Abstract
Current treatments for neurodegenerative diseases and neural injuries face major challenges, primarily due to the diminished regenerative capacity of neurons in the mammalian CNS as they mature. Here, we investigated the role of Ezh2, a histone methyltransferase, in regulating mammalian axon regeneration. We found that Ezh2 declined in the mouse nervous system during maturation but was upregulated in adult dorsal root ganglion neurons following peripheral nerve injury to facilitate spontaneous axon regeneration. In addition, overexpression of Ezh2 in retinal ganglion cells in the CNS promoted optic nerve regeneration via both histone methylation–dependent and –independent mechanisms. Further investigation revealed that Ezh2 fostered axon regeneration by orchestrating the transcriptional silencing of genes governing synaptic function and those inhibiting axon regeneration, while concurrently activating various factors that support axon regeneration. Notably, we demonstrated that GABA transporter 2, encoded by Slc6a13, acted downstream of Ezh2 to control axon regeneration. Overall, our study underscores the potential of modulating chromatin accessibility as a promising strategy for promoting CNS axon regeneration.
Keywords: Neuroscience, Ophthalmology
Keywords: Epigenetics, Neurodegeneration, Neurodevelopment
<p>Ezh2 fosters optic nerve regeneration by regulating multiple categories of target genes in retinal ganglion cells.</p>
Introduction
Axon degeneration and neuronal cell death are common consequences of neurodegenerative diseases and neural injuries. Unfortunately, current clinical therapeutics for neural injuries and neurodegenerative diseases still fall short of success. In the mammalian CNS, the inability of mature neurons to regenerate axons after injury or neurodegeneration results in poor functional recovery and permanent disabilities. Therefore, understanding why mature neurons in the mammalian CNS cannot regrow axons has been a longstanding challenge for the field. Research over the past decades has revealed that the low intrinsic axon growth competency of mature CNS neurons (1–3), together with extrinsic inhibitory molecules (4–6), are the major contributors to unsatisfactory regenerative outcomes. During development, young neurons are intrinsically competent in axon growth to establish neural circuits, whereas adult neurons possess poor axon growth ability to maintain circuit stability. Moreover, the inhibitory extracellular environment also limits unnecessary axon sprouting, acting as another factor to stabilize the neural circuits (7).
During maturation, the cellular state of neurons changes from favoring to limiting axon growth, likely regulated by modifications of the epigenomic and subsequent transcriptomic landscapes in neurons. Unlike CNS neurons, the axon regeneration ability of neurons in the peripheral nervous system (PNS) can be reactivated upon peripheral nerve injury by initiating a transcription-dependent regenerative response (8, 9). Recent studies demonstrated that such a response also involves massive changes in the epigenome and transcriptome of PNS neurons (10–12). It has been revealed that nerve injuries induce a common developmental-like transcriptional program in sensory neurons (12–14). Similar reversal to an embryonic transcriptomic state also occurs in mature corticospinal neurons at an early stage following spinal cord injury, although it cannot be sustained (15). Thus, it is vital to unveil the epigenomic changes that occur during neuronal maturation and PNS axon regeneration. The knowledge gained may be useful for epigenetically remodeling the transcriptomic landscape of mature CNS neurons and enhancing their axon regeneration ability.
In this study, we investigated the role of enhancer of zeste homolog 2 (Ezh2) histone methyltransferase in mammalian axon regeneration. Ezh2 is the catalytic core of the polycomb repressive complex 2 (PRC2), which catalyzes trimethylation of lysine 27 on histone H3 (H3K27me3). H3K27me3 condenses nearby chromatin to downregulate transcription (16). Although most studies focus on the role of Ezh2 as a histone methyltransferase, a number of studies clearly showed that Ezh2 can also methylate nonhistone substrates (17, 18) or exert methylation-independent functions (19–21), suggesting its versatility.
Here, we showed that Ezh2 loss-of-function impaired spontaneous axon regeneration of dorsal root ganglion (DRG) neurons. In addition, overexpression of Ezh2 in retinal ganglion cells (RGCs) promoted optic nerve regeneration in both histone methylation-dependent and -independent manners. Mechanistic exploration revealed that Ezh2 orchestrated mammalian axon regeneration by targeting both the intrinsic regenerative ability and the extrinsic hostile environment.
Results
Ezh2 is developmentally downregulated in the nervous system and upregulated in adult DRG neurons following peripheral nerve injury.
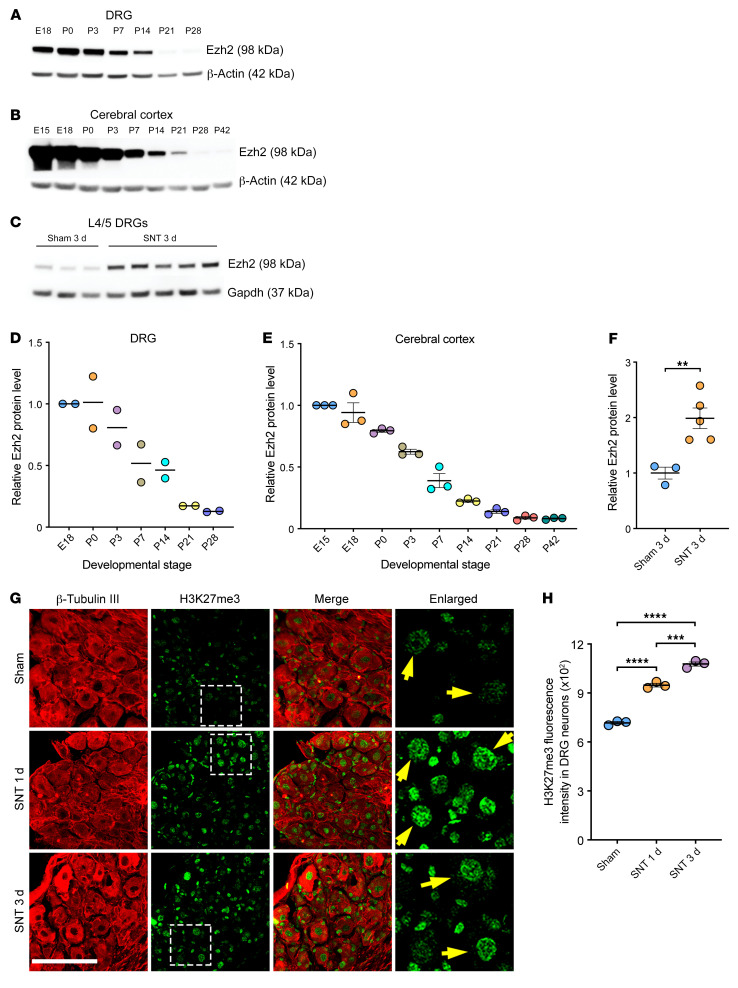
To evaluate how Ezh2 expression was regulated during neural development, we first examined Ezh2 protein levels in the mouse DRG and cerebral cortex at different developmental stages. We found that Ezh2 was abundantly expressed in DRGs and the cortex at the late embryonic stage, remained high during the first several postnatal days, and then gradually declined to become hardly detectable at 3 to 4 weeks after birth (Figure 1, A, B, D, and E). DRG neurons extend a single axon that bifurcates into 2 branches, a peripheral branch that readily regenerates upon injury in a transcription-dependent manner (8), and a central branch lacking the spontaneous regenerative ability. Sensory axons in the mouse sciatic nerve are primarily comprised of peripheral branches of lumbar 4 and 5 (L4/5) DRG neurons. A sharp increase of Ezh2 in L4/5 DRGs was detected 3 days after sciatic nerve transection (Figure 1, C and F). Other PRC2 subunits were not discernibly altered (Supplemental Figure 1, A–E; supplemental material available online with this article; https://doi.org/10.1172/JCI163145DS1). Immunofluorescence of DRG sections showed significantly increased neuronal H3K27me3 following the injury (Figure 1, G and H), suggesting cell-autonomous upregulation of Ezh2 in DRG neurons. These results were consistent with a previous study showing increased Ezh2 and H3K27me3 in the DRG after spinal nerve ligation (22), which also injures peripheral axons of DRG neurons. During development, neurons lose axon growth capacities after reaching their targets, correlating with the decline of Ezh2. On the other hand, the upregulation of Ezh2 in DRG neurons after peripheral nerve injury accompanies the robust regenerative response, suggesting that Ezh2 might facilitate axon regeneration.
Figure 1. Ezh2 is developmentally downregulated in the nervous system and upregulated in DRG neurons following peripheral nerve injury.
(A and B) Representative immunoblotting showing that Ezh2 is developmentally downregulated in the DRG (A) and cerebral cortex (B). (C) Immunoblotting showing that Ezh2 is significantly increased in L4/5 DRGs 3 days after sciatic nerve transection. (D) Quantification of relative protein levels of Ezh2 in A (n = 2 independent experiments). (E) Quantification of relative protein levels of Ezh2 in B (1-way ANOVA followed by Tukey’s multiple comparisons; P 0.0001; n = 3 independent experiments). (F) Quantification of relative protein levels of Ezh2 in C (unpaired, 2-tailed t test; P = 0.0092; n = 3 for sham, n = 5 for sciatic nerve transection). (G) Representative immunofluorescence of DRG sections showing increased H3K27me3 levels in nuclei of DRG neurons 1 or 3 days after sciatic nerve transection. DRG sections were stained with anti-H3K27me3 (green) and anti-β-tubulin III (red). The rightmost column displays enlarged images of the areas outlined in white, dashed boxes. Yellow arrows indicate H3K27me3 in nuclei of DRG neurons. Scale bars: 100 μm, 30 μm for enlarged images. (H) Quantification of fluorescence intensity of H3K27me3 immunoreactivity in DRG neurons in G (1-way ANOVA followed by Tukey’s multiple comparisons; P 0.0001; n = 3 mice for all). SNT, sciatic nerve transection. **P 0.01, ***P 0.001, ****P 0.0001.
Upregulation of Ezh2 contributes to spontaneous axon regeneration of DRG neurons in vitro and in vivo.
To test our hypothesis, we first investigated if Ezh2 loss-of-function would impair regenerative axon growth of cultured DRG neurons. Using in vitro electroporation (23), siRNAs targeting Ezh2 mRNA (siEzh2) were transfected into DRG neurons. Control neurons were electroporated with nontargeting siRNAs (siNT). Immunoblotting confirmed that Ezh2 was efficiently knocked down 3 days after the electroporation (Supplemental Figure 1, F and H). Thus, on the fourth day, we replated the neurons and cultured them for another 24 hours, as described in our earlier study (9). The results showed that Ezh2 knockdown significantly reduced regenerative axon growth by approximately 25% (Supplemental Figure 1, J and L). To rule out the possibility that the phenotype was caused by off-target effects of the siRNAs, we crossed Ezh2fl/fl mice with Advillin-Cre mice (24) to generate Advillin-Cre;Ezh2fl/fl mice, in which Ezh2 was specifically deleted in sensory neurons. We performed a peripheral nerve conditioning lesion in Advillin-Cre;Ezh2fl/fl and Ezh2fl/fl (control) mice and waited for 3 days, and then cultured L4/5 DRG neurons for 24 hours. Successful knockout of Ezh2 was confirmed by immunoblotting of protein extracted from the cultured cells (Supplemental Figure 1, G and I). The remaining Ezh2 signal likely came from nonneuronal cells in the culture. We found that Ezh2 deletion significantly impaired regenerative axon growth of conditioning lesioned DRG neurons by approximately 20% (Supplemental Figure 1, K and M). These results demonstrated that Ezh2 supported regenerative axon growth of sensory neurons in vitro.
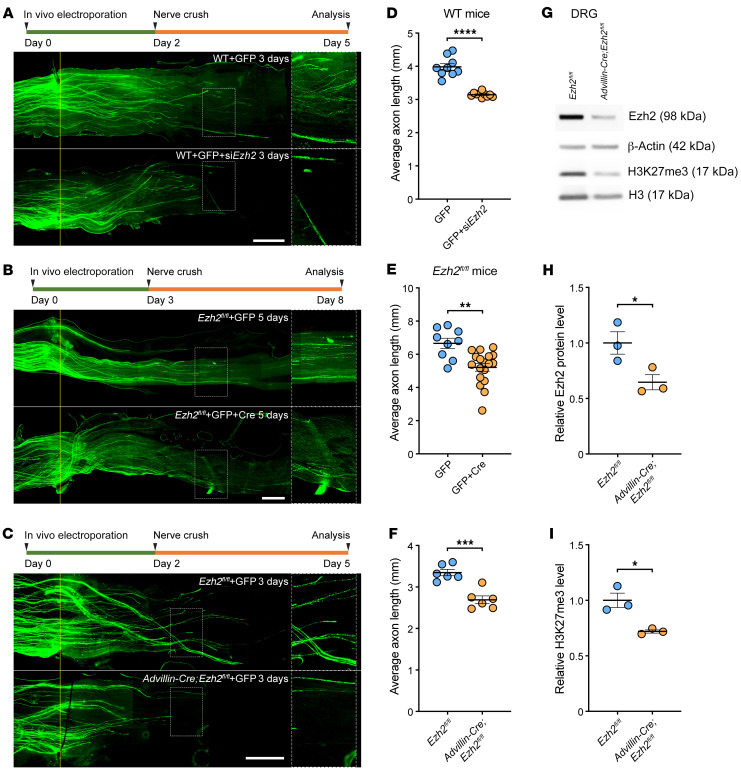
To further explore if Ezh2 was also required for axon regeneration of DRG neurons in vivo, we knocked down Ezh2 in L4/5 DRGs by in vivo electroporation of siEzh2, a technique widely used in our previous studies (25, 26). CMV-GFP plasmid was simultaneously electroporated to label the axons. Control mice were electroporated solely with the CMV-GFP plasmid, as our previous study demonstrated that electroporation of siNT had no impact on axon regeneration of sensory neurons (26). Two days after the electroporation, the sciatic nerve was crushed. After 3 days, we found that Ezh2 knockdown in DRGs significantly impaired axon regeneration of sensory neurons in vivo by approximately 20% (Figure 2, A and D). To rule out off-target effects of the siRNAs, we electroporated CMV-Cre and CMV-GFP plasmids into L4/5 DRGs of Ezh2fl/fl mice to knockout Ezh2. Ezh2fl/fl mice electroporated with the CMV-GFP plasmid only were the control group. To allow sufficient time for Cre-mediated recombination, the sciatic nerve was crushed 3 days later. Five days after the crush, we found that axon regeneration was significantly impaired by Ezh2 knockout (Figure 2, B and E). To further rule out the possibility that the observed phenotype resulted from Ezh2 loss-of-function in nonneuronal cells in the DRG, we electroporated the CMV-GFP plasmid in Advillin-Cre;Ezh2fl/fl and Ezh2fl/fl (control) mice; 2 days later, we crushed the sciatic nerve. Three days after the crush, we found that specific deletion of Ezh2 in sensory neurons significantly reduced axon regeneration in vivo by approximately 20% (Figure 2, C and F). Successful knockout of Ezh2 and decrease of H3K27me3 in DRG neurons of Advillin-Cre;Ezh2fl/fl mice were confirmed by immunoblotting (Figure 2, G–I). Collectively, these results demonstrated that Ezh2 upregulation contributed to spontaneous axon regeneration of DRG neurons triggered by peripheral nerve injury both in vitro and in vivo.
Figure 2. Upregulation of Ezh2 contributes to spontaneous axon regeneration of DRG neurons in vivo.
(A–C) Top: Experimental timeline. Bottom: Representative images of sciatic nerves showing that Ezh2 knockdown (A) or knockout (B) in L4/5 DRGs or sensory neuron-specific knockout of Ezh2 (C) impairs spontaneous axon regeneration of DRG neurons in vivo. The right column displays enlarged images of the areas in white, dashed boxes on the left. The crush sites are aligned with the yellow line. Scale bars: 1 mm, 0.5 mm for enlarged images. (D) Quantification of lengths of regenerating axons in A (unpaired, 2-tailed t test; P 0.0001; n = 9 mice for control, n = 10 mice for Ezh2 knockdown). (E) Quantification of lengths of regenerating axons in B (unpaired 2-tailed t test; P = 0.0011; n = 9 mice for control, n = 18 mice for Ezh2 knockout). (F) Quantification of lengths of regenerating axons in C (unpaired 2-tailed t test; P = 0.0003; n = 6 mice for both). (G) Representative immunoblotting showing successful knockout of Ezh2 and downregulation of H3K27me3 in DRG neurons of Advillin-Cre;Ezh2fl/fl mice. (H) Quantification of relative protein levels of Ezh2 in G (unpaired 2-tailed t test P = 0.0436; n = 3 independent experiments). (I) Quantification of relative levels of H3K27me3 in G (unpaired 2-tailed t test; P = 0.0137; n = 3 independent experiments). siEzh2, siRNAs targeting Ezh2 mRNA. *P 0.05, **P 0.01, ***P 0.001, ****P 0.0001.
Ezh2 overexpression enhances optic nerve regeneration via both histone methylation–dependent and –independent mechanisms.
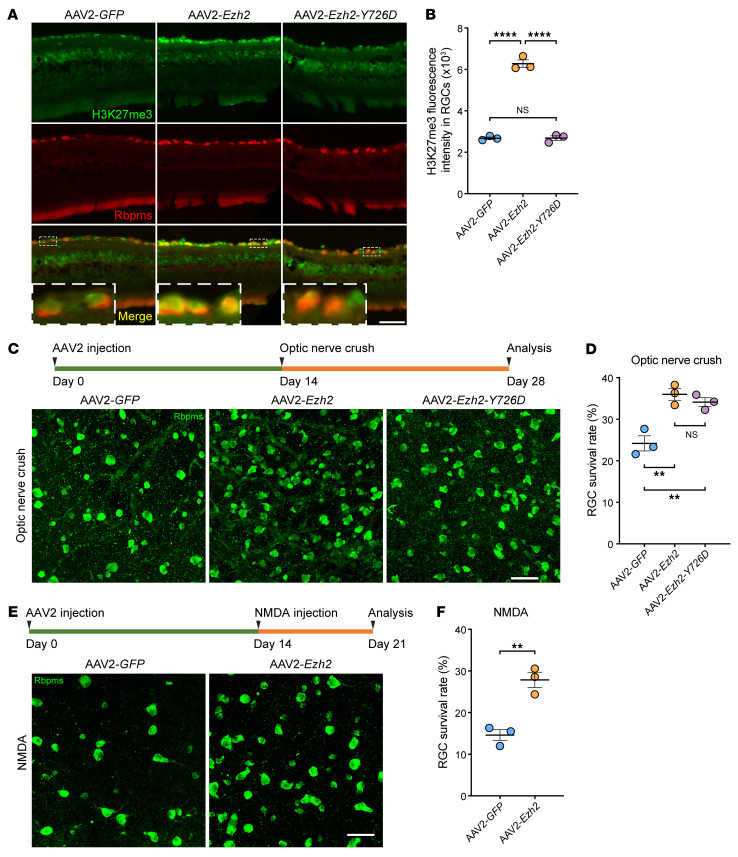
Since upregulation of Ezh2 contributed to axon regeneration of regenerative DRG neurons, we questioned whether forced overexpression of Ezh2 would similarly promote axon regeneration in nonregenerative adult RGCs. We first examined if Ezh2 and other PRC2 subunits in RGCs were changed by optic nerve crush (ONC). By analyzing a single-cell RNA-Seq data set of RGCs (27), we found that their mRNA levels remained relatively stable (Supplemental Figure 2, A and B), consistent with the nonregenerative characteristic of RGCs. We then overexpressed Ezh2 in RGCs by intravitreal injection of AAV2-Ezh2. Control mice were injected with AAV2-GFP. Previously, we showed that this approach could successfully transduce approximately 90% of RGCs (26, 28). Successful overexpression of Ezh2 2 weeks after virus injection was confirmed by immunoblotting of whole retinas or RGCs enriched from dissociated retinal cells by FACS (Supplemental Figure 3, A–D). In addition, immunofluorescence of retinal sections showed that H3K27me3 levels in RGCs consequently increased (Figure 3, A and B). Therefore, the optic nerve was crushed 2 weeks after virus injection. Ezh2 overexpression improved RGC survival by approximately 50% 2 weeks after the ONC (Figure 3, C and D and Supplemental Figure 3E). In a different retinal injury model induced by intravitreal injection of N-methyl-D-aspartate (NMDA) (29), Ezh2 overexpression almost doubled RGC survival 1 week after the excitotoxic injury to the retina (Figure 3, E and F), suggesting that Ezh2 could protect RGCs against various types of injury.
Figure 3. Ezh2 overexpression enhances RGC survival after ONC or excitotoxic injury.
(A) Representative immunofluorescence of retinal sections showing increased H3K27me3 levels in RGCs 2 weeks after intravitreal injection of AAV2-Ezh2, but not AAV2-Ezh2-Y726D. Retinal sections were stained with anti-H3K27me3 (green) and anti-Rbpms (red). Insets display enlarged images of RGCs in white, dashed boxes. Scale bars: 50 μm, 10 μm for enlarged images. (B) Quantification of fluorescence intensity of H3K27me3 immunoreactivity in RGCs in A (1-way ANOVA followed by Tukey’s multiple comparisons; P 0.0001; n = 3 mice for all; at least 150 RGCs from 10–12 nonadjacent sections were analyzed for each mouse). (C) Top: Experimental timeline. Bottom: Representative immunofluorescence of whole-mount retinas showing that overexpression of Ezh2 or Ezh2-Y726D improves RGC survival 2 weeks after optic nerve crush. Whole-mount retinas were stained with anti-Rbpms (green). Scale bar: 50 μm. (D) Quantification of RGC survival rate 2 weeks after ONC in C (1-way ANOVA followed by Tukey’s multiple comparisons; P = 0.0025; n = 3 mice for all; 6–9 fields were analyzed for each mouse). (E) Top: Experimental timeline. Bottom: Representative immunofluorescence of whole-mount retinas showing that overexpression of Ezh2 improves RGC survival 1 week after NMDA-induced excitotoxic injury. Whole-mount retinas were stained with anti-Rbpms (green). Scale bar: 50 μm. (F) Quantification of RGC survival rate 1 week after NMDA-induced excitotoxic injury in E (unpaired 2-tailed t test; P = 0.0042; n = 3 mice for both; 6–8 fields were analyzed for each mouse). **P 0.01, ****P 0.0001.
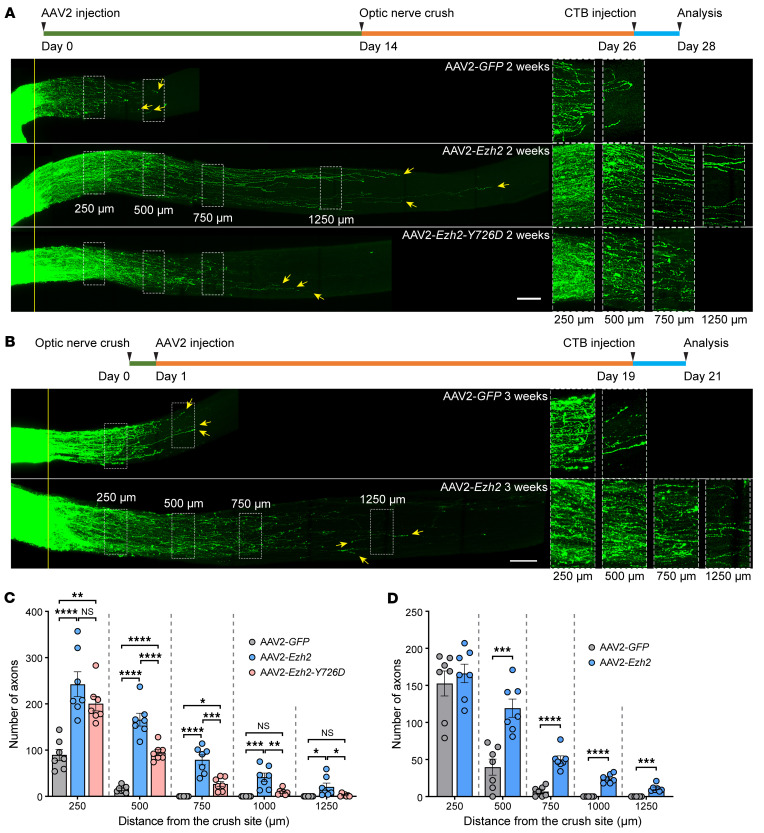
Optic nerve regeneration was also assessed 2 weeks after the ONC. Regenerating axons were labeled by Alexa Fluor–conjugated cholera toxin subunit B (CTB) intravitreally injected 2 days before tissue harvesting. Optic nerves were tissue-cleared and imaged as previously described (26, 28). Compared to the control group, where only a limited number of axons crossed the crush site, overexpression of Ezh2 induced markedly enhanced optic nerve regeneration (Figure 4, A and C). Some axons grew over 1,250 μm in 2 weeks.
Figure 4. Ezh2 overexpression enhances optic nerve regeneration via both histone methylation-dependent and -independent mechanisms.
(A and B) Top: Experimental timeline. Bottom: Representative images of optic nerves showing that pre- (A) or postinjury (B) overexpression of Ezh2 induces strong optic nerve regeneration 2 (A) or 3 weeks (B) after optic nerve crush. Preinjury overexpression of Ezh2-Y726D also modestly promotes optic nerve regeneration (A). Columns on the right display enlarged images of the areas in the white, dashed boxes on the left, showing axons at 250, 500, 750, and 1,250 μm distal to the crush sites, which are aligned with the yellow line. Yellow arrows indicate longest axons in each nerve. Scale bars: 100 μm, 50 μm for enlarged images. (C) Quantification of optic nerve regeneration in A (1-way ANOVA followed by Tukey’s multiple comparisons; P 0.0001 at 250, 500, 750, and 1,000 μm, P = 0.0126 at 1,250 μm; n = 7 mice for all). (D) Quantification of optic nerve regeneration in B (unpaired 2-tailed t test; P = 0.5305 at 250 μm, P = 0.0004 at 500 μm, P 0.0001 at 750 and 1,000 μm, P = 0.0003 at 1,250 μm; n = 7 mice for both). *P 0.05, **P 0.01, ***P 0.001, ****P 0.0001.
To investigate if the histone methyltransferase activity of Ezh2 was required for promoting optic nerve regeneration and RGC survival, we overexpressed a mutant form of Ezh2 with the 726th amino acid mutated from a tyrosine to an aspartic acid (Ezh2-Y726D) in RGCs (Supplemental Figure 3, A and C). Previous studies reported that this single amino acid mutation eliminated the methyltransferase activity of human and mouse Ezh2 (30, 31). Immunostaining of retinal sections confirmed that overexpression of Ezh2-Y726D did not increase H3K27me3 levels in RGCs (Figure 3, A and B). Surprisingly, this catalytically dead Ezh2 mutant exhibited comparable ability to enhance RGC survival as WT Ezh2 at 2 weeks after the ONC (Figure 3, C and D and Supplemental Figure 3E), suggesting a histone methylation–independent neuroprotective role of Ezh2. Moreover, Ezh2-Y726D also induced optic nerve regeneration, albeit to a much lesser extent (Figure 4, A and C), indicating that both histone methylation–dependent and –independent mechanisms contributed to promoting axon regeneration.
To explore the translational potential of Ezh2 gain-of-function in CNS axon regeneration, we tested if postinjury overexpression of Ezh2 in RGCs could also promote optic nerve regeneration. AAV2-GFP or AAV2-Ezh2 was intravitreally injected 1 day after the ONC. 3 weeks later, we found that postinjury Ezh2 overexpression still evidently promoted optic nerve regeneration (Figure 4, B and D), albeit weaker than that induced by preinjury overexpression of Ezh2. Specifically, more regenerating axons could be observed at 500–1,250 μm from the crush site after Ezh2 overexpression. However, the numbers of regenerating axons at 250 μm from the crush site were equivalent between the 2 conditions. This was likely caused by the extended regeneration period in the control group (3 weeks in Figure 4, B and D versus 2 weeks in Figure 4, A and C) and delayed Ezh2 expression in RGCs. These results demonstrated the translational potential of Ezh2 gain-of-function for enhancing axon regeneration in the CNS.
Ezh2 modifies the RGC transcriptome to regulate multiple categories of target genes.
To gain mechanistic insights into how Ezh2 supports RGC axon regeneration, we profiled the transcriptomic and epigenomic changes in RGCs induced by Ezh2 or Ezh2-Y726D overexpression with RNA-Seq and assay for transposase-accessible chromatin with sequencing (ATAC-Seq). We intravitreally injected AAV2-GFP (control), AAV2-Ezh2, or AAV2-Ezh2-Y726D and crushed the optic nerve after 2 weeks. 3 days after the ONC, RGCs were enriched from dissociated retinal cells by FACS to construct RNA-Seq and ATAC-Seq libraries. All sequencing libraries exhibited high quality (Supplemental Figure 4, A–F and Supplemental Figure 5, A–S), except for 1 RNA-Seq library from the Ezh2 overexpression condition, which was excluded from subsequent data analysis based on results of principle component analysis (PCA) and hierarchical clustering (Supplemental Figure 4, B and C). Chromatin accessibility at the promoter region moderately correlated with RNA expression within each condition (Supplemental Figure 6, A–D), suggesting consistency between the RNA-Seq and ATAC-Seq.
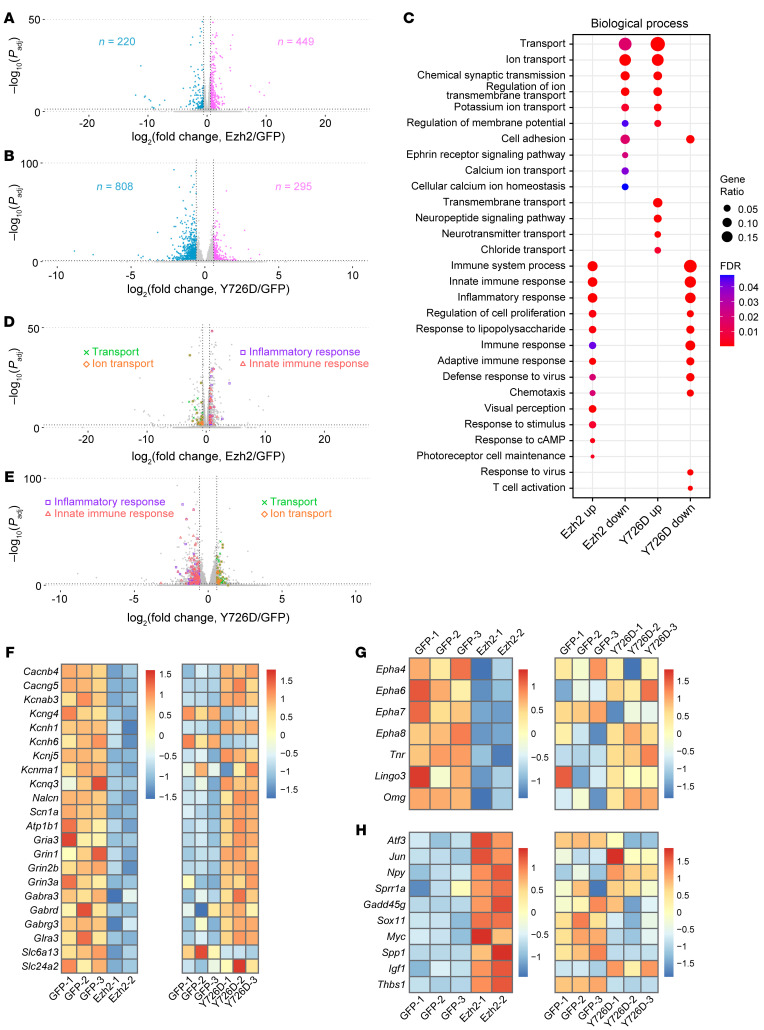
We identified 669 differentially expressed genes (DEGs) in the RNA-Seq between control and Ezh2 overexpression conditions at the threshold of adjusted P less than 0.05 and fold change greater than 1.5 (Figure 5A, Supplemental Figure 4G, and Supplemental Table 1). Surprisingly, despite being a catalytically dead form of Ezh2, Ezh2-Y726D overexpression resulted in more DEGs (1,103, Figure 5B and Supplemental Table 1). This was consistent with our ATAC-Seq results, in which considerably more differentially accessible regions were found after Ezh2-Y726D overexpression (Supplemental Table 2), suggesting that unknown transcriptional regulatory activities of Ezh2, independent of the methyltransferase function, remain to be discovered. We then examined how the 669 DEGs induced by Ezh2 overexpression were regulated by Ezh2-Y726D overexpression. Although not all of them were significantly regulated by Ezh2-Y726D, most showed opposite patterns of regulation after Ezh2 or Ezh2-Y726D overexpression (Supplemental Figure 4G). Indeed, among the 236 common DEGs regulated by both Ezh2 and Ezh2-Y726D, 203 (86%) changed in opposite directions (Supplemental Figure 4H and Supplemental Table 1). Gene ontology (GO) analysis of the DEGs further revealed that Ezh2 and Ezh2-Y726D inversely modified the RGC transcriptome (Figure 5C and Supplemental Table 3). Specifically, Ezh2 overexpression downregulated a series of ion transport and synaptic transmission-related genes and upregulated many immune response programs, both of which were oppositely regulated by Ezh2-Y726D overexpression (Figure 5, C–E). Similarly, in the ATAC-Seq, a large number of GO terms (immune response genes) were shared between genes whose promoter regions became more open after Ezh2 overexpression and those whose promoter regions became more closed after Ezh2-Y726D overexpression (Supplemental Figure 6E and Supplemental Table 4). These results implied that Ezh2-Y726D might inhibit functions of endogenous Ezh2 in a dominant-negative manner.
Figure 5. Ezh2 modifies the RGC transcriptome to regulate multiple categories of target genes.
(A and B) Volcano plots showing differences in gene expression between control and Ezh2 overexpression conditions (A) or between control and Ezh2-Y726D overexpression conditions (B). Note that 12 genes with –log10 (Padj) 50 and 3 genes with –log10(Padj) 100 are not plotted in A and B, respectively. (C) GO analysis of DEGs induced by Ezh2 or Ezh2-Y726D overexpression. A subset of most significantly enriched GO terms in the biological process category are shown here. (D and E) Volcano plots described in A and B with DEGs in 4 enriched GO terms labeled. (F and G) Heatmaps of mRNA levels of neuronal excitability and synaptic transmission regulators (F) and axon regeneration inhibitory factors (G) downregulated by Ezh2 overexpression in the control versus Ezh2 overexpression RNA-Seq (left) and the control versus Ezh2-Y726D overexpression RNA-Seq (right). (H) Heatmaps of mRNA levels of axon regeneration positive regulators upregulated by Ezh2 overexpression in the control versus Ezh2 overexpression RNA-Seq (left) and the control versus Ezh2-Y726D overexpression RNA-Seq (right). Note that the control versus. Ezh2 overexpression RNA-Seq and the control versus Ezh2-Y726D overexpression RNA-Seq were performed separately. Therefore, control (GFP) libraries in one RNA-Seq are independent of those in the other.
Because Ezh2 primarily functions to repress gene transcription through H3K27me3, we first focused on genes downregulated by Ezh2 overexpression. GO analysis showed that Ezh2 overexpression decreased transcription of many genes coding for ion channels and transporters as well as neurotransmitter receptors and transporters (Figure 5, C, D, and F), which are all important regulators of neuronal excitability and synaptic transmission. Except for a few genes, most of them were upregulated by Ezh2-Y726D (Figure 5F), consistent with its dominant-negative role. Because neuronal excitability and synaptic transmission are fundamental biological functions of mature neurons, these results suggested that Ezh2 might turn mature RGCs to a developmental-like state at the transcriptomic level, favoring axon regeneration.
Close examination of the downregulated genes further revealed that Ezh2 suppressed transcription of multiple axon regeneration inhibitory factors or their receptors, including ephrin receptors (encoded by Epha4, 6, 7, and 8; note that Epha4 was not among the 669 DEGs but had an adjusted P value under 0.05), tenascin-R (encoded by Tnr), Lingo3, and oligodendrocyte myelin glycoprotein (OMgp; encoded by Omg) (Figure 5G). The available single-cell RNA-Seq data set of RGCs (27) or immunostaining of retinal sections confirmed their expression in RGCs (Supplemental Figure 7, A–H). Moreover, Ezh2-Y726D did not transcriptionally downregulate these genes (Figure 5G), suggesting H3K27me3-dependent regulation.
In addition to suppressing the above genes, Ezh2 overexpression also upregulated many positive regulators of axon regeneration (Figure 5H). Among them, Atf3, Jun, Npy, Sprr1a, Gadd45g, and Sox11 are well-known regeneration-associated genes (32–37). Others, including Myc, Spp1 (which codes for osteopontin), Igf1, and Thbs1 (which codes for thrombospondin-1), have been well documented to promote CNS axon regeneration (38–42). In contrast, Ezh2-Y726D overexpression downregulated many of these genes (Figure 5H), indicating that their upregulation by Ezh2 overexpression was largely H3K27me3 dependent. Since H3K27me3 is associated with transcriptional suppression of its targets, these changes were likely secondary to increased H3K27me3.
Ezh2 overexpression enhances optic nerve regeneration by downregulating GABA transporter 2.
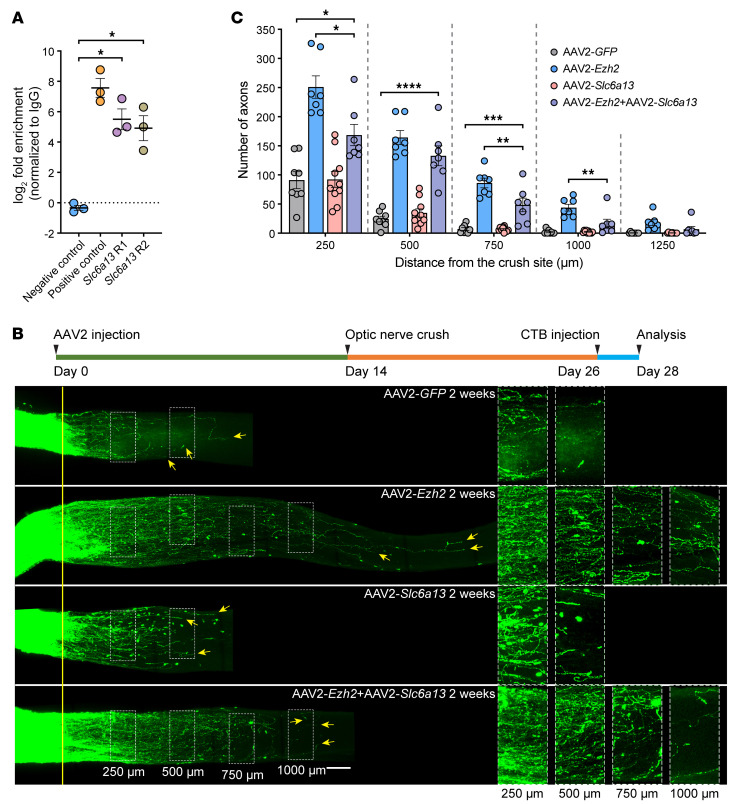
To determine if genes regulated by Ezh2 functionally acted downstream to regulate axon regeneration, we first examined the function of Slc6a13 (which codes for GABA transporter 2 [Gat2]), one of the most significantly downregulated genes after Ezh2 overexpression (see Figure 5F and Supplemental Table 1). We verified that Slc6a13 was broadly expressed by RGCs (Supplemental Figure 7, I and J). Additionally, employing the cleavage under targets and tagmentation (CUTTag) method (43) followed by quantitative PCR (qPCR), we showed H3K27me3 enrichment at Slc6a13 promoter region (Figure 6A), indicating its transcription could be directly regulated by Ezh2 via H3K27me3. Functionally, when Slc6a13 was overexpressed along with Ezh2 in RGCs by intravitreal injection of AAV2-Slc6a13, the strong optic nerve regeneration stimulated by Ezh2 overexpression was partially blocked, whereas Slc6a13 overexpression per se had no effect (Figure 6, B and C). These results demonstrated that Slc6a13 downregulation was required, at least in part, by Ezh2 to enhance optic nerve regeneration, and suggested a pivotal role of extracellular GABA levels in regulating axon regeneration, in line with insights from several prior studies (44–46). Slc6a13 overexpression also had a mild but significant inhibitory effect on regenerative axon growth of DRG neurons (Supplemental Figure 8, A and B), indicating its broadly consistent role in inhibiting axon regeneration.
Figure 6. Ezh2 overexpression enhances optic nerve regeneration by transcriptionally suppressing Slc6a13.
(A) CUTTag followed by qPCR showing H3K27me3 enrichment in the promoter region of Slc6a13 (paired 2-tailed t test; P = 0.0161 between negative control and Slc6a13 R1, P = 0.0306 between negative control and Slc6a13 R2; n = 3 independent experiments). (B) Top: Experimental timeline. Bottom: Representative images of optic nerves showing that Slc6a13 overexpression partially blocks Ezh2 overexpression–induced optic nerve regeneration. Columns on the right display enlarged images of the areas in the white, dashed boxes on the left, showing axons at 250, 500, 750, and 1,000 μm distal to the crush sites, which are aligned with the yellow line. Yellow arrows indicate longest axons in each nerve. Scale bars: 100 μm, 50 μm for enlarged images. (C) Quantification of optic nerve regeneration in B (1-way ANOVA followed by Tukey’s multiple comparisons; P 0.0001 at 250, 500, 750, and 1,000 μm, P = 0.0010 at 1,250 μm; n = 8 mice for control, n = 10 mice for Slc6a13 overexpression, n = 7 mice for others). *P 0.05, **P 0.01, ***P 0.001, ****P 0.0001.
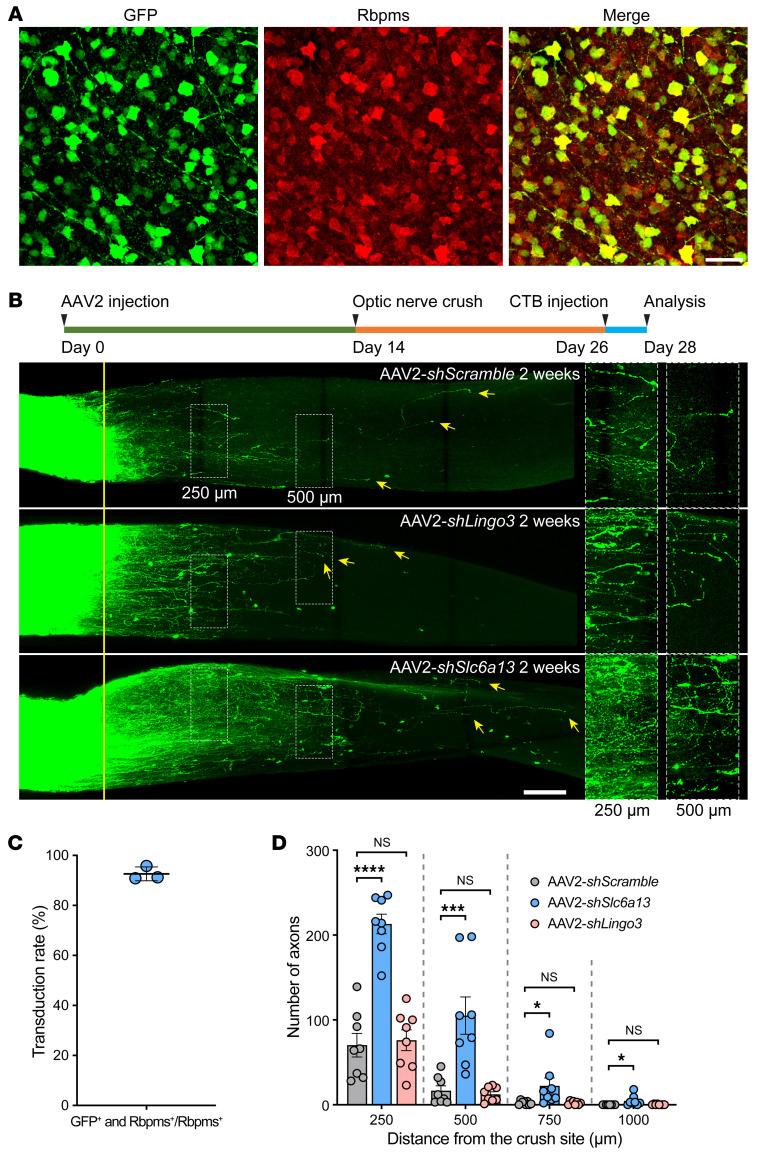
Based on these results, we wondered if downregulation of Slc6a13 could sufficiently induce optic nerve regeneration. We knocked down Slc6a13 in RGCs by intravitreal injection of AAV2 vectors encoding an shRNA against Slc6a13 mRNA. The transduction efficiency in RGCs was 92.65% ± 2.743% (Figure 7, A and C). We found that Slc6a13 loss-of-function effectively induced optic nerve regeneration (Figure 7, B and D). Together, these results demonstrated that Slc6a13 was a key downstream target of Ezh2 that mediates axon regeneration.
Figure 7. Slc6a13 loss-of-function promotes optic nerve regeneration.
(A) Representative immunofluorescence of whole-mount retinas showing high transduction efficiency of AAV2-shSlc6a13-EGFP in RGCs by intravitreal injection. Whole-mount retinas were stained with anti-GFP (green) and anti-Rbpms (red). Scale bar: 50 μm. (B) Top: Experimental timeline. Bottom: Representative images of optic nerves showing that knockdown of Slc6a13, but not Lingo3, modestly promotes optic nerve regeneration 2 weeks after optic nerve crush. Columns on the right display enlarged images of the areas in the white, dashed boxes on the left, showing axons at 250 and 500 μm distal to the crush sites, which are aligned with the yellow line. Yellow arrows indicate longest axons in each nerve. Scale bars: 100 μm, 50 μm for enlarged images. (C) Quantification of the percentage of GFP-positive RGCs in A. The average transduction rate was 92.65% ± 2.743% (n = 3 mice; 8 fields were analyzed for each mouse). Data represent mean ± SD. (D) Quantification of optic nerve regeneration in B (1-way ANOVA followed by Tukey’s multiple comparisons; P 0.0001 at 250 and 500 μm, P = 0.0248 at 750 μm, P = 0.0263 at 1,000 μm; n = 8 mice for all). *P 0.05, ***P 0.001, ****P 0.0001.
Ezh2 overexpression enhances optic nerve regeneration by suppressing major axon regeneration inhibitory signaling.
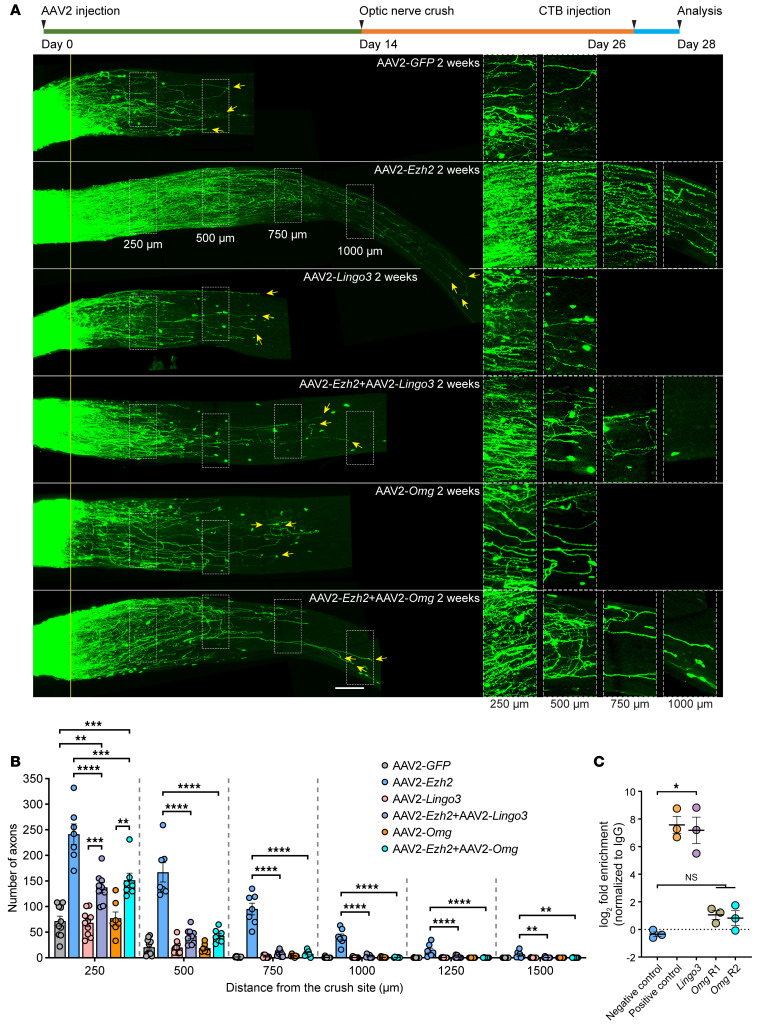
Next, we tested if downregulation of axon regeneration inhibitory signaling contributed to Ezh2 overexpression–induced optic nerve regeneration. Overexpression of Omg or Lingo3, per se, had no effect on optic nerve regeneration, but almost completely blocked Ezh2 overexpression–induced regeneration (Figure 8, A and B). Only at 250 μm from the crush site were more axons observed in the cooverexpression groups than in the control group. CUTTag followed by qPCR revealed H3K27me3 binding at the Lingo3 promoter region (Figure 8C), suggesting H3K27me3-mediated transcriptional repression of Lingo3. Unlike Slc6a13, Lingo3 loss-of-function did not promote optic nerve regeneration (Figure 7, B and D). In contrast, no binding of the Omg promoter region by H3K27me3 was detected (Figure 8C), suggesting that Omg downregulation might be a secondary effect of elevated H3K27me3. Collectively, these results suggested that downregulation of Lingo3 and Omg contributed to optic nerve regeneration induced by Ezh2 overexpression, and that Ezh2 might be a key suppressor of signaling pathways that impede CNS axon regeneration (see Discussion).
Figure 8. Ezh2 overexpression enhances optic nerve regeneration by transcriptionally suppressing Lingo3 and Omg.
(A) Top: Experimental timeline. Bottom: Representative images of optic nerves showing that Lingo3 or Omg overexpression almost completely blocks Ezh2 overexpression-induced optic nerve regeneration. Columns on the right display enlarged images of the areas in white, dashed boxes on the left, showing axons at 250, 500, 750, and 1,000 μm distal to the crush sites, which are aligned with the yellow line. Yellow arrows indicate longest axons in each nerve. Scale bars: 100 μm, 50 μm for enlarged images. (B) Quantification of optic nerve regeneration in A (1-way ANOVA followed by Tukey’s multiple comparisons; P 0.0001 at 250, 500, 750, 1,000, and 1,250 μm, P = 0.0003 at 1,500 μm; n = 10 mice for control and Lingo3 overexpression, n = 9 mice for Ezh2 and Lingo3 cooverexpression, n = 7 mice for others). (C) CUTTag followed by qPCR showing H3K27me3 enrichment in the promoter region of Lingo3, but not that of Omg (paired 2-tailed t test; P = 0.0197 between negative control and Lingo3 R1, P = 0.1010 between negative control and Omg R1, P = 0.2329 between negative control and Omg R2; n = 3 independent experiments). Note that the negative control and positive control are identical to those in Figure 6A. *P 0.05, **P 0.01, ***P 0.001, ****P 0.0001.
Ezh2 activates multiple axon regeneration enhancing pathways.
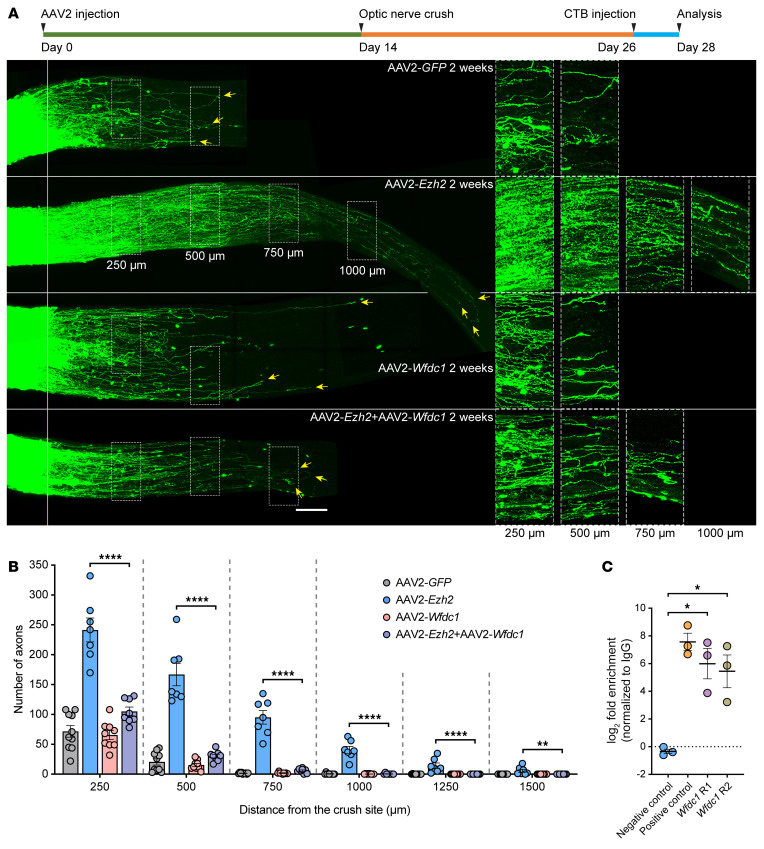
Ezh2 overexpression in RGCs resulted in upregulation of osteopontin (encoded by Spp1, see Figure 5H and Supplemental Table 1), which selectively promotes axon regeneration of αRGCs (40). In addition to retinal repair, increased osteopontin expression was also shown to underlie the enhanced tissue repair induced by knocking out Wfdc1 (47), known as a tumor suppressor (48) and a wound repair inhibitor (47). Interestingly, mRNA levels of Wfdc1 in RGCs were also reduced by Ezh2 in our RNA-Seq (see Supplemental Table 1), suggesting that it might regulate Ezh2-induced optic nerve regeneration via osteopontin. Indeed, overexpression of Wfdc1 completely blocked the optic nerve regeneration induced by Ezh2 overexpression (Figure 9, A and B), suggesting that Wfdc1 was a strong axon regeneration inhibitor transcriptionally suppressed by Ezh2. CUTTag followed by qPCR showed that the Wfdc1 promoter region was bound by H3K27me3 (Figure 9C), again suggesting histone methylation–dependent regulation by Ezh2. These results provided a potential mechanism of how Ezh2 indirectly upregulated axon regeneration–enhancing factors.
Figure 9. Ezh2 overexpression enhances optic nerve regeneration by transcriptionally suppressing Wfdc1.
(A) Top: Experimental timeline. Bottom: Representative images of optic nerves showing that Wfdc1 overexpression completely blocks Ezh2 overexpression–induced optic nerve regeneration. Columns on the right display enlarged images of the areas in the white, dashed boxes on the left, showing axons at 250, 500, 750, and 1,000 μm distal to the crush sites, which are aligned with the yellow line. Yellow arrows indicate longest axons in each nerve. Scale bars: 100 μm, 50 μm for enlarged images. (B) Quantification of optic nerve regeneration in A (1-way ANOVA followed by Tukey’s multiple comparisons; P 0.0001 at 250, 500, 750, 1,000, and 1,250 μm, P = 0.0015 at 1,500 μm; n = 10 mice for control, n = 7 mice for Ezh2 overexpression, n = 9 mice for Wfdc1 overexpression, n = 8 mice for Ezh2 and Wfdc1 cooverexpression). Note that the control and Ezh2 overexpression conditions are identical to those in Figure 8B. (C) CUTTag followed by qPCR showing H3K27me3 enrichment in the promoter region of Wfdc1 (paired 2-tailed t test; P = 0.0357 between negative control and Wfdc1 R1, P = 0.0478 between negative control and Wfdc1 R2; n = 3 independent experiments). Note that the negative control and positive control are identical to those in Figure 6A. *P 0.05, **P 0.01, ***P 0.001, ****P 0.0001.
Among other upregulated genes (Supplemental Table 1), Ascl1 and Neurog2 are important regulators of neurogenesis and axon guidance during development and direct reprogramming factors converting glial cells into neurons (49). Moreover, Ascl1, Neurog2, and Ezh2 were identified as key factors driving neuronal differentiation in a Crispr-based screening (50), suggesting similar or cooperative functions. Interestingly, Ascl1 was shown to support PNS axon regeneration in mice (51) and CNS axon regeneration in zebrafish and rats (52). We therefore investigated if Neurog2 could also regulate optic nerve regeneration. Overexpression of Neurog2 in RGCs had little effect on optic nerve regeneration (Supplemental Figure 9, A and B), suggesting that distinct mechanisms mediate neurogenesis and axon regeneration.
Ezh2 overexpression does not alter the epigenetic aging clock of RGCs.
A recent study discovered that ONC increased the DNA methylation age of RGCs, and that polycistronic expression of reprogramming factors Oct4, Sox2, and Klf4 counteracted the aging effect of ONC and promoted optic nerve regeneration (53). Ezh2 is also critical for efficient cellular reprogramming (54, 55) and was shown to participate in shaping the aging epigenome (56). Moreover, we showed that Ezh2 overexpression specifically silenced transcription of many genes functionally involved in synaptic activities of mature neurons, in some way turning adult RGCs to a developmental-like state at the transcriptomic level. We thus wondered if Ezh2 overexpression could also reverse the aging effect of ONC on the RGC epigenome. We intravitreally injected AAV2-GFP, AAV2-Ezh2, or AAV2-Ezh2-Y726D into mice of exactly the same age, performed ONC 2 weeks after the injection, and extracted DNA from FACS-enriched RGCs 3 days after the ONC. Uninjured groups only received AAV2 injection but did not undergo ONC. Reduced representation bisulfite sequencing (RRBS) libraries were constructed from RGC DNA and sequenced to obtain the DNA methylation landscape. A predictive PCA model (53) was used to estimate changes in the DNA methylation age of RGCs. The results confirmed that ONC accelerated epigenetic aging of RGCs, but neither WT Ezh2 nor Ezh2-Y726D could reverse the changes (Supplemental Figure 10A). Consistently, our RNA-Seq did not detect significant changes in mRNA levels of most 5mC DNA methyltransferases or demethylases (Supplemental Figure 10, B and C). These results indicated that Ezh2 overexpression was not able to rejuvenate mature RGCs epigenetically.
Collectively, our study not only revealed a role of Ezh2 in coordinating axon regeneration via regulation of multiple key regenerative pathways, but also identified chromatin accessibility as a promising target to promote CNS axon regeneration.
Discussion
Developmental decline of axon regeneration ability with transcriptomic changes regulated by Ezh2.
Axon regeneration ability of mammalian neurons declines as they mature. While PNS neurons can reactivate their regenerative ability upon peripheral axonal injury, most adult CNS neurons permanently lose their ability to regenerate axons. Given that every single cell in an organism has completely the same genome, and so does a neuron in different states (e.g., developmental versus mature or healthy versus injured), it is conceivable that the tuning of the axon regeneration ability in neurons is largely regulated by changes in the epigenomic and transcriptomic landscapes. Here we found that Ezh2, an epigenetic regulator that controls chromatin accessibility and gene transcription via histone methylation, was developmentally downregulated in both the PNS and CNS and could be upregulated in PNS neurons by peripheral nerve injury. Thus, Ezh2 levels in the nervous system and the axon growth and regeneration potential of neurons rise and fall in parallel. Indeed, we found that Ezh2 loss-of-function impaired spontaneous axon regeneration of mature PNS neurons, while Ezh2 gain-of-function promoted axon regeneration in nonregenerative adult CNS neurons. Mechanistic exploration revealed that Ezh2 overexpression in RGCs suppressed transcription of a large number of genes regulating synaptic transmission and neuronal excitability, which are housekeeping functions of mature neurons. Therefore, our study suggested that Ezh2 upregulation might turn mature neurons into a developmental-like cellular state at the transcriptomic level to empower them with stronger axon regeneration ability. In support of this, several previous studies also found a negative correlation between synaptic functions and axon regeneration ability (44, 57–59). On the other hand, Ezh2 overexpression also resulted in upregulation of many factors known to enhance axon regeneration, some of which are highly expressed in developing neurons (60–62).
When the epigenetic aging biomarker, the DNA methylation clock, was examined in RGCs (53), we confirmed that optic nerve injury significantly accelerated RGC epigenetic aging. However, Ezh2 overexpression was not able to reverse it. These results indicated that physiological aging at the transcriptomic level could be uncoupled from the DNA methylation aging clock. The discrepancy between DNA methylation–based aging clock and transcriptomic landscape–based cellular state was not surprising. A recent study (63) of naked mole rats (NMRs), which live an exceptionally long life and are considered a nonaging mammal, showed a normal aging progress in many tissues at the epigenetic level without significant overlap with age-related transcriptomic changes. Interestingly, cell reprogramming was able to rejuvenate the DNA methylation clock of NMR cells, consistent with a recent study in which partial reprogramming of RGCs with 3 reprogramming factors, Oct4, Sox2, and Klf4, reversed the DNA methylation aging induced by optic nerve injury (53). Collectively, we think that Ezh2 overexpression in mature RGCs switched their transcriptomic landscape to a developmental-like state with stronger axon growth ability.
Ezh2 is a master suppressor of CNS axon regeneration inhibitory signaling.
Here, we demonstrated that Ezh2 overexpression transcriptionally silenced OMgp (1 of the 3 major MAIs), Lingo3, tenascin-R, and several ephrin receptors. OMgp and other MAIs (MAG and Nogo) act through the Nogo receptor 1 (NgR1) complex or PirB to inhibit axon growth (64–66). Lingo3 is a paralog of Lingo1 that codes for a critical component of the NgR1 complex (67), which blocks axon regeneration via RhoA when activated by MAIs and CSPGs (68). Lingo1 loss-of-function also promotes axon regeneration and neuronal survival in various CNS injury and disease models (69–71). Additionally, Lingo family receptors can form heteromers with one another in the mouse brain (72), strongly suggesting functional overlap between the paralogs. Tenascin-R, an extracellular matrix molecule, is a repulsive guidance cue in zebrafish during development (73) and an inhibitor of mouse optic nerve regeneration (74). Ephrin receptors are chemorepellent axon guidance molecules that can cause growth cone collapse when activated by their ligands, ephrins (75). MAIs, CSPGs, and repulsive axon guidance cues are 3 major classes of extracellular axon regeneration inhibitors in the mature CNS (76). Ezh2 downregulated transcription of regeneration inhibitors or their receptors associated with all 3 classes. Functionally, we found that overexpression of Omg or Lingo3 blocked optic nerve regeneration induced by Ezh2 overexpression to a great extent. Thus, Ezh2 might be a master suppressor of CNS axon regeneration inhibitory signaling pathways. Notably, our study indicated that, besides glial cells, neurons per se could also contribute to the production of extracellular CNS regeneration inhibitors, such as OMgp and tenascin-R. It would be interesting for future studies to investigate the mechanisms of neuron-secreted axon regeneration inhibitors.
Ezh2 enhances optic nerve regeneration via both methylation-dependent and -independent pathways.
Most previous studies of Ezh2 focused on H3K27me3-mediated transcriptional repression in various biological processes. Evidence has been emerging, however, to suggest that Ezh2 also has activities unrelated to protein methylation. For example, Ezh2 can transactivate the androgen receptor by directly binding to its promoter region (20). Similarly, a ternary complex of Ezh2, RelA, and RelB bind to promoters of Il6 and Tnf to enhance their transcription (19). Furthermore, Ezh2 can even regulate protein translation via interacting with fibrillarin and controlling rRNA methylation, completely independent of its methyltransferase function (77). In our study, an Ezh2 mutant lacking the methyltransferase activity, Ezh2-Y726D, was still able to modestly promote optic nerve regeneration, clearly indicating that methyltransferase-independent activities of Ezh2 also contributed to the enhanced axon regeneration.
In the current study, we did not further investigate these methyltransferase-independent mechanisms. Our RNA-Seq results implied that Ezh2-Y726D acted in a dominant-negative manner of WT Ezh2 to control gene transcription. Such results appeared perplexing, as Ezh2-Y726D still promoted RGC survival and optic nerve regeneration. A likely explanation is that the overall effect of Ezh2-Y726D overexpression was to enhance the maturation state of RGCs, which was unlikely to further reduce the already very low intrinsic axon growth ability. However, Ezh2-Y726D did regulate some genes in the same way as Ezh2. For example, both Ezh2 and Ezh2-Y726D overexpression significantly upregulated Jun, Npy, and Igf1, all of which have been shown to support axon regeneration (78–80). Likewise, both Ezh2 and Ezh2-Y726D downregulated mRNA levels of corticotropin releasing hormone binding protein (encoded by Crhbp) and Slc6a13 (see Figure 5F and Supplemental Table 1). Crhbp is selectively expressed in RGC subtypes susceptible to ONC, and its loss-of-function significantly promotes RGC survival and optic nerve regeneration (27). Similarly, our current study showed that knocking down Slc6a13 induced optic nerve regeneration (see Figure 7, B and D). Upregulation of axon regeneration enhancers and downregulation of Crhbp and Slc6a13 might be methylation-independent mechanisms by which Ezh2-Y726D modestly promoted optic nerve regeneration. Future studies are needed to further explore the roles noncanonical pathways of Ezh2 play in mammalian axon regeneration.
Although AAV2 also transduces other retinal cells besides RGCs, our data indicated that Ezh2 overexpression–induced increase of H3K27me3 was mostly observed in cells within the ganglion cell layer of the retina (see Figure 3A), which contains mainly RGCs and some displaced amacrine cells. However, we cannot exclude the possibility that non-RGC-autonomous factors also contributed to the optic nerve regeneration observed here (80). Future studies using Vglut2-Cre mice to restrict Ezh2 expression in RGCs would provide a clearer answer.
Methods
Mice.
Adult C57BL/6J mice (6–10 weeks old) of both sexes were used unless otherwise stated. The Ezh2fl/fl (stock no. 015499-UNC) mouse strain was obtained from the Mutant Mouse Resource and Research Center (MMRRC) at University of North Carolina at Chapel Hill, an NIH-funded strain repository, and was donated to the MMRRC by Alexander Tarakhovsky (the Rockefeller University [New York, New York, USA]). The Advillin-Cre mouse line (the Jackson Laboratory) stock no. 032536) was a gift from Fan Wang at Duke University in Durham, North Carolina, and was crossed with Ezh2fl/fl to obtain Advillin-Cre;Ezh2fl/fl conditional knockout mice. Because female Advillin-Cre mice have weak Cre expression in oocytes, only male Advillin-Cretg/+;Ezh2fl/fl mice and female Ezh2fl/fl mice were used for breeding. Therefore, the resulting offspring was either heterozygous or negative for Advillin-Cre. Both male and female Advillin-Cretg/+;Ezh2fl/fl mice were used for experiments. Genotypes of the mice were determined by PCR using primers and programs provided by the MMRRC and the Jackson Laboratory. All mouse surgeries were performed under anesthesia induced by i.p. injection of ketamine (100 mg/kg) and xylazine (10 mg/kg) diluted in sterile saline. Details of surgeries are described below.
Immunoblotting.
Total protein was extracted from mouse DRGs, cultured DRG cells, retinas, or FACS-enriched RGCs using the RIPA buffer (Sigma-Aldrich) containing the protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitor cocktail (Sigma-Aldrich). Identical amounts of total protein from each condition were separated by SDS-PAGE on 4-to-12% Bis-Tris gels and transferred onto polyvinylidene difluoride membranes. Membranes were blocked with TBST containing 5% blotting-grade blocker (Bio-Rad), incubated in primary antibodies against target molecules overnight at 4°C, washed 4 times (5, 5, 10, and 10 minutes) with TBST, incubated in corresponding HRP-linked secondary antibodies (1:2,000; Cell Signaling Technology 7074 or 7076) for 1 hour at room temperature, and washed 4 times (5, 5, 10, and 10 minutes) again with TBST. All antibodies were diluted with TBST containing 5% blotting-grade blocker. Primary antibodies used for immunoblotting in this study include rabbit anti-Ezh2 (1:1,000, Cell Signaling Technology 5246), rabbit anti-H3 (1:1,000, Cell Signaling Technology 9715), mouse anti-H3K27me3 (1:10,000, Sigma-Aldrich 05-1951), rabbit anti-Ezh1 (1:2,000, Sigma-Aldrich ABE281), rabbit anti-Suz12 (1:1,000, Cell Signaling Technology 3737), rabbit anti-Eed (1: 1,000, Sigma-Aldrich 09-774), rabbit anti-Rbap46/48 (1:1,000, Active Motif 39199), mouse anti-β-actin (1:10,000, Sigma-Aldrich A1978), and mouse anti-Gapdh (1:20,000, Sigma-Aldrich G8795). See complete unedited blots in the supplemental material.
In vivo DRG electroporation.
Under anesthesia, a small dorsolateral laminectomy was performed on the left side to expose left L4/5 DRGs. Using a pulled glass micropipette (World Precision Instruments) connected to a Picospritzer III (pressure: 20 psi, pulse duration: 6 ms, Parker Hannifin), 1 μL plasmid vectors (2 μg/μL) and/or siEzh2 (100 μM, Horizon Discovery, see Supplemental Table 5 for target sequences) containing 0.05% fast green FCF (Sigma-Aldrich) were injected into each DRG. After injection, in vivo electroporation was performed by applying 5 electric pulses (voltage: 35 V, pulse duration: 15 ms, pulse interval: 950 ms) using a platinum tweezertrode (BTX) powered by an ECM 830 Electro Square Porator (BTX). The wound was then closed with sutures.
Sciatic nerve crush or transection.
Under anesthesia, sciatic nerves were exposed right below the pelvis and crushed with Dumont #5 forceps (Fine Science Tools) for 15 seconds or cut with scissors, and the wound was closed by sutures. In sham surgeries, sciatic nerves were only exposed but not injured. Both sciatic nerve transection and sciatic nerve crush result in axotomy of all axons in the sciatic nerve. Sciatic nerve crush was performed in in vivo DRG neuron axon regeneration experiments and was only done on the left side. The crush site was marked with 10-0 nylon epineural sutures that are identifiable during dissection and imaging data analysis. Sciatic nerve transection was performed in other experiments to ensure completeness of the axotomy and was done bilaterally.
Analysis of in vivo DRG neuron axon regeneration.
Three or 5 days after sciatic nerve crush, mice were anesthetized and transcardially perfused with PBS followed by 4% PFA. Sciatic nerve segments (proximal end: 5 mm proximal to the crush site; distal end: the point where the sciatic nerve branches into 3 nerves) were dissected and postfixed in 4% PFA overnight at 4°C. The next day, nerve segments were mounted in Fluoroshield (Sigma-Aldrich) onto microscope slides, covered with coverslips, and flattened by applying a heavy weight on coverslips. Tiled fluorescent images of whole-mount nerve segments were obtained with a Zeiss inverted fluorescence microscope controlled by the AxioVision software using a 5× objective. Nerve segments were imaged from approximately 1 mm proximal to the crush site to approximately 0.5 mm distal to the end of the longest axon. Using the built-in “measure/curve spline” function of the AxioVision software, GFP-labeled axons were manually traced from the crush site to axonal tips to determine the lengths. The mean length of all axons traced in 1 nerve segment was used as the average axon length of this nerve. Nerves whose epineural sutures were missing or with less than 10 identifiable GFP-labeled axons were excluded from data analysis. Measurement was done by experimenters blinded to experimental conditions. Nerve images were put on a black background when figures were generated.
ONC and regeneration.
Intravitreal virus injection, ONC, and RGC axon labeling were performed as previously described (28). Briefly, under anesthesia, 1.5 μL AAV2 virus (approximately 1 × 1013 genome copies/mL) was injected into the vitreous humor with a Hamilton syringe (33-gauge needle). The position and direction of the needle were well controlled to avoid injury to the lens. Two weeks after the virus injection, under anesthesia, a small incision was made in the skin right behind the eye and the conjunctiva was incised to expose the extraocular muscles. The muscles were pushed aside with forceps to expose the optic nerve, and the optic nerve was crushed with Dumont #5 forceps (Fine Science Tools) for 5 seconds at approximately 0.5 mm behind the optic disc. Care was taken to avoid damage to the ophthalmic artery. For the postinjury treatment model, ONC was done 1 day before virus injection. To label RGC axons in the optic nerve, under anesthesia, 1.5 μL Alexa Fluor 555 or 647–conjugated CTB (1 μg/μL, Thermo Fisher Scientific C22843 or C34778) was injected into the vitreous humor with a Hamilton syringe (33-gauge needle) 2 days before tissue harvesting. Mice with lens injury, hemorrhage, or incomplete crush evidenced by continuous CTB labeling through the chiasm were excluded from data analysis.
Analysis of optic nerve regeneration.
Z-stacked (step size: 2 μm) and tiled fluorescent images of tissue-cleared whole-mount optic nerves were obtained with a Zeiss LSM 800 confocal microscope using a 20× objective. Optic nerves were imaged from approximately 0.5 mm proximal to the crush site to approximately 0.5 mm distal to the end of the longest axon. To quantify the number of regenerating axons in each optic nerve, every 8 consecutive planes were Z-projected in maximum intensity to generate a series of Z-projection images of 16-μm-thick optical sections. At each 250-μm interval from the crush site, the number of CTB-labeled axons was counted in each Z-projection image and summed over all optical sections. Nerve images were put on a black background when figures were generated.
NMDA-induced excitotoxicity model.
Under anesthesia, 1.5 μL AAV2 virus (approximately 1 × 1013 genome copies/mL) was injected into the vitreous humor with a Hamilton syringe (33-gauge needle). Two weeks after the virus injection, under anesthesia, 1.5 μL NMDA (20 mM, Sigma-Aldrich) was injected into the vitreous humor with a Hamilton syringe (33-gauge needle). The position and direction of the needle were well controlled to avoid injury to the lens.
RGC enrichment.
Retinas were dissected from euthanized mice, digested with papain (Thermo Fisher Scientific 88285) containing 0.005% DNase (Worthington) at 37°C for 8 minutes, washed 3 times with HBSS, and dissociated into cell suspension by trituration in NeuroBasal medium containing 1% BSA. Cells were filtered with a 40 μm cell strainer, pelleted by centrifugation (500g for 5 minutes at room temperature), resuspended in NeuroBasal medium containing 1% BSA, blocked with rat anti-mouse CD16/CD32 (1:50, BD Biosciences 553141) for 5 minutes on ice, and labeled with PE-conjugated rat anti-CD90.2 (1:100, Thermo Fisher Scientific 12-0902-81) for 30 minutes on ice. After that, cells were washed twice with HBSS containing 1% BSA, pelleted by centrifugation (500g for 5 minutes at room temperature), and again resuspended in Neurobasal medium containing 1% BSA. Propidium iodide (PI) or DAPI was mixed with the cell suspension to label dead cells 2 minutes before cells were loaded into a Beckman Coulter MoFlo Legacy Cell Sorter. CD90.2-positive and PI or DAPI–negative cells were sorted into NeuroBasal medium containing 1% BSA with a 70 μm nozzle.
CUTTag and qPCR.
ChIP-Seq libraries were constructed from FACS-enriched RGCs (100,000 cells for each library) using rabbit anti-H3K27me3 (1:50, Active Motif 39155) or normal rabbit IgG (1:50, Sigma-Aldrich NI01) and the CUTTag-IT Assay Kit (Active Motif) following the manufacture’s manual. Identical amounts of DNA from each library were used in qPCR to determine the enrichment of H3K23me3 in the promoter region of each gene. Two pairs of primers were designed for each gene. One pair for Lingo3 promoter region resulted in no amplification and was excluded from data analysis. Sequences of primers used in qPCR are in Supplemental Table 5. Positive control (71020) and negative control primers (71013) were purchased from Active Motif. Fold enrichment of H3K27me3 binding was determined using the ddCt method and normalized to IgG. All qPCR experiments were done in triplicate.
Immunofluorescence of whole-mount retinas.
Retinas were dissected from transcardially perfused mice and postfixed in 4% PFA overnight at 4°C. On the next day, retinas were postfixed in ice-cold methanol for 20 minutes, washed 3 times for 5 minutes each time with PBS, radially cut into a petal shape, and blocked with PBST (1%) containing 10% goat serum for 1 hour at room temperature. After blocking, retinas were incubated in primary antibodies against target molecules overnight at 4°C, washed 4 times for 15 minutes each time with PBST (0.3%), incubated in corresponding Alexa Fluor–conjugated secondary antibodies (1:500, Thermo Fisher Scientific) for 2 hours at room temperature, and washed 4 times for 15 minutes each time again with PBST (0.3%). All antibodies were diluted with PBST (1%) containing 10% goat serum. Retinas were flat-mounted in Fluoroshield (Sigma-Aldrich) onto microscope slides and covered by coverslips. Fluorescent images of whole-mount retinas were obtained with a Zeiss LSM 800 confocal microscope using a 20× objective.
Analysis of RGC survival rate.
To quantify RGC survival rate, mice were transcardially perfused 2 weeks after ONC or 1 week after NMDA injection and both retinas of each mouse were dissected. Retinas were stained with guinea pig anti-Rbpms (1:100, Sigma-Aldrich ABN1376) following the steps described above (see Immunofluorescence of whole-mount retinas). Randomly, 6–9 fields were taken from the peripheral regions of each retina. For each mouse, RGC survival rate was calculated by dividing the average number of Rbpms-positive cells in 1 field in the injured retina by that in the uninjured retina. Only cells in the ganglion cell layer were counted.
Analysis of RGC transduction efficiency.
To quantify RGC transduction efficiency, mice were transcardially perfused 2 weeks after intravitreal injection of AAV2-shSlc6a13-EGFP. Retinas were stained with guinea pig anti-Rbpms (1:100, PhosphoSolutions 1832-RBPMS) and chicken anti-GFP (1:100, Thermo Fisher Scientific A10262) following the steps described above (see Immunofluorescence of whole-mount retinas). Eight fields were randomly taken from the peripheral regions of each retina. For each mouse, RGC transduction efficiency was calculated by dividing the total number of GFP and Rbpms double-positive cells in all fields by the total number of Rbpms-positive cells in all fields. Only cells in the ganglion cell layer were counted.
Immunofluorescence of retinal sections.
Fixed retinas were immersed in 30% sucrose overnight at 4°C. On the next day, retinas were embedded in OCT compound, frozen, and cut into 10 μm sections with a cryostat. Sections were transferred onto microscope slides and warmed on a slide warmer for 1 hour at 37°C. Sections on slides were rinsed once in PBS, soaked in 100°C citrate buffer (pH 6) for 15 minutes, cooled down in the buffer to room temperature, washed 2 times for 5 minutes each time with PBS, and blocked with PBST (0.3%) containing 10% goat serum for 1 hour at room temperature. After blocking, sections were incubated in primary antibodies against target molecules overnight at 4°C, washed 4 times (5, 5, 10, and 10 minutes) with PBST (0.3%), incubated in corresponding Alexa Fluor–conjugated secondary antibodies (1:500, Thermo Fisher Scientific) for 1 hour at room temperature, and washed 4 times (5, 5, 10, and 10 minutes) again with PBST (0.3%). All antibodies were diluted with PBST (0.3%) containing 10% goat serum. Sections were mounted in Fluoroshield (Sigma-Aldrich) and covered by coverslips. Fluorescent images of retinal sections were obtained with a Zeiss inverted fluorescence microscope controlled by the AxioVision software using a 20× objective.
Analysis of H3K27me3 levels in RGCs.
To analyze H3K27me3 levels in RGCs, retinas were dissected from transcardially perfused mice 2 weeks after intravitreal injection of AAV2-GFP, AAV2-Ezh2, or AAV2-Ezh2-Y726D and sectioned with a cryostat. Retinal sections were stained with guinea pig anti-Rbpms (1:500, Sigma-Aldrich ABN1376) and mouse anti-H3K27me3 (1:100, Sigma-Aldrich 05-1951) following the steps described above (see Immunofluorescence of retinal sections).
To quantify H3K27me3 levels in RGCs, fluorescence intensity of H3K27me3 immunoreactivity of at least 150 Rbpms-positive cells from 10–12 nonadjacent retinal sections acquired with identical imaging configurations was analyzed for each retina. Fluorescence intensity was measured using the “outline spline” function of the AxioVision software and the background fluorescence intensity was subtracted.
Statistics.
Statistical analysis was done with GraphPad Prism 10 and the significance level was set as P 0.05. Data represent mean ± SEM unless otherwise stated. For comparisons between 2 conditions, 2-tailed unpaired or paired t test was used. For comparisons among 3 or more conditions, 1-way ANOVA followed by Tukey’s multiple comparisons was used to determine the statistical significance. All details of statistics, including tests used, P values, and sample sizes, are described in figure legends. P values of posthoc analyses are illustrated in figures. A P value less than 0.05 was considered significant.
Study approval.
Protocols for animal experiments in this study were approved by the Animal Care and Use Committee of Johns Hopkins University.
Data availability.
Raw and processed sequencing data are available in the Gene Expression Omnibus (GSE247320). Values for all data points in graphs are reported in the Supporting Data Values file.
Author contributions
XWW, CML, and FQZ conceived the study and designed the project; XWW, SGY, and RYW performed the experiments. MWH, XL, and JQ analyzed the sequencing data. XWW and CZ performed data analysis. JJJ, ARK, and AJO helped with data analysis. YG helped with manuscript revision. XWW and FQZ wrote the manuscript with contributions from all authors. The order among cofirst authors was assigned based on contributions.
Supplementary Material
Acknowledgments
We acknowledge Michele Pucak and Abigail Bush from the Multiphoton Imaging Core (supported by P30NS050274) of the Department of Neuroscience at Johns Hopkins University School of Medicine and Hao Zhang from the Flow Cytometry Cell Sorting Core Facility at Johns Hopkins University Bloomberg School of Public Health for their help in confocal microscopy and FACS sorting. We appreciate the Johns Hopkins Single Cell and Transcriptomics Core for RNA-Seq and ATAC-Seq experiments. FQZ is supported by grants from the NIH (R01NS085176, R01EY027347, R01EY030883, and R01EY031779), the Craig H. Neilsen Foundation (259450), and the BrightFocus Foundation (G2017037). XWW and JQ are supported by the NIH (K99EY031742 and R01EY029548).
Version 1. 11/28/2023
In-Press Preview
Version 2. 02/01/2024
Electronic publication
Footnotes
FQZ’s present address is: Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2023, Wang et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2024;134(3):e163145. https://doi.org/10.1172/JCI163145.
Contributor Information
Xue-Wei Wang, Email: xwang137@jhmi.edu.
Shu-Guang Yang, Email: lionamms@hotmail.com.
Ming-Wen Hu, Email: mhu10@jhmi.edu.
Rui-Ying Wang, Email: 77276533@qq.com.
Chi Zhang, Email: czhan110@jhmi.edu.
Anish R. Kosanam, Email: akosana1@jhu.edu.
Arinze J. Ochuba, Email: aochuba1@jh.edu.
Jing-Jing Jiang, Email: jjj_sj@163.com.
Ximei Luo, Email: luoximei@uestc.edu.cn.
Yun Guan, Email: Yguan1@jhmi.edu.
Jiang Qian, Email: jiang.qian@jhmi.edu.
Chang-Mei Liu, Email: liuchm@ioz.ac.cn.
Feng-Quan Zhou, Email: fengquan@yahoo.com.
References
- 1.He Z, Jin Y. Intrinsic control of axon regeneration. Neuron. 2016;90(3):437–451. doi: 10.1016/j.neuron.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Curcio M, Bradke F. Axon regeneration in the central nervous system: facing the challenges from the inside. Annu Rev Cell Dev Biol. 2018;34:495–521. doi: 10.1146/annurev-cellbio-100617-062508. [DOI] [PubMed] [Google Scholar]
- 3.Mahar M, Cavalli V. Intrinsic mechanisms of neuronal axon regeneration. Nat Rev Neurosci. 2018;19(6):323–337. doi: 10.1038/s41583-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nogueira-Rodrigues J, et al. Rewired glycosylation activity promotes scarless regeneration and functional recovery in spiny mice after complete spinal cord transection. Dev Cell. 2021;57(47):440–450. doi: 10.1016/j.devcel.2021.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Geoffroy CG, Zheng B. Myelin-associated inhibitors in axonal growth after CNS injury. Curr Opin Neurobiol. 2014;27:31–38. doi: 10.1016/j.conb.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwab ME, Strittmatter SM. Nogo limits neural plasticity and recovery from injury. Curr Opin Neurobiol. 2014;27:53–60. doi: 10.1016/j.conb.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006;7(8):617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith DS, Skene JH. A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J Neurosci. 1997;17(2):646–658. doi: 10.1523/JNEUROSCI.17-02-00646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saijilafu, et al. PI3K-GSK3 signalling regulates mammalian axon regeneration by inducing the expression of Smad1. Nat Commun. 2013;4:2690. doi: 10.1038/ncomms3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weng YL, et al. Epigenetic regulation of axonal regenerative capacity. Epigenomics. 2016;8(10):1429–1442. doi: 10.2217/epi-2016-0058. [DOI] [PubMed] [Google Scholar]
- 11.Palmisano I, et al. Epigenomic signatures underpin the axonal regenerative ability of dorsal root ganglia sensory neurons. Nat Neurosci. 2019;22(11):1913–1924. doi: 10.1038/s41593-019-0490-4. [DOI] [PubMed] [Google Scholar]
- 12.Renthal W, et al. Transcriptional reprogramming of distinct peripheral sensory neuron subtypes after axonal injury. Neuron. 2020;108(1):128–144. doi: 10.1016/j.neuron.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandran V, et al. A systems-level analysis of the peripheral nerve intrinsic axonal growth program. Neuron. 2016;89(5):956–970. doi: 10.1016/j.neuron.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou FQ, et al. Neurotrophins support regenerative axon assembly over CSPGs by an ECM-integrin-independent mechanism. J Cell Sci. 2006;119(pt 13):2787–2796. doi: 10.1242/jcs.03016. [DOI] [PubMed] [Google Scholar]
- 15.Poplawski GHD, et al. Injured adult neurons regress to an embryonic transcriptional growth state. Nature. 2020;581(7806):77–82. doi: 10.1038/s41586-020-2200-5. [DOI] [PubMed] [Google Scholar]
- 16.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasanthakumar A, et al. A non-canonical function of Ezh2 preserves immune homeostasis. EMBO Rep. 2017;18(4):619–631. doi: 10.15252/embr.201643237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu KX, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338(6113):1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee ST, et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, et al. Polycomb- and methylation-independent roles of EZH2 as a transcription activator. Cell Rep. 2018;25(10):2808–2820. doi: 10.1016/j.celrep.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi B, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27(14):5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laumet G, et al. G9a is essential for epigenetic silencing of K(+) channel genes in acute-to-chronic pain transition. Nat Neurosci. 2015;18(12):1746–1755. doi: 10.1038/nn.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou FQ, et al. NGF-induced axon growth is mediated by localized inactivation of GSK-3beta and functions of the microtubule plus end binding protein APC. Neuron. 2004;42(6):897–912. doi: 10.1016/j.neuron.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X, et al. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci U S A. 2010;107(20):9424–9429. doi: 10.1073/pnas.0914725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saijilafu, et al. Genetic dissection of axon regeneration via in vivo electroporation of adult mouse sensory neurons. Nat Commun. 2011;2:543. doi: 10.1038/ncomms1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang XW, et al. Lin28 signaling supports mammalian PNS and CNS axon regeneration. Cell Rep. 2018;24(10):2540–2552. doi: 10.1016/j.celrep.2018.07.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran NM, et al. Single-cell profiles of retinal ganglion cells differing in resilience to injury reveal neuroprotective genes. Neuron. 2019;104(6):1039–1055. doi: 10.1016/j.neuron.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang XW, et al. Knocking out non-muscle myosin II in retinal ganglion cells promotes long-distance optic nerve regeneration. Cell Rep. 2020;31(3):107537. doi: 10.1016/j.celrep.2020.107537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X, et al. Preservation of vision after CaMKII-mediated protection of retinal ganglion cells. Cell. 2021;184(16):4299–4314. doi: 10.1016/j.cell.2021.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42(8):722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 31.Lavarone E, et al. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat Commun. 2019;10(1):1679. doi: 10.1038/s41467-019-09624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsujino H, et al. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: A novel neuronal marker of nerve injury. Mol Cell Neurosci. 2000;15(2):170–182. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins R, Hunt SP. Long-term increase in the levels of c-jun mRNA and jun protein-like immunoreactivity in motor and sensory neurons following axon damage. Neurosci Lett. 1991;129(1):107–110. doi: 10.1016/0304-3940(91)90731-8. [DOI] [PubMed] [Google Scholar]
- 34.Bonilla IE, et al. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci. 2002;22(4):1303–1315. doi: 10.1523/JNEUROSCI.22-04-01303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Befort K, et al. Selective up-regulation of the growth arrest DNA damage-inducible gene Gadd45 alpha in sensory and motor neurons after peripheral nerve injury. Eur J Neurosci. 2003;18(4):911–922. doi: 10.1046/j.1460-9568.2003.02827.x. [DOI] [PubMed] [Google Scholar]
- 36.Tanabe K, et al. Fibroblast growth factor-inducible-14 is induced in axotomized neurons and promotes neurite outgrowth. J Neurosci. 2003;23(29):9675–9686. doi: 10.1523/JNEUROSCI.23-29-09675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakisaka S, et al. Increased neuropeptide Y (NPY)-like immunoreactivity in rat sensory neurons following peripheral axotomy. Neurosci Lett. 1991;124(2):200–203. doi: 10.1016/0304-3940(91)90093-9. [DOI] [PubMed] [Google Scholar]
- 38.Bray ER, et al. Thrombospondin-1 mediates axon regeneration in retinal ganglion cells. Neuron. 2019;103(4):642–657. doi: 10.1016/j.neuron.2019.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belin S, et al. Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics. Neuron. 2015;86(4):1000–1014. doi: 10.1016/j.neuron.2015.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan X, et al. Subtype-specific regeneration of retinal ganglion cells following axotomy: effects of osteopontin and mTOR signaling. Neuron. 2015;85(6):1244–1256. doi: 10.1016/j.neuron.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian C, Zhou FQ. Updates and challenges of axon regeneration in the mammalian central nervous system. J Mol Cell Biol. 2020;12(10):798–806. doi: 10.1093/jmcb/mjaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang SG, et al. Strategies to promote long-distance optic nerve regeneration. Front Cell Neurosci. 2020;14:119. doi: 10.3389/fncel.2020.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaya-Okur HS, et al. CUTTag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10(1):1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hilton BJ, et al. An active vesicle priming machinery suppresses axon regeneration upon adult CNS injury. Neuron. 2022;110(1):51–69. doi: 10.1016/j.neuron.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li F, et al. Glial metabolic rewiring promotes axon regeneration and functional recovery in the central nervous system. Cell Metab. 2020;32(5):767–785. doi: 10.1016/j.cmet.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romaus-Sanjurjo D, et al. GABA promotes survival and axonal regeneration in identifiable descending neurons after spinal cord injury in larval lampreys. Cell Death Dis. 2018;9(6):663. doi: 10.1038/s41419-018-0704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ressler SJ, et al. WFDC1 is a key modulator of inflammatory and wound repair responses. Am J Pathol. 2014;184(11):2951–2964. doi: 10.1016/j.ajpath.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu S, et al. Molecular structure, gene expression and functional role of WFDC1 in angiogenesis and cancer. Cell Biochem Funct. 2021;39(5):588–595. doi: 10.1002/cbf.3624. [DOI] [PubMed] [Google Scholar]
- 49.Dennis DJ, et al. bHLH transcription factors in neural development, disease, and reprogramming. Brain Res. 2019;1705:48–65. doi: 10.1016/j.brainres.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, et al. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell. 2018;23(5):758–771. doi: 10.1016/j.stem.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lisi V, et al. Enhanced neuronal regeneration in the CAST/Ei mouse strain is linked to expression of differentiation markers after Injury. Cell Rep. 2017;20(5):1136–1147. doi: 10.1016/j.celrep.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams RR, et al. MASH1/Ascl1a leads to GAP43 expression and axon regeneration in the adult CNS. PLoS One. 2015;10(3):e0118918. doi: 10.1371/journal.pone.0118918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Y, et al. Reprogramming to recover youthful epigenetic information and restore vision. Nature. 2020;588(7836):124–129. doi: 10.1038/s41586-020-2975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Onder TT, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483(7391):598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding XL, et al. The polycomb protein Ezh2 impacts on induced pluripotent stem cell generation. Stem Cells Dev. 2014;23(9):931–940. doi: 10.1089/scd.2013.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mozhui K, Pandey AK. Conserved effect of aging on DNA methylation and association with EZH2 polycomb protein in mice and humans. Mech Ageing Dev. 2017;162:27–37. doi: 10.1016/j.mad.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sekine Y, et al. Functional genome-wide screen identifies pathways restricting central nervous system axonal regeneration. Cell Rep. 2018;23(2):415–428. doi: 10.1016/j.celrep.2018.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Norsworthy MW, et al. Sox11 Expression promotes regeneration of some retinal ganglion cell types but kills others. Neuron. 2017;94(6):1112–1120. doi: 10.1016/j.neuron.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z, et al. KLF6 and STAT3 co-occupy regulatory DNA and functionally synergize to promote axon growth in CNS neurons. Sci Rep. 2018;8(1):12565. doi: 10.1038/s41598-018-31101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42(3):425–428. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaytseva O, et al. MYC in brain development and cancer. Int J Mol Sci. 2020;21(20):7742. doi: 10.3390/ijms21207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costales J, Kolevzon A. The therapeutic potential of insulin-like growth factor-1 in central nervous system disorders. Neurosci Biobehav Rev. 2016;63:207–222. doi: 10.1016/j.neubiorev.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horvath S, et al. DNA methylation clocks tick in naked mole rats but queens age more slowly than nonbreeders. Nat Aging. 2022;2(1):46–59. doi: 10.1038/s43587-021-00152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang KC, et al. Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature. 2002;417(6892):941–944. doi: 10.1038/nature00867. [DOI] [PubMed] [Google Scholar]
- 65.Atwal JK, et al. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science. 2008;322(5903):967–970. doi: 10.1126/science.1161151. [DOI] [PubMed] [Google Scholar]
- 66.Fournier AE, et al. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature. 2001;409(6818):341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- 67.Mi S, et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7(3):221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- 68.Dickendesher TL, et al. NgR1 and NgR3 are receptors for chondroitin sulfate proteoglycans. Nat Neurosci. 2012;15(5):703–712. doi: 10.1038/nn.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ji B, et al. LINGO-1 antagonist promotes functional recovery and axonal sprouting after spinal cord injury. Mol Cell Neurosci. 2006;33(3):311–320. doi: 10.1016/j.mcn.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 70.Fu QL, et al. Blocking LINGO-1 function promotes retinal ganglion cell survival following ocular hypertension and optic nerve transection. Invest Ophthalmol Vis Sci. 2008;49(3):975–985. doi: 10.1167/iovs.07-1199. [DOI] [PubMed] [Google Scholar]
- 71.Inoue H, et al. Inhibition of the leucine-rich repeat protein LINGO-1 enhances survival, structure, and function of dopaminergic neurons in Parkinson’s disease models. Proc Natl Acad Sci U S A. 2007;104(36):14430–14435. doi: 10.1073/pnas.0700901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guillemain A, et al. LINGO family receptors are differentially expressed in the mouse brain and form native multimeric complexes. FASEB J. 2020;34(10):13641–13653. doi: 10.1096/fj.202000826R. [DOI] [PubMed] [Google Scholar]
- 73.Becker CG, et al. Tenascin-R as a repellent guidance molecule for developing optic axons in zebrafish. J Neurosci. 2003;23(15):6232–6237. doi: 10.1523/JNEUROSCI.23-15-06232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Becker T, et al. Tenascin-R inhibits regrowth of optic fibers in vitro and persists in the optic nerve of mice after injury. Glia. 2000;29(4):330–346. doi: 10.1002/(SICI)1098-1136(20000215)29:4330::AID-GLIA43.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 75.Taylor H, et al. Ephs and ephrins. Curr Biol. 2017;27(3):R90–R95. doi: 10.1016/j.cub.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 76.Giger RJ, et al. Guidance molecules in axon regeneration. Cold Spring Harb Perspect Biol. 2010;2(7):a001867. doi: 10.1101/cshperspect.a001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yi Y, et al. A PRC2-independent function for EZH2 in regulating rRNA 2’-O methylation and IRES-dependent translation. Nat Cell Biol. 2021;23(4):341–354. doi: 10.1038/s41556-021-00653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raivich G, et al. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43(1):57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 79.Cui C, et al. Adequate expression of neuropeptide Y is essential for the recovery of zebrafish motor function following spinal cord injury. Exp Neurol. 2021;345:113831. doi: 10.1016/j.expneurol.2021.113831. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y, et al. Elevating growth factor responsiveness and axon regeneration by modulating presynaptic inputs. Neuron. 2019;103(1):39–51. doi: 10.1016/j.neuron.2019.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed sequencing data are available in the Gene Expression Omnibus (GSE247320). Values for all data points in graphs are reported in the Supporting Data Values file.