Abstract
The cysteine proteinases referred to as gingipains R (gingipain R1 and gingipain R2) and gingipain K produced by Porphyromonas gingivalis are virulence factors of this periodontal pathogen which likely act by interrupting host defense mechanisms and by participating in the penetration and destruction of host connective tissue. To examine the effect of immunization with gingipains R on the ability of P. gingivalis to colonize and invade in the mouse chamber model, BALB/c mice were immunized intraperitoneally with the 95-kDa gingipain R1, the 50-kDa gingipain R2, or multiple antigenic peptide (MAP)-conjugated gingipain R-derived peptides and then challenged with P. gingivalis. Immunization of mice with the 95-kDa gingipain R1, the 50-kDa gingipain R2, or a peptide derived from the N-terminal sequence of the catalytic domain of gingipains R (peptide A) followed by challenge with P. gingivalis A7436 resulted in protection from P. gingivalis invasion. In contrast, immunization with peptides corresponding to either a sequence encompassing the catalytic cysteine residue of gingipains R (peptide B) or an identical sequence within the catalytic domains of gingipain R1 and gingipain K (peptide C), followed by challenge with P. gingivalis, did not protect animals, nor did immunization with a peptide corresponding to sequences within the adhesion/hemagglutinin domain of gingipain R1 (peptide D) which have been shown to be directly involved in the hemagglutinin activity of gingipain R1. However, the immunoglobulin G (IgG) titer obtained following immunization with peptide D was comparable to that obtained following immunization with the N-terminal peptide (peptide A). Competitive enzyme-linked immunosorbent assays, using either the 95-kDa gingipain R1 or gingipain K as the competing soluble antigen, indicated that 42 and 53% of the antibodies induced by immunization with heat-killed bacteria recognize gingipain R1 and gingipain K, respectively; however, even at very high concentrations, the 50-kDa gingipain R2 did not hinder IgG binding to P. gingivalis. These results indicate that antibodies directed to the amino-terminal region of the catalytic domain of gingipains R are capable of inducing a protective immune response against P. gingivalis infection in the mouse chamber model.
Porphyromonas gingivalis has been implicated as a major etiological agent in periodontitis (6), which becomes a chronic state typified by extensive tissue degradation, with eventual bone erosion and tooth loss (53). A number of virulence factors have been implicated in the pathogenicity of P. gingivalis, with the cysteine proteinases having received considerable attention due to their ability to activate and/or degrade a broad range of host proteins (42). The cysteine proteinases referred to as gingipains cleave synthetic and natural substrates after arginine or lysine residues. Recent in vitro data support the role of gingipains R in the inflammatory process by the generation of C5a and the activation of prokallikrein, with subsequent bradykinin release from kininogen (7, 43). This cascade of events induced by gingipains R has been postulated to contribute to the pathogenic potential of P. gingivalis in vivo (20, 21, 54). Gingipain K may also be involved in disease progression, primarily through binding and degradation of fibrinogen and fibronectin (7, 40, 48).
Arginine-specific gingipains are encoded by two related genes, while lysine-specific gingipains are encoded by only a single genetic locus (reviewed in reference 12). For gingipain R1 and gingipain K, the translated part of each gene, rgp1 and kgp, respectively, encodes a prepropeptide, a catalytic domain, and a hemagglutinin domain, and the initial polyprotein is apparently subject to posttranslational processing. The major forms of high-molecular-mass gingipains, gingipain R1 and gingipain K, released by P. gingivalis HG66 are purified as noncovalent complexes of the catalytic domain with several polypeptide chains (GP44, GP19, GP17, and GP27) derived from the nascent hemagglutinin domain via putative proteolytic processing after one lysine and three arginine residues (39). The hemagglutinin domains of gingipain R1 and gingipain K are nearly identical in primary sequence in each enzyme. The catalytic domains of both gingipain R1 and gingipain K share only limited identity (27%) scattered throughout the polypeptide chain, except for an identical 30-amino-acid residue fragment located in the C-terminal part of the molecule. In contrast to gingipain R1, gingipain R2 is expressed as a preproenzyme missing the majority of hemagglutinin domain but otherwise closely related to the rgp1 gene product (29). The N-terminal two-thirds of the primary structure of mature gingipain R2, containing the active-site cysteine residue and putative histidine residue of the catalytic dyad, is nearly identical with the catalytic domain of gingipain R1, but the two structures differ considerably at the C terminus (31).
In P. gingivalis W50, an alternative proteolytic processing of the initial polypeptide encoded by the prpR1 gene, a gene basically identical to rgp1, apparently leads to assembly of a slightly different noncovalent complex composed of the catalytic (a-chain) and the hemagglutinin (b-chain) domains (2). Posttranslational modification of the catalytic domain can give rise to three biochemically distinct enzymes, a dimer and two catalytic monomers, one unmodified and one lipopolysaccharide modified (45). In this regard, the 95-kDa gingipain R1 is an equivalent of the dimer, while the 50-kDa gingipain R2 is analogous to unmodified catalytic monomer, a product of homologous gene designated as prpR2 in strain W50 (45).
Antibodies specific for gingipains are produced in patients with adult periodontitis patients, with the majority being reactive with antigenic determinants within the hemagglutinin/adhesion domain of gingipain R1 (22). However, the function of these antibodies has not been elucidated. Patients with a history of destructive periodontal disease frequently demonstrate an elevated immunoglobulin G (IgG) response to P. gingivalis; however, these antibodies are apparently ineffective at limiting continued disease progression (16, 28, 51, 55). Indeed, in several animal studies, induction of an immune response to P. gingivalis antigens has been demonstrated to actually exacerbate disease (reviewed in reference 26). In this report, we describe animal experiments which were designed to determine the protective effect of P. gingivalis-specific antibodies produced against functionally defined peptide fragments derived from the catalytic and hemagglutinin/adhesion domains of the 95-kDa gingipain R1.
MATERIALS AND METHODS
Bacteria.
P. gingivalis A7436 is the virulent strain used in our previous studies in the mouse chamber model (13, 14). Strain A7436 expresses gingipains R1 and R2 and contains the rgp1 and rgp2 genes as detected by enzymatic activity and Northern blot analysis, respectively (unpublished data). P. gingivalis A7436 was typically maintained on anaerobic blood agar plates (Difco) and grown for 2 to 3 days under anaerobic conditions as previously described (14). For mouse challenge studies, P. gingivalis cultures were grown for 18 h in anaerobic broth MIC (Difco), washed, and adjusted to a final cell density of 1010 CFU/ml. Mice were infected with approximately 109 CFU of P. gingivalis A7436 directly into subcutaneous chambers implanted in mice as described below. Exact CFU used for each experiment was determined by serially diluting the inoculum and plating on anaerobic blood agar followed by incubation for 7 days under anaerobic conditions.
Antigen preparation.
Whole-cell antigens used for immunization were prepared by centrifugation of P. gingivalis cultures for 10 min at 10,000 × g at room temperature and resuspended in 1/10 the original volume in anaerobic broth. Bacterial cells (approximately 108 CFU, as determined by serially diluting cultures prior to heat inactivation) were heated to 95°C for 10 min; the heat-treated preparations were plated on anaerobic blood agar and incubated for 7 days under anaerobic conditions to confirm effective killing. Gingipains were purified from strain HG66 (39), and multiple antigenic peptide (MAP)-conjugated peptides were synthesized (49) at the University of Georgia Core Facility (Athens, Ga.). The amount of antigen used for immunization was quantified by dry weight for the MAP-conjugated peptides and by bicinchoninic acid protein assay kit (Sigma Chemical Co., St. Louis, Mo.) for the purified gingipains.
Experimental animals.
Female BALB/c mice 4 to 6 weeks of age (Charles River Laboratories) were used for these experiments. The total number of mice used for each group is indicated in Table 1. Approval to conduct these studies was obtained from the Animal Use Committee at Morehouse School of Medicine (Atlanta, Ga.). Mice were immunized by injection of each immunogen (50 μg/mouse in Freund’s complete adjuvant) in subcutaneous chambers implanted in mice (13). Animals immunized with heat-killed P. gingivalis received an initial immunization corresponding to approximately 108 CFU (as determined prior to heat inactivation). Control mice were immunized with Freund’s adjuvant only. Seven days after primary immunization, mice were boosted (10 times) at 3-day intervals with the 95-kDa gingipain R1, the 50-kDa gingipain R2, or MAP-conjugated peptides (50 μg/mouse in Freund’s incomplete adjuvant). Animals immunized with P. gingivalis were boosted (10 times) at 3-day intervals with heat-killed P. gingivalis corresponding to approximately 102 CFU (as determined prior to heat inactivation). At 14, 21, and 28 days postimmunization, chamber fluid was removed with a hypodermic needle and syringe to assess the IgG response to P. gingivalis and gingipains.
TABLE 1.
Pathological course of P. gingivalis infection in immunized mice
| Group | Total no. of mice | Lesionsa | Deathsb | Cachexiac |
|---|---|---|---|---|
| Nonimmunized | 22 | 14/22 | 14/22 | +++++ |
| Peptide A | 32 | 1/32 | 0/32 | + |
| Scrambled peptide Ad | 8 | 5/8 | 5/8 | ++++ |
| Peptide D | 8 | 6/8 | 6/8 | +++ |
| Scrambled peptide Dd | 8 | 4/8 | 0/8 | ++++ |
| Whole cells | 24 | 0/24 | 0/24 | + |
| Gingipain R1 | 22 | 0/22 | 0/22 | ++ |
| Gingipain R2 | 24 | 0/24 | 0/22 | + |
Number of mice with secondary lesions on the ventral abdomen/total of mice tested as detected on day 7.
Number of dead mice/total number of mice tested by day 7.
Cachexia was determined over the 30-day period, and results are presented as detected on day 7. By day 30, no cachexia was observed in any of the surviving animals. Cachexia was defined as ruffled hair, hunched bodies, and weight loss and was scored on a scale from +++++ (severe cachexia) to − (no cachexia).
Scrambled peptides corresponding to sequences within peptide A or peptide D were used as negative controls.
Forty-nine days after the first immunization, mice were challenged by inoculation of approximately 109 CFU of P. gingivalis A7436 directly into chambers. Mice were examined daily for secondary lesions and health status. Severe cachexia was defined as ruffled hair, hunched bodies, and weight loss. Chamber fluid was removed from each implanted chamber at 1 to 7 days postchallenge for bacteriological culturing and immunological analysis. All surviving animals were sacrificed 30 days postchallenge, and sera were separated from blood obtained by cardiac puncture.
Chamber fluid analysis.
Aliquots of fluid from each chamber (50 μl) were streaked for isolation onto anaerobic blood agar plates and cultured at 37°C for 7 days under anaerobic conditions (14). Results were expressed as the number of positive cultures obtained from chamber samples/total number of chambers sampled. Samples from individual mice were also pooled for each group and serially diluted. Samples were then plated on anaerobic blood agar plates and cultured at 37°C under anaerobic conditions for 7 days. Results are expressed as CFU/milliliter for each group.
IgG specific for gingipains and whole cells was quantitated by an enzyme-linked immunosorbent assay (ELISA) (10) and expressed as a dilution factor of chamber fluid at which there was 50% maximal OD540 reading calculated from sigmoidal curves obtained in the ELISA. For competitive ELISA, chamber fluid from mice immunized with heat-killed P. gingivalis (1:10,000) was preincubated with increasing concentrations of gingipains as competing antigens, before the mixture was added to a microtitration plate coated with whole P. gingivalis cells. Chamber fluid was mixed with buffer alone as a control. The amount of antibody specifically bound to bacterial surface antigens was determined by subsequent binding of peroxidase-labeled goat anti-mouse IgG antibodies and expressed as residual IgG binding to P. gingivalis surface antigens.
For Western blot analysis, purified gingipains and samples of P. gingivalis vesicles and membranes (41) were boiled, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose. The nitrocellulose membranes were transiently stained with Ponceau S, the positions of molecular weight markers, gingipain R2, and polypeptide chains constituting the gingipain R1 complex were marked, and the membranes were incubated in chamber fluid obtained from mice immunized with the 95-kDa gingipain R1, the 50-kDa gingipain R2, peptides, or heat-killed whole cells. Alkaline phosphatase-labeled goat anti-mouse IgG was then added, and the blots were developed.
RESULTS
Virulence of P. gingivalis in immunized mice.
Female BALB/c mice were immunized with either the 95-kDa gingipain R1, the 50-kDa gingipain R2, or MAP-conjugated gingipain R-derived peptides (Fig. 1), by direct injection into stainless steel chambers implanted subcutaneously, and subsequently challenged by injection of live P. gingivalis into chambers (13). Nonimmunized animals, or animals immunized with a scrambled peptide A control and challenged with P. gingivalis, developed ulcerated necrotic lesions on their abdomens and exhibited severe cachexia with ruffled hair, hunched bodies, and weight loss; 14 of 22 and 5 of 8 mice, respectively, died (Table 1). In contrast, animals immunized with MAP-conjugated peptide A, corresponding to the N terminus of the catalytic domain of gingipains R (Fig. 1), followed by challenge with P. gingivalis were protected from abscess formation and death (Table 1). Similar results were obtained for animals that had been immunized with either whole P. gingivalis cells, the 95-kDa gingipain R1, or the 50-kDa gingipain R2. However, immunization with a peptide corresponding to sequences within the adhesion/hemagglutinin domain of gingipain R1 (peptide D) (Fig. 1), which has been shown to be directly involved in the hemagglutinin activity of this gingipain (5), followed by challenge with P. gingivalis, did not protect animals (Table 1). Similar results were obtained following challenge with P. gingivalis in mice immunized with peptides corresponding to either a sequence encompassing the catalytic cysteine residue of gingipains R (peptide B) or a homologous sequence within the catalytic domains of gingipains R and gingipain K (peptide C) (data not shown). Immunization with either peptide A, the 95-kDa gingipain R1, the 50-kDa gingipain R2, or P. gingivalis whole cells, followed by challenge with live bacteria, resulted in fewer animals from which P. gingivalis was cultured compared to the nonimmunized group. Following challenge with P. gingivalis, viable organisms were cultured from chamber fluid obtained from 20 of 22 nonimmunized mice up to the time of death (Table 2). Viable organisms were also cultured from animals immunized with peptides B, C, and D (Table 2 and data not shown). Following challenge with live bacteria, in the nonimmunized group, P. gingivalis levels increased relative to the initial inoculum (108 to 1012 CFU) throughout the course of the experiments (Table 2). In animals that were immunized with peptide A, peptide D, gingipain R1, gingipain R2, or whole cells, followed by challenge with P. gingivalis, we observed an initial increase in P. gingivalis CFU in those chambers that were positive for P. gingivalis; however, by day 7, the CFUs had decreased to <106 (Table 2). These results indicate that immunization with a peptide corresponding to the N-terminal catalytic domain of gingipains R or with active proteinases or whole bacteria can limit the ability of P. gingivalis to invade in the mouse chamber model.
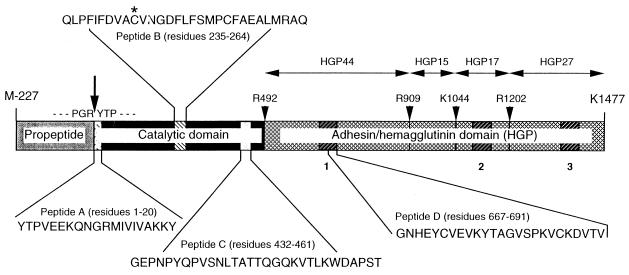
FIG. 1.
Structure of pro-gingipain R1, with indicated location of peptides used for immunization. The initial transcript of the rgp1 gene consists of a propeptide, a catalytic domain, and an adhesion/hemagglutinin domain (38). During translocation onto the P. gingivalis surface, the polyprotein undergoes proteolytic processing, resulting in the formation of the mature 95-kDa gingipain R1, either in membrane-bound or soluble forms consisting of a noncovalent complex of catalytic polypeptide and fragments of the adhesion/hemagglutinin domain (41). The adhesion/hemagglutinin domain is conveniently divided into subdomains (HGPs) of 44, 15, 17, and 27 kDa, according to putative proteolytic processing after one lysine and three arginine residues (arrowheads). The hemagglutination active site (peptide D) (5) is a part of a triplicate amino acid sequence repeat present in the HGP44, HGP17, and HGP27 subdomains. The triplicate repeats of the 50-amino-acid sequence within the adhesion/hemagglutinin domain are represented by hatched boxes numbered beneath the structure. Gingipain R2 is also translated as a proenzyme, nearly identical in sequence to the catalytic domain of gingipain R1 but missing the entire adhesion/hemagglutinin domain. The structure of the gingipain K initial polyprotein is similar to that of gingipain R1, with the adhesion/hemagglutinin domain being virtually identical. The gingipain K initial transcript is apparently subject to posttranslational processing by gingipains R (34). The catalytic domains of both gingipains share only limited identity (27%) scattered throughout the polypeptide chain, except for an identical 30-amino-acid residue fragment (peptide C). The cleavage of the propeptide releasing the N-terminal sequence of active gingipains R is shown by an arrow. Arrowheads indicate putative proteolytic processing sites leading to assembly of the soluble or membrane-bound enzyme (molecular mass 95 of kDa) in the form of a noncovalent complex of the catalytic domain with indicated, active fragments of the adhesion/hemagglutinin domain.
TABLE 2.
Recovery of P. gingivalis from chamber fluid following challengea
| Group | No. of mice from which P. gingivalis was cultured/total no. sampled on indicated day following challenge
|
|||
|---|---|---|---|---|
| 1 | 2 | 5 | 7 | |
| Nonimmunized | 21/22 (1.4 × 1012c) | 20/22 (1.1 × 1012) | 20/22 (2.4 × 1012) | 0b |
| Peptide A | 23/32 (7.2 × 1011) | 21/32 (1.9 × 1010) | 19/32 (9.8 × 108) | 19/32 (<106) |
| Scrambled peptide Ad | 8/8 (6.7 × 1010) | 8/8 (4.8 × 1010) | 7/8 (2.0 × 1010) | 7/8 (5.6 × 108) |
| Peptide D | 8/8 (1.0 × 1011) | 8/8 (8.2 × 1010) | 8/8 (2.2 × 108) | 6/8 (<106) |
| Scrambled peptide Dd | 8/8 (8.6 × 1010) | 7/8 (6.0 × 1010) | 6/8 (2.6 × 108) | 6/8 (6.7 × 108) |
| Whole cells | 17/24 (7.4 × 1011) | 11/24 (8.8 × 1011) | 9/24 (4.6 × 108) | 6/24 (<106) |
| Gingipain R1 | 12/24 (2 × 1012) | 9/24 (8 × 109) | 4/24 (<106) | 3/24 (<106) |
| Gingipain R2 | 15/22 (6.1 × 1011) | 9/22 (1.8 × 1011) | 7/22 (1.2 × 1010) | 6/22 (<106) |
Aliquots of fluid from each chamber (50 μl) were streaked for isolation onto anaerobic blood agar plates and cultured at 37°C for 7 days under anaerobic conditions.
The six surviving animals in this group that were positive for P. gingivalis were sacrificed on day 7 due to severe cachexia.
CFU obtained from chamber fluid as detected in samples pooled from individual mice.
Scrambled peptides corresponding to sequences within peptide A or peptide D were used as negative controls.
IgG response to P. gingivalis.
Immunization with the N-terminal peptide induced a moderate IgG response to the 95-kDa gingipain R1 and the 50-kDa gingipain R2 as detected in chamber fluid samples (Table 3). The absence of a response to whole cells may be due to the lack of exposure of this epitope on cell surfaces so that the N-terminal sequence of the membrane-associated gingipain R1 catalytic domain is not available for antibody binding (41). The IgG response obtained following immunization with peptide fragment D, representing sequences within the adhesion/hemagglutinin domain of the 95-kDa gingipain R1, was comparable to that induced by the N-terminal peptide; however, protection against P. gingivalis challenge was not observed when this peptide was used as an immunogen (Tables 1 and 2). Immunization with the 95-kDa gingipain R1 induced a high IgG titer to all antigens examined except for the 50-kDa gingipain R2 (Table 3). The low titer to the 50-kDa gingipain R2 may be due to the absence of the highly immunogenic adhesion/hemagglutinin domain in this enzyme (24). Indeed, immunization with whole cells induced a good response to the 95-kDa gingipain R1 and gingipain K, with essentially no binding to the 50-kDa gingipain R2. For all immunization groups, postchallenge serum IgG titers were higher than the chamber fluid IgG titers obtained 3 weeks postimmunization but prior to challenge and reflected the effect of challenge with P. gingivalis (data not shown).
TABLE 3.
ELISA analysis of chamber fluid from mice immunized with gingipains R, peptide fragments of gingipains, and whole bacteriaa
| Immunogen | Antibody titerb (mean ± SD)
|
|||
|---|---|---|---|---|
| Gingipain R1 | Gingipain R2 | Gingipain K | P. gingivalis | |
| Gingipain R1 | 200,000 ± 28,000 | 6,600 ± 1,440 | 105,000 ± 13,500 | 13,000 ± 1,100 |
| Gingipain R2 | 3,600 ± 510 | 2,800 ± 415 | NQ | 400 ± 28 |
| Peptide A | 710 ± 52 | 145 ± 8 | NQ | NQ |
| Scrambled peptide A | ND | ND | ND | ND |
| Peptide D | 210 ± 21 | ND | 290 ± 17 | 50 ± 4 |
| Scrambled peptide D | ND | ND | ND | ND |
| P. gingivalis | 22,000 ± 2,500 | 760 ± 48 | 20,000 ± 2,600 | 12,000 ± 980 |
Microplates were coated with purified gingipains (1.0 μg/ml) or whole P. gingivalis cells (23), nonspecific binding sites were blocked with bovine serum albumin, and plates were incubated with serial dilutions of chamber fluid. Chamber fluid was obtained at 28 days postimmunization and prior to challenge with live P. gingivalis. Chamber fluid samples were pooled from individual mice in each group. Quantity of antibody bound to immobilized antigen was determined with peroxidase-labeled goat anti-mouse IgG.
Expressed as a dilution factor of chamber fluid at which there was 50% of maximal OD540 reading calculated from sigmoidal curves obtained in the ELISA. NQ, not quantitated (IgG binding detectable but too low to be quantitated); ND, not detected (IgG binding at the lowest [fivefold] chamber fluid dilution).
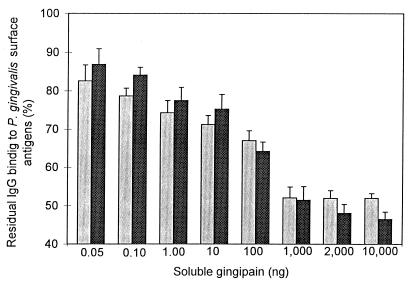
Competitive ELISAs, using either the 95-kDa gingipain R1 or gingipain K as the competing soluble antigen, indicated that 42 and 53% of the antibodies induced by immunization with heat-killed bacteria recognize the 95-kDa gingipain R1 and gingipain K, respectively (Fig. 2). However, even at very high concentrations, the 50-kDa gingipain R2 did not hinder IgG binding to P. gingivalis (Fig. 2, legend). These observations were also confirmed by Western blot analysis (Fig. 3D; see below) and indicate that sequences within the noncatalytic/hemagglutinin domains of the 95-kDa gingipain R1 and gingipain K are responsible for approximately 50% of the induced IgG response.
FIG. 2.
Competitive ELISA. Chamber fluid from mice immunized with heat-killed P. gingivalis was preincubated with increasing concentration of the 95-kDa gingipain R1 (light bars) and gingipain K (dark bars) as competing antigens before the mixture was added to a microtitration plate coated with whole P. gingivalis cells. The amount of antibody specifically bound to bacterial surface antigens was determined by subsequent binding of peroxidase-labeled goat anti-mouse IgG antibodies. When the 50-kDa gingipain R2 was used as the competing antigen, levels of residual IgG binding to P. gingivalis were as follows: 0.05 ng, 102.9%; 0.10 ng, 98.3%; 1.0 ng, 96.8%; 10 ng, 93.8%, 100 ng, 100.2%; 2,000 ng, 91.9%; and 10,000 ng, 96.8%.
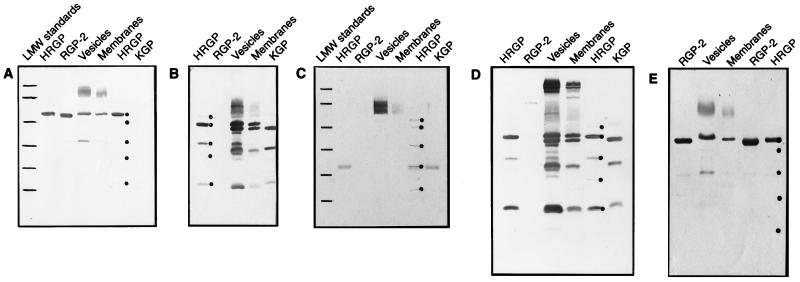
FIG. 3.
Western blot analysis of chamber fluid samples. Purified gingipains (95-kDa gingipain R1 [HRGP]), (50-kDa gingipain R2 [RGP-2]), and (gingipain K [KGP]) and samples of P. gingivalis vesicles and membranes (1.0 μg of purified gingipains or 5 μg of protein in vesicles and membrane fractions) were boiled, resolved by SDS-PAGE, and transferred to nitrocellulose. The nitrocellulose was transiently stained with Ponceau S; the positions of RGP-2 and polypeptide chains constituting the HRGP complex were then marked (dots to the right of an appropriate lane) and incubated in chamber fluid obtained from mice immunized with either the N-terminal peptide of the catalytic domain of RGPs (peptide A; 1,000-fold dilution) (A), HRGP (1,000-fold dilution) (B), the peptide derived from sequences within the adhesion/hemagglutinin domain of HRGP (peptide D; 100-fold dilution) (C), heat-killed P. gingivalis (1,000-fold dilution) (D), or RGP-2 (1,000-fold dilution) (E). Alkaline phosphatase-labeled goat anti-mouse IgG was then added, and blots were developed. Low-molecular weight (LMW) standards were loaded in the gels depicted in panels A and C. The molecular mass values for the markers from top to bottom are denoted by lines and correspond to 94, 67, 43, 30, 21, and 14 kDa.
Western blot analysis.
Chamber fluid from mice immunized with the N-terminal peptide of the catalytic domain of gingipains R reacted with the catalytic domain (50 kDa) of gingipain R1, with gingipains R present in vesicles and bacterial membrane fractions, and with the 50-kDa gingipain R2 (Fig. 3A). A similar pattern was observed when chamber fluid from animals immunized with whole gingipain R2 was used (Fig. 3E). The lack of reactivity with gingipain K is in agreement with antibody specificity results (Table 3). Although the adhesion domain-derived peptide induced a poor IgG response as detected by ELISA, we found reactivity to several proteins by Western blot analysis (Fig. 3C). The 50-kDa gingipain R2 was not recognized by this antibody due to the lack of an adhesion domain. However, reactivity could be detected with the 27-kDa domain of the 95-kDa gingipain R1 and gingipain K, as well as proteins migrating in the range of 60 to 70 kDa in vesicle and membrane preparations. Significantly, the putative sequences of the adhesion domains present in the 44- and 17-kDa subunits (Fig. 1) did not bind antibody (Fig. 3C). Immunization with the 95-kDa gingipain R1 resulted in antibodies with specificity predominantly directed against the 44-kDa adhesion/hemagglutinin domain of gingipain R1 and the 42-kDa domain of gingipain K (Fig. 3B). These domains were also recognized in vesicle and membrane preparations. Additional protein bands recognized by this antiserum included the 32- and 17-kDa proteins in gingipain K, as well as their apparent equivalents in vesicles and membranes. However, the 95-kDa gingipain R1 catalytic domain was only weakly recognized, and the 50-kDa gingipain R2 was not recognized at all. These results are in agreement with previous studies in which the catalytic domains of gingipains R were poorly recognized in antisera obtained from rabbits or chickens immunized with the entire gingipain R1 molecule (12).
Immunization with heat-killed bacteria resulted in antibodies (Fig. 3D) with specificity similar to that induced by immunization with the 95-kDa gingipain R1. In addition to polypeptides composing the 95-kDa gingipain R1 complex, high-molecular-weight proteins were also detected in vesicles and membranes. However, we could not detect any reactivity in Western blot analysis to the catalytic domain of either the 95-kDa gingipain R1 or the 50-kDa gingipain R2, results in agreement with those obtained with mice immunized with the 95-kDa gingipain R1 (Fig. 3B) and consistent with data obtained by ELISA in which antibodies generated following immunization with heat-killed P. gingivalis demonstrated a very low titer against the 50-kDa gingipain R2. Our results also indicate that antisera obtained from mice immunized with P. gingivalis A7436 whole cells react with gingipains and their fragments obtained from P. gingivalis HG66 (Fig. 3D). Thus, it appears that antigenic conversation of the gingipains exists among different P. gingivalis strains. Furthermore, antisera raised to gingipains isolated from P. gingivalis strain HG66 recognize gingipains from a large panel of P. gingivalis strains (27).
DISCUSSION
Previous studies have established a role for gingipains R in the ability of P. gingivalis to colonize and invade in several animal models (11, 23, 31). In a previous study we found that inhibition of the enzymatic activity of the P. gingivalis cysteine proteinases including gingipains R severely limits the ability of P. gingivalis to colonize and invade in the mouse chamber model (12). A P. gingivalis rgpA rgpB double mutant has recently been shown to exhibit a marked reduction in virulence, indicating a major role for gingipains in the virulence potential of P. gingivalis (31). In the present study, we found that immunization of mice with the 95-kDa gingipain R1, the 50-kDa gingipain R2, or a peptide derived from the N-terminal sequence of the catalytic domain of gingipains R (peptide A) resulted in protection from P. gingivalis invasion. The protection observed following neutralization of these enzymes is most likely due to the ability of these enzymes to contribute to the virulence potential of P. gingivalis in a multifactorial manner. It is known that gingipains R may act as processing proteinases responsible for self-maturation as well as for maturation of gingipain K, fimbrillin, and a 75-kDa outer membrane protein (30, 35). Since fimbrillin, gingipains R, and gingipain K have been documented in the virulence potential of P. gingivalis (24, 25, 32, 50), gingipains R may participate in a central role in the pathogenesis of periodontal disease via production of pathophysiologically significant proteins.
The ability to modulate the colonization or pathogenicity of P. gingivalis has been demonstrated in several animal models using whole-cell and component vaccines. Whole-cell and polysaccharide protein conjugate vaccines have been demonstrated to protect against the invasive capabilities of P. gingivalis in mice (13, 47). Immunization of rats with purified P. gingivalis fimbriae and fimbrial peptides also has been shown to protect against periodontal destruction induced by infection with P. gingivalis in a rat periodontitis model (25). There is also evidence that immunization can influence the colonization of P. gingivalis in several primate models (3, 8, 19). However, in several animal models, induction of an immune response to P. gingivalis antigens has been demonstrated to actually exacerbate disease (reviewed in reference 26). Thus, it is critical that an immune response be generated to antigens which can function in a protective capacity. Indeed, it has been documented that although patients with a history of destructive periodontal disease frequently demonstrate an elevated IgG antibody response to P. gingivalis, these antibodies are apparently ineffective at limiting continued disease progression (9, 16, 26, 46, 51, 55).
The results reported in this study indicate that in mice the major IgG response is targeted to sequences within the adhesion/hemagglutinin domain of the 95-kDa gingipain R1 and gingipain K. It has recently been demonstrated that antibodies specific for P. gingivalis gingipains R were produced in patients with adult periodontitis, with the majority of the antibodies reactive with antigenic determinants within the hemagglutinin/adhesion domain of the 95-kDa gingipain R1 (22). We have also observed that patients with severe, untreated periodontitis mount a strong IgG response to this same domain (12). The strong reactivity against this part of gingipains may result from the fact that sequences within the hemagglutinin/adhesion domain are present in at least two other P. gingivalis proteins. These sequences are repeated four times in the hagA gene product (18) and once in a TonB-dependent protein encoded by the tla gene (1). It is possible that such a specific response to sequences within the adhesion/hemagglutinin domain of gingipains mounted in patients with periodontitis actually serve to divert the immune response from other protective antigens. In the mouse model, antibodies with this specificity may function to limit the invasion of P. gingivalis. However, in human subjects, where the local inflammatory response can lead to bone and tissue destruction within the periodontal ligament, the production of antibodies may function to aggravate local tissue damage.
Recent studies have demonstrated that the inhibition of hemagglutination in vivo by administration of a monoclonal antibody specific for the hemagglutinin domain of the prpRI gene (P. gingivalis W50 equivalent of the rgp1 gene) product results in the inhibition of P. gingivalis colonization (5). In the present study, we found that immunization with a peptide derived from sequences within the hemagglutinin domain did not induce a protective immunological response against P. gingivalis; however, the peptide used for our immunization studies did not contain a FEED motif. This sequence is present in several microbial adhesins and has been demonstrated to be directly responsible for fibronectin binding (36, 44, 52); the absence of the FEED motif in the peptide used in our studies could explain the lack of a protective response. Thus, in future studies it will be critical to examine peptides derived from other regions of the hemagglutinin domain.
Although immunization with the entire gingipains R induced an IgG response to the cell surface enzyme in intact bacteria, those antibodies generated following immunization with the N-terminal peptide were unable to recognize the mature protein in whole cells, suggesting that this epitope was not exposed in whole cells. Rabbit antisera generated to the N-terminal portion of the catalytic domain of the 95-kDa gingipain R1 and the 50-kDa gingipain R2 also did not recognize gingipain R1 in membrane or vesicle preparations unless samples were dissociated by boiling (41), again suggesting that this epitope is not exposed in whole cells or vesicles. The protective effect induced following immunization with the N-terminal peptide may result from IgG binding within the sequence of progingipain transiently expressed on the cell surface. We postulate that this binding interferes with the processing and or folding of progingipain and in this way hinders the pathogenic potency of P. gingivalis.
In summary, immunization with a MAP-conjugated peptide corresponding to the N-terminal domain of gingipains R results in the production of antibodies which afford protection of mice from P. gingivalis infection in the mouse chamber model. These findings support the hypothesis that inhibition of the maturation and/or catalytic activity of gingipains Rs can inhibit invasion of P. gingivalis in mice. Future studies are aimed at further defining specific domains of gingipains R which function in inducing a protective response in a periodontitis model of infection.
ACKNOWLEDGMENTS
This study was supported by Public Health Service (PHS) grants DE09161 from the National Institute of Dental Research and RR03034 from the National Center for Research Resources to C.A.G., by PHS grant DE 09761 from the National Institute of Dental Research to J.T., and by grant P204A019 from Committee of Scientific Research (KBN, Poland) to J.P.
REFERENCES
- 1.Aduse-Opoku J, Slaney J M, Rangarajan M, Muir J, Young K A, Curtis M A. The Tla protein of Porphyromonas gingivalis W50: a homolog of the RI protease precursor (PrpRI) is an outer membrane receptor required for growth on low levels of hemin. J Bacteriol. 1997;179:4778–4788. doi: 10.1128/jb.179.15.4778-4788.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aduse-Opoku J, Muir J, Slaney J M, Rangarajan M, Curtis M A. Characterization, genetic analysis, and expression of a protease antigen (PrpR1) of Porphyromonas gingivalis W50. Infect Immun. 1995;63:4744–4754. doi: 10.1128/iai.63.12.4744-4754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson D M, Ebersole J L, Novak M J. Functional properties of nonhuman primate antibody to Porphyromonas gingivalis. Infect Immun. 1995;63:3245–3552. doi: 10.1128/iai.63.9.3245-3252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkocy-Gallagher G A, Han N, Patti J M, Whitlock J, Progulske-Fox A, Lantz M S. Analysis of the prtP gene encoding porphypain, a cysteine proteinase of Porphyromonas gingivalis. J Bacteriol. 1996;178:2734–2741. doi: 10.1128/jb.178.10.2734-2741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis M A, Aduse-Opoku J, Slaney J M, Rangarajan M, Booth V, Cridland J, Shepard P. Characterization of an adherence and antigenic determinant of the ArgI protease of Porphyromonas gingivalis which is present on multiple gene products. Infect Immun. 1996;64:2532–2539. doi: 10.1128/iai.64.7.2532-2539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutler C W, Kalmar J R, Genco C A. Pathogenic strategies of the oral anaerobe Porphyromonas gingivalis. Trends Microbiol. 1995;3:45–51. doi: 10.1016/s0966-842x(00)88874-5. [DOI] [PubMed] [Google Scholar]
- 7.Discipio R G, Daffern P J, Kawahara M, Pike R, Travis J, Hugli T E. Cleavage of human complement component C5 by cysteine proteinases from Porphyromonas gingivalis. Prior oxidation of C5 augments proteinase digestion of C5. Immunology. 1996;87:660–667. doi: 10.1046/j.1365-2567.1996.478594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebersole J L, Brunsvold M, Steffensen B, Wood R, Holt S C. Effects of immunization with Porphyromonas gingivalis and Prevotella intermedia on progression of ligature-induced periodontitis in the nonhuman primate Macaca fascularis. Infect Immun. 1991;59:3351–3359. doi: 10.1128/iai.59.10.3351-3359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebersole J L, Taubman M A, Smith D J, Frey D E, Haffajee A D, Socransky S S. Human serum antibody responses to oral microorganisms. IV. Correlation with homologous infection. Oral Microbiol Immunol. 1987;2:53–59. doi: 10.1111/j.1399-302x.1987.tb00290.x. [DOI] [PubMed] [Google Scholar]
- 10.Ebersole J L, Frey D E, Taubman M A, Smith D J, Socransky S S, Tanner A C R. Serological identification of oral Bacteroides spp. by enzyme-linked immunosorbent assay. J Clin Microbiol. 1984;19:639–644. doi: 10.1128/jcm.19.5.639-644.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher H M, Schenkein H A, Morgan R M, Bailey K A, Berry C R, Macrina F L. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genco, C. A., J. Potempa, J. Mikolajczyk-Pawlinska, and J. Travis. Role of gingipains R in Porphyromonas gingivalis pathogenesis. Clin. Infect. Dis., in press. [DOI] [PubMed]
- 13.Genco C A, Kapczynski D R, Cutler C W, Arko R J, Arnold R R. Influence of immunization on Porphyromonas gingivalis colonization and invasion in the mouse chamber model. Infect Immun. 1992;60:1447–1454. doi: 10.1128/iai.60.4.1447-1454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genco C A, Cutler C W, Kapczynski D R, Maloney K H, Arnold R R. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect Immun. 1991;59:1255–1263. doi: 10.1128/iai.59.4.1255-1263.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goulbourne P A, Ellen R P. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J Bacteriol. 1991;173:5266–5274. doi: 10.1128/jb.173.17.5266-5274.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunsolley J C, Tew J G, Gooss C, Marshall D R, Burmeister J A, Schenkein H A. Serum antibodies to periodontal bacteria. J Periodontol. 1990;61:412–419. doi: 10.1902/jop.1990.61.7.412. [DOI] [PubMed] [Google Scholar]
- 17.Hamada N, Watanabe K, Sasakawa C, Yoshikawa M, Yoshimura F, Umemoto T. Construction and characterization of a fimA mutant of Porphyromonas gingivalis. Infect Immun. 1994;62:1696–1704. doi: 10.1128/iai.62.5.1696-1704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han N, Whitlock J, Progulske-Fox A. The hemagglutinin gene (hagA) of Porphyromonas gingivalis 381 contains four large, contiguous, direct repeats. Infect Immun. 1996;64:4000–4007. doi: 10.1128/iai.64.10.4000-4007.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holt S C, Brunsvold M, Jones A, Wood R, Ebersole J L. Cell envelope and cell wall immunization of Macaca fascularis: effect on the progression of ligature-induced periodontitis. Oral Microbiol Immunol. 1995;10:321–333. doi: 10.1111/j.1399-302x.1995.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 20.Imamura T, Potempa J, Pike R E, Travis J. Dependence of vascular permeability enhancement on cysteine proteinases in vesicles of Porphyromonas gingivalis. Infect Immun. 1995;63:1999–2003. doi: 10.1128/iai.63.5.1999-2003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imamura T, Pike R N, Potempa J, Travis J. Pathogenesis of periodontitis: a major arginine-specific cysteine proteinase from Porphyromonas gingivalis induces vascular permeability enhancement through activation of the kallikrein/kinin pathway. J Clin Invest. 1994;94:361–367. doi: 10.1172/JCI117330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly C G, Booth V, Kendal H, Slaney J M, Curtis M A, Lehner T. The relationship between colonization and hemagglutination inhibiting and B cell epitopes of Porphyromonas gingivalis. Clin Exp Immunol. 1997;110:285–291. doi: 10.1111/j.1365-2249.1997.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kesavalu L, Holt S C, Ebersole J L. Trypsin-like protease activity of Porphyromonas gingivalis as a potential virulence factor in a murine lesion model. Microb Pathog. 1996;20:1–10. doi: 10.1006/mpat.1996.0001. [DOI] [PubMed] [Google Scholar]
- 24.Kontani M, Ono H, Shibata H, Okamura Y, Tanaka T, Fujiwara T, Kimura S, Hamada S. Cysteine protease of Porphyromonas gingivalis 381 enhances binding of fimbriae to cultures human fibroblasts and matrix proteins. Infect Immun. 1996;64:756–762. doi: 10.1128/iai.64.3.756-762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malek R, Fisher G, Caleca A, Stinson M, Van Oss C J, Lee J-Y, Cho M-I, Genco R J, Evans R T, Dyer D W. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994;176:1052–1059. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McArthur W P, Clark W B. Specific antibodies and their potential role in periodontal diseases. J Periodontol. 1993;64:807–818. doi: 10.1902/jop.1993.64.8s.807. [DOI] [PubMed] [Google Scholar]
- 27.Mikolajczyk-Pawlinska J, Kordula T, Pavloff N, Pemberton P A, Kiefer M C, Travis J, Potempa J. Genetic variations of Porphyromonas gingivalis genes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biol Chem. 1998;379:205–211. doi: 10.1515/bchm.1998.379.2.205. [DOI] [PubMed] [Google Scholar]
- 28.Naito Y, Okuda K, Takazoe I. Detection of specific antibody in adult human periodontitis sera to surface antigens of Bacteroides gingivalis. Infect Immun. 1987;55:832–834. doi: 10.1128/iai.55.3.832-834.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama K. Domain-specific rearrangement between the two arg-gingipain encoding genes in Porphyromonas gingivalis: possible involvement of nonreciprocal recombination. Microbiol Immunol. 1997;41:185–196. doi: 10.1111/j.1348-0421.1997.tb01189.x. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama K M, Yoshimura F, Kadowaki T, Yamamoto K. Involvement of arginine-specific cysteine proteinases (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J Bacteriol. 1996;178:2818–2824. doi: 10.1128/jb.178.10.2818-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. Construction and characterization of arginine-specific cysteine proteinase (arg-gingipain)-deficient mutants of Porphyromonas gingivalis. J Biol Chem. 1995;270:23619–23626. doi: 10.1074/jbc.270.40.23619. [DOI] [PubMed] [Google Scholar]
- 32.Njoroge T, Genco R J, Sojar H T, Hamada N, Genco C A. A role for fimbriae in the invasion of oral epithelial cells by Porphyromonas gingivalis. Infect Immun. 1997;65:1980–1984. doi: 10.1128/iai.65.5.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogawa T, Uchida H, Hamada S. Porphyromonas gingivalis fimbriae and their synthetic peptides induce pro inflammatory cytokines in human peripheral blood monocyte cultures. FEMS Microbiol Lett. 1994;116:237–242. doi: 10.1111/j.1574-6968.1994.tb06707.x. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto K, Kadowaki T, Nakayama K, Yamamoto K. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine proteinase (Lys-gingipain) in Porphyromonas gingivalis: structural relationship with the arginine-specific cysteine proteinase (arg-gingipain) J Biochem. 1996;120:398–406. doi: 10.1093/oxfordjournals.jbchem.a021426. [DOI] [PubMed] [Google Scholar]
- 35.Onoe T, Hoover C I, Nakayama K, Ideka T, Nakamura H, Yoshimura F. Identification of Porphyromonas gingivalis prefimbrillin possessing a long leader peptide: possible involvement of trypsin-like protease in fimbrillin maturation. Microb Pathog. 1995;19:351–364. doi: 10.1016/s0882-4010(96)80006-4. [DOI] [PubMed] [Google Scholar]
- 36.Patti J M, Allen B L, McGavin M J, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 37.Pavloff N, Pemberton P A, Potempa J, Chen W C A, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P. Molecular cloning and characterization of Porphyromonas gingivalis Lys-gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. J Biol Chem. 1997;272:1595–1600. doi: 10.1074/jbc.272.3.1595. [DOI] [PubMed] [Google Scholar]
- 38.Pavloff N, Potempa J, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and structural characterization of the arg-gingipain proteinase of Porphyromonas gingivalis. J Biol Chem. 1995;270:1007–1010. doi: 10.1074/jbc.270.3.1007. [DOI] [PubMed] [Google Scholar]
- 39.Pike R, McGraw W, Potempa J, Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 40.Pike R N, Potempa J, McGraw W, Coetzer H T, Travis J. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J Bacteriol. 1996;178:2876–2882. doi: 10.1128/jb.178.10.2876-2882.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Potempa J, Pike R, Travis J. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect Immun. 1995;63:1176–1182. doi: 10.1128/iai.63.4.1176-1182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potempa J, Pavloff N, Travis J. Porphyromonas gingivalis: a proteinase/gene accounting audit. Trends Microbiol. 1995;3:430–434. doi: 10.1016/s0966-842x(00)88996-9. [DOI] [PubMed] [Google Scholar]
- 43.Potempa J, Pike R, Travis J. Host and Porphyromonas gingivalis proteinases (gingipains) in periodontitis: a biochemical model of infection and tissue destruction. Perspect Drug Discovery Design. 1995;2:445–458. [Google Scholar]
- 44.Progulske-Fox A, Tumwasorn S, Lepine G, Whitlock J, Savett D, Ferretti J J, Banas J A. The cloning, expression and sequence analysis of a second Porphyromonas gingivalis gene that codes for a protein involved in hemagglutination. Oral Microbiol Immunol. 1995;10:311–318. doi: 10.1111/j.1399-302x.1995.tb00160.x. [DOI] [PubMed] [Google Scholar]
- 45.Rangarajan M, Aduse-Opoku J, Slaney J M, Young K J, Curtis M A. The prpR1 and prpR2 arginine-specific protease genes of Porphyromonas gingivalis W50 produce five biochemically distinct enzymes. Mol Microbiol. 1995;23:955–965. doi: 10.1046/j.1365-2958.1997.2831647.x. [DOI] [PubMed] [Google Scholar]
- 46.Schenck K, Michaelson T E. IgG subclass distribution of serum antibodies against lipopolysaccharide from Bacteroides gingivalis periodontal health and disease. Acta Pathol Microbiol Immunol Scand. 1987;95:41–46. doi: 10.1111/j.1699-0463.1987.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 47.Schifferle R E, Chen P B, Davern L B, Aguirre A, Genco R J, Levine M J. Modification of experimental Porphyromonas gingivalis murine infection by immunization with a polysaccharide-protein conjugate. Oral Microbiol Immunol. 1993;8:266–271. doi: 10.1111/j.1399-302x.1993.tb00572.x. [DOI] [PubMed] [Google Scholar]
- 48.Scott C F, Whitlaker E J, Hammond B F, Colman R W. Purification and characterization of a potent 70-kDa thiol lysyl-proteinase (Lys-gingivain) from Porphyromonas gingivalis that cleaves kininogen and fibrinogen. J Biol Chem. 1993;268:7935–7942. [PubMed] [Google Scholar]
- 49.Tam J P. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc Natl Acad Sci USA. 1988;85:5409–5413. doi: 10.1073/pnas.85.15.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tokuda M, Duncan M, Cho M-I, Kuramitsu H K. Role of Porphyromonas gingivalis protease activity in colonization of oral surfaces. Infect Immun. 1996;64:4067–4073. doi: 10.1128/iai.64.10.4067-4073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turner D W, Dai G J, Merrell B, Robinson J P. Serum and gingival tissue antibody levels to oral microbial antigens in human chronic adult periodontitis. Microbios. 1989;60:133–140. [PubMed] [Google Scholar]
- 52.Westerlund B, Korhonen T K. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]
- 53.Williams D M, Hughes F J, Odell E W, Farthing P M. Pathology of periodontal disease. Oxford, England: Oxford University Press; 1992. [Google Scholar]
- 54.Wingrove J A, DiScipio R G, Chen Z, Potempa J, Travis J, Hugli T E. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas gingivalis. J Biol Chem. 1992;267:18902–18907. [PubMed] [Google Scholar]
- 55.Yoshimura F, Sugano T, Kawanami M, Kato H, Suziki T. Detection of specific antibodies against fimbriae and membrane proteins from the oral anaerobe Bacteroides gingivalis in patients with periodontal disease. Microbiol Immunol. 1987;31:935–957. doi: 10.1111/j.1348-0421.1987.tb03154.x. [DOI] [PubMed] [Google Scholar]