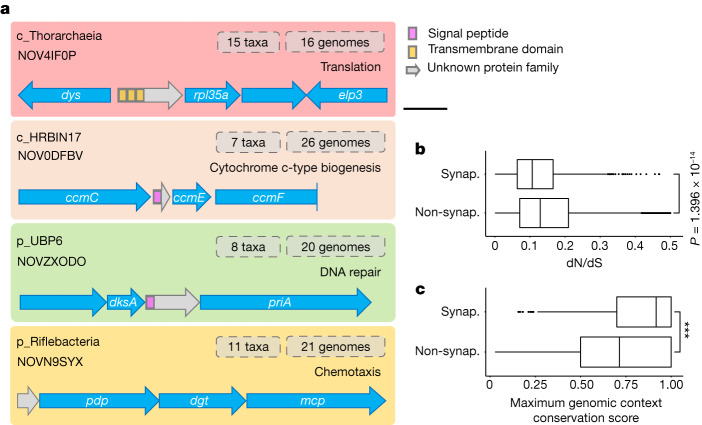
Fig. 4. FESNov synapomorphic gene families found at high-level taxonomic rank.
a, Schematic overview of four phylum- and class-level synapomorphic FESNov gene families located in conserved gene clusters involved in relevant cellular processes. Gene names correspond to: dys (K00809), deoxyhypusine synthase; rpl35a (K02917), large subunit ribosomal protein L35Ae; elp3 (K07739), elongation protein 3 homolog; ccmC (K02195), haem exporter protein C; ccmE (K02197), cytochrome c-type biogenesis protein; ccmF (K02198), cytochrome c-type biogenesis protein; dksA (K06204), DnaK suppressor protein; priA (K04066), primosomal protein N’; pdp (K00756), pyrimidine-nucleoside phosphorylase; dgt (K01129), dGTPase; mcp (K03406), methyl-accepting chemotaxis protein. Scale bar, 500 base pairs. b, Comparison of dN/dS values between synapomorphic (synap., n = 1,034) and non-synapomorphic (non-synap., n = 403,051) FESNov gene families. c, Comparison of genomic context conservation between synap. (n = 1,034) and non-synap. (n = 402,987) FESNov gene families. b,c, Data are represented as boxplots in which the middle line is the median, the lower and upper hinges correspond to the first and third quartiles, the upper whisker extends from the hinge to the highest value no further than 1.5 × interquartile range (IQR) from the hinge and the lower whisker extends from the hinge to the lowest value no further than 1.5 × IQR of the hinge; data beyond the end of the whiskers are outlying points that are plotted individually. b,c, P values by two-sided Wilcoxon test are indicated (***P < 1 × 10−15).