Abstract
Background.
In electronic health records, patterns of missing laboratory test results could capture patients’ course of disease as well as reflect clinician’s concerns or worries for possible conditions. These patterns are often understudied and overlooked. This study aims to identify informative patterns of missingness among laboratory data collected across 15 healthcare system sites in three countries for COVID-19 inpatients.
Methods.
We collected and analyzed demographic, diagnosis, and laboratory data for 69,939 patients with positive COVID-19 PCR tests across three countries from 1 January 2020 through 30 September 2021. We analyzed missing laboratory measurements across sites, missingness stratification by demographic variables, temporal trends of missingness, correlations between labs based on missingness indicators over time, and clustering of groups of labs based on their missingness/ordering pattern.
Results.
With these analyses, we identified mapping issues faced in seven out of 15 sites. We also identified nuances in data collection and variable definition for the various sites. Temporal trend analyses may support the use of laboratory test result missingness patterns in identifying severe COVID-19 patients. Lastly, using missingness patterns, we determined relationships between various labs that reflect clinical behaviors.
Conclusion.
In this work, we use computational approaches to relate missingness patterns to hospital treatment capacity and highlight the heterogeneity of looking at COVID-19 over time and at multiple sites, where there might be different phases, policies, etc. Changes in missingness could suggest a change in a patient’s condition, and patterns of missingness among laboratory measurements could potentially identify clinical outcomes. This allows sites to consider missing data as informative to analyses and help researchers identify which sites are better poised to study particular questions.
Keywords: Missing data, Electronic health records, COVID-19, Laboratory tests, Multi-site health data
Graphical Abstract
Introduction
The increasing availability of electronic health record (EHR) data has led to the burgeoning use of these data in various domains, including the identification of disease phenotypes and the clinical course of disease. Most recently, the EHR has been used as a rich source of data for characterizing the trajectory of the Coronavirus Disease (COVID-19) that is caused by the SARS-CoV-2 virus. However, it is commonly acknowledged that EHR data often require rigorous processing before they are of usable quality, thereby presenting considerable challenges to those using these data for research, quality improvement, or disease surveillance. Issues such as data availability [1–3], data recording or format inconsistencies [4,5], temporal changes in data policies [5,6], poorly standardized free-text [1,6], lack of interoperability between EHR systems [6,7], healthcare processes that can bias inferences made from EHR data [8]and diagnostic coding errors [1] all impair the usability of EHR data. Moreover, the most frequently reported barrier to EHR usability is missing data or data which are expected to be in the record but are not [3,4,6,9–13].
There is an important difference in the way that clinicians and informaticians tend to view missing data. To a clinician, data is considered missing if a laboratory test was supposed to be conducted and its value recorded, but for some known or unknown reason, it is absent from the medical record. Therefore, if a test is measured and recorded once per week according to protocol, and this is carried out without issues, there is no missing data for this particular test. On the other hand, if an informatician is carrying out a time series analysis, and their specified time window is daily measurements, this indicates that a laboratory test that is not measured on a given day will be considered missing, despite the fact that the correct ordering frequency is being carried out by the clinician. This study was originally motivated by the desire to carry out a time series analysis; but with laboratory tests collected at different frequencies, the issue of missing data and how to deal with it needed to be addressed. As we delved into this, we realized that missing data itself could be leveraged to learn more about EHRs, nuances across sites, ordering patterns, and relationships between the labs themselves.
Much has been written about missing data as a principal contributing factor to poor data quality in the clinical record [1]. Data are missing in the EHR for two principal reasons. First, a laboratory test might have been ordered, but the result is missing from the record. Although important for ascertaining the quality of reporting systems, characterizing this type of missing data is difficult without access to clinical notes and ordering systems in the EHR. The second reason missing data is that a laboratory test was never ordered, or where a test was ordered and resulted, but for some reason was not resulted for some time or ever again during a hospital stay, and thus a result would not be expected in the EHR. Such missing data should not be considered as a direct measure of EHR data quality, since there are many factors, often clinical, that determine when and if a test result is absent from the record. We focus here on the latter type of missing data, and we propose that the absence of data can be informative, and that patterns of these missing data can be considered as informative missingness.
Thus, our research question is “What can we learn from patterns of missingness in EHR data that could amplify our understanding of the pathophysiology, severity, trajectory, and outcomes of disease?”
Patterns of Data Missingness
Missing data is typically characterized according to three commonly accepted missingness patterns. The first is missing completely at random (MCAR). In this pattern, the missingness of a variable is not associated with any observed or unobserved variables, including the variable itself. An example would be where responses to a survey question about smoking status is not present on some proportion of respondents because the question was asked (or not) in a truly random fashion; in other words, there is no nonrandom pattern of missingness. In the second pattern, missing at random (MAR), the missingness of a variable is associated with the value of another observed variable. For example, responses to a question on smoking status are dependent on one’s occupation, resulting in a missing value for smoking when a respondent notes that their occupation is in health care. Finally, when data are missing in the EHR, it is often missing not at random (MNAR) [14]. A variable may be missing because of the value of the variable itself. For example, a smoker may be less likely to answer a survey question about smoking status because they are a heavy smoker. Note that it is not known whether or not the respondent is, in fact, a smoker, and that would not be known because they did not answer the question. In other words, the probability of determining if a respondent is a smoker is depending on the value of the smoking question. For this reason, such missing data are nonignorable, which implies that such data violate assumptions for imputation and need to be considered (encoded) explicitly as missing data for purposes of imputation.
However, we contend that there is a special case of MNAR, where the missing data are informative, in this case indicating a clinician’s assumption and a decision not to order a test subsequent to the previous one. This type of missingness has been referred to as structurally missing data [15], in that there is a logical, non-random reason the data are missing. However, we refer to this pattern as informative-missing not at random (I-MNAR). In this pattern, the missingness of a variable is dependent on the value of the variable, like MNAR, but may also be influenced by the value of other variables as well, whether they are observed or unobserved. It is a pattern commonly seen in the EHR, where once a normal laboratory result is obtained, no further assays of the same type are present in the record. The absent results indicate that the decision not to order the test after the normal result was likely due to the normal value itself, but it could be that the values of other variables (such as other laboratory tests or clinical assessments) are taken into account during the decision-making process. Thus, the absence of laboratory results after a given result is informative, perhaps about the severity of the disease, the availability of the test, practice guidelines, or clinician preferences. An example of this is in [16], where the recording of rheumatoid factor test results in the EHR was found to be missing when a test result was negative. In other words, a test was not ordered because it was assumed that the test would be negative based on a prior result of the test.
Informative Missing-Not-At Random in EHR Studies
The I-MNAR pattern has been investigated in the literature, albeit under a different nomenclature, typically referred to simply as “informative missingness”. For example, in [17] it was noted that missing data are often correlated with a target variable, such as outcome. Informative missing data has been identified in genotype analysis and genetic association studies [18–23], longitudinal cohort studies [24,25], meta-analyses [26–31], exposure assessment in case-control studies [32], and particularly in studies using EHR data [33].
The goal of this study was to identify patterns of missing laboratory tests that might suggest levels of disease severity or other factors, such as patient sex or hospital characteristics that could influence the availability of laboratory data in the EHR. From this study, we hope to determine if these missing data fit an I-MNAR pattern and interpret this informativeness. Accordingly, we focus here specifically on the laboratory data patterns found in our examination of EHR data in an international federated data consortium.
After this introduction, we describe our methodology for defining the sample population, the variable set, and the analytic methods used to quantify and describe missing values. Next, we present the results as characterization of the distribution of missing data by laboratory test, stratified by sex, hospital site, and disease severity, across different time periods from admission through 60 days thereafter. We also present the results of an analysis that seeks to investigate the patterns of missingness with regard to pairs of laboratory tests. Finally, we consider a topic model analysis that clusters groups of labs together based on missingness patterns.
Statement of significance
Problem
Missing data is pervasive in the electronic health record (EHR).
What is already known
Missing data in EHRs are difficult to remediate through source data or imputation. In addition, missing data of all types are often used to reflect the quality of EHR data.
What this paper adds
This study focuses on the informativeness of missing data, thereby providing new insights into the nature of EHR data, hospital treatment capacity, relation to clinical outcomes, and opportunities for applying novel statistical and machine learning methods to temporal pattern estimation in the presence of missing data.
Methods
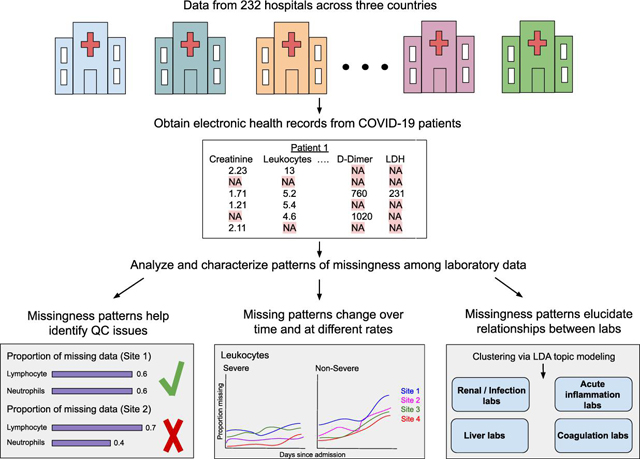
For an overview of our workflow, please see the graphical abstract corresponding to this paper.
Study Population and Setting
COVID-19 was and continues to be a pressing global health crisis that is still poorly understood. Due to a general lack of guidelines for COVID-19 (as compared to other diseases) and the rapidly evolving phenotype and treatment regimens for COVID-19 we imagined there would be many interesting missing data patterns in this setting as clinicians investigated how to diagnose and treat it. In addition, the COVID-19 pandemic presented a unique opportunity of collaboration between many diverse and international sites that led to the formation of the Consortium for the Clinical Characterization of COVID-19 by the EHR (4CE) consortium. It was through the efforts of this consortium where the common data model allowed for analysis across multiple institutions across the world.
We leveraged the resources of the international 4CE consortium (4CE). The 4CE uses a federated data model and predictive analytics framework in a hub-and-spoke configuration. Specifically, all 4CE-contributing academic medical centers (spokes) query and standardize EHR data elements using a COVID ontology, and apply analysis locally to their COVID datasets, and then provide aggregate statistics to a coordinating academic medical center (hub). This agile, rapid, and privacy-preserving data sharing approach has efficiently and effectively supported several COVID studies over the last two years [34–40]. For sites with multiple hospitals, we assume that practice patterns are similar across the hospitals within those sites. There are many benefits to analyzing multiple sites across the world– it enables us to be able to look at a broader population across multiple countries and ensure the results are generalizable. We keep in mind the difference in practices and focus on how the missing patterns might bring out these differences/similarities across systems and their role in providing additional insight to the relationship between labs, disease severity, and test ordering patterns that may reflect adherence to clinical guidelines.
This retrospective observational study of EHR was reviewed and approved by the ethics and institutional review boards for all participating 4CE contributing sites.
We analyzed laboratory test ordering patterns in EHR data for 69,939 patients from 232 hospitals across three countries from the 4CE consortium for the period of 1 January 2020 through 30 September 2021. Fifteen 4CE consortium contributing sites are described in Table 1. Most sites in 4CE use the i2b2 common data model, ensuring consistent mapping of the variables. While there are sites which are not on i2b2, quality control is performed after data extraction to ensure that the correct mappings are used.
Table 1.
4CE contributing sites.
| 4CE contributing site | Location | Number of Hospitals | Number of Beds | Inpatient discharges/year | Ratio of Female: Male | Ratio of Severe: Non-Severe |
|---|---|---|---|---|---|---|
| Boston Children’s Hospital | Boston, Massachusetts, United States | 1 | 404 | 28,000 | 1.03 | 0.30 |
| Beth Israel Deaconess Medical Center | Boston, Massachusetts, United States | 1 | 673 | 40,752 | 1.02 | 1.14 |
| Bordeaux University Hospital | Bordeaux, France | 3 | 2,676 | 130,033 | 0.76 | 2.23 |
| ASST Papa GGiovanni XXIII Bergamo | Bergamo, Italy | 1 | 1,080 | 45,000 | 0.60 | 0.36 |
| Istituti Clinici Scientifici Maugeri | Pavia, Lumezzane, and Milano, Italy | 3 | 775 | 12,344 | 1.10 | 0.06 |
| Mass General Brigham (Partners Healthcare) | Boston, Massachusetts, United States | 10 | 3,418 | 163,521 | 0.94 | 0.58 |
| Northwestern University | Evanston, Illinois, United States | 5 | 193 | 15,748 | 0.97 | 0.61 |
| University of Pennsylvania | Philadelphia, Pennsylvania, United States | 5 | 2,469 | 118,188 | 1.13 | 0.33 |
| University of Michigan | Ann Arbor, Michigan, United States | 3 | 1,000 | 49,008 | 0.98 | 1.66 |
| University of Pittsburgh | Pittsburgh, Pennsylvania, United States | 39 | 8,085 | 369,300 | 0.98 | 0.36 |
| Department of Veteran Affairs (1) | North Atlantic District, United States | 49 | 3,594 | 151,075 | 0.05 | 0.87 |
| Department of Veteran Affairs (2) | Southwest District, United States | 29 | 3,115 | 156,315 | 0.07 | 0.843 |
| Department of Veteran Affairs (3) | Midwest District, United States | 39 | 2,686 | 145,468 | 0.06 | 0.59 |
| Department of Veteran Affairs (4) | Continental District, United States | 24 | 2,110 | 113,260 | 0.07 | 1.06 |
| Department of Veteran Affairs (5) | Pacific District, United States | 29 | 2,296 | 114,569 | 0.06 | 0.86 |
The inclusion criteria include a positive COVID-19 polymerase chain reaction (PCR) test on or during admission to the inpatient setting. Only data from a patient’s first COVID-19 admission was considered; subsequent admissions were not included in the analysis. We collected results from 16 laboratory tests conducted over the entire admission. We selected these labs because (1) their abnormal values have been associated with worse outcomes among COVID patients in the literature and in our own 4CE mortality risk prediction models; (2) they reflect acute pathophysiology of COVID-19 patients as markers of cardiac, renal, hepatic, and immune dysfunction; (3) the labs were mappable across sites with an identifiable LOINC code. The clinical significance of these tests and common ordering practice is described in [41–43].
Quantifying missingness: definitions
A missing laboratory test is logged when no results for the test are available in the EHR for a patient within a time point, usually within a day. Thus, we assume that a missing test result serves as a proxy for a test that wasn’t ordered. The number of missing results per patient will hence be the total number of days without a given laboratory test result for the period of hospital admission. Note that this is oftentimes not how missing data are defined in a clinical setting. Even though it could be routine practice not to order a given laboratory test each day, we still want to capture this information on a per-day basis to understand the ordering patterns of the various labs.
Quantifying missingness across selected indicators
We first investigated the overall number of missing results and the proportion of missing values for each site and lab. The proportion missing for a given patient is defined as the total number of hospital days with no results for a given laboratory test divided by the total number of days admitted. We characterized the degree of missingness across variables of severity, sex, and time. We defined COVID severity by applying an EHR-based algorithm that defines severe patients based on the blood gas results (partial pressure of carbon dioxide or partial pressure of oxygen), medications (sedatives/anesthetics or treatment for shock), diagnoses (acute respiratory distress syndrome or ventilator-associated pneumonia), and procedures (endotracheal tube insertion or invasive mechanical ventilation) [35]. A patient with one or more of the aforementioned data elements was noted as severe; otherwise, the patient was assigned as non-severe. We assess for differences in quantiles of missingness between male and female patients and levels of severity.
We also investigated the proportion of missing laboratory values over different time intervals to capture the differences in patterns of laboratory missingness. To identify initial differences in missingness during the early days of hospital admission, we plotted on a heatmap the difference in proportion missing during the first 3 days of admission between severe and non-severe patients.
To identify changes in trends of missingness over time, we first modeled the rates of change of proportion missing over time for severe and non-severe patients separately and then plotted on a heatmap the difference in the rates of change. We obtained rates of change by fitting linear models across days since admission and obtaining the beta coefficients. We examined three different time intervals: 0–10 days, 0–30 days, and 0–60 days to capture patients with short, medium, and long term hospital stays. For 0–10 days and 0–30 days, linear models were fit across the proportion missing on each hospital day following admission for all patients. For 0–60 days, the models were fit over three-day periods.
Patterns of missingness shared between pairs of laboratory tests
We were interested in the temporal relationships between the various labs based on their missingness. For each lab, we compiled a list of missing indicators for each patient on each day since admission. We then calculated the Spearman correlation between two laboratory test pairs at a given time point based on the missing indicators. We repeated this out to 30 days after admission. Then, for each laboratory test pair, we fit a linear model across the Spearman correlation values across time points. A positive slope indicates that tests were not initially ordered or missing together and they become more concordant as the hospital admission continues; a negative slope indicates that tests are initially ordered or missing together and lose their concordance over time. Both of these dynamics could reveal biological and clinical mechanisms at work.
Patterns of missingness shared between groups of laboratory tests
Identifying relationships between variables based on missingness and ordering patterns
We also characterized the relationships between the labs themselves based on their missingness patterns. To this end, we employed Latent Dirichlet Allocation (LDA) topic modeling to identify similar labs based on their ordering and missingness patterns.
Topic modeling, or the discovery of hidden semantic structures in a text body, can help us identify some of the relationships between the labs based on their missingness patterns. Our “topics” would be groups or clusters of similar labs. To this end, we employed Latent Dirichlet Allocation (LDA) topic modeling.
LDA topic modeling is a generative probabilistic approach applied to collections of data, which in practice is typically text corpora [42]. In a natural language processing setting, documents are represented as random mixtures over latent topics, and each topic is characterized by a distribution over the words in the documents. The topic probabilities provide a representation of a document. In our case, the “words” are the individual labs, each “document” corresponds to each patient– the sum of indicators (whether or not a lab was ordered) for each lab across 30 days. The “topics” are the groups / clusters of similar labs based on those patterns. In essence, it is identifying which labs tend to co-occur, or be missing together. For more information on LDA topic modeling, see [42].
We first identified the optimal number of topics for our input data; and then LDA learns the probabilities that a given word belongs to a topic.
We used four metrics to determine the optimal number of topics to learn from the data; in order to accomplish this, we evaluated a range of two to eight topics. We assessed which of these maximized:
the held-out likelihood, which provides a measure of how predictive the model is on unseen documents;
the semantic coherence, which captures the tendency of a topic’s high probability words to co-occur in the same document;
the lower bound on the marginal likelihood;
and which minimized the residuals.
Because every laboratory test has a non-zero probability of belonging to a given topic, we determine a cut-off based on the sum of the cumulative probabilities in decreasing order. If the difference between 1 and the sum of the cumulative probabilities is <= 0.05, then only the labs that have been summed up to that cutoff are determined to describe the topic or make up the majority of the probability. In essence, the labs that make up a topic should be responsible for about 95% of the probability mass.
For each site, we generate a list of topics and the labs that describe that given topic based on their cumulative sum cut-off. From there, we are interested in groups of laboratory tests that intersect topics frequently across sites. We looked for the largest unique combination of labs that intersect at least nine times across all the topics from all the sites. Nine intersections was chosen empirically because it allowed for four unique groups of labs across the 15 sites. From there, we identified other lab tests that might intersect with a given established unique group at least six times to show the heterogeneity between topics.
Results
Quantifying missingness of laboratory test across sites
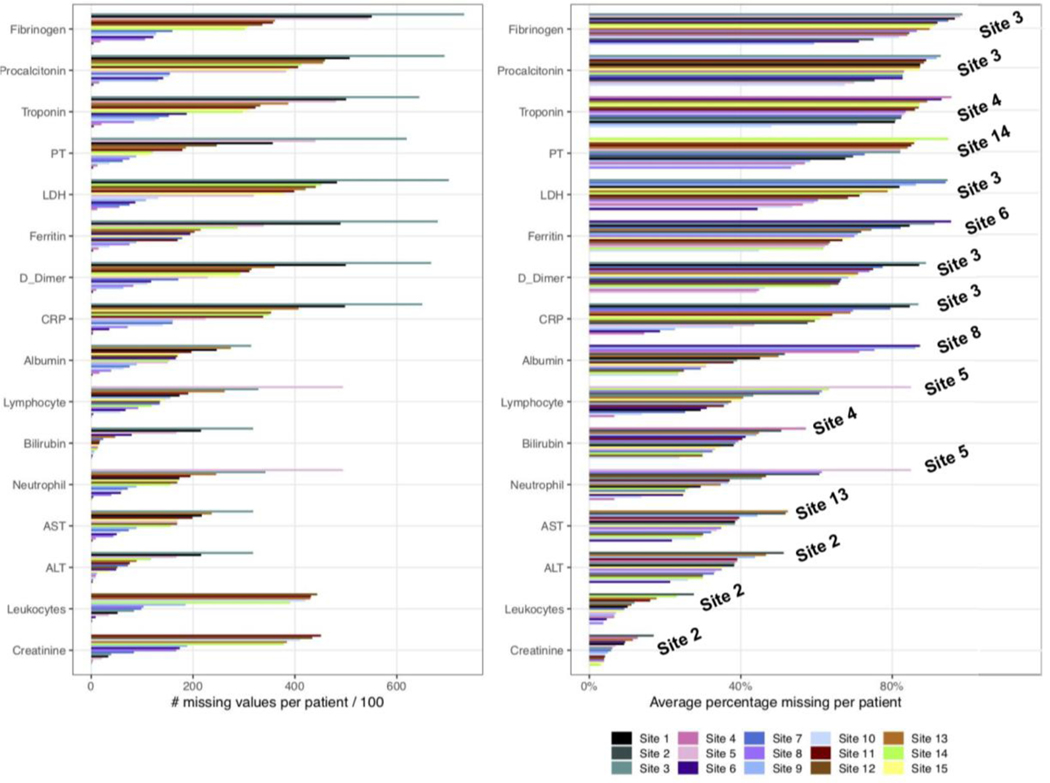
Figure 1 shows that the missingness across labs varies widely across sites and lab measures. The larger variability in the number of missing values per patient Figure 1 (A) compared to the average proportion missing per patient Figure 1 (B) across sites is possibly due to differences in the distribution of the length of stay and sample size at particular sites. Nonetheless, the number of missing values per patient still informs the exact number of missing tests and is useful for identifying sites with more valid values which might be required for some analyses. To account for the length of admission, the proportion missing (Figure 1 (B)) is normalized against the total number of admitted days for each patient. Generally, creatinine and leukocytes show the lowest number and proportion missing compared to all other tests. This is closely followed by bilirubin, albumin, ALT, and AST, which have a considerably lower number of average missing values per patient and a lower proportion missing (throughout patient admission) across most of the sites. Lymphocytes and neutrophils show a similar level of missingness as the above labs except for a visibly higher measure of missingness in Site 5.
Figure 1: Summary of missingness for lab measures across all sites.
(A) Number of missing labs is calculated for each patient before it is averaged over the total number of patients. (B) Proportion missing is calculated by taking into account the total number of days with no measurements of labs divided by the total number of admitted days for each patient. The proportion missing is averaged across all patients. The sites with the highest percentage missing are labeled on the right within the plot.
Quantifying missingness across sex and severity
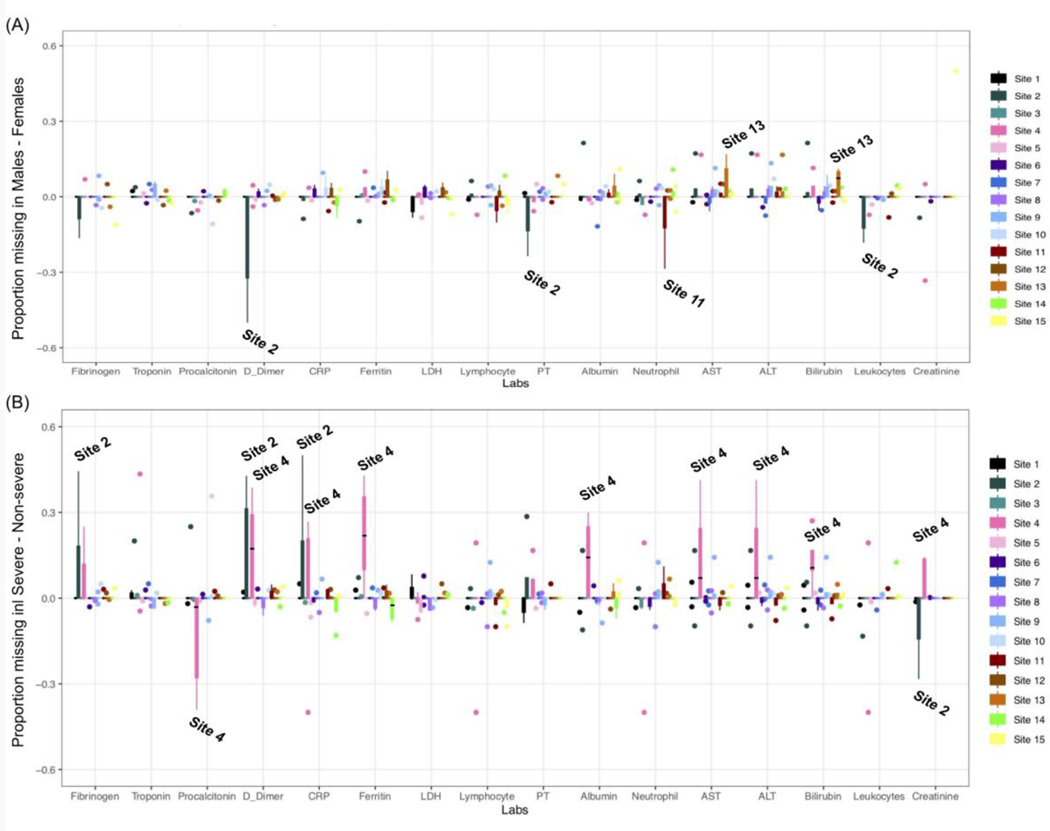
Figure 2(A) shows the difference in proportion missing between males and females for each lab. Generally, across all labs, the difference in missingness by sex is varied in both directions across all sites. We observe the largest difference in missingness between males and females at Site 2, with some female patients having much more missingness for D-dimer, PT, and leukocytes. We also observe some slight deviations at some of the sites with more missingness among females in neutrophils at Site 12 and more missingness among males at Site 13 in AST and bilirubin. It is important to note that Site 2 has the smallest sample size (n=162), and Sites 11–15 have very few female patients (ranging from 5.2–6.4 percent). Thus deviations are likely a result of variability in the data. Beyond these findings, we observe that most sites are well-balanced across the different sexes with regard to missingness in the data.
Figure 2: Difference in the quantiles of proportion missing across labs between (A) males and females as well as between (B) severe and non-severe patients.
Sites with the largest deviation in the proportion missing are labeled in the plots. For (A), sites with more missingness in the positive direction have more missingness in male populations. For (B), sites with more missingness in the positive direction have more missingness in severe populations.
Next, we quantified the difference in proportion missing between severe and non-severe patients across the entire cohort in Figure 2(B). We hypothesized for this experiment that there would be a higher proportion missing in non-severe patients. We observe in Figure 2(B) that most deviations occur at Site 2 and Site 4. We note more missing data in severe patients at Site 2 for fibrinogen, D-dimer, and CRP, and more missing procalcitonin results in non-severe patients. These attributes could be explained by the fact that Site 2 has a smaller sample size and is a pediatric hospital, hence the current severity definition might not be suited for its patients. At Site 4, we observe more missing data in severe patients for D-dimer, CRP, ferritin, albumin, AST, ALT, bilirubin, and creatinine while we observe more missing procalcitonin data in non-severe patients. Site 4 does not have an intensive care unit (ICU) which could explain the higher proportion of missingness in severe patients. Other than these two sites, the remaining sites seem to be well-balanced across levels of severity with regard to missingness in the data over the whole admission period.
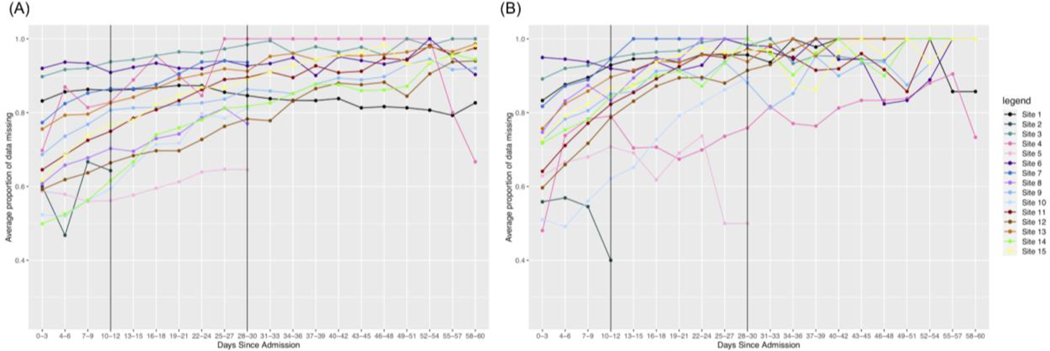
Quantifying missingness over course of admission
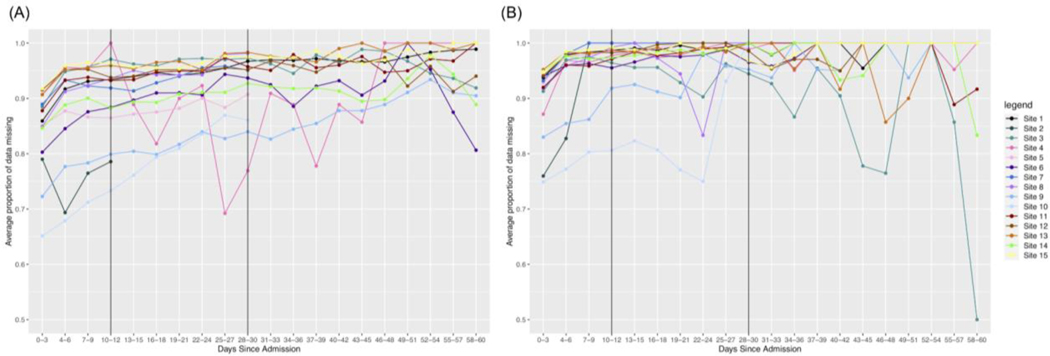
After quantifying overall missingness in severe vs. non-severe patients, we were also interested in the temporal dynamics of this missingness. We observe that for all labs, missingness increases faster over time in non-severe patients as opposed to severe patients. We present the results for three labs with varying amounts of initial missingness below.
Troponin overall has more missingness than all other labs. The range for possible missingness proportions is limited to [0,1] and initially we observe more missingness in the non-severe group as opposed to the severe group. Thus, many sites hit a maximum proportion of missingness much faster in the non-severe group, making the rate of change smaller than it would be for severe patients. We also see more variability in the non-severe group out to later days in the hospital admission.
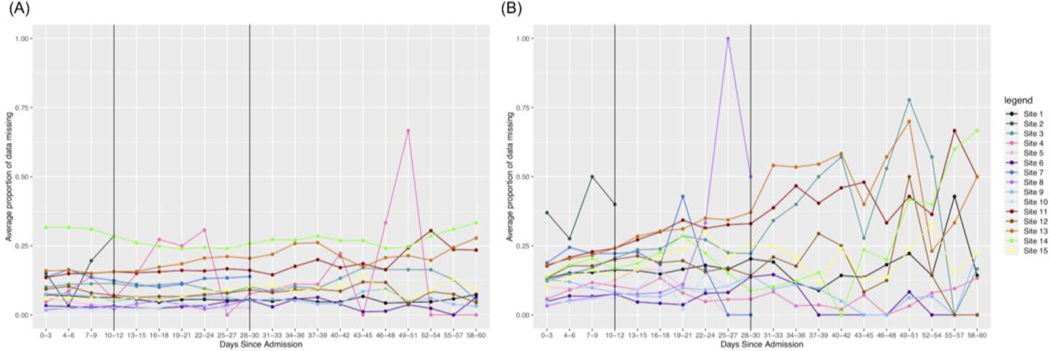
Another trend is that a test may initially have a higher rate of change in non-severe patients early on, but the rate of change is higher in severe patients out to 60 days. We see that this is the case for Ferritin, as shown in Figure 4. Initially, we see a stronger increase in missingness in the first 10 days for non-severe patients and this remains to be the case for many sites in 0–30 days. However, again, patients in the non-severe group reach the maximum level of missingness faster than patients in the severe group. Thus, because the severe patients take more time to reach the maximum level of missingness, many sites will have an overall greater rate of change in the severe group.
Figure 4. Changes in missing Ferritin results over time.
(A) severe patients and (B) non-severe patients. Since the range of proportions is limited to [0,1], and non-severe patients initially have more missingness than severe patients, we see that the rate of change across all time is higher for severe patients.
Lastly, some labs might have a consistently larger rate of change across time intervals for non-severe patients. We see in Table S1 that this is the case for Leukocytes, where this lab has a consistently lower amount of missingness compared to the other labs. Because the upper bound for missingness is further away from the initial amounts of missingness, we see that in non-severe patients it increases more quickly and is more pronounced overall.
Based on the trends we observe in the temporal line plots, we conclude that overall, there is more missingness in non-severe patients as opposed to severe patients over time across many of the labs in our dataset. This is what we might expect to see because clinicians might not test non-severe patients as heavily over time as severe patients who could be having more issues as their stay continues.
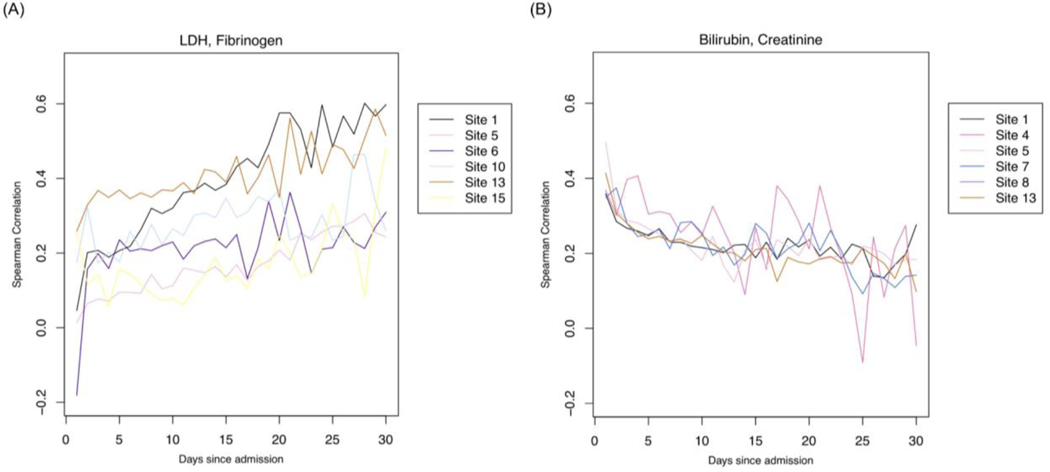
Patterns of missingness shared between pairs of laboratory tests
In identifying relationships in missingness across labs, we investigated laboratory test pairs that show a change in correlation across admission days (Figure 6). We then shortlisted test pairs with either a significant positive or negative change in correlation over admission days (Figure 6). Only test pairs that show reproducible correlations across sites are shortlisted. To obtain the rate of change, we fit linear models across the Spearman correlation values over time (Figure 6 (A) & (B)). Sites were included for a pair of the slope = |0.3| and standard error <= 0.25.
Figure 6. Correlations of pairs of laboratory tests.
Pairs of labs with (A) positive or (B) negative correlations through admission. Labs with position correlations are more strongly correlated in their missingness during later parts of admission; Labs with a negative association are more strongly correlated in their missingness during the earlier parts of admission.
In Table 2, we observe several lab pairs that become positively associated during the later parts of admission. Notable lab pairs indicate suspicion of infection, coagulopathy including cardiac involvement, liver involvement, severe COVID-19 outcomes as well as rule out of differential diagnoses such as bacterial pneumonia.
Table 2.
Lab pairs that are more strongly associated during later parts of admission and their implications
| Pair of Labs | # of Sites | Median Slope (Min, Max) | Implication |
|---|---|---|---|
| LDH, Fibrinogen | 6 | 0.591 (0.439, 1.608) | Suspicion of infection |
| Fibrinogen, D-dimer | 6 | 0.629 (0.4, 1.843) | Suspicion of coagulopathy |
| Fibrinogen, Procalcitonin | 7 | 0.489 (0.341, 1.6) | Suspicion of coagulopathy with cardiac involvement |
| LDH, Albumin | 6 | 0.5725 (0.33, 0.918) | Suspicion of severe COVID-related outcome |
| LDH, Bilirubin | 6 | 0.6685 (0.347, 0.882) | Suspicion of liver involvement |
| LDH, ALT | 6 | 0.572 (0.348, 0.698) | Suspicion of liver involvement |
| LDH, AST | 6 | 0.541 (0.342, 0.688) | Suspicion of liver involvement |
| Procalcitonin, Ferritin | 7 | 0.698 (0.364, 1.787) | Differential diagnosis- COVID-pneumonia vs. bacterial pneumonia |
| Procalcitonin, D-dimer | 6 | 0.7325 (0.657, 1.7) | Suspicion of severe COVID-related outcome |
In Table 3, we observe several lab pairs that become strongly associated during early admission. Notable lab pairs indicate suspicion of infection with or without kidney or liver involvement.
Table 3.
Lab pairs that are more strongly associated during earlier parts of admission and their implications
| Pair of Labs | # of Sites | Median Slope (Min, Max) | Implication |
|---|---|---|---|
| Bilirubin, Creatinine | 6 | −0.5065 (−0.809, −0.311) | Suspicion of kidney involvement |
| ALT, Creatinine | 6 | −0.499 (−0.74, −0.317) | Suspicion of severe COVID-related outcome due to liver and kidney involvement |
| Neutrophil, Creatinine | 6 | −0.44 (−1, −0.31) | Suspicion of infection with kidney involvement |
| Lymphocyte, Creatinine | 6 | −0.5535 (−0.994, −0.307) | Suspicion of infection with kidney involvement |
Patterns of missingness shared between groups of laboratory tests
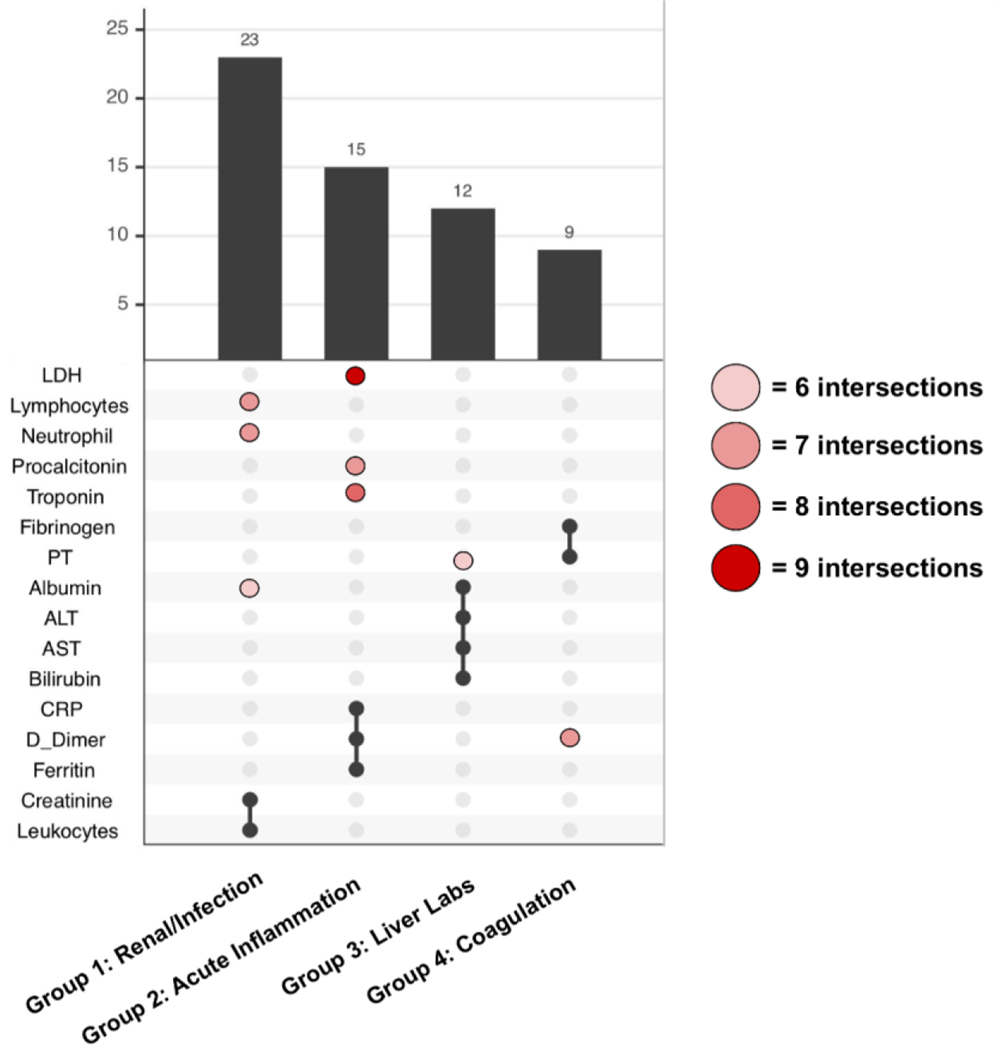
To identify groups of labs that share patterns in missingness, topic modeling was done to arrive at a common set of labs across sites (Figure 7(A)). From 15 sites, we end up with 91 topics derived from LDA. We include unique groups of labs with at least nine intersections across topics in an pSet plot. Based on these criteria, we obtained four unique groups that are reproducible across sites presented in Figure 7(A). The black points represent the labs that are consistent across all topics found across sites while the pink and red ones represent labs that are only found in some but not all of the topics across sites (Figure 7(A)). We find that overall, the groups of intersections represent groups of labs that measure for similar issues in a COVID-19 setting, and thus are related to one another. Group 1 consists of labs that are commonly ordered together on a daily basis, measuring for renal issues and infection. Group 2 are likely tests ordered individually rather than as a group. They all are labs that have at times been thought to help prognosticate potential severity of COVID. Group 3 represents tests that are ordered together to assess liver function, and Group 4 represents coagulation studies that may or may not be ordered as a group.
Figure 7: Grouped labs with similar patterns of missingness.
Prevalence of the top four groups of labs from topic modeling analyses across sites. The red/pink points show slight deviations in group membership of labs across different sites.
Discussion
Through characterizing and exploring laboratory test missingness patterns across a multi-national COVID-19 study, we identified several insights and opportunities [1].
1. Missingness patterns can indicate responses to hospital treatment capacity
During the global pandemic, several academic medical centers were not prepared for the large influx of patients with intensive care needs. Many created makeshift ICUs. Furthermore, some facilities may have implemented new triaging procedures. For example, the higher proportion of missing data in the initial three days of admission in Site 4 across almost all the labs could be attributed to the lack of an ICU in Site 4. Because patients with serious conditions might be in the process of having a transfer being arranged, it could explain the higher proportion of missing labs in the severe patients from Site 4.
2. Missingness patterns can change over time and at different rates
Although all patient groups exhibit some proportion of non-reported labs, not all patient groups consistently demonstrate the same rate of non-reported labs over time. This finding echoes one study that found that counting lab measurements from an arbitrary, rather than clock, time is superior in attaining a stable time series [45]. Patients among the non-severe group reach the maximum level of non-ordered labs faster than patients among the severe group. For short-stay patients, we observe this change for labs like troponin. For ferritin, we observe a higher rate of change in non-severe patients. Intuitively, for non-severe patients, clinicians may become less concerned about a patient’s condition, leading to reduced ordering rates. As a corollary, the absence of test ordering can indicate improvements in the patient’s health.
3. Missingness patterns could be predictive of clinical outcomes
Missingness patterns in healthcare data also carries a signal within itself. Previous work has shown that missing data can indicate a variety of issues: access to healthcare, the health status of patients, all of which can impact disease models [53]. In this setting, because the reporting patterns between labs are cross-correlated (groups of labs with shared reporting (and non-reporting) patterns as shown in topic modeling Figure 7), the removal of one test’s missing values from a model could potentially affect model performance. Furthermore, using two different data-driven methods -- spearman correlations over time as well as topic models -- we observed common clinical themes among lab pairs and sets including coagulation, infection with renal involvement, and liver involvement. For future work, it would be interesting to see how the collective missingness in the groups identified by LDA topic modeling is associated with certain comorbidities such as thrombotic events, neurological outcomes, and acute respiratory distress. We also plan to incorporate concepts from causal inference, such as propensity score matching to ensure that the associations found between missingness and comorbidities are reliable.
4. Computational approaches for addressing ordering patterns
Missingness patterns could inform which variables would benefit from imputation for future studies. A number of approaches to dealing with informative missingness have been reported. These include using a Monte Carlo Expectation-Maximization simulation series that incorporates within-subject autocorrelation with a latent autoregressive process for longitudinal binary data [46], a Bayesian shrinkage approach to improve estimation of sparse patterns [47], the use of an informative ordering pattern odds ratio [48–50]. Another approach could extend the PopKLD algorithm for high-throughput phenotyping to deal with informative missingness [51]. This could be especially valuable in our continued work in temporal pattern estimation in the face of informative missing data that will investigate these methods in the context of these COVID consortium data [52].
This study has several limitations.
Order sets were not investigated. The patterns of missingness for laboratory and other tests and procedures were likely influenced by standing order sets. Furthermore, order sets are likely to change over time as knowledge about COVID and therapies for it evolve over time. We plan to correlate patterns of missingness with the content of order sets and how they change over the course of the pandemic.
Missingness was not correlated with secular trends of the pandemic. As the pandemic has evolved, there have been a number of irregular cycles in the epidemic curve, with marked changes in disease incidence associated with the Delta and Omicron variants. These changes are likely associated with corresponding changes in order sets and clinical practice. We will investigate patterns of laboratory test missingness as they may reflect clinical practice patterns which in turn may reflect the undulation in the incidence of COVID over time.
Type of patient care unit was not captured. It is possible that test ordering patterns could be influenced by the type of unit a patient was on at given points in time. For example, order sets may be used less frequently in intensive care units than in medical units, even those dedicated to COVID. Our future work will include an examination of “unit effect” in a temporal context, which will be important as patients move between acute and less-acute care settings in a given hospitalization.
Focus on missing test values, as opposed to missing orders. We assumed that a missing laboratory test result indicated that the test was not ordered. While a reasonable assumption, a more accurate indicator of ordering behavior would be to capture orders in addition to test results. We will investigate the feasibility of obtaining these data from the EHR in future work.
Severity definitions might change over time. When patients stay in the hospital for long periods of time, it is possible that their severity changes. Thus, there are limitations in looking at Figure 4 due to the fact that patients initially labeled as non-severe that remain in the hospital out to 60 days might not actually be non-severe at that point.
Conclusion
In this study, we investigated and demonstrated how characterization of missing data patterns in EHRs, particularly lab results, could support various steps in scientific study ranging from hypothesis generation to inferential analyses. Our key takeaway is that missingness patterns can relate to hospital treatment capacity and reflect heterogeneity when looking at the disease over time and at multiple sites, where there might be different phases, policies. Changes in missingness could potentially reflect a change in patient condition and suggest correlations to clinical outcomes like coagulation, infection with renal involvement, and liver involvement. Furthermore, missing data patterns will enable consortia to identify which sites are better poised to study particular questions and potentially inform the use of imputation methods for addressing these challenges. Finally, our results may provide insights into some of the biological relationships between labs in EHRs data for COVID-19 patients.
Supplementary Material
Figure 3. Changes in missing Troponin results over time.
(A) severe patients and (B) non-severe patients. Since the range of proportions is limited to [0,1], and non-severe patients initially have more missingness than severe patients, we see that the rate of change across all time is higher for severe patients.
Figure 5. Changes in missing Leukocytes over time.
(A) severe patients and (B) non-severe patients. Since the range of proportions is limited to [0,1], and non-severe patients initially have more missingness than severe patients, we see that the rate of change across all time is higher for non-severe patients.
Acknowledgements
MM is supported by National Center for Advancing Translational Sciences (NCATS) UL1 TR001857. DAH is supported by NCATS UL1TR002240. WY is supported by National Institutes of Health (NIH) T32HD040128. BJA is supported by National Heart, Lung, and Blood Institute (NHLBI) U24 HL148865. WGL is supported by National Library of Medicine (NLM) R00LM012926. YL is supported by R01LM013337. SNM is supported by NCATS 5UL1TR001857-05 and National Human Genome Research Institute (NHGRI) 5R01HG009174-04. GSO is supported by NIH grants U24CA210967 and P30ES017885. LPP is supported by NCATS CTSA Award UL1TR002366. SV is supported by NCATS UL1TR001857. GMW is supported by NCATS UL1TR002541, NCATS UL1TR000005, NLM R01LM013345, and NHGRI 3U01HG008685-05S2. ZX is supported by National Institute of Neurological Disorders and Stroke (NINDS) R01NS098023 and R01NS124882. QL is supported by National Institute of General Medical Sciences (NIGMS) R01GM124111 and NIH National Institute on Aging RF1AG063481. DLM is supported by NCATS UL1-TR001878. JHH is supported by NCATS UL1-TR001878.
Footnotes
CRediT Author Statement
Tan: Conceptualization, Methodology, Formal analysis, Validation, Resources, Software, Data curation, Writing – Original Draft, Writing – Review and Editing, Visualization, Approval; Getzen: Conceptualization, Methodology, Formal analysis, Validation, Resources, Software, Data curation, Writing – Original Draft, Writing – Review and Editing, Visualization, Approval; Hutch: Data Curation, Writing – Review and Editing, Visualization, Approval; Strasser: Formal analysis, Writing – Review and Editing, Validation, Visualization, Approval; Gutiérrez-Sacristán: Data Curation, Formal analysis, Writing – Review and Editing; Le: Conceptualization, Methodology, Software, Formal analysis, Writing – Original Draft, Writing – Review and Editing, Visualization, Approval; Dagliati: Conceptualization, Methodology, Software, Formal analysis, Writing – Original Draft, Writing – Review and Editing, Visualization, Approval; Morris: Data Curation, Writing – Review and Editing, Approval; Hanauer: Data Curation, Writing – Review and Editing, Approval; Moal: Data Curation, Writing – Review and Editing, Approval; Bonzel: Writing – Review and Editing, Approval; Yuan: Writing – Review and Editing, Approval; Chiudinelli: Data Curation, Approval; Das: Formal analysis, Approval; Zhang: Data Curation, Formal analysis, Writing – Review and Editing, Approval; Aronow: Data Curation, Formal analysis, Writing – Review and Editing, Visualization, Approval; Avillach: Data Curation, Formal analysis, Writing – Review and Editing, Visualization, Approval; Brat: Conceptualization, Methodology, Formal analysis, Writing – Review and Editing, Visualization, Approval; Cai: Conceptualization, Methodology, Formal analysis, Writing – Review and Editing, Visualization, Approval; Hong: Conceptualization, Formal analysis, Writing – Review and Editing, Visualization, Approval; La Cava: Formal analysis, Writing – Review and Editing, Visualization, Approval; Loh: Data Curation, Approval; Murphy: Conceptualization, Data curation, Writing – Review and Editing, Visualization, Approval; Ngiam: Data curation, Writing – Review and Editing, Approval; Omenn: Conceptualization, Writing – Review and Editing, Visualization, Approval; Patel: Data curation, Formal analysis, Writing – Review and Editing, Approval; Samayamuthu: Data curation, Writing – Review and Editing, Approval; Shriver: Data curation, Writing – Review and Editing, Approval; Shakeri Hossein Abad: Formal analysis, Writing – Review and Editing, Visualization, Approval; Tan BWL: Data curation, Writing – Review and Editing, Approval; Tan BWQ: Conceptualization, Formal analysis, Writing – Review and Editing, Approval; Visweswaran: Data curation, Writing – Review and Editing, Approval; Wang: Formal analysis, Writing – Review and Editing, Approval; Weber: Data curation, Writing – Review and Editing, Approval; Xia: Data curation, Writing – Review and Editing, Approval; Verdy: Data curation, Writing – Review and Editing, Approval; Long: Conceptualization, Methodology, Formal analysis, Validation, Resources, Data curation, Writing – Original Draft, Writing – Review and Editing, Visualization, Supervision, Approval; Mowery: Conceptualization, Methodology, Formal analysis, Validation, Resources, Data curation, Writing – Original Draft, Writing – Review and Editing, Visualization, Supervision, Approval; Holmes: Conceptualization, Methodology, Formal analysis, Validation, Resources, Data curation, Writing – Original Draft, Writing – Review and Editing, Visualization, Supervision, Approval.
References
- 1.Denny JC. Chapter 13: Mining electronic health records in the genomics era. PLoS Comput Biol. 2012;8: e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush RA, Connelly CD, Pérez A, Barlow H, Chiang GJ. Extracting autism spectrum disorder data from the electronic health record. Appl Clin Inform. 2017;8: 731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apte M, Neidell M, Furuya EY, Caplan D, Glied S, Larson E. Using electronically available inpatient hospital data for research. Clin Transl Sci. 2011;4: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dittmar MS, Zimmermann S, Creutzenberg M, Bele S, Bitzinger D, Lunz D, et al. Evaluation of comprehensiveness and reliability of electronic health records concerning resuscitation efforts within academic intensive care units: a retrospective chart analysis. BMC Emerg Med. 2021;21: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farmer R, Mathur R, Bhaskaran K, Eastwood SV, Chaturvedi N, Smeeth L. Promises and pitfalls of electronic health record analysis. Diabetologia. 2018;61: 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayley KB, Belnap T, Savitz L, Masica AL, Shah N, Fleming NS. Challenges in using electronic health record data for CER: experience of 4 learning organizations and solutions applied. Med Care. 2013;51: S80–6. [DOI] [PubMed] [Google Scholar]
- 7.Samal L, Dykes PC, Greenberg JO, Hasan O, Venkatesh AK, Volk LA, et al. Care coordination gaps due to lack of interoperability in the United States: a qualitative study and literature review. BMC Health Serv Res. 2016;16: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agniel D, Kohane IS, Weber GM. Biases in electronic health record data due to processes within the healthcare system: retrospective observational study. BMJ. 2018. p. k1479. doi: 10.1136/bmj.k1479 [DOI] [PMC free article] [PubMed]
- 9.Aerts H, Kalra D, Sáez C, Ramírez-Anguita JM, Mayer M-A, Garcia-Gomez JM, et al. Quality of Hospital Electronic Health Record (EHR) Data Based on the International Consortium for Health Outcomes Measurement (ICHOM) in Heart Failure: Pilot Data Quality Assessment Study. JMIR Med Inform. 2021;9: e27842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argalious MY, Dalton JE, Sreenivasalu T, O’Hara J, Sessler DI. The association of preoperative statin use and acute kidney injury after noncardiac surgery. Anesth Analg. 2013;117: 916–923. [DOI] [PubMed] [Google Scholar]
- 11.Chang C, Deng Y, Jiang X, Long Q. Multiple imputation for analysis of incomplete data in distributed health data networks. Nat Commun. 2020;11: 5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman SS, Davlyatov G, Hall AG. Toward Understanding the Value of Missing Social Determinants of Health Data in Care Transition Planning. Appl Clin Inform. 2020;11: 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gianfrancesco MA, Goldstein ND. A narrative review on the validity of electronic health record-based research in epidemiology. BMC Med Res Methodol. 2021;21: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. Journal of the American Medical Informatics Association. 2013. pp. 117–121. doi: 10.1136/amiajnl-2012-001145 [DOI] [PMC free article] [PubMed]
- 15.Petrazzini BO, Naya H, Lopez-Bello F, Vazquez G, Spangenberg L. Evaluation of different approaches for missing data imputation on features associated to genomic data. BioData Min. 2021;14: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sammon CJ, Miller A, Mahtani KR, Holt TA, McHugh NJ, Luqmani RA, et al. Missing laboratory test data in electronic general practice records: analysis of rheumatoid factor recording in the clinical practice research datalink. Pharmacoepidemiol Drug Saf. 2015;24: 504–509. [DOI] [PubMed] [Google Scholar]
- 17.Che Z, Purushotham S, Cho K, Sontag D, Liu Y. Recurrent Neural Networks for Multivariate Time Series with Missing Values. Sci Rep. 2018;8: 6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen AS, Collins JS, Rathouz PJ, Selander CL, Satten GA. Bootstrap calibration of TRANSMIT for informative missingness of parental genotype data. BMC Genet. 2003;4 Suppl 1: S39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen AS, Rathouz PJ, Satten GA. Informative missingness in genetic association studies: case-parent designs. Am J Hum Genet. 2003;72: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James I, McKinnon E, Gaudieri S, Morahan G, Diabetes Genetics Consortium. Missingness in the T1DGC MHC fine-mapping SNP data: association with HLA genotype and potential influence on genetic association studies. Diabetes Obes Metab. 2009;11 Suppl 1: 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kujala M, Nevalainen J. A case study of normalization, missing data and variable selection methods in lipidomics. Stat Med. 2015;34: 59–73. [DOI] [PubMed] [Google Scholar]
- 22.Lin W-Y, Liu N. Reducing bias of allele frequency estimates by modeling SNP genotype data with informative missingness. Front Genet. 2012;3: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu SH, Erion G, Novitsky V, De Gruttola V. Viral Genetic Linkage Analysis in the Presence of Missing Data. PLoS One. 2015;10: e0135469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butera NM, Zeng D, Green Howard A, Gordon-Larsen P, Cai J. A doubly robust method to handle missing multilevel outcome data with application to the China Health and Nutrition Survey. Stat Med. 2022;41: 769–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu MC, Follmann DA. Use of summary measures to adjust for informative missingness in repeated measures data with random effects. Biometrics. 1999;55: 75–84. [DOI] [PubMed] [Google Scholar]
- 26.Chaimani A, Mavridis D, Higgins JPT, Salanti G, White IR. Allowing for informative missingness in aggregate data meta-analysis with continuous or binary outcomes: Extensions to metamiss. Stata J. 2018;18: 716–740. [PMC free article] [PubMed] [Google Scholar]
- 27.Harris RG, Batterham M, Neale EP, Ferreira I. Impact of missing outcome data in meta-analyses of lifestyle interventions during pregnancy to reduce postpartum weight retention: An overview of systematic reviews with meta-analyses and additional sensitivity analyses. Obes Rev. 2021;22: e13318. [DOI] [PubMed] [Google Scholar]
- 28.Kahale LA, Khamis AM, Diab B, Chang Y, Lopes LC, Agarwal A, et al. Potential impact of missing outcome data on treatment effects in systematic reviews: imputation study. BMJ. 2020. p. m2898. doi: 10.1136/bmj.m2898 [DOI] [PMC free article] [PubMed]
- 29.Mavridis D, Salanti G, Furukawa TA, Cipriani A, Chaimani A, White IR. Allowing for uncertainty due to missing and LOCF imputed outcomes in meta-analysis. Stat Med. 2019;38: 720–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mavridis D, White IR, Higgins JPT, Cipriani A, Salanti G. Allowing for uncertainty due to missing continuous outcome data in pairwise and network meta-analysis. Stat Med. 2015;34: 721–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White IR, Higgins JPT, Wood AM. Allowing for uncertainty due to missing data in meta-analysis--part 1: two-stage methods. Stat Med. 2008;27: 711–727. [DOI] [PubMed] [Google Scholar]
- 32.Lyles RH, Allen AS, Dana Flanders W, Kupper LL, Christensen DL. Inference for case-control studies when exposure status is both informatively missing and misclassified. Statistics in Medicine. 2006. pp. 4065–4080. doi: 10.1002/sim.2500 [DOI] [PubMed]
- 33.Groenwold RHH. Informative missingness in electronic health record systems: the curse of knowing. Diagn Progn Res. 2020;4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brat GA, Weber GM, Gehlenborg N, Avillach P, Palmer NP, Chiovato L, et al. International electronic health record-derived COVID-19 clinical course profiles: the 4CE consortium. NPJ Digit Med. 2020;3: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klann JG, Weber GM, Estiri H, Moal B, Avillach P, Hong C, et al. Validation of an Internationally Derived Patient Severity Phenotype to Support COVID-19 Analytics from Electronic Health Record Data. J Am Med Inform Assoc. 2021. doi: 10.1093/jamia/ocab018 [DOI] [PMC free article] [PubMed]
- 36.Weber GM, Hong C, Palmer NP, Avillach P, Murphy SN, Gutiérrez-Sacristán A, et al. International Comparisons of Harmonized Laboratory Value Trajectories to Predict Severe COVID-19: Leveraging the 4CE Collaborative Across 342 Hospitals and 6 Countries: A Retrospective Cohort Study. medRxiv. 2021. doi: 10.1101/2020.12.16.20247684 [DOI]
- 37.Le TT, Gutiérrez-Sacristán A, Son J, Hong C, South AM, Beaulieu-Jones BK, et al. Multinational characterization of neurological phenotypes in patients hospitalized with COVID-19. Sci Rep. 2021;11: 20238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bourgeois FT, Gutiérrez-Sacristán A, Keller MS, Liu M, Hong C, Bonzel C-L, et al. International Analysis of Electronic Health Records of Children and Youth Hospitalized With COVID-19 Infection in 6 Countries. JAMA Netw Open. 2021;4: e2112596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Estiri H, Strasser ZH, Brat GA, Semenov YR, Consortium for Characterization of COVID-19 by EHR (4CE), Patel CJ, et al. Evolving phenotypes of non-hospitalized patients that indicate long COVID. BMC Med. 2021;19: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan BWL, Tan BWQ, Tan ALM, Schriver ER, Gutiérrez-Sacristán A, Das P, et al. Long-term kidney function recovery and mortality after COVID-19-associated acute kidney injury: An international multi-centre observational cohort study. eClinicalMedicine. 2023;55. doi: 10.1016/j.eclinm.2022.101724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudolf JW, Dighe AS, Coley CM, Kamis IK, Wertheim BM, Wright DE, et al. Analysis of Daily Laboratory Orders at a Large Urban Academic Center: A Multifaceted Approach to Changing Test Ordering Patterns. Am J Clin Pathol. 2017;148: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Website. Available: https://shmpublications.onlinelibrary.wiley.com/doi/full/10.1002/jhm.2354?casa_token=Tqb8-Vv7sbgAAAAA%3AT5YCnn2VadQTjLM83pk6_zI2fmn9nzKPCGzzf_KE8SRDoSa1ZyCioazf0eCn6nxV3fuV2bul6FD0DpE
- 43.Journal of Hospital Medicine. [cited 19 Apr 2022]. doi: 10.1002/(ISSN)1553-5606 [DOI]
- 44.Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. J Mach Learn Res. 2003;3: 993–1022. [Google Scholar]
- 45.Hripcsak G, Albers DJ, Perotte A. Parameterizing time in electronic health record studies. Journal of the American Medical Informatics Association. 2015. pp. 794–804. doi: 10.1093/jamia/ocu051 [DOI] [PMC free article] [PubMed]
- 46.Albert PS, Follmann DA, Wang SA, Suh EB. A latent autoregressive model for longitudinal binary data subject to informative missingness. Biometrics. 2002;58: 631–642. [DOI] [PubMed] [Google Scholar]
- 47.Gaskins JT, Daniels MJ, Marcus BH. Bayesian methods for nonignorable dropout in joint models in smoking cessation studies. J Am Stat Assoc. 2016;111: 1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Higgins JPT, White IR, Wood AM. Imputation methods for missing outcome data in meta-analysis of clinical trials. Clin Trials. 2008;5: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spineli LM. An empirical comparison of Bayesian modelling strategies for missing binary outcome data in network meta-analysis. BMC Med Res Methodol. 2019;19: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spineli LM, Kalyvas C, Pateras K. Participants’ outcomes gone missing within a network of interventions: Bayesian modeling strategies. Stat Med. 2019;38: 3861–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albers DJ, Elhadad N, Claassen J, Perotte R, Goldstein A, Hripcsak G. Estimating summary statistics for electronic health record laboratory data for use in high-throughput phenotyping algorithms. Journal of Biomedical Informatics. 2018. pp. 87–101. doi: 10.1016/j.jbi.2018.01.004 [DOI] [PMC free article] [PubMed]
- 52.Weber GM, Zhang HG, L’Yi S, Bonzel C-L, Hong C, Avillach P, et al. International Changes in COVID-19 Clinical Trajectories Across 315 Hospitals and 6 Countries: Retrospective Cohort Study. J Med Internet Res. 2021;23: e31400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Getzen E, Ungar L, Mowery D, Jiang X, Long Q. Mining for Equitable Health: Assessing the Impact of Missing Data in Electronic Health Records. Journal of Biomedical Informatics. 2023;104269. doi: 10.1016/j.jbi.2022.104269. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.