Abstract
Goat production is an important source of livelihood and food. Goats may serve as reservoir of surra affecting livestock production. Here, forty-two free-roaming goats from Cavite, Philippines were screened using two primer sets, Trypanosoma brucei minisatellite chromosome for initial detection and the internal transcribed spacer 1 (ITS-1) to determine phylogeny. Initial PCR detection showed that 19/42 (45%) goats were positive, much higher than the rate previously reported in goats from Cebu (34%). The infectivity rate was higher in male (56%) than in female (42%) and the rate was higher in young ≤1 year old (100%) than in adult >1 year old (43%). Phylogenetic analysis of the ITS-1 sequences between T. evansi goat samples and other isolates indicate potential interspecies transmission.
Keywords: Cavite, goat, internal transcribed spacer-1, PCR, Trypanosoma evansi
Surra, caused by Trypanosoma evansi is endemic in Central and South America, Northern Africa and Southeast Asia including the Philippines [17]. The infection causes anemia, lethargy and wasting that result in decreased production and impair performance in the animal which can lead to severe economic losses. This hemoflagellate parasite can infect a wide range of animal species including horses, cattle and goats [3, 9, 14]. In the Philippines, goat is considered to be an important livestock animal, valued at $210.7 million nationally and contributes to about 3.4% of the total livestock industry of the country. While annual economic losses attributed to trypanosomosis amounts to $770,000 [13].
Surra is mainly transmitted by tabanid flies. These flies are intermittent feeders and can contribute to the efficient transmission of surra across different animal species [2, 14]. Although goats are generally considered trypanotolerant, their role and function as reservoir of infection has not been clearly substantiated especially in the Philippines [4, 5, 8].
Blood samples from 42 free-roaming goats were collected from Naic, Cavite, Philippines. Briefly, 2 mL of blood was collected via venipuncture of the jugular vein. The blood was placed in a 3 mL tube containing ethylenediaminetetraacetic acid (EDTA) as the anticoagulant (Surgitech, Quezon city, Philippines). Sample collection was conducted in accordance with Philippines’ law on animal welfare and by virtue of permit No. 2019-001 issued by the Institutional Animal Care and Use Committee (IACUC) of Cavite State University. Total genomic DNA from the samples were extracted using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) following manufacturers protocol. All DNA samples were stored in −30°C freezer prior to PCR.
Two rounds of PCR were done in this study, The first PCR amplified the Trypanosoma brucei minisatellite chromosome repeat sequence a ~164 bp repeat of minisatellite chromosome and the internal transcribed spacer 1 (ITS-1) region approximately ~480 bp in size. These primers have previously been used successfully for the molecular detection and characterization of T. evansi [5, 11, 16, 19]. The primers used for the first PCR (TBR1 and 2) were as follows; forward (5′-CGA ATG AAT ATT AAA CAA TGC GCA G-3′) and reverse (5′-AGA ACC ATT TAT TAG CTT TGT TGC-3′), respectively [10]. The optimized PCR conditions used for the amplification of this gene was done as previously described [18]. Positive samples from the initial PCR were then subjected to the second PCR for amplifying ITS-1 sequence. The primers used were as follows; forward (5′-CCG GAA GTT CAC CGA TAT TG-3′) and reverse (5′-TTG CTG CGT TCT TCA ACG AA-3′), respectively [12].
The second PCR amplifying the ITS-1 region were cloned into TOPO vector (Invitrogen, Carlsbad, CA, USA) and sequenced using BigDyeTM Terminator V3.1 Sequencing Kit (Thermo Fisher Scientific, Tokyo, Japan) with ABI Prism 3130 sequencing machine as previously described [8]. The M13 forward (5′-CTG GCC GTC GTT TTA C-3′) and reverse (5′-CAG GAA ACA GCT ATG AC-3′) primers were used to assemble the forward and reverse sequences of the insert. The ITS-1 sequence showing the highest identity with the sequence obtained from T. evansi in goats was retrieved from National Center for Biotechnology Information (NCBI) following a Basic Local Alignment Search Tool (BLAST). In addition, identities between ITS-1 sequences from T. evansi in goat samples were determined by generating an identity matrix using Genetyx software (Genetyx Corp., Tokyo, Japan). A phylogenetic tree was assembled using MEGA X software [7]. Statistical analysis using Fisher’s exact test to compare the differences between the infection rate found in males and females were done using GraphPad QuickCalcs (Dotmatics, San Diego, CA, USA).
This study, showed that 19 out of the 42 (45%) goats were PCR-positive in the first PCR for TBR1 and TBR2 sequence as shown in Table 1. This rate was much higher than the prevalence (34%) reported in a recent study for goats in Cebu, Philippines [5]. The result indicates high prevalence of T. evansi infection in goats in Cavite since almost half of the samples were PCR positive. Therefore, it is suggestive that increased infectivity rate in goats, that are generally considered trypanotolerant [11], may perpetuate the parasite infection. Action toward control of the infection in goats must be taken in order to prevent cross infection between goats and other animals [6]. We likewise observed a higher detection rate in males at 56% than in females 42% although the difference is not statistically significant. It may be attributed to the small sample size used in this study. The infection rate between goats ≤1 year old was higher as compared to goats >1 year old (Table 1). Similar to male goats, the sample size of young goats is few such that it may not be able to capture the actual prevalence rate in the population [15].
Table 1. Prevalence of Trypanosoma evansi in goats in Cavite, Philippines based on PCR with Trypanosoma brucei minisatellite chromosome.
| Variable | Total (n) |
Detection Rate (%) |
Positives (n) |
P-value | |
|---|---|---|---|---|---|
| Sex | |||||
| Male | 9 | 56 | 5 | 0.71 | |
| Female | 33 | 42 | 14 | ||
| Age group | |||||
| ≤1 year | 2 | 100 | 2 | 0.20 | |
| >1 year | 40 | 43 | 17 | ||
In addition, phylogenetic analysis revealed that the ITS-1 sequences were highly identical to the sequences from the parasite originating from a wide range of hosts including camel, water buffalo and horses as seen in Fig. 1. This further suggests that a potential active transmission between domestic animals is a common occurrence contributing to the genetic diversity of the parasite [1, 4, 8, 17, 18]. Further, complete ITS-1 sequences from 6 selected samples namely: S2, S6, S7, S9, S10 and S14 were analyzed from the total of 14 positive samples. The sequences obtained in this study were closely related to those previously described from different geographic locations in the Philippines [5, 19]. However, in contrast to the previous findings [5, 19], we found in this study that the ITS-1 sequences of the parasite obtained from goat samples were highly variable as seen in Fig. 1. It is possible that several genotypes may exist in a single host as these animals may be infected several times throughout their lifetime [4]. The sequence obtained from goat S6 and goat S2 samples showed the highest identity of 99.8% while those obtained from goat S14 and goat S9 samples showed the lowest identity of 33.8%. These results suggested a diversity in ITS-1 sequence of T. evansi in the goat population examined in this study. However, it should be noted that the ITS-1 sequence is not related to parasite virulence.
Fig. 1.
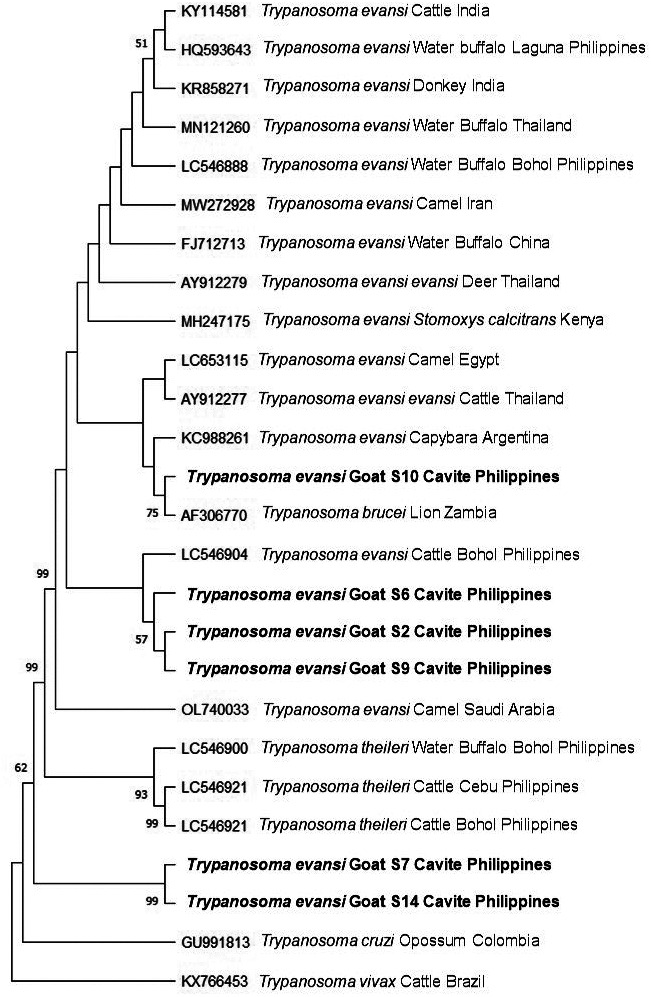
Phylogenetic analysis of Trypanosoma evansi based on the internal transcribed spacer-1 sequences obtained from goat samples in this study indicate similarities with parasite isolated from a wide range of animal hosts and in different geographic locations. This further supports potential interspecies transmission of surra. The maximum likelihood tree was constructed using the Tamura-3 model. The phylogeny test was run using the bootstrap analysis with 1,000 iterations. The sequences obtained from the current study are shown in bold. T. vivax was designated the outgroup.
Considering the high prevalence rate of surra observed in goats in this study, and its proximity to equine, cattle and buffalo farms, it is highly likely that the transmission of this disease may be perpetuated by goats potentially serving as reservoir host as previously observed [4]. As the goat industry contributes greatly to the economy and food security of the Philippines, further studies are needed to determine the extent and impact of surra on goats [4, 14]. Likewise, additional studies on the infection dynamics between goats enrolling more samples and other domestic animals should be done in order to clarify parasite transmission. Therefore, careful consideration, revisit and redesign of control measures against surra in the Philippines incorporating goats as important livestock animals may help in the control of surra.
CONFLICT OF INTEREST
The authors declare no conflicts of interest in this study.
Acknowledgments
We thank Ms. Daisy Kaye de Asis and the students from the College of Veterinary Medicine and Biomedical Sciences of Cavite State University for their assistance in collecting blood samples from goats. To all the farmers of Naic, Cavite who allowed us to collect blood samples from their animals and most especially to the Office of the Provincial Veterinarian of Cavite headed by Dr. May M. Magno. To Dr. Gladys L. Credo for supporting us in the field sampling. This study was supported in part by the Cavite State University Research Grant and by the National Research Center for Protozoan Diseases–Obihiro University of Agriculture and Veterinary Medicine. We also thank the Japan Society for the Promotion of Science (P23084).
REFERENCES
- 1.Amer S, Ryu O, Tada C, Fukuda Y, Inoue N, Nakai Y. 2011. Molecular identification and phylogenetic analysis of Trypanosoma evansi from dromedary camels (Camelus dromedarius) in Egypt, a pilot study. Acta Trop 117: 39–46. doi: 10.1016/j.actatropica.2010.09.010 [DOI] [PubMed] [Google Scholar]
- 2.Baldacchino F, Desquesnes M, Mihok S, Foil LD, Duvallet G, Jittapalapong S. 2014. Tabanids: neglected subjects of research, but important vectors of disease agents! Infect Genet Evol 28: 596–615. doi: 10.1016/j.meegid.2014.03.029 [DOI] [PubMed] [Google Scholar]
- 3.Dargantes AP, Mercado RT, Dobson RJ, Reid SA. 2009. Estimating the impact of Trypanosoma evansi infection (surra) on buffalo population dynamics in southern Philippines using data from cross-sectional surveys. Int J Parasitol 39: 1109–1114. doi: 10.1016/j.ijpara.2009.02.012 [DOI] [PubMed] [Google Scholar]
- 4.Desquesnes M, Dargantes A, Lai DH, Lun ZR, Holzmuller P, Jittapalapong S. 2013. Trypanosoma evansi and surra: a review and perspectives on transmission, epidemiology and control, impact, and zoonotic aspects. BioMed Res Int 2013: 321237. doi: 10.1155/2013/321237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elata A, Galon EM, Moumouni PFA, Ybanez RHD, Mossaad E, Salces CB, Bajenting GP, Ybanez AP, Xuan X, Inoue N, Suganuma K. 2020. First molecular detection and identification of Trypanosoma evansi in goats from Cebu, Philippines using a PCR-based assay. Vet Parasitol Reg Stud Rep 21: 100414. [DOI] [PubMed] [Google Scholar]
- 6.Gutierrez C, Corbera JA, Morales M, Büscher P. 2006. Trypanosomosis in goats: current status. Ann N Y Acad Sci 1081: 300–310. doi: 10.1196/annals.1373.040 [DOI] [PubMed] [Google Scholar]
- 7.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macalanda AMC, Angeles JMM, Moendeg KJ, Dang AT, Higuchi L, Inoue N, Xuan X, Kirinoki M, Chigusa Y, Leonardo LR, Villacorte EA, Rivera PT, Goto Y, Kawazu SI. 2018. Evaluation of Schistosoma japonicum thioredoxin peroxidase-1 as a potential circulating antigen target for the diagnosis of Asian schistosomiasis. J Vet Med Sci 80: 156–163. doi: 10.1292/jvms.17-0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mekata H, Konnai S, Witola WH, Inoue N, Onuma M, Ohashi K. 2009. Molecular detection of trypanosomes in cattle in South America and genetic diversity of Trypanosoma evansi based on expression-site-associated gene 6. Infect Genet Evol 9: 1301–1305. doi: 10.1016/j.meegid.2009.07.009 [DOI] [PubMed] [Google Scholar]
- 10.Moser DR, Cook GA, Ochs DE, Bailey CP, McKane MR, Donelson JE. 1989. Detection of Trypanosoma congolense and Trypanosoma brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitology 99: 57–66. doi: 10.1017/S0031182000061023 [DOI] [PubMed] [Google Scholar]
- 11.Musinguzi SP, Suganuma K, Asada M, Laohasinnarong D, Sivakumar T, Yokoyama N, Namangala B, Sugimoto C, Suzuki Y, Xuan X, Inoue N. 2017. A PCR-based survey of animal African trypanosomosis and selected piroplasm parasites of cattle and goats in Zambia. J Vet Med Sci 78: 1819–1824. doi: 10.1292/jvms.16-0240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC, Dávila AM. 2005. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res 95: 186–192. doi: 10.1007/s00436-004-1267-5 [DOI] [PubMed] [Google Scholar]
- 13.Philippine Statistics Authority. 2022. Philippine Statistics Authority Annual Situation Report. https://psa.gov.ph/sites/default/files/1_Goat%20Annual%20Situation%20Report_ONSedits_v2_ONS-signed.pdf [accessed on August 19, 2022].
- 14.Reid SA. 2002. Trypanosoma evansi control and containment in Australasia. Trends Parasitol 18: 219–224. doi: 10.1016/S1471-4922(02)02250-X [DOI] [PubMed] [Google Scholar]
- 15.Reynolds L, Adediran S. 1994. Composition of village goat herds in southwest Nigeria. Small Rumin Res 13: 49–53. doi: 10.1016/0921-4488(94)90030-2 [DOI] [Google Scholar]
- 16.Rjeibi MR, Ben Hamida T, Dalgatova Z, Mahjoub T, Rejeb A, Dridi W, Gharbi M. 2015. First report of surra (Trypanosoma evansi infection) in a Tunisian dog. Parasite 22: 3. doi: 10.1051/parasite/2015004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sánchez E, Perrone T, Recchimuzzi G, Cardozo I, Biteau N, Aso PM, Mijares A, Baltz T, Berthier D, Balzano-Nogueira L, Gonzatti MI. 2015. Molecular characterization and classification of Trypanosoma spp. Venezuelan isolates based on microsatellite markers and kinetoplast maxicircle genes. Parasit Vectors 8: 536. doi: 10.1186/s13071-015-1129-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suganuma K, Acosta TJ, Valinotti MFR, Sanchez AR, Mossaad E, Elata A, Inoue N. 2022. First molecular survey of animal trypanosomes in Paraguayan horses. Vet Parasitol Reg Stud Rep 27: 100664. [DOI] [PubMed] [Google Scholar]
- 19.Villareal MV, Mingala CN, Rivera WL. 2013. Molecular characterization of Trypanosoma evansi isolates from water buffaloes (Bubalus bubalis) in the Philippines. Acta Parasitol 58: 6–12. doi: 10.2478/s11686-013-0110-5 [DOI] [PubMed] [Google Scholar]


