Warm D., Schroer J., Sinning A. Gabaergic interneurons in early brain development: Conducting and orchestrated by cortical network activity. Front. Mol. Neurosci. 2021;14 doi: 10.3389/fnmol.2021.807969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Akin O., Bajar B.T., Keles M.F., Frye M.A., Zipursky S.L. Cell-type-specific patterned stimulus-independent neuronal activity in the Drosophila visual system during synapse formation. Neuron. 2019;101:894–904.e5. doi: 10.1016/j.neuron.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ponce-Alvarez A., Jouary A., Privat M., Deco G., Sumbre G. Whole-brain neuronal activity displays crackling noise dynamics. Neuron. 2018;100:1446–1459.e6. doi: 10.1016/j.neuron.2018.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Blankenship A.G., Feller M.B. Mechanisms underlying spontaneous patterned activity in developing neural circuits. Nat. Rev. Neurosci. 2010;11:18–29. doi: 10.1038/nrn2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Choi B.J., Chen Y.D., Desplan C. Building a circuit through correlated spontaneous neuronal activity in the developing vertebrate and invertebrate visual systems. Genes Dev. 2021;35:677–691. doi: 10.1101/gad.348241.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ge X., Zhang K., Gribizis A., Hamodi A.S., Sabino A.M., Crair M.C. Retinal waves prime visual motion detection by simulating future optic flow. Science. 2021;373:abd0830. doi: 10.1126/science.abd0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Martini F.J., Guillamon-Vivancos T., Moreno-Juan V., Valdeolmillos M., Lopez-Bendito G. Spontaneous activity in developing thalamic and cortical sensory networks. Neuron. 2021;109:2519–2534. doi: 10.1016/j.neuron.2021.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guillamon-Vivancos T., Anibal-Martinez M., Puche-Aroca L., Moreno-Bravo J.A., Valdeolmillos M., Martini F.J., Lopez-Bendito G. Input-dependent segregation of visual and somatosensory circuits in the mouse superior colliculus. Science. 2022;377:845–850. doi: 10.1126/science.abq2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tiriac A., Bistrong K., Pitcher M.N., Tworig J.M., Feller M.B. The influence of spontaneous and visual activity on the development of direction selectivity maps in mouse retina. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2021.110225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Akin O., Zipursky S.L. Activity regulates brain development in the fly. Curr. Opin. Genet. Dev. 2020;65:8–13. doi: 10.1016/j.gde.2020.04.005. [DOI] [PubMed] [Google Scholar]
- 146.Bajar B.T., Phi N.T., Isaacman-Beck J., Reichl J., Randhawa H., Akin O. A discrete neuronal population coordinates brain-wide developmental activity. Nature. 2022;602:639–646. doi: 10.1038/s41586-022-04406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nakashima A., Ihara N., Shigeta M., Kiyonari H., Ikegaya Y., Takeuchi H. Structured spike series specify gene expression patterns for olfactory circuit formation. Science. 2019;365:eaaw5030. doi: 10.1126/science.aaw5030. [DOI] [PubMed] [Google Scholar]
- 148.Cheng S., Butrus S., Tan L., Xu R., Sagireddy S., Trachtenberg J.T., Shekhar K., Zipursky S.L. Vision-dependent specification of cell types and function in the developing cortex. Cell. 2022;185:311–327.e24. doi: 10.1016/j.cell.2021.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hejnol A., Rentzsch F. Neural nets. Curr. Biol. 2015;25:R782–R786. doi: 10.1016/j.cub.2015.08.001. [DOI] [PubMed] [Google Scholar]