Abstract
Tra1 is an essential coactivator protein of the yeast SAGA and NuA4 acetyltransferase complexes that regulate gene expression through multiple mechanisms including the acetylation of histone proteins. Tra1 is a pseudokinase of the PIKK family characterized by a C-terminal PI3K domain with no known kinase activity. However, mutations of specific arginine residues to glutamine in the PI3K domains (an allele termed tra1Q3) result in reduced growth and increased sensitivity to multiple stresses. In the opportunistic fungal pathogen Candida albicans, the tra1Q3 allele reduces pathogenicity and increases sensitivity to the echinocandin antifungal drug caspofungin, which disrupts the fungal cell wall. Here, we found that compromised Tra1 function, in contrast to what is seen with caspofungin, increases tolerance to the azole class of antifungal drugs, which inhibits ergosterol synthesis. In C. albicans, tra1Q3 increases the expression of genes linked to azole resistance, such as ERG11 and CDR1. CDR1 encodes a multidrug ABC transporter associated with efflux of multiple xenobiotics, including azoles. Consequently, cells carrying tra1Q3 show reduced intracellular accumulation of fluconazole. In contrast, a tra1Q3 Saccharomyces cerevisiae strain displayed opposite phenotypes: decreased tolerance to azole, decreased expression of the efflux pump PDR5, and increased intracellular accumulation of fluconazole. Therefore, our data provide evidence that Tra1 differentially regulates the antifungal response across yeast species.
Keywords: Tra1, SAGA complex, azole resistance, fungal pathogen, ergosterol, Candida albicans, CDR1
Introduction
Fungal infections represent a major public health concern with over a billion infections each year resulting in more than 1.5 million deaths (Bongomin et al. 2017). Members of the Candida genus, including Candida albicans, are opportunistic pathogens that can cause a wide range of severe infections in susceptible populations, such as the elderly or immunocompromised individuals (Pappas et al. 2018). Candida species represent the leading cause of fungal infection–related deaths worldwide (Rokas 2022). Treatment of Candida infection (candidiasis) is unfortunately limited to 4 major classes of antifungal drugs: echinocandins, azoles, polyenes, and flucytosines. Echinocandins, such as caspofungin, target the fungal cell wall by inhibiting the synthesis of the carbohydrate β-1,3-glucan (Balashov et al. 2006; Lee et al. 2012), while azoles, such as fluconazole and miconazole, inhibit the synthesis of ergosterol, thereby compromising the lipid composition of fungal membranes (Kuznetsov 2021). Azoles specifically inhibit the 14α-demethylase, encoded by the C. albicans ERG11 gene, in the ergosterol synthesis pathway. Consequently, mutations in ERG11 are one of the main sources of acquired azole resistance (Wu et al. 2017; Kaur and Nobile 2022).
Tra1 is an essential component of the SAGA and NuA4 complexes that regulate the acetylation of both histone and nonhistone substrates (Grant et al. 1997; Clarke et al. 1999; Steunou et al. 2014; Downey 2021) in all eukaryotic cells, including fungi (Saleh et al. 1998; Grant et al. 1998; Allard et al. 1999). Recently, Tra1 was shown to be essential for viability in C. albicans (Razzaq et al. 2021; Rashid et al. 2022). Tra1 is essential due to its function in NuA4. This is highlighted in the fission yeast Schizosaccharomyces pombe where the SAGA-incorporated Tra1 is dispensable for viability while conversely, the Nua4-localized Tra2 is essential (Helmlinger et al. 2011). Many components of the SAGA and NuA4 complexes regulate different aspects of the C. albicans antifungal response and pathogenicity (Laprade et al. 2002; Lu et al. 2008; Sellam et al. 2009; Askew et al. 2009; Chang et al. 2015; Shivarathri et al. 2019; Razzaq et al. 2021; Rashid et al. 2022). However, the functions of Tra1 in C. albicans remain poorly understood.
Tra1 is a member of the phosphoinositide 3-kinase-related kinase (PIKK) family, but unlike other family members such as Tor1, it does not possess any detectable kinase activity due to the lack of specific kinase motifs (McMahon et al. 1998; Saleh et al. 1998; Helmlinger et al. 2011). Despite its pseudokinase nature, we have previously shown that mutation of key arginine residues to glutamine in the Tra1 PI3K domain, an allele termed tra1Q3, is associated with increased sensitivity to various stress conditions, including cell wall stress, protein misfolding, and high temperature in the budding yeast Saccharomyces cerevisiae (Berg et al. 2018). tra1Q3 mutants also display increased sensitivity to acid stress, impaired growth in respiratory conditions, and reduced chronological lifespan (Bari et al. 2022). Similar to their S. cerevisiae counterparts, C. albicans tra1Q3 mutants display increased sensitivity to cell wall stress induced by caspofungin, as well as reduced biofilm formation and pathogenicity (Razzaq et al. 2021). However, the impact of Tra1 on C. albicans resistance to other classes of antifungal drugs remains unknown. Hence, here we tested the effect of the tra1Q3 mutation to modulate sensitivity to other antifungal compounds.
Materials and methods
Reagents
Fluconazole, amphotericin B, miconazole, FK506, rhodamine 6G (R6G), and caspofungin were from MilliporeSigma. Propidium iodide (PI) was from Thermo Fisher Scientific.
Yeast strains and growth conditions
The tra1 mutant strains used in this study were previously described (Berg et al. 2018; Razzaq et al. 2021) and are listed in Supplementary Table 1. Both S. cerevisiae and C. albicans strains were cultured in YPD (2% Bacto peptone, 1% yeast extract, and 2% glucose) unless noted. Cells were grown in liquid YPD overnight at 30°C with shaking. The next day, cells were diluted at 1:10 ratio and then incubated for 2 h at 30°C with shaking. Cell growth on agar plates was measured as previously described (Petropavlovskiy et al. 2020).
Fluorescent fluconazole probe uptake
The fluorescent fluconazole probe (RB510) was added to log phase cells cultured at 30°C with shaking to a final concentration of 1 µg/mL as previously described (Benhamou et al. 2017). Next, the cells were incubated in the dark for 60 min at 30°C with shaking. Then, cells were washed with phosphate-buffered saline (PBS). The mean fluorescence intensity of the probe was measured with a BD FacsCelesta flow cytometer. Data were collected from 30,000 cells per time point using a 561-nm yellow–green laser. Mean fluorescent intensity was calculated using FlowJo. No gates were applied.
Cell viability assay
Saccharomyces cerevisiae cells were grown in liquid SC media lacking leucine overnight at 30°C with shaking. The next day, cells were diluted at 1:5 ratio and then incubated for 5 h at 30°C with shaking until the log phase in 50-mL culture flasks. Next, cells were equalized to OD600 = 0.80, treated or not with 20 µg/mL of fluconazole, and incubated with shaking for 1–24 h at 30°C. After each time point, 1 mL of cells was pelleted by centrifugation and resuspended in a final volume of 1000 µL PBS. For positive control, 1 sample of each strain was boiled for 15 min at 100°C. Five hundred microliters of cell suspension was stained with 2.5 µL of 1 mg/mL PI solution, and cells were incubated in the dark for 10 min at room temperature as previously described (Chadwick et al. 2016). Data were collected from 30,000 cells per time point using a BD FACSCelesta flow cytometer (BD Biosciences) equipped with a 561-nm yellow–green laser. Analysis was performed using FlowJo.
Gene expression analysis
Total RNA was isolated using the MasterPure Yeast RNA purification kit (Lucigen) according to the manufacturer’s instructions. cDNA was synthesized from 2.5 µg total RNA using the SuperScript IV VILO Master Mix (Thermo Fisher Scientific) according to the manufacturer’s instructions. qRT-PCR for ERG11 and CDR1/PDR5 together with ACT1/TDH3 as housekeeping gene was amplified from the synthesized cDNA using primers listed in Supplementary Table 2 with a QuantStudio 3 real-time PCR system using the ΔΔCT method (Thermo Fisher Scientific).
Efflux of R6G
To measure C. albicans drug efflux capacity, R6G efflux was measured by fluorescence assay with whole cells. Candida albicans were grown in liquid YPD overnight at 30°C with shaking in 10-mL culture tubes. First, cells were diluted at a 1:10 ratio and then incubated for 2 h at 30°C with shaking until the log phase. Next, cells were pelleted by centrifugation, washed with 5 mL PBS (pH 7), resuspended in 2 mL PBS, and incubated for 1 h at 30°C with shaking in PBS to energy-deprived cells. R6G was added at a concentration of 4 µM, and the incubation continued for 1 h, thus facilitating R6G accumulation. After this incubation, cells were sedimented by centrifugation, washed with PBS, and resuspended in a final volume of 200 µL PBS. Fifty microliters of individual strains was diluted in 150 µL PBS and aliquoted in a 96-well microtiter plate, which was placed in a BioTek Cytation5 Cell Imaging Multimode Reader (Agilent) with temperature control set at 30°C. Baseline emission of fluorescence (excitation wavelength: 584 nm; emission wavelength: 625 nm) and OD600 was recorded for 0, 2, and 4 min. Glucose (2% final concentration) was next added to each strain to initiate R6G efflux. As a negative control, no glucose was added to separate aliquots of each strain. Data points were recorded in triplicate for 60 min at 2-min intervals. Data were plotted as the ratio of fluorescence value/OD600 data point.
Results and discussion
Compromise of Tra1 function differentially impacts azole resistance across yeast species
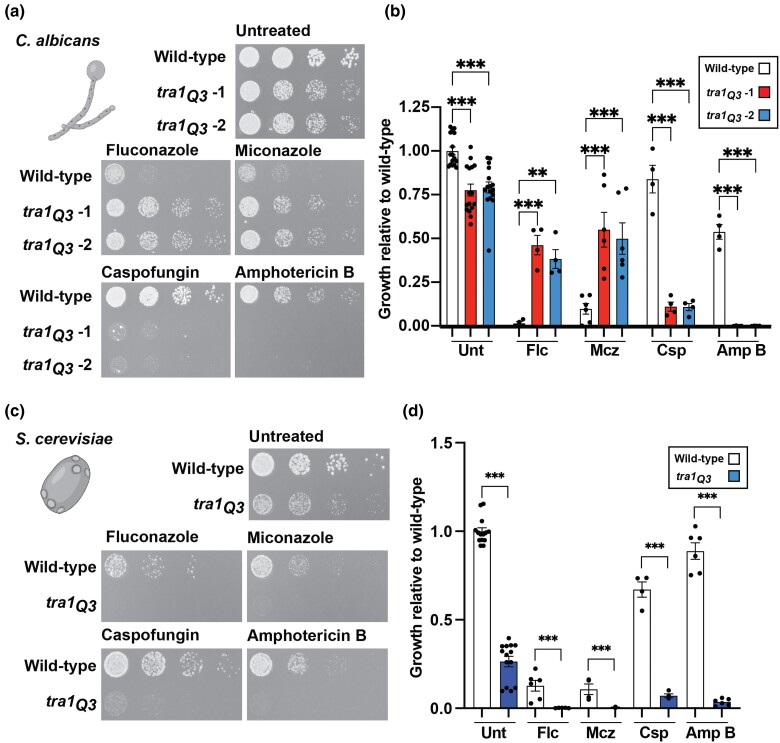
To test the effect of the tra1Q3 allele on the C. albicans antifungal resistance, wild-type cells and cells carrying the mutations were spotted on agar plates containing the echinocandin caspofungin, the azoles miconazole and fluconazole, or amphotericin B (Fig. 1a and b). While caspofungin disrupts the fungal cell wall by inhibiting the β-(1,3)-D-glucan synthase, both azoles and amphotericin B affect fungal membranes. Azoles inhibit ergosterol synthesis by inhibiting the cytochrome P450 enzyme 14α-demethylase (Hitchcock 1991; Wu et al. 2017). Polyenes, such as amphotericin B, disrupt membrane integrity by directly binding to ergosterol (Nett et al. 2010). As previously observed (Razzaq et al. 2021), 2 independently generated tra1Q3 mutants display increased sensitivity to caspofungin. Similarly, we observed increased sensitivity to amphotericin B. Unexpectedly, tra1Q3 cells showed increased tolerance to the azoles miconazole and fluconazole compared to wild type. While both azoles and amphotericin B affect fungal membrane integrity, they do so via very distinct mechanisms; mechanisms for the development of antifungal resistance to these drugs are also very different (Lee et al. 2020). Interestingly, deleting other SAGA complex components such as SPT7, SPT8, and ADA2 sensitizes C. albicans to both caspofungin and fluconazole (Bruno et al. 2006; Sellam et al. 2009; Rashid et al. 2022), whereas deleting other components, such as NGG1 and UPB8, has no effect on antifungal drug resistance (Rashid et al. 2022). Therefore, different SAGA components and/or modules may differentially contribute to antifungal drug resistance. Moreover, due to its incorporation in both SAGA and NuA4, Tra1 has additional roles that may contribute to the phenotypes observed here. Indeed, while the role for NuA4 in hyphal growth has been characterized (Lu et al. 2008), how it regulates antifungal drug resistance remains unclear.
Fig. 1.
tra1Q3 results in differential resistance to azoles across yeast species. a) Wild-type and tra1Q3 C. albicans cells were spotted on YPD agar plates at 30°C without treatment or containing either 20 µg/mL fluconazole (Flc), 0.5 µg/mL miconazole (Mcz), 0.05 µg/mL caspofungin (Csp), or 0.5 µg/mL amphotericin B (Amp B). b) Quantification of the growth relative to untreated wild type is shown in the bar graph. The second dilution was used for quantification. Data are presented ± SEM n ≥ 4 and ***P < 0.0003; **P < 0.01. c) Wild-type and tra1Q3 S. cerevisiae cells were spotted on YPD agar plates at 30°C without treatment or containing either 20 µg/mL Flc, 0.3 µg/mL Mcz, 0.10 µg/mL Csp, or 0.5 µg/mL Amp B. d) Quantification of the growth relative to untreated wild type is shown in the bar graph. Second dilution was used for quantification. Data are presented ± SEM n ≥ 4 and ***P < 0.0003. Yeast illustrations were generated using biorender.com.
In contrast, in S. cerevisiae, tra1Q3 cells are hypersensitive to caspofungin, amphotericin B, and azoles (Fig. 1c and d). This is consistent with previous results showing that loss of Tra1 function in budding yeast sensitizes cells to multiple stresses such as heat shock, protein misfolding, ageing, cell wall perturbation, and DNA damage (Mutiu et al. 2007; Hoke et al. 2008, 2010; Berg et al. 2018; Cheung and Díaz-Santín 2019; Jiang et al. 2019; Bari et al. 2022). The differential sensitivity to azoles between C. albicans and S. cerevisiae suggests that differences exist between the genes impacted by Tra1 across yeast species.
Tra1 regulates the expression of genes associated with azole resistance
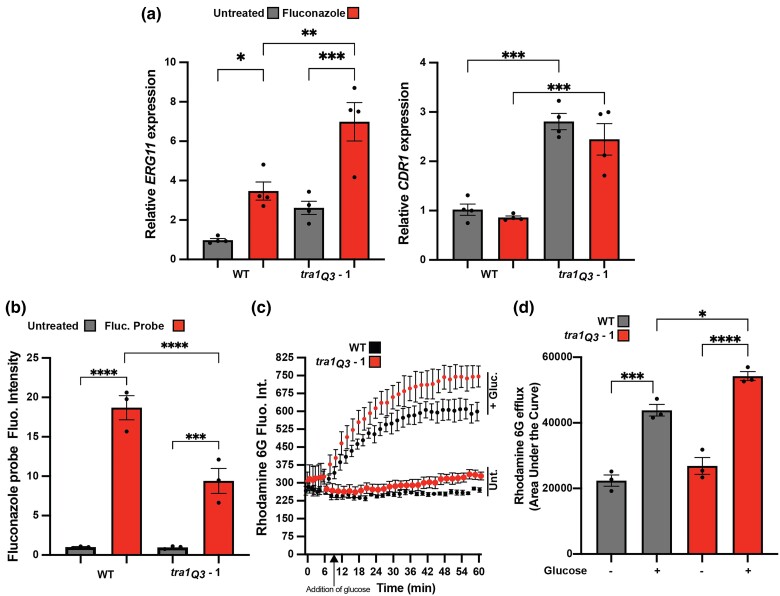
Given the decreased azole susceptibility in tra1Q3 cells, we next assessed the expression of genes previously associated with azole resistance in C. albicans. Specifically, we addressed the expression of ERG11 and CDR1. Mutations in ERG11 resulting in overexpression or loss of drug affinity are associated with azole resistance in multiple C. albicans clinical isolates (Franz et al. 1998; MacPherson et al. 2005; Flowers et al. 2015). Cdr1 is a member of the ATP-binding cassette transporter family associated with multidrug resistance (Dogra et al. 1999; Holmes et al. 2008). Cdr1 is known for its role in fluconazole efflux and acquired multidrug resistance in clinical isolates of C. albicans (Holmes et al. 2008). As shown in Fig. 2a, there was a significant increase in the expression of CDR1 and ERG11 in tra1Q3 cells relative to wild type both in untreated conditions (both 2.8-fold) and upon the addition of fluconazole (2.7- and 2.1-fold, respectively). In the past, we have shown, in budding yeast, that components of the SAGA complex can act as repressors of transcription under different conditions (Ricci et al. 2002).
Fig. 2.
Candida albicans tra1Q3 mutants display phenotypes associated with azole resistance. a) tra1Q3 cells show increased expression of genes linked to azole resistance. Wild-type and tra1Q3 C. albicans cells were incubated with 20 µg/mL fluconazole for 1 h and the expression of ERG11 and CDR1 was assessed by qRT-PCR. n = 4. b) tra1Q3 cells show reduced accumulation of intracellular fluconazole. Wild-type and tra1Q3 C. albicans cells were incubated with a fluorescent fluconazole probe for 1 h, and mean fluorescent intensity of intracellular fluconazole was assessed by flow cytometry. n = 3. c) R6G efflux is increased in tra1Q3 cells. Energy-depleted cells were incubated with R6G for 1 h and then left untreated or treated with glucose to induce efflux. Mean rhodamine 6D fluorescence was monitored over time. n = 3. d) Quantification of the area under the curve calculated from R6G release assays is shown in the bar graph. n = 3. Means are shown ± SEM ****P < 0.0001; ***P < 0.0003; **P < 0.01; *P < 0.05.
In light of the increased expression of CDR1 in tra1Q3 cells in C. albicans, we investigated whether compromising Tra1 function affects drug efflux and intracellular bioavailability. To do so, we took advantage of a fluorescent probe that allows the real-time imaging of azole uptake in fungal cells (Benhamou et al. 2017). We found that intracellular accumulation of fluorescently tagged azole is significantly reduced by 50% in tra1Q3 cells (Fig. 2b). Since CDR1 expression has been linked to increased efflux of fluconazole in fungi (Hernáez et al. 1998; Kim et al. 2019), we tested whether tra1Q3 C. albicans have higher drug efflux capacity using the well-characterized Cdr1 efflux substrate R6G (Maesaki et al. 1999). Our findings support that there is increased efflux of R6G in tra1Q3 cells (Fig. 2c and d), suggesting that elevated expression of CDR1 contributes to greater azole tolerance. This upregulation of efflux pumps may explain the unique azole resistance of tra1Q3. Upregulation of efflux pumps is a well-established mechanism of azole resistance but has not been linked to resistance of amphotericin B or caspofungin (Lee et al. 2020).
In response to azole, C. albicans activates the calcineurin pathway, which is essential for virulence (Blankenship et al. 2003; Juvvadi et al. 2014). Inhibiting calcineurin-dependent signaling with FK506 increases susceptibility to azole in C. albicans (Cruz et al. 2002; Uppuluri et al. 2008; Khodavaisy et al. 2023). Thus, we tested whether FK506 alleviates the azole resistance observed in tra1Q3 cells. Indeed, we found that azole tolerance in tra1Q3 cells is suppressed by treatment with the calcineurin inhibitor (Supplementary Fig. 1). Together, our findings suggest that while some cellular mechanisms associated with azole resistance, such as ERG11 and CDR1 expression, are increased in tra1Q3 cells, drug tolerance still requires a functional calcineurin pathway. This is in agreement with previous studies showing that the expression of CDR1 and ergosterol biosynthesis genes is independent of calcineurin signaling (Cruz et al. 2002; Jia et al. 2016).
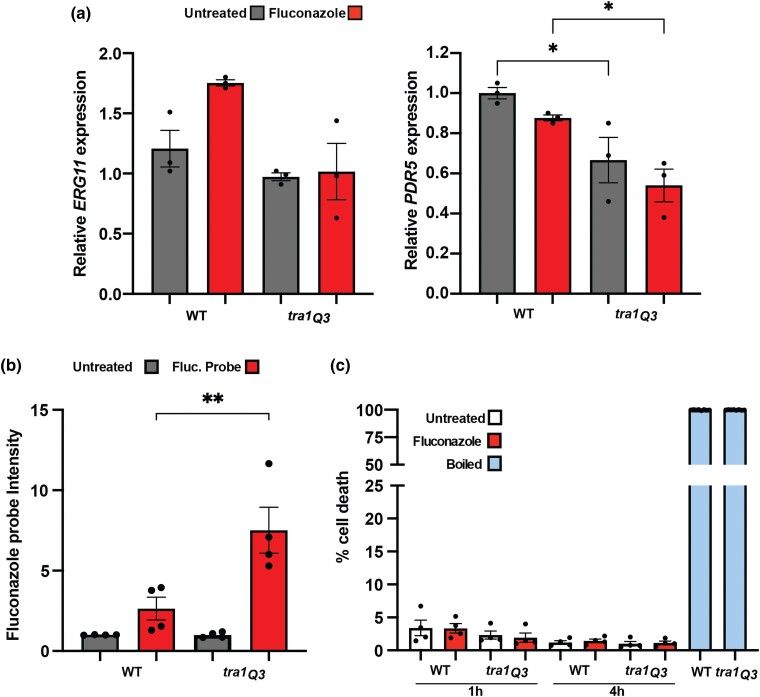
Next, we investigated the effect of tra1Q3 on azole resistance mechanisms in budding yeast given the differences between C. albicans and S. cerevisiae (Fig. 1). In S. cerevisiae, SAGA regulates gene expression in response to changes in sterol content (Dewhurst-Maridor et al. 2017). NuA4 is also linked to sterol metabolism, and eaf1Δ cells display increased accumulation of ergosterol esters (Pham et al. 2022). Unlike C. albicans, S. cerevisiae tra1Q3 cells did not demonstrate significant changes in ERG11 expression, as compared to the wild type (Fig. 3a). This difference with our C. albicans data is consistent with a potential rewiring of the role of Tra1 in the transcription of sterol genes between species. Indeed, rewiring of SAGA functions between S. cerevisiae and C. albicans has been previously reported. Spt3 negatively regulates filamentous growth (a key virulence trait) in C. albicans but has an opposite role in S. cerevisiae (Laprade et al. 2002). Moreover, Sch9 has been shown to play a critical role in chromosome segregation in C. albicans, a function absent in budding yeast (Varshney et al. 2015). Functional rewiring between the 2 species is not surprising given that the phylogenetic distance between S. cerevisiae and C. albicans is approximately the same as between humans and sponges (Shen et al. 2018).
Fig. 3.
Saccharomyces cerevisiae tra1Q3 mutants display phenotypes associated with increased azole sensitivity. a) tra1Q3 cells show decreased expression of genes linked to azole resistance. Wild-type and tra1Q3 C. albicans cells were incubated with 20 µg/mL fluconazole for 1 h and the expression of ERG11 and PDR5 was assessed by qRT-PCR. n = 3. b) tra1Q3 cells show increased accumulation of intracellular fluconazole. Wild-type and tra1Q3 cells were incubated with a fluorescent fluconazole probe for 1 h, and mean fluorescent intensity of intracellular fluconazole was assessed by flow cytometry. n = 4. c) tra1Q3 cells do not show increased cell death upon fluconazole treatment. Wild-type and tra1Q3 S. cerevisiae cells were incubated with 20 µg/mL fluconazole for 1 and 4 h and stained with PI to assess cell viability. Boiled cells are shown as positive control. The percentage of PI-positive cells was assessed by flow cytometry and presented in a bar graph. n = 4. All data are shown ± SEM *P < 0.05 **P<0.01.
Since the tra1Q3 cells showed increased expression of CDR1 in C. albicans, we investigated its impact on the expression of the PDR5 ABC transporter (the CDR1 homolog) in S. cerevisiae. We found reduced expression of PDR5 in tra1Q3 cells (Fig. 3a). This is consistent with the role of SAGA in the regulation of PDR5 expression (Gao et al. 2004) and reduced global transcription of SAGA-regulated genes previously observed in the tra1Q3 mutant (Berg et al. 2018). Consistent with reduced PDR5 expression, intracellular accumulation of fluorescently labeled fluconazole increased in tra1Q3 cells (Fig. 3b).
In contrast to other antifungal drugs, such as amphotericin B, which behave in a fungicidal manner, azoles are fungistatic, thus inducing minimal cell death in multiple yeast species (Manavathu et al. 1998). For this reason, we investigated whether the decreased growth observed in the tra1Q3 S. cerevisiae cells treated with fluconazole was associated with changes in cell death. Wild-type and tra1Q3 S. cerevisiae cells were treated with fluconazole and stained with PI at different time intervals for up to 24 h (Fig. 3c). No significant differences were observed, indicating that the effect of fluconazole remains fungistatic within the tra1Q3 cells. Finally, treatment of S. cerevisiae with the calcineurin inhibitor FK506 sensitized cells to fluconazole (Supplementary Fig. 2a). While tra1Q3 cells were not inherently sensitive to the inhibitor, the cells displayed a synthetic negative interaction when crossed with a cnb1Δ mutant (Supplementary Fig. 2b), which encodes a calcineurin subunit (Cyert and Thorner 1992). We previously observed a similar phenotype with a distinct Tra1 mutant (Hoke et al. 2008). These results suggest that Tra1 and calcineurin, like in C. albicans, function within distinct signaling pathways.
Conclusions and perspectives
While biochemical and structural studies have extensively characterized Tra1 functions in model yeast, its specific impact for gene expression in fungal pathogens such as C. albicans is poorly understood. How compromise of Tra1 function leads to the upregulation of genes linked to azole resistance will require more detailed investigations to define genome-wide occupancy of coactivator complexes and their role in activation, repression, and maintenance of transcription under different conditions.
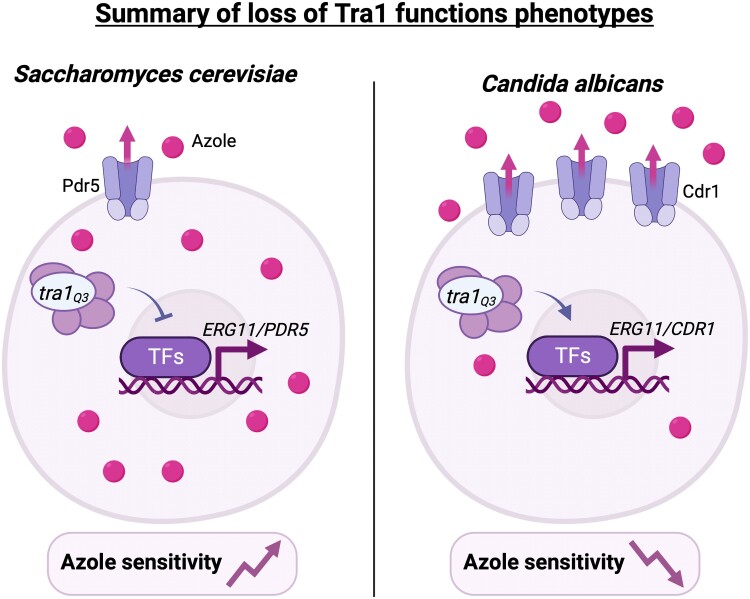
Here, we also show that negatively impacting Tra1 function has opposite effects with regard to azole tolerance between S. cerevisiae and C. albicans (Fig. 4). This highlights the evolutionary diversity of the control of the antifungal response by Tra1-containing complexes. Future investigations should aim at defining the breadth of Tra1 functions across fungi. Nakaseomyces (Candida) glabrata is the second most common cause of candidiasis (McCarty et al. 2021) but is evolutionarily more closely related to S. cerevisiae than other pathogenic Candida species. Nakaseomyces glabrata is highly dependent upon the upregulation of CDR1 and CDR2 in response to antifungal stress (Sanglard et al. 2009) and thus could serve as a compelling comparison to assess this divergent role of Tra1 among yeast species. Finally, similar to other members of the PIKK family, such as Tor, Tra1 should be druggable. However, our data suggest that its potential as a candidate for combinational therapy with antifungal drugs would have to be considered carefully.
Fig. 4.
Summary of tra1Q3 phenotypes associated with azole treatment in S. cerevisiae and C. albicans. In S. cerevisiae, compromise of Tra1 function associated with the tra1Q3 allele results in decreased expression of genes associated with azole resistance such as ERG11 and PDR5. Consequently, tra1Q3 cells show increased accumulation of intracellular azole. In C. albicans, tra1Q3 is linked to the increased expression of ERG11 and CDR1, increased efflux of azole, and consequently, increased drug resistance. Created with Biorender.com.
Supplementary Material
Acknowledgments
The authors thank Micha Fridman (University of Tel-Aviv) for the fluorescent fluconazole probe. We also thank Matthew Berg for his help in the early stages of the project.
Contributor Information
Gabriela Marsiglio Nunes Librais, Department of Anatomy and Cell Biology, The University of Western Ontario, London, Ontario, N6A 5C1, Canada.
Yuwei Jiang, Department of Anatomy and Cell Biology, The University of Western Ontario, London, Ontario, N6A 5C1, Canada.
Iqra Razzaq, Department of Molecular and Cellular Biology, University of Guelph, Guelph, Ontario N1G 2W1, Canada.
Christopher J Brandl, Department of Biochemistry, The University of Western Ontario, London, Ontario, N6A 5C1, Canada.
Rebecca S Shapiro, Department of Molecular and Cellular Biology, University of Guelph, Guelph, Ontario N1G 2W1, Canada.
Patrick Lajoie, Department of Anatomy and Cell Biology, The University of Western Ontario, London, Ontario, N6A 5C1, Canada.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at G3 online.
Funding
This study was supported by a Canadian Institutes of Health Research (CIHR) Project Grant (PJT 168882), a Natural Sciences and Engineering Research Council (NSERC) Discovery Grant (RGPIN-2022-05267), and a John R Evans Leader Fund from the Canadian Foundation for Innovation (CFI) Grant (65183) to PL. RSS is supported by a Canadian Institutes offor Health Research Project Grant (PJT 162195) and a NSERC Discovery Grant (RGPIN-2018-4914). CJB was supported by a NSERC Discovery Grant (RGPIN-2015-04394). YJ was supported by a Masters to PhD Transfer Scholarship from the Dean of the Schulich School of Medicine and Dentistry at Western University.
Literature cited
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Côté J. 1999. Nua4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18(18):5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew C, Sellam A, Epp E, Hogues H, Mullick A, Nantel A, Whiteway M. 2009. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog. 5(10):e1000612. doi: 10.1371/journal.ppat.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashov SV, Park S, Perlin DS. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob Agents Chemother. 50(6):2058–2063. doi: 10.1128/AAC.01653-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari KA, Berg MD, Genereaux J, Brandl CJ, Lajoie P. 2022. Tra1 controls the transcriptional landscape of the aging cell. G3 (Bethesda) 13(1):jkac287. doi: 10.1093/g3journal/jkac287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamou RI, Bibi M, Steinbuch KB, Engel H, Levin M, Roichman Y, Berman J, Fridman M. 2017. Real-time imaging of the azole class of antifungal drugs in live Candida cells. ACS Chem Biol. 12(7):1769–1777. doi: 10.1021/acschembio.7b00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MD, Genereaux J, Karagiannis J, Brandl CJ. 2018. The pseudokinase domain of tra1 is required for nuclear localization and incorporation into the SAGA and NuA4 complexes. G3 (Bethesda) 8(6):1943–1957. doi: 10.1534/g3.118.200288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JR, Wormley FL, Boyce MK, Schell WA, Filler SG, Perfect JR, Heitman J. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot Cell. 2(3):422–430. doi: 10.1128/EC.2.3.422-430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel). 3(4):57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno VM, Kalachikov S, Subaran R, Nobile CJ, Kyratsous C, Mitchell AP. 2006. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2(3):e21. doi: 10.1371/journal.ppat.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick SR, Pananos AD, Di Gregorio SE, Park AE, Etedali-Zadeh P, Duennwald ML, Lajoie P. 2016. A toolbox for rapid quantitative assessment of chronological lifespan and survival in Saccharomyces cerevisiae. Traffic 17(6):689–703. doi: 10.1111/tra.12391. [DOI] [PubMed] [Google Scholar]
- Chang P, Fan X, Chen J. 2015. Function and subcellular localization of Gcn5, a histone acetyltransferase in Candida albicans. Fungal Genet Biol. 81:132–141. doi: 10.1016/j.fgb.2015.01.011. [DOI] [PubMed] [Google Scholar]
- Cheung ACM, Díaz-Santín LM. 2019. Share and share alike: the role of Tra1 from the SAGA and NuA4 coactivator complexes. Transcription 10(1):37–43. doi: 10.1080/21541264.2018.1530936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 19(4):2515–2526. doi: 10.1128/MCB.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, Perfect JR, McCusker JH, Heitman J. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21(4):546–559. doi: 10.1093/emboj/21.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS, Thorner J. 1992. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol Cell Biol. 12(8):3460–3469. doi: 10.1128/mcb.12.8.3460-3469.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhurst-Maridor G, Abegg D, David FPA, Rougemont J, Scott CC, Adibekian A, Riezman H. 2017. The SAGA complex, together with transcription factors and the endocytic protein Rvs167p, coordinates the reprofiling of gene expression in response to changes in sterol composition in. Mol Biol Cell. 28(20):2637–2649. doi: 10.1091/mbc.e17-03-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra S, Krishnamurthy S, Gupta V, Dixit BL, Gupta CM, Sanglard D, Prasad R. 1999. Asymmetric distribution of phosphatidylethanolamine in C. albicans: possible mediation by CDR1, a multidrug transporter belonging to ATP binding cassette (ABC) superfamily. Yeast 15(2):111–121. doi:doi:. [DOI] [PubMed] [Google Scholar]
- Downey M. 2021. Non-histone protein acetylation by the evolutionarily conserved GCN5 and PCAF acetyltransferases. Biochim Biophys Acta Gene Regul Mech. 1864(2):194608. doi: 10.1016/j.bbagrm.2020.194608. [DOI] [PubMed] [Google Scholar]
- Flowers SA, Colón B, Whaley SG, Schuler MA, Rogers PD. 2015. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother. 59(1):450–460. doi: 10.1128/AAC.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz R, Kelly SL, Lamb DC, Kelly DE, Ruhnke M, Morschhäuser J. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob Agents Chemother. 42(12):3065–3072. doi: 10.1128/AAC.42.12.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Wang L, Milgrom E, Shen W-CW. 2004. On the mechanism of constitutive Pdr1 activator-mediated PDR5 transcription in Saccharomyces cerevisiae: evidence for enhanced recruitment of coactivators and altered nucleosome structures. J Biol Chem. 279(41):42677–42686. doi: 10.1074/jbc.M406363200. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11(13):1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Yates JR 3rd, Workman JL. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 2(6):863–867. doi: 10.1016/S1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- Helmlinger D, Marguerat S, Villén J, Swaney DL, Gygi SP, Bähler J, Winston F. 2011. Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. EMBO J. 30(14):2843–2852. doi: 10.1038/emboj.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernáez ML, Gil C, Pla J, Nombela C. 1998. Induced expression of the Candida albicans multidrug resistance gene CDR1 in response to fluconazole and other antifungals. Yeast 14(6):517–526. doi:. [DOI] [PubMed] [Google Scholar]
- Hitchcock CA. 1991. Cytochrome P-450-dependent 14 alpha-sterol demethylase of Candida albicans and its interaction with azole antifungals. Biochem Soc Trans. 19(3):782–787. doi: 10.1042/bst0190782. [DOI] [PubMed] [Google Scholar]
- Hoke SMT, Guzzo J, Andrews B, Brandl CJ. 2008. Systematic genetic array analysis links the Saccharomyces cerevisiae SAGA/SLIK and NuA4 component Tra1 to multiple cellular processes. BMC Genet. 9(1):46. doi: 10.1186/1471-2156-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoke SMT, Irina Mutiu A, Genereaux J, Kvas S, Buck M, Yu M, Gloor GB, Brandl CJ. 2010. Mutational analysis of the C-terminal FATC domain of Saccharomyces cerevisiae Tra1. Curr Genet. 56(5):447–465. doi: 10.1007/s00294-010-0313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AR, Lin Y-H, Niimi K, Lamping E, Keniya M, Niimi M, Tanabe K, Monk BC, Cannon RD. 2008. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrob Agents Chemother. 52(11):3851–3862. doi: 10.1128/AAC.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Zhang H, Li C, Li G, Liu X, Wei J. 2016. The calcineruin inhibitor cyclosporine a synergistically enhances the susceptibility of Candida albicans biofilms to fluconazole by multiple mechanisms. BMC Microbiol. 16(1):113. doi: 10.1186/s12866-016-0728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Berg MD, Genereaux J, Ahmed K, Duennwald ML, Brandl CJ, Lajoie P. 2019. Sfp1 links TORC1 and cell growth regulation to the yeast SAGA-complex component Tra1 in response to polyQ proteotoxicity. Traffic 20(4):267–283. doi: 10.1111/tra.12637. [DOI] [PubMed] [Google Scholar]
- Juvvadi PR, Lamoth F, Steinbach WJ. 2014. Calcineurin as a multifunctional regulator: unraveling novel functions in fungal stress responses, hyphal growth, drug resistance, and pathogenesis. Fungal Biol Rev. 28(2–3):56–69. doi: 10.1016/j.fbr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Nobile CJ. 2022. Antifungal drug-resistance mechanisms in Candida biofilms. Curr Opin Microbiol. 71:102237. doi: 10.1016/j.mib.2022.102237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodavaisy S, Gharehbolagh SA, Abdorahimi M, Rezaie S, Ahmadikia K, Badali H, Meis JF, Mahmoudi S. 2023. In vitro combination of antifungal drugs with tacrolimus (FK506) holds promise against clinical Candida species, including Candida auris. Med Mycol. 61(7):myad069. doi: 10.1093/mmy/myad069. [DOI] [PubMed] [Google Scholar]
- Kim SH, Iyer KR, Pardeshi L, Muñoz JF, Robbins N, Cuomo CA, Wong KH, Cowen LE. 2019. Genetic analysis of implicates Hsp90 in morphogenesis and azole tolerance and cdr1 in azole resistance. mBio 10(1):e02529-18. doi: 10.1128/mBio.02529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov AE. 2021. Azoles—Synthesis, Properties, Applications and Perspectives. London, UK: IntechOpen. p. 1–146. [Google Scholar]
- Laprade L, Boyartchuk VL, Dietrich WF, Winston F. 2002. Spt3 plays opposite roles in filamentous growth in Saccharomyces cerevisiae and Candida albicans and is required for C. albicans virulence. Genetics 161(2):509–519. doi: 10.1093/genetics/161.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Maccallum DM, Jacobsen MD, Walker LA, Odds FC, Gow NA, Munro CA. 2012. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob Agents Chemother. 56(1):208–217. doi: 10.1128/AAC.00683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Puumala E, Robbins N, Cowen LE. 2020. Antifungal drug resistance: molecular mechanisms in Candida albicans and beyond. Chem Rev. 121(6):3390–3411. doi: 10.1021/acs.chemrev.0c00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Su C, Mao X, Raniga PP, Liu H, Chen J. 2008. Efg1-mediated recruitment of NuA4 to promoters is required for hypha-specific Swi/Snf binding and activation in Candida albicans. Mol Biol Cell. 19(10):4260–4272. doi: 10.1091/mbc.e08-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson S, Akache B, Weber S, De Deken X, Raymond M, Turcotte B. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob Agents Chemother. 49(5):1745–1752. doi: 10.1128/AAC.49.5.1745-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maesaki S, Marichal P, Vanden Bossche H, Sanglard D, Kohno S. 1999. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J Antimicrob Chemother. 44(1):27–31. doi: 10.1093/jac/44.1.27. [DOI] [PubMed] [Google Scholar]
- Manavathu EK, Cutright JL, Chandrasekar PH. 1998. Organism-dependent fungicidal activities of azoles. Antimicrob Agents Chemother. 42(11):3018–3021. doi: 10.1128/AAC.42.11.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty TP, White CM, Pappas PG. 2021. Candidemia and invasive candidiasis. Infect Dis Clin North Am. 35(2):389–413. doi: 10.1016/j.idc.2021.03.007. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94(3):363–374. doi: 10.1016/S0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- Mutiu AI, Hoke SMT, Genereaux J, Hannam C, MacKenzie K, Jobin-Robitaille O, Guzzo J, Côté J, Andrews B, Haniford DB, et al. 2007. Structure/function analysis of the phosphatidylinositol-3-kinase domain of yeast tra1. Genetics 177(1):151–166. doi: 10.1534/genetics.107.074476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Crawford K, Marchillo K, Andes DR. 2010. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob Agents Chemother. 54(8):3505–3508. doi: 10.1128/AAC.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. 2018. Invasive candidiasis. Nat Rev Dis Primers. 4(1):18026. doi: 10.1038/nrdp.2018.26. [DOI] [PubMed] [Google Scholar]
- Petropavlovskiy AA, Tauro MG, Lajoie P, Duennwald ML. 2020. A quantitative imaging-based protocol for yeast growth and survival on agar plates. STAR Protoc. 1(3):100182. doi: 10.1016/j.xpro.2020.100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T, Walden E, Huard S, Pezacki J, Fullerton MD, Baetz K. 2022. Fine-tuning acetyl-CoA carboxylase 1 activity through localization: functional genomics reveals a role for the lysine acetyltransferase NuA4 and sphingolipid metabolism in regulating Acc1 activity and localization. Genetics 221(4):iyac086. doi: 10.1093/genetics/iyac086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid S, Correia-Mesquita TO, Godoy P, Omran RP, Whiteway M. 2022. SAGA complex subunits in differentially regulate filamentation, invasiveness, and biofilm formation. Front Cell Infect Microbiol. 12:764711. doi: 10.3389/fcimb.2022.764711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaq I, Berg MD, Jiang Y, Genereaux J, Uthayakumar D, Kim GH, Agyare-Tabbi M, Halder V, Brandl CJ, Lajoie P, et al. 2021. The SAGA and NuA4 component Tra1 regulates Candida albicans drug resistance and pathogenesis. Genetics 219(2):iyab131. doi: 10.1093/genetics/iyab131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci AR, Genereaux J, Brandl CJ. 2002. Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol Cell Biol. 22(12):4033–4042. doi: 10.1128/MCB.22.12.4033-4042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A. 2022. Evolution of the human pathogenic lifestyle in fungi. Nat Microbiol. 7(5):607–619. doi: 10.1038/s41564-022-01112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Schieltz D, Ting N, McMahon SB, Litchfield DW, Yates JR 3rd, Beys-da-Silva WO. 1998. Tra1p is a component of the yeast Ada.Spt transcriptional regulatory complexes. J Biol Chem. 273(41):26559–26565. doi: 10.1074/jbc.273.41.26559. [DOI] [PubMed] [Google Scholar]
- Sanglard D, Coste A, Ferrari S. 2009. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 9(7):1029–1050. doi: 10.1111/j.1567-1364.2009.00578.x. [DOI] [PubMed] [Google Scholar]
- Sellam A, Askew C, Epp E, Lavoie H, Whiteway M, Nantel A. 2009. Genome-wide mapping of the coactivator Ada2p yields insight into the functional roles of SAGA/ADA complex in Candida albicans. Mol Biol Cell. 20(9):2389–2400. doi: 10.1091/mbc.e08-11-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X-X, Opulente DA, Kominek J, Zhou X, Steenwyk JL, Buh KV, Haase MAB, Wisecaver JH, Wang M, Doering DT, et al. 2018. Tempo and mode of genome evolution in the budding yeast subphylum. Cell 175(6):1533–1545.e20. doi: 10.1016/j.cell.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivarathri R, Tscherner M, Zwolanek F, Singh NK, Chauhan N, Kuchler K. 2019. The fungal histone acetyl transferase Gcn5 controls virulence of the human pathogen Candida albicans through multiple pathways. Sci Rep. 9(1):9445. doi: 10.1038/s41598-019-45817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steunou A-L, Rossetto D, Côté J. 2014. Regulating chromatin by histone acetylation. In: Workman J, Abmayr S, editors. Fundamentals of Chromatin. New York, NY: Springer. p. 147–212. [Google Scholar]
- Uppuluri P, Nett J, Heitman J, Andes D. 2008. Synergistic effect of calcineurin inhibitors and fluconazole against Candida albicans biofilms. Antimicrob Agents Chemother. 52(3):1127–1132. doi: 10.1128/AAC.01397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney N, Schaekel A, Singha R, Chakraborty T, van Wijlick L, Ernst JF, Sanyal K. 2015. A surprising role for the Sch9 protein kinase in chromosome segregation in Candida albicans. Genetics 199(3):671–674. doi: 10.1534/genetics.114.173542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Gao N, Li C, Gao J, Ying C. 2017. A newly identified amino acid substitution T123I in the 14α-demethylase (Erg11p) of Candida albicans confers azole resistance. FEMS Yeast Res. 17(3):1–6. doi: 10.1093/femsyr/fox012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at G3 online.