Abstract
In Staphylococcus epidermidis and Staphylococcus aureus, a number of cell wall- and cytoplasmic membrane-associated lipoproteins are induced in response to iron starvation. To gain insights into the molecular basis of iron-dependent gene regulation in the staphylococci, we sequenced the DNA upstream of the 3-kb S. epidermidis sitABC operon, which Northern blot analysis indicates is transcriptionally regulated by the growth medium iron content. We identified two DNA sequences which are homologous to elements of the Corynebacterium diphtheriae DtxR regulon, which controls, in response to iron stress, for example, production of diphtheria toxin, siderophore, and a heme oxygenase. Upstream of the sitABC operon and divergently transcribed lies a 645-bp open reading frame (ORF), which codes for a polypeptide of approximately 25 kDa with homology to the DtxR family of metal-dependent repressor proteins. This ORF has been designated SirR (staphylococcal iron regulator repressor). Within the sitABC promoter/operator region, we also located a region of dyad symmetry overlapping the transcriptional start of sitABC which shows high homology to the DtxR operator consensus sequence, suggesting that this region, termed the Sir box, is the SirR-binding site. The SirR protein was overexpressed, purified, and used in DNA mobility shift assays; SirR retarded the migration of a synthetic oligonucleotide based on the Sir box in a metal (Fe2+ or Mn2+)-dependent manner, providing confirmatory evidence that this motif is the SirR-binding site. Furthermore, Southern blot analysis of staphylococcal chromosomal DNA with the synthetic Sir box as a probe confirmed that there are at least five Sir boxes in the S. epidermidis genome and at least three in the genome of S. aureus, suggesting that SirR controls the expression of multiple target genes. Using a monospecific polyclonal antibody raised against SirR to probe Western blots of whole-cell lysates of S. aureus, S. carnosus, S. epidermidis, S. hominis, S. cohnii, S. lugdunensis, and S. haemolyticus, we identified an approximately 25-kDa cross-reactive protein in each of the staphylococcal species examined. Taken together, these data suggest that SirR functions as a divalent metal cation-dependent transcriptional repressor which is widespread among the staphylococci.
Staphylococcus aureus is well recognized as a human pathogen responsible for a variety of pyogenic and toxin-related infections (7). In contrast, coagulase-negative staphylococci such as Staphylococcus epidermidis have emerged more recently as a major medical problem with the widespread use of implanted medical devices (7). Both organisms are now frequent pathogens in hospitals and account for much morbidity and mortality. S. epidermidis is much less biologically active than S. aureus being almost devoid of conventional exotoxins but possessing a marked capacity to adhere to and form biofilms on the surfaces of implanted medical devices (7). However, the acquisition of essential nutrients to facilitate growth in host tissues is a problem common for all bacterial pathogens, and therefore relevant, to both S. aureus and S. epidermidis infections. Such growth is critical to the establishment of infection and depends in part on the ability of the pathogen to scavenge nutrients such as iron (44, 46). Although there is an abundance of iron in the extracellular body fluids, the free ionic iron concentration (10−18 M), due to the presence of the iron-binding glycoproteins transferrin (in serum) and lactoferrin (on mucosal surfaces and in polymorphonuclear leukocytes), is far too low to support the growth of bacteria such as the staphylococci (44, 46). Furthermore, in a number of bacterial pathogens, this lack of readily available iron constitutes a major environmental signal which coordinately controls the expression of a number of virulence and metabolic genes unrelated to iron acquisition (21).
To grow in host tissues, staphylococci must therefore acquire iron. While there is a considerable amount of information on the iron-sequestering systems of gram-negative bacteria and their contribution to virulence (46, 48), there is comparatively little information on the staphylococci (44). Although they produce and use siderophores (low-molecular-mass iron chelators), the genes and gene products involved in their regulation, synthesis, export, or import are unknown (44). Previously, we identified a number of iron-repressible S. aureus and S. epidermidis cell wall- and cytoplasmic membrane-associated proteins which are expressed during growth in vivo during both human (37, 45) and experimental animal (23, 25) infections. These include a 42-kDa cell wall protein which functions as a receptor for human transferrin (24, 26) and a 32-kDa cytoplasmic membrane-associated lipoprotein (6, 23, 37). The gene encoding this lipoprotein has recently been cloned from S. epidermidis and shown to be a component of a translationally coupled, iron-regulated operon which consists of three genes (sitABC), the products of which constitute an ABC transporter (6). Although the function and contribution of the sit operon to growth in vivo and to the pathogenesis of staphylococcal infection are not known, this operon does show significant homology to a family of streptococcal ABC operons involved in adherence and genetic competence and which are essential for virulence (10). Since SitC is not exposed at the staphylococcal cell surface (6), it is unlikely to function as an adhesin and is probably involved in the acquisition of metal ions. Furthermore, the mechanism by which the sit operon is regulated via the growth medium iron content is not known.
In gram-negative bacteria such as Escherichia coli, the ferric uptake regulator (Fur) protein is responsible for the iron-dependent transcriptional regulation of genes involved in the biosynthesis and transport of siderophores such as aerobactin and enterobactin and in the regulation of virulence determinants such as the enterotoxin Stx1 (3, 21, 48). Since the fur locus was first identified in Salmonella typhimurium (12) and extensively characterized in E. coli (21), numerous other Fur homologs have been found in gram-negative bacteria such as Vibrio cholerae (21), Pseudomonas aeruginosa (30), Yersinia pestis (38), and Neisseria meningitidis (43). In these bacteria, the coordinate regulation of multiple genes by iron depends on Fur functioning as an iron-responsive, DNA-binding repressor protein. The 17-kDa Fur protein functions as a dimer and in the presence of Fe2+ binds to a consensus sequence termed the Fur box located within the promoter region of the target genes (3). However, when iron levels are low, Fur does not bind and the genes are expressed (21).
Although Fur homologs have been identified in both Bacillus subtilis (5) and S. epidermidis (19), by far the most intensively investigated iron-dependent repressor in gram-positive bacteria is DtxR (41). This protein was first identified as repressor of diphtheria toxin synthesis in Corynebacterium diphtheriae (41). Although DtxR is a functional homolog of Fur, it shares no amino acid homology and belongs to a newly emerging family of iron-dependent repressors. Apart from C. diphtheriae, DtxR homologs have now been identified in Streptomyces spp. (16), Brevibacterium lactofermentum (28), and mycobacteria (11) and also in the spirochete Treponema pallidum (17), where they regulate genes encoding iron transport systems, heme oxygenase, and virulence determinants and genes involved in protecting bacteria from oxidative stress (11, 16, 17, 28, 34, 41). This finding suggests that the growing family of DtxR homologs play a central role in regulating the adaptation, survival, and virulence of gram-positive pathogens. In this report, we describe the identification and characterization of a novel DtxR homolog, SirR, from S. epidermidis, which is located immediately upstream of the sitABC operon and which is common to both S. aureus and the coagulase-negative staphylococci.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
S. aureus, S. epidermidis, Staphylococcus hominis, Staphylococcus cohnii, Staphylococcus lugdunensis, and Staphylococcus haemolyticus clinical isolates were obtained from University Hospital, Nottingham, United Kingdom. Staphylococcus carnosus TM300 was a gift from F. Götz (Tübingen, Germany). Strains were maintained by regular subculture on horse blood agar. For broth culture, strains were grown statically for 18 h at 37°C in RPMI 1640 tissue culture medium containing 2 mg of NaHCO3 per ml as described before (6). RPMI 1640 medium was depleted of iron by batch incubation with Chelex 100 (Bio-Rad Laboratories) as described previously (18). Cultures were incubated in 5% CO2 in air; where indicated, the medium was supplemented with 20 μM Fe2(SO4)3 to produce iron-rich growth conditions. Where the effects of different metal ions on gene expression were examined, metal salts (MgSO4, MnCl2, CuSO4, CoCl2, NiCl2, and ZnCl2) were added to iron-depleted RPMI 1640 at 20 μM. E. coli strains were routinely cultured at 37°C in Luria-Bertani (LB) broth or LB agar containing appropriate antibiotics. Plasmid pW32 was derived from a λ-Zap II library and contains a 5.4-kb EcoRI fragment of S. epidermidis genomic DNA which contains the sitABC operon as previously described (6).
SDS-PAGE and immunoblotting.
Staphylococci were digested with lysostaphin (80 μg/μl; Sigma) in phosphate-buffered saline (pH 7.4) for 30 min at 37°C prior to solubilization by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for 5 min, and proteins were separated by SDS-PAGE using a 10% polyacrylamide gel (1). For immunoblotting, polypeptides were transferred to BioTrace NT membrane (Gelman) followed by blocking, incubation with primary antibody (1/500 dilution overnight for polyclonal antisera or 1/2 dilution overnight for monoclonal antibody) and conjugate (1/2,000 dilution of anti-rabbit, anti-rat, or anti-mouse peroxidase conjugate for 4 h), and finally detection of bound antibody (1).
Overexpression and partial purification of SirR.
Recombinant plasmids were recovered by alkaline lysis and subjected to restriction analysis (2). Inserts from positive clones were sequenced with an ABI automated DNA sequencer. DNA and protein sequence analyses were performed with LASERGENE software (DNAstar Inc.). To produce sufficient amounts of SirR to enable in vitro studies, the SirR coding region was amplified by PCR incorporating NcoI and HindIII restriction enzyme sites in the primers. Following digestion, the sirR fragment was inserted into expression vector pTRC99 (Pharmacia) and used to transform E. coli JM107. E. coli carrying the correct construct was grown to an A600 of 0.5 prior to addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.5 mM to induce expression of sirR. Once expression was confirmed by SDS-PAGE, lysates from E. coli overexpressing sirR were subjected to fractionation by fast protein liquid chromatography (FPLC) on a MonoQ column (Pharmacia) over a gradient of 0 to 1 M NaCl. Fractions shown to contain SirR by gel retardation assays, using the 382-bp PCR-derived probe described below, were further purified by affinity purification on Ni-nitrilotriacetic acid resin (Qiagen) as described by Schmitt and Holmes (36).
Gel retardation assay.
Gel retardation assays were performed essentially as described in the protocol for the Boehringer Mannheim digoxigenin gel shift kit. Briefly DNA probes containing the putative Sir box were end labeled with digoxigenin-11-ddUTP (Boehringer Mannheim) by using terminal transferase (Promega) according to the manufacturers’ instructions. Two different DNA probes were used: a 382-bp PCR product (oligonucleotide primer sequences, 5′-TTCACTTACTGATGGTGG-3′ and 5′-CTTTGAGAAGAGATGATT-3′) containing the 5′ coding regions of sitA and sirR and the complete intergenic region; and a synthetic oligonucleotide probe containing the Sir box (oligonucleotide sequences, 5′-TAAAATAAATTAGGTTAACCTAAACTTTTTATTA-3′ and 5′-TAATAAAAAGTTTAGGTTAACCTAATTTATTTTA-3′). The end-labeled fragments were incubated for 15 min at room temperature with partially purified SirR or FPLC fractions in 20 μl (total volume) in buffer containing 20 mM Tris HCl (pH 7), 5 mM MgCl2, 40 mM KCl, 2 mM dithiothreitol, 10% (vol/vol) glycerol, 1 μg of poly(dI-dC), and 5 μg of bovine serum albumin. Freshly prepared FeSO4 or MnSO4 was added at 125 μM. In some experiments, the divalent metal ion chelator EDTA at a concentration of 0.1 mM was also added to the reaction mixture. Samples were immediately electrophoresed at 100 V on a 5% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA buffer (2) and then electroblotted onto a positively charged nylon membrane. The digoxigenin-labeled probe was detected by using anti-digoxigenin-alkaline phosphatase conjugate and the luminogenic substrate CDPstar (Boehringer Mannheim). The signal was captured with a Berthold Luminograph LB980 or by exposure to X-ray film.
Production of antibodies.
A monospecific polyclonal antiserum to SirR was raised in adult female New Zealand White rabbits. Total-cell protein from E. coli overexpressing sirR was subjected to SDS-PAGE; after electrophoresis, the Coomassie blue-stained band corresponding to SirR was excised from the gel and electroeluted, and the denatured protein was used for immunization. A mouse monoclonal antibody to the S. epidermidis 32-kDa lipoprotein, SitC, was produced as described before (6).
Southern and Northern blot analyses.
Staphylococcal chromosomal DNA was digested with restriction endonucleases, electrophoresed, and transferred to a Hybond N+ membrane. The blot was probed with digoxigenin-labeled Sir box oligonucleotides as described above and visualized with CDPstar. A 3-kb EcoRV-EcoRI fragment from pW32 containing sitABC and most of sirR was labeled with digoxigenin and used as the probe for Northern blot analysis. Total RNA was extracted from S. epidermidis 901 grown for 18 h under iron-rich or iron-restricted conditions in RPMI 1640, using a Qiagen RNeasy total RNA kit. Northern blotting was performed as described by Ausubel et al. (2).
Identification of transcriptional start site.
The transcriptional start of the sitABC operon was mapped by primer extension incorporating [35S]dATP, using avian myeloblastosis virus reverse transcriptase (Promega) and RNA template prepared from S. epidermidis 901 grown under iron-restricted conditions, essentially as described by Ausubel et al. (2). Manual DNA sequencing for transcriptional mapping was carried out by using Sequenase version 2.0 according to the manufacturer’s instructions.
Nucleotide sequence accession number.
The sequence of sirR is available in the GenBank database under accession no. X99128.
RESULTS
Identification of DNA coding for SirR and its putative operator site.
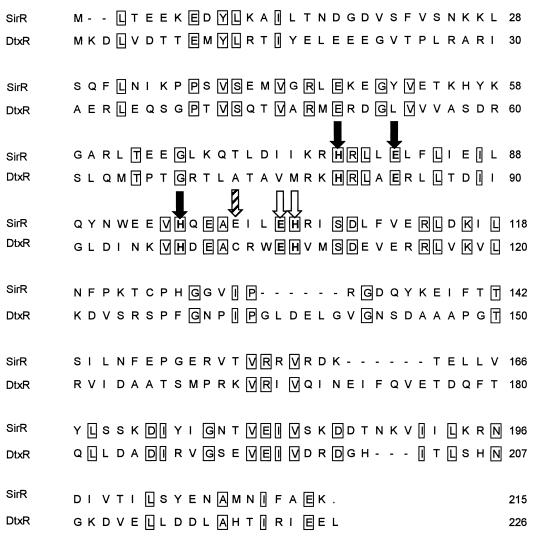
Plasmid pW32 was derived from a λ-Zap II genomic library and contains a 5.4-kb EcoRI fragment of S. epidermidis DNA which includes sitABC operon (6). DNA sequence analysis of pW32 plasmid indicated that upstream of the sitABC operon, and divergently transcribed, lies a 645-bp open reading frame that codes for a polypeptide of approximately 25 kDa with homology to the DtxR family of metal-dependent repressor proteins (Fig. 1 and Table 1). This open reading frame has been designated sirR (staphylococcal iron regulator repressor). Alignment of the deduced SirR protein sequence with the DtxR family reveals that it is most closely related (38% identical) to TroR from the spirochete T. pallidum, followed by IdeR (33%) found in the mycobacteria (Table 1). Furthermore, SirR, like TroR, is located adjacent to a putative ABC transporter system (17) with homology to the streptococcal family of multifunctional ABC operons involved in adherence and genetic competence (10). Although this staphylococcal gene product exhibits only 29% identity with DtxR, the metal coordination sites are conserved (Fig. 1).
FIG. 1.
Comparison of the deduced amino acid sequences of SirR and DtxR. Identical amino acid residues are boxed. Solid arrows indicate residues identified from the DtxR crystal structure as metal coordination sites (M1), the open arrows indicate metal-binding site 2 (M2), and the hatched arrow indicates the cysteine residue (C102) positioned within M2 which is replaced by a glutamate residue in SirR and TroR.
TABLE 1.
Sequence homologies of SirR with other DtxR homologs
| Accession no. | % Identity | Organism | Protein |
|---|---|---|---|
| X99128 | 100 | Staphylococcus epidermidis | SirR |
| U55214 | 38 | Treponema pallidum | TroR |
| Z96072 | 33 | Mycobacterium tuberculosis | IdeR |
| U14190 | 32 | M. smegmatis | IdeR |
| P33120 | 29 | Corynebacterium diphtheriae | DtxR |
| JC4513 | 28 | Streptomyces pilosus | DesR |
| JC4512 | 26 | Streptomyces lividans | DesR |
DNA sequence analysis of the region 5′ to the sitABC operon and transcriptional start analysis of this operon have identified a promoter sequence with similarity to E. coli ς70 promoters and a region of dyad symmetry overlapping the transcriptional start of sitABC termed the Sir box (Fig. 2A). This box shows high homology to the DtxR operator consensus sequence (20), suggesting that this region is SirR-binding site, and so it has been designated the Sir box. To map the transcriptional start site of the sitABC operon, RNA was prepared from S. epidermidis 901 grown in iron-restricted RPMI 1640 medium. Primer extension analysis indicated that the transcriptional start site was 38 nucleotides upstream of the TTG translational start and overlaps the Sir box (Fig. 2).
FIG. 2.
(A) Nucleotide sequence of the S. epidermidis sitABC operon promoter region. The −10 and −35 promoter sequences are underlined; the putative ribosome-binding site (RBS) and translation start codon of sitA are shown in bold. The vertical arrow indicates the transcriptional start of the sitABC operon, and the Sir box is indicated by convergent horizontal arrows. (B) Comparison of the S. epidermidis Sir box nucleotide sequence with sequences of known DtxR- and DesR-binding sites in C. diphtheriae and S. lividans. The 19-bp consensus sequence for DtxR derived by Lee et al. (20) is also shown.
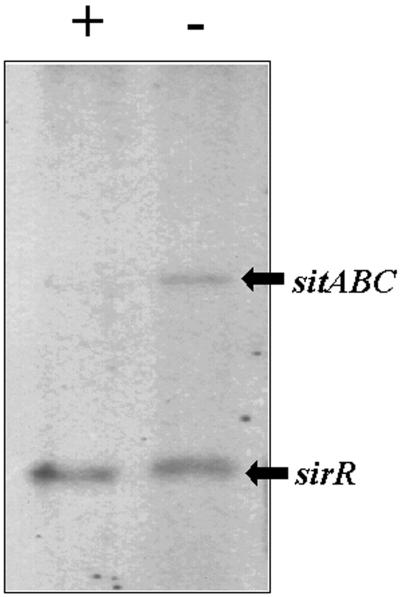
When grown in an iron-rich growth medium, the 32-kDa SitC lipoprotein is not expressed. To determine whether the sit operon is regulated by iron at the transcriptional level, RNA was prepared from S. epidermidis 901 grown in RPMI 1640 under iron-rich and iron-restricted conditions. Northern blot analysis using a DNA probe which hybridizes to both sirR and the sitABC operon revealed that expression of the 3-kb sitABC operon in response to iron is controlled at the transcriptional level (Fig. 3). In addition, Fig. 3 shows that the 0.7-kb sirR transcript is monocistronic and not influenced by the iron content of the growth medium.
FIG. 3.
Northern blot analysis of S. epidermidis sirR and sitABC transcripts. Total RNA was isolated from S. epidermidis 901 grown in iron-sufficient (+) or iron-deficient (−) RPMI 1640 medium. The 2.7-kb sitABC transcript and the 0.7-kb sirR transcript are indicated by arrows.
SitC responds to divalent metal ions other than iron.
The production of diphtheria toxin is known to be repressed in a DtxR-dependent manner by iron and various divalent metal cations, including Co2+, Mn2+, and Ni2+ (15, 39). To investigate whether the sitABC operon is similarly controlled by metal cations, we used a monoclonal antibody raised against SitC (6) to probe immunoblots of the whole-cell proteins of S. epidermidis 901 grown in iron-depleted RPMI 1640 supplemented with different divalent metal cations. Figure 4 shows that SitC production in S. epidermidis is fully repressed by the presence of iron and Mn2+ but unaffected by the presence of Co2+, Cu2+, Ni2+, Zn2+, or Mg2+ in the growth medium.
FIG. 4.
Immunoblot analysis of SitC production in S. epidermidis grown in iron-restricted RPMI 1640 or supplemented with cobalt, copper, iron, magnesium, manganese, nickel, or zinc (added at 20 μM). Whole-cell proteins prepared by lysostaphin digestion of staphylococcal suspensions adjusted to the same optical density were subjected to SDS-PAGE, immunoblotted, and probed with a monoclonal antibody to SitC. Lane 1, no addition; lane 2, Co2+; lane 3, Cu2+; lane 4, Fe2+; lane 5, Mg2+; lane 6, Mn2+; lane 7, Ni2+; lane 8, Zn2+.
DNA gel retardation assays.
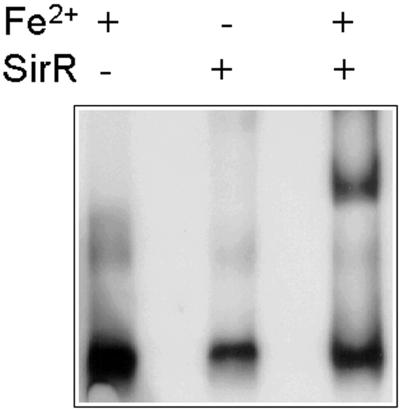
To demonstrate that SirR is capable of binding to the Sir box in a metal ion-dependent manner, we partially purified the protein by FPLC and Ni-nitrilotriacetic acid affinity chromatography. In DNA gel retardation assays using a synthetic oligonucleotide based on the Sir box sequence, SirR retarded movement of the oligonucleotide in the presence of Fe2+ (Fig. 5) or Mn2+, and this retardation could be abolished by the inclusion of the divalent metal ion chelator EDTA in the buffer. Furthermore, binding of SirR to the labeled Sir box oligonucleotide could be abolished by competition with the unlabeled Sir box but not by nonspecific DNA (data not shown).
FIG. 5.
DNA gel mobility shift assay of synthetic Sir box (labeled with digoxigenin) with partially purified SirR in the presence (+) or absence (−) of Fe2+. The Sir box oligonucleotide is clearly retarded only in the presence of both SirR and Fe2+.
SirR is a global regulator.
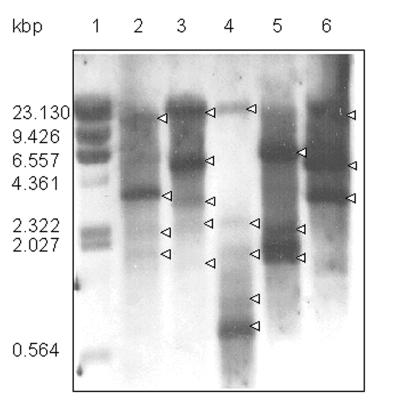
To determine whether SirR is present in other staphylococci, we subjected whole-cell lysates of S. aureus, S. carnosus, S. epidermidis, S. hominis, S. cohnii, S. lugdunensis, and S. haemolyticus to Western blotting using monospecific anti-SirR antibodies. The presence of an immunoreactive polypeptide of ca. 25 kDa, corresponding to SirR, from each of the strains tested (Fig. 6) indicates that SirR is common to the staphylococci. Southern blotting of S. epidermidis and S. aureus chromosomal DNA using the synthetic Sir box used for the gel retardation studies as a probe confirms that there are at least five Sir boxes in the S. epidermidis genome and at least three Sir boxes in the genome of S. aureus. This finding provides evidence that the SirR regulon may contain a number of operons expressed only by staphylococci growing under iron restriction (Fig. 7).
FIG. 6.
Immunoblot analysis of SirR production in coagulase-negative staphylococci grown in iron-restricted RPMI 1640 medium. Whole-cell proteins prepared by lysostaphin digestion were subjected to SDS-PAGE, immunoblotted, and probed with a monospecific polyclonal antibody to SirR. Lane 1, S. epidermidis; lane 2, S. cohnii; lane 3, S. hominis; lane 4, S. carnosus; lane 5, S. lugdunensis; lane 6, S. warneri; lane 7, S. aureus.
FIG. 7.
Southern blot analysis showing the presence of multiple Sir boxes in genomic DNA prepared from S. epidermidis 901 and S. aureus BB restricted with MunI (lanes 2 and 5), HindIII (lanes 3 and 6), and SalI (lane 4). Lane 1, λ-HindIII marker; lanes 2 to 4, S. epidermidis; lanes 5 and 6, S. aureus. Strongly hybridizing bands are indicated by arrowheads.
DISCUSSION
In both S. aureus and S. epidermidis, a number of cell wall- and cytoplasmic membrane-associated proteins, the production of which depends on the growth medium iron content, have been identified (6, 24, 37, 45). Of these, we have recently cloned and sequenced an iron-regulated 32-kDa lipoprotein, SitC, which is encoded by a component of the sitABC operon (6). Analysis of DNA sequence 5′ to sitABC failed to identify a region with any homology to a Fur box, indicating that sitABC expression is unlikely to be modulated by the Fur-like protein recently identified in S. epidermidis (19). However, in the present report, we have provided evidence suggesting that the iron-dependent regulation of the sit operon may be mediated via SirR, a protein related to the DtxR family of metal-dependent transcriptional repressors, the target for which is a 19-bp palindrome termed the Sir box. Since S. epidermidis contains a Fur homolog, this raises the intriguing possibility that staphylococci possess two distinct families of metal-dependent repressor proteins.
The S. epidermidis Fur-like protein described by Heidrich et al. (19) is located upstream of a putative superoxide dismutase, and both genes contain a sequence motif with low similarity to Fur boxes but no Sir box. This staphylococcal fur gene therefore may, like the E. coli homolog, bind to its own promoter and autoregulate its expression (8). The lack of an additional Sir box within the sirR operator indicates that sirR is, however, unlikely to be autoregulated. Although the staphylococcal Fur protein is unable to complement an E. coli fur mutant, the E. coli Fur protein does recognize weakly the Fur box in front of the S. epidermidis fur gene (19). Whether this staphylococcal Fur-like protein is functional and regulates the superoxide dismutase gene and other target genes has yet to be established. In this context, it is perhaps worth noting that Fur boxes overlapping numerous B. subtilis genes, as well as three different Fur homologs, have been identified (5).
Transcriptional mapping of the S. epidermidis sit operon indicates that the Sir box is located within the promoter/operator of the sit operon. DNA mobility shift analysis using a synthetic Sir box oligonucleotide confirmed that SirR bound in a metal-dependent manner, indicating that this motif is indeed the SirR target. The Sir box is closely related to the C. diphtheriae DtxR-binding site, conforming to the DtxR box consensus sequence and sharing some 12 of 19 identical nucleotides with the DtxR-binding site located within the promoter/operator of the diphtheria toxin gene (20). In C. diphtheriae, at least six other genes are regulated by DtxR and contain DtxR-binding sites (20, 34). These include hmuO, the product of which is a heme oxygenase required for the utilization of heme and hemoglobin (34), and irp1, which codes for an iron-regulated 38-kDa protein (34). While the function of IRP1 is not known, it may be functionally homologous to SitC since both proteins are lipoproteins anchored to the outer surface of the cytoplasmic membrane, they contain similar signal peptidase II cleavage sites, and both are probably involved in metal ion transport (6, 34). Whether irp1, like sitC, is part of an operon coding for an ABC transporter is not known, but it does show homology with the siderophore receptor FhuD from B. subtilis. It is possible that in the staphylococci, SirR regulates multiple genes, a hypothesis supported by (i) immunoblot data revealing an antigenically conserved 25-kDa protein in the staphylococci and (ii) Southern blot analysis which revealed a number of Sir boxes in the genomes of both S. epidermidis and S. aureus, indicating that SirR may, in common with DtxR and Fur, be a pleiotropic regulator of gene expression in these gram-positive pathogens.
The DNA target for DtxR was identified by DNase I footprinting analysis as a 27-bp interrupted palindrome which overlaps the basal promoter of the operons that it controls (13, 36, 39), while in vitro affinity selection has identified the minimal DtxR operator to be a 9-bp palindrome separated by a single base pair (40). In addition, binding of DtxR to its operator site has been shown to occur in the presence of divalent metal ions (e.g., Mn2+ and Co2+) other than Fe2+ (35, 39). Since the cloning and sequencing of dtxR (4), mutational analysis has identified specific residues which are important for the function of DtxR in terms of its interaction with DNA (e.g., R47) or divalent metal binding (e.g., H98, C102, and H106), and subsequent crystallographic studies have placed these functional assays in the context of a high-resolution crystal structure (9, 29, 31–33, 40, 47). From this work, four domains necessary for the regulatory functions of DtxR have been identified. These are an N-terminal, helix-turn-helix DNA-binding domain, two metal ion-binding sites, and a dimerization/stabilization domain (9, 29, 31–33). From secondary structure predictions and a comparison of the amino acid sequences of SirR and DtxR, SirR appears to contain three N-terminal helices at positions 1 to 16 (helix 1), 27 to 33 (helix 2), and 38 to 49 (helix 3) similar to those found in DtxR (31), with residues highly conserved in the DNA recognition helix (helix 3 at G38 to R50 in DtxR) of other DtxR homologs (i.e., P39, V41, S42, V45, R47, and E49) also being conserved in SirR. Metal-binding site 1 of DtxR has metal coordination sites at H79, E83, and H98, residues which are conserved in SirR. The DtxR metal-binding site 2 has metal coordination sites at E105 and H106, both of which are retained in SirR. In DtxR, C102 has been shown to be important for activity since it resides in metal-binding site 2 (42), although it does not directly appear to provide a metal-binding ligand (31). In SirR, residue C102 is replaced by a glutamate residue, a conservative substitution also found in TroR, the DtxR homolog from T. pallidum. In contrast to TroR, SirR, like the other DtxR homologs so far described, also contains a region homologous to the DtxR dimerization domain located in helix 5 of DtxR (31).
Using DNA gel retardation studies, we have provided in vitro evidence to support the function of SirR as a metal-dependent repressor in that SirR binds to a synthetic Sir box only in the presence of divalent metal ions such as Fe2+ and Mn2+. These data are further supported by in vivo data, as Western blot analysis shows that Mn2+ as well as Fe2+ can modulate SitC production. Given that the sit operon is related to a family of streptococcal ABC transporters found to be essential for virulence and competence and which also transport Mn2+ or Zn2+ ions (10), it is possible that SirR functions in vivo as a Mn2+ rather than an Fe2+-dependent repressor. In E. coli, Mn2+, Fe2+, and Co2+ are all capable of repressing in vivo expression of transcriptional fusions to the promoters of Fur-regulated genes (21). However, in E. coli, the Fur target genes are involved in the biosynthesis and transport of Fe3+ chelators such as aerobactin which have only a low affinity for divalent metal cations such as Fe2+ (27). Determination of the nature of the ligand transported via the SitABC transporter in S. epidermidis requires the construction of defined mutants. Such mutants will enable us to determine whether SirR is involved in the regulation of genes involved in the uptake of iron or other metal cations, an important consideration given the existence of Fur-like proteins in the staphylococci.
ACKNOWLEDGMENT
This work was supported by program grant G9219778 from the Medical Research Council to P.W.
REFERENCES
- 1.Arbuthnott J P, Arbuthnott E, Arbuthnott A D J, Pike W J, Cockayne A. Investigation of microbial growth in vivo: evaluation of a novel in vivo chamber implant system. FEMS Microbiol Lett. 1992;100:75–80. doi: 10.1111/j.1574-6968.1992.tb14022.x. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1994. [Google Scholar]
- 3.Bagg A, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd J, Oza M N, Murphy J R. Molecular-cloning and DNA-sequence analysis of a diphtheria toxin iron-dependent regulatory element (DtxR) from Corynebacterium diphtheriae. Proc Natl Acad Sci USA. 1990;87:5968–5972. doi: 10.1073/pnas.87.15.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bsat N, Helmann J D. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Regulation of Bacillus subtilis iron uptake genes by Fur protein, abstr. H-193. [Google Scholar]
- 6.Cockayne A, Hill P J, Powell N B L, Bishop K, Sims C M, Williams P. Infect. Immun. submitted. 1998. Molecular cloning of a 32kDa lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crossley K B, Archer G L. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. [Google Scholar]
- 8.de Lorenzo V, Herrero M, Giovannini F, Neilands J B. Fur (ferric uptake regulation) protein and CAP (catabolite-activator protein) modulate transcription of fur gene in Escherichia coli. Eur J Biochem. 1988;173:537–546. doi: 10.1111/j.1432-1033.1988.tb14032.x. [DOI] [PubMed] [Google Scholar]
- 9.Ding X, Zeng H, Schiering N, Ringe D, Murphy J R. Identification of the primary metal ion-activation sites of the diphtheria, tox repressor by X-ray crystallography and site-directed mutational analysis. Nat Struct Biol. 1996;3:382–387. doi: 10.1038/nsb0496-382. [DOI] [PubMed] [Google Scholar]
- 10.Dintilhac A, Alloing G, Granadel C, Claverys J-P. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 11.Dussurget O, Rodriguez M, Smith I. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol Microbiol. 1996;22:535–544. doi: 10.1046/j.1365-2958.1996.1461511.x. [DOI] [PubMed] [Google Scholar]
- 12.Ernst J F, Bennett R L, Rothfield L I. Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J Bacteriol. 1978;135:928–934. doi: 10.1128/jb.135.3.928-934.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fourel G, Phalipon A, Kaczorek M. Evidence for direct regulation of diphtheria toxin gene transcription by an Fe2+-dependent DNA-binding repressor, DtxR, in Corynebacterium diphtheriae. Infect Immun. 1989;57:3221–3225. doi: 10.1128/iai.57.10.3221-3225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis R T, Booth J W, Becker R R. Uptake of iron from hemoglobin and haptoglobin-hemoglobin complex by hemolytic bacteria. Int J Biochem. 1995;17:767–773. doi: 10.1016/0020-711x(85)90262-9. [DOI] [PubMed] [Google Scholar]
- 15.Groman N B, Judge K. Effect of metal ions on diphtheria toxin production. Infect Immun. 1979;26:1065–1070. doi: 10.1128/iai.26.3.1065-1070.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunter-Seeboth K, Schupp T. Cloning and sequence-analysis of the Corynebacterium-diphtheriae dtxR homolog from Streptomyces lividans and S. pilosus encoding a putative iron repressor protein. Gene. 1995;166:117–119. doi: 10.1016/0378-1119(95)00628-7. [DOI] [PubMed] [Google Scholar]
- 17.Hardham J M, Stamm L V, Porcella S F, Frye J G, Barnes N Y, Howell J K. Identification and transcriptional analysis of a Treponema pallidum operon encoding a putative ABC transport system, an iron-activated repressor protein homolog, and a glycolytic pathway enzyme homolog. Gene. 1997;197:47–64. doi: 10.1016/s0378-1119(97)00234-5. [DOI] [PubMed] [Google Scholar]
- 18.Hasan A A, Holland J, Smith A, Williams P. Elemental iron does repress transferrin, haemopexin and haemoglobin receptor expression in Haemophilus influenzae. FEMS Microbiol Lett. 1997;150:19–26. doi: 10.1111/j.1574-6968.1997.tb10344.x. [DOI] [PubMed] [Google Scholar]
- 19.Heidrich C, Hantke K, Bierbaum G, Sahl H G. Identification and analysis of a gene encoding a Fur-like protein of Staphylococcus epidermidis. FEMS Microbiol Lett. 1996;140:253–259. doi: 10.1111/j.1574-6968.1996.tb08345.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee J H, Wang T, Ault K, Liu J, Schmitt M P, Holmes R K. Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect Immun. 1997;65:4273–4280. doi: 10.1128/iai.65.10.4273-4280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litwin C M, Boyko S A, Calderwood S B. Cloning, sequencing, and transcriptional regulation of the Vibrio cholerae fur gene. J Bacteriol. 1992;174:1879–1903. doi: 10.1128/jb.174.6.1897-1903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modun B, Williams P, Pike W J, Cockayne A, Arbuthnott J P, Finch R, Denyer S P. Cell envelope proteins of Staphylococcus epidermidis grown in vivo in peritoneal chamber implant. Infect Immun. 1992;60:2551–2553. doi: 10.1128/iai.60.6.2551-2553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Modun B, Kendall D, Williams P. Staphylococci express a receptor for human transferrin: identification of a 42-kilodalton cell wall transferrin-binding protein. Infect Immun. 1994;62:3850–3858. doi: 10.1128/iai.62.9.3850-3858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modun B, Cockayne A, Finch R G, Williams P. The Staphylococcus aureus and Staphylococcus epidermidis transferrin-binding proteins are expressed in vivo during infection. Microbiology. 1998;144:1005–1012. doi: 10.1099/00221287-144-4-1005. [DOI] [PubMed] [Google Scholar]
- 26.Modun B, Evans R W, Joannou C L, Williams P. Receptor-mediated recognition and uptake of iron from human transferrin by Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 1998;66:3591–3596. doi: 10.1128/iai.66.8.3591-3596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neilands J B. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 28.Oguiza J A, Tao X, Marcos A T, Martin J F, Murphy J R. Molecular cloning, DNA sequence analysis, and characterization of the Corynebacterium diphtheriae dtxR homolog from Brevibacterium lactofermentum. J Bacteriol. 1995;177:465–467. doi: 10.1128/jb.177.2.465-467.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pohl E, Qiu X Y, Must L M, Holmes R K, Hol W G Y. Comparison of high-resolution structures of the diphtheria toxin repressor in complex with cobalt and zinc at the cation-anion binding site. Protein Sci. 1997;6:1114–1118. doi: 10.1002/pro.5560060519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prince R W, Cox C D, Vasil M L. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J Bacteriol. 1993;175:2589–2598. doi: 10.1128/jb.175.9.2589-2598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu X Y, Pohl E, Holmes R K, Hol W J G. High-resolution structure of the diphtheria-toxin repressor complexed with cobalt and manganese reveals an SH3-like 3rd domain and suggests a possible role of phosphate as co-corepressor. Biochemistry. 1996;35:12292–12302. doi: 10.1021/bi960861d. [DOI] [PubMed] [Google Scholar]
- 32.Qiu X Y, Verlinde C L M J, Zhang S P, Schmitt M P, Holmes R K, Hol W G J. Three-dimensional structure of the diphtheria-toxin repressor in complex with divalent-cation co-repressors. Structure. 1995;3:87–100. doi: 10.1016/s0969-2126(01)00137-x. [DOI] [PubMed] [Google Scholar]
- 33.Schiering N, Tao X, Murphy J R, Petsko G A, Ringe D. Crystallization and preliminary-X-ray studies of the diphtheria tox repressor from Corynebacterium diphtheriae. J Mol Biol. 1994;244:654–656. doi: 10.1006/jmbi.1994.1760. [DOI] [PubMed] [Google Scholar]
- 34.Schmitt M P. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect Immun. 1997;65:4634–4641. doi: 10.1128/iai.65.11.4634-4641.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt M P, Twiddy E M, Holmes R K. Purification and characterization of the diphtheria-toxin repressor. Proc Natl Acad Sci USA. 1992;89:7576–7580. doi: 10.1073/pnas.89.16.7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt M P, Holmes R K. Analysis of diphtheria-toxin repressor-operator interactions and characterization of a mutant repressor with decreased binding-activity for divalent metals. Mol Microbiol. 1993;9:173–181. doi: 10.1111/j.1365-2958.1993.tb01679.x. [DOI] [PubMed] [Google Scholar]
- 37.Smith D G E, Wilcox M H, Williams P, Finch R G, Denyer S P. Characterization of the cell envelope proteins of Staphylococcus epidermidis cultured in human peritoneal dialysate. Infect Immun. 1991;59:617–624. doi: 10.1128/iai.59.2.617-624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staggs T M, Perry R D. Identification and cloning of a fur regulatory gene in Yersinia pestis. J Bacteriol. 1991;173:417–425. doi: 10.1128/jb.173.2.417-425.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao X, Boyd J, Murphy J R. Specific binding of the diphtheria tox regulatory element DtxR to the tox operator requires divalent heavy-metal ions and a 9-base-pair interrupted palindromic sequence. Proc Natl Acad Sci USA. 1992;89:5897–5901. doi: 10.1073/pnas.89.13.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao X, Murphy J R. Determination of the minimal essential nucleotide-sequence for diphtheria tox repressor binding by in-vitro affinity selection. Proc Natl Acad Sci USA. 1994;91:9646–9650. doi: 10.1073/pnas.91.20.9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao X, Schiering N, Zeng H Y, Ringe D, Murphy J R. Iron, DtxR and the regulation of diphtheria-toxin expression. Mol Microbiol. 1994;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 42.Tao X, Murphy J R. Cysteine-102 is positioned in the metal-binding activation site of the Corynebacterium diphtheriae regulatory element DtxR. Proc Natl Acad Sci USA. 1993;90:8524–8528. doi: 10.1073/pnas.90.18.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas C E, Sparling P F. Identification and cloning of a fur homologue from Neisseria meningitidis. Mol Microbiol. 1994;11:725–737. doi: 10.1111/j.1365-2958.1994.tb00350.x. [DOI] [PubMed] [Google Scholar]
- 44.Trivier D, Courcol R J. Iron depletion and virulence in Staphylococcus aureus. FEMS Microbiol Lett. 1996;141:117–127. doi: 10.1111/j.1574-6968.1996.tb08373.x. [DOI] [PubMed] [Google Scholar]
- 45.Wilcox M H, Williams P, Smith D G E, Finch R G, Denyer S P. Variation in the expression of cell envelope proteins of coagulase-negative staphylococci cultured under iron-restricted conditions in human peritoneal dialysate. J Gen Microbiol. 1991;137:2561–2570. doi: 10.1099/00221287-137-11-2561. [DOI] [PubMed] [Google Scholar]
- 46.Williams P, Griffiths E. Bacterial transferrin receptors—structure, function and contribution to virulence. Med Microbiol Immunol. 1992;181:301–322. doi: 10.1007/BF00191543. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z O, Schmitt M P, Holmes R K. Characterization of mutations that inactivate the diphtheria toxin repressor gene (dtxR) Infect Immun. 1994;62:1600–1608. doi: 10.1128/iai.62.5.1600-1608.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wooldridge K G, Williams P H. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 49.Wooldridge K G, Williams P H, Ketley J M. Iron-responsive genetic regulation in Campylobacter jejuni: cloning and characterization of a fur homolog. J Bacteriol. 1994;176:5852–5856. doi: 10.1128/jb.176.18.5852-5856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]