Abstract
Transcriptional activation of target genes is essential for TP53-mediated tumour suppression, though the roles of the diverse TP53-activated target genes in tumour suppression remains poorly understood. Knockdown of ZMAT3, an RNA-binding zinc-finger protein involved in regulating alternative splicing, in haematopoietic cells by shRNA caused leukaemia only with the concomitant absence of the PUMA and p21, the critical effectors of TRP53-mediated apoptosis and cell cycle arrest respectively. We were interested to further investigate the role of ZMAT3 in tumour suppression beyond the haematopoietic system. Therefore, we generated Zmat3 knockout and compound gene knockout mice, lacking Zmat3 and p21, Zmat3 and Puma or all three genes. Puma−/−p21−/−Zmat3−/− triple knockout mice developed tumours at a significantly higher frequency compared to wild-type, Puma−/−Zmat3−/− or p21−/−Zmat3−/−deficient mice. Interestingly, we observed that the triple knockout and Puma−/−Zmat3−/− double deficient animals succumbed to lymphoma, while p21−/−Zmat3−/− animals developed mainly solid cancers. This analysis suggests that in addition to ZMAT3 loss, additional TRP53-regulated processes must be disabled simultaneously for TRP53-mediated tumour suppression to fail. Our findings reveal that the absence of different TRP53 regulated tumour suppressive processes changes the tumour spectrum, indicating that different TRP53 tumour suppressive pathways are more critical in different tissues.
Subject terms: Cancer models, Cancer genetics, Cancer
Introduction
The tumour suppressor TP53 (mouse TRP53, often referred to as p53) is a frequently mutated gene in human cancer [1–3]. Although TP53-mediated transcriptional regulation of diverse cellular responses is known to be critical for its ability to prevent the development of cancer, the role of many TP53 target genes in tumour suppression remains unclear [2, 4]. Recent studies in mice sought to identify the mechanisms that are critical for TRP53-mediated tumour suppression, using sensitised shRNA gene knock-down and CRISPR/Cas9 knockout screens in cells in culture and even in vivo [5, 6]. Amongst the hits identified in these screens, the RNA binding protein ZMAT3 [7, 8] was found to be a potent tumour suppressor. ZMAT3 (also known as WIG-1) is an RNA-binding zinc-finger protein that is involved in regulating alternative splicing and as such is expressed in a broad range of tissues [6]. ZMAT3 was reported to control splicing of mRNAs, including those encoding the negative regulators of TRP53 function, MDM2 and MDM4, several other splicing factors (e.g., HNRNPDL, DHX9) and the cell adhesion and stem cell marker CD44 [6, 9]. ZMAT3 has been described to be under the direct transcriptional control of TRP53 in various cell types [5, 6, 8, 10], indicating its broad role for tumour suppression in diverse tissues. The absence of TRP53 causes a reduction in Zmat3 expression in a range of human carcinomas (e.g., breast, lung) and high levels of Zmat3 expression in malignant cells predict increased patient survival in certain cancers [6]. Moreover, an analysis of the Project Achilles CRISPR/Cas9 gene knockout screening data in human cancer cells revealed that ZMAT3 inactivation enhances proliferation of malignant cells with wild-type (wt) TP53. This further supports a role for ZMAT3 in tumour suppression acting downstream of TP53 [6]. Of note, CRISPR/Cas9-mediated loss of ZMAT3 enhanced tumorigenesis in autochthonous mouse models of lung adenocarcinoma and hepatocellular carcinoma [6]. However, unlike loss of TRP53, the absence of ZMAT3 did not have marked impact on the rate of tumour development or severity of malignant disease in the context of murine c-MYC-driven lymphomagenesis or mutant KrasG12D-driven lung adenocarcinoma development [11]. This indicates that the relative importance of ZMAT3 in TRP53-mediated tumour suppression may vary depending on cell type and/or the context of other oncogenic drivers present in the emerging neoplastic cell population. In vivo studies have shown that shRNA-mediated silencing of ZMAT3 in haematopoietic stem/progenitor cells (HSPCs) caused development of leukaemia/lymphoma in transplant recipient mice only when PUMA and p21, the critical effectors of TRP53-mediated apoptosis [12, 13] and cell cycle arrest [14] respectively, were also absent [5]. A limitation of these studies was that ZMAT3, PUMA and p21 were removed only in the haematopoietic compartment. Therefore, it remains unclear what impact the absence of ZMAT3 either on its own, or in conjunction with the loss of TRP53 induced apoptosis and cell cycle arrest/cell senescence might have in other tissues. To address this question, we generated mice lacking ZMAT3, PUMA and p21. We found that these triple knockout (TKO) mice spontaneously developed malignancy at a considerably higher frequency compared to wt controls as well as Puma−/−Zmat3−/− and p21−/−Zmat3−/− double knockout (DKO) mice. Most of the tumours from the TKO mice were of haematopoietic cell origin. These findings demonstrate that this combination of defects in TRP53-activated cellular responses, even if present in all cells, predominantly causes leukaemia/lymphoma. They also reaffirm that defects in multiple TRP53-activated cellular responses must collude to drive tumorigenesis.
Results
Adult pre-neoplastic Puma−/−p21−/−Zmat3−/− mice have only minor abnormalities in their haematopoietic cell populations
To investigate the impact of combined loss of ZMAT3, PUMA and p21 in TRP53-mediated tumour suppression we crossed Zmat3−/− with Puma−/−p21−/− mice to obtain Puma−/−Zmat3−/−, p21−/−Zmat3−/− and Puma−/−p21−/−Zmat3−/− animals (Fig. 1A). Puma−/−Zmat3−/−, p21−/−Zmat3−/− and Puma−/−p21−/−Zmat3−/− offspring were born at the expected Mendelian ratios of inheritance observed for heterozygous crosses of each allele within the colony (Table S1), as well as from Puma+/-Zmat3+/- and p21+/-Zmat3+/- di-hybrid inter-crosses (Table S2). The DKO and TKO mutant mice reached adulthood without any notable defects.
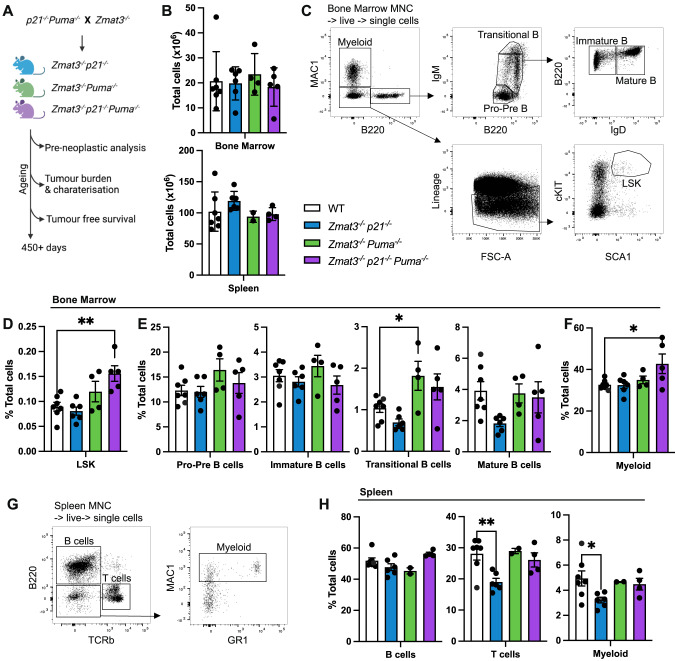
Fig. 1. Lymphoid organ analysis of young adult mice of the indicated genotypes.
A Schematic for tumour development study. Mouse cohorts were analysed for pre-neoplastic phenotypes and monitored for tumour-free survival. B–H Single-cell suspensions were prepared from spleen, thymus, peripheral blood and bone marrow of Puma−/−p21−/−Zmat3−/− (N = 5), Puma−/−Zmat3−/− (N = 2–4), p21−/−Zmat3−/− (N = 6), and wt (N = 7) mice and the indicated haematopoietic cell subsets were examined by immunostaining and FACS analysis. B Total cell counts for bone marrow (1 femur, top) and whole spleen (bottom). C Representative FACS plots from wt mice that indicate gating strategy to identify the cell populations of interest in the bone marrow: myeloid (MAC1+), LSK (Lineage-SCA1+cKIT+) haematopoietic stem/progenitor cells (HSPCs) and B lymphoid cells; pro-pre (B220loIgM-), immature (B220+IgMlo IgD-), transitional (B220+IgMhi) and mature (B220+IgMmedIgD+) B lymphoid cells. The lineage marker antibody cocktail included antibodies against NK1.1, TER119, Ly6G, F4/80, CD2, CD4 and CD8. D–F Percentages of the indicated cell subsets in the bone marrow from mice of the indicated genotypes. G Representative FACS plots of wt mice indicate gating strategy to identify cell populations in the spleen: B cells (B220+), T cells (TCRβ+) and myeloid cells (MAC1+B220-TCRβ-). H Percentages of the indicated cell subsets in the spleen of mice of the indicated genotypes. Data represent mean ± SEM. Statistical significance was calculated by one-way ANOVA *p < 0.05. N number of mice, MNC mono-nuclear cells as determined by forward/side scatter. For data of analysis of single knockout control mice, Puma−/− (N = 7), p21−/−(N = 8), Zmat3−/− (N = 6), refer to Fig. S1.
Previous reports have demonstrated important individual and overlapping roles of ZMAT3, PUMA and p21 in lymphoid cells [5, 12, 14]. Therefore, we hypothesised that Puma−/−p21−/−Zmat3−/− TKO mice may display a marked pre-leukaemic phenotype with a pronounced expansion of certain haematopoietic cell populations. We therefore determined the overall composition of the haematopoietic system of the TKO, and both DKO mice by immunostaining and fluorescence-activated cell sorter (FACS) analysis at 8–12 weeks of age. The cellularity of the different lymphoid organs (e.g. spleen, bone marrow, thymus) in the DKO and TKO mice were comparable to those in wt controls (Figs. 1B, S1A), as were the white blood cell (WBC) counts in peripheral blood (PB) (Fig. S1B). T cell development in the thymus appeared normal with the overall distribution of the double negative (DN, CD4-CD8- progenitors), double-positive (DP, CD4+CD8+ immature) and single positive CD4+ and CD8+ (mature) thymocytes comparable between the DKO as well as TKO mice with those seen in wt controls (Fig. S1C, D). In the bone marrow, we observed a small but significant increase in the immature stem and multi-potent progenitor population (termed LSK, defined by lack of any mature lineage markers and expression of both SCA1 and cKIT) in the Puma−/−p21−/−Zmat3−/− TKO mice compared to wt controls (Fig. 1C, D). However, this did not result in any differences in B-cell development in the bone marrow, with only a small, yet significant increase in myeloid cells observed (Figs. 1E, F, S1E, F). The numbers of all mature cell types in the spleen that we tested were comparable between DKO as well as TKO mice with those seen in wt controls, with comparable frequencies of B cells (B220+), T cells (TCRβ+) and myeloid cells (MAC1+) (Figs. 1G, H, S1G–I). In addition, histological analysis of major organs revealed no lesions of significance in young adult DKO and TKO mice (Fig. S2). Overall, these findings demonstrate that the combined absence of ZMAT3, PUMA and p21 does not cause marked defects in pre-cancerous mice.
Cells from Puma−/−p21−/−Zmat3−/− mice are resistant to TRP53-mediated apoptosis triggered by DNA damage
The BH3-only protein PUMA is critical for apoptosis induced by stress stimuli that activate TRP53, such as DNA damage [12, 13, 15]. To verify that cells from TKO mice were, as expected, resistant to TRP53-mediated apoptosis, we exposed thymocytes from Puma−/−p21−/−Zmat3−/− as well as Puma−/−Zmat3−/−, p21−/−Zmat3−/−, Zmat3−/−, p21−/−, Puma−/− and wt mice to apoptosis inducing agents in vitro and their survival was then measured by flow cytometric analysis. The survival of Puma−/−p21−/−Zmat3−/− TKO thymocytes was comparable to that of DKO and wt thymocytes when either left untreated (DMSO control), treated with ionomycin or starved of serum (growth factor deprivation) (Fig. 2A–C). As expected, thymocytes from TKO and Puma−/−Zmat3−/− DKO mice were resistant to etoposide, a cytotoxic stimulus that kills these cells via a TRP53-dependent process. At 24 h, there was less than 40% survival of wt thymocytes but more than 80% survival of the TKO, Puma−/−Zmat3−/− DKO and Puma−/− thymocytes (Fig. 2D). To confirm that this survival advantage was due to failure to induce apoptosis owing to the absence of PUMA, we performed cleaved caspase-3 (CC3) staining, a marker for cells that have initiated apoptosis, after etoposide treatment, followed by flow cytometric analysis. At 24 h after treatment a drastic decrease in CC3 staining in Puma−/−p21−/−Zmat3−/− as well as Puma−/−Zmat3−/− and Puma−/− thymocytes was observed compared to wt thymocytes (Fig. 2E). These data show that cells from TKO mice are resistant to TRP53-driven apoptosis triggered by DNA damage due to the absence of PUMA, but are normally sensitive to several TRP53-independent apoptotic stimuli.
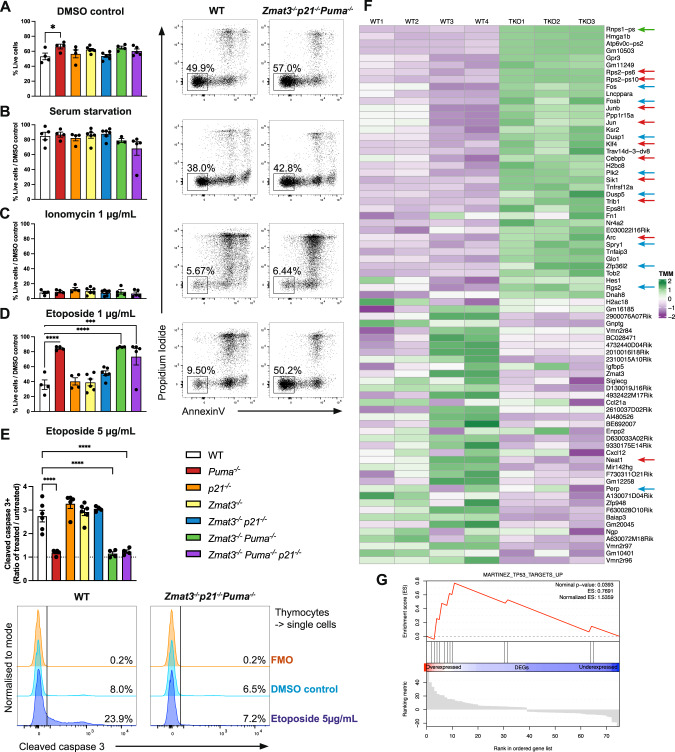
Fig. 2. Impact of combined loss of ZMAT3, PUMA and p21 on thymocyte transcriptional landscape and survival upon treatment with various apoptotic stimuli.
Thymocytes from mice of the indicated genotypes were isolated and either (A) left untreated (DMSO control), (B) treated with 1 µg/mL ionomycin, (C) exposed to serum deprivation in culture or (D) treated with 1 µg/mL etoposide. Cell viability was assessed after 24 h by Annexin-V/PI staining and FACS analysis. Data are presented as mean ± SEM of Annexin-V-PI- population (live cells) (A) and live cells relative to DMSO treated control samples (B–D). Representative FACS plots are shown to the right of each graph. E Thymocytes were treated with 5 µg/mL etoposide for 6 h, fixed, followed by intracellular staining for activated (i.e. cleaved) caspase-3 (CC3) and then subjected to FACS analysis. Data are presented as ratio of CC3+ cells in treated vs untreated (DMSO control) thymocytes with representative histograms shown below. FMO = fluorescence minus one staining control. Statistical significance was calculated by one-way ANOVA *p = 0.035, ***p = 0.0009, ****p < 0.0001. F, G RNAseq analysis of isolated thymocytes from 8–12 week old wt (N = 4) and Puma−/−p21−/−Zmat3−/− TKO (N = 3) mice. F Heat map showing all significant differentially expressed (DE) genes between thymocytes from wt and TKO mice. Each column represents data from an individual mouse. Data are colour coded by gene expression Z-scores. Arrows indicate genes that regulate TRP53 and are reported to compromise its tumour suppression function (red), known TRP53 target genes (blue) and splicing regulators (green). G Gene-set enrichment analysis (GSEA) on DE genes between thymocytes from wt and TKO mice shows significant overlap with the Mouse Gene Set: MARTINEZ_TP53_TARGETS_UP [65].
ZMAT3 impacts expression of genes involved in TRP53 mediated regulation of the haematopoietic compartment
Given the importance of ZMAT3 as an RNA binding protein we performed RNA-seq analysis on cells from young adult (8–12 weeks old) age-matched Puma−/−p21−/−Zmat3−/−, Zmat3−/− and wt control mice. Comparison of gene expression profiles of Puma−/−p21−/−Zmat3−/− and wt thymocytes revealed minor changes in the transcriptional landscape and identified 75 significantly differentially expressed (DE) genes (Figs. 2F, G, S3A, B, Table S3). GO term annotation uncovered various categories related to mechanisms that cells use to control gene expression and maintain cellular proliferation, differentiation, or cell adhesion (Fig. S3A). Notably, the most statistically DE genes in Puma−/−p21−/−Zmat3−/− cells included genes that regulate TP53 and compromise its tumour suppression function (Fig. 2G, red arrows), such as Rps2 - regulator of TRP53-MDM2 signalling [16], Klf4 [17], Trb1 [18], Neat1 [19], Cebpb [20], Sik1 [21], Jun [22], Arc [23], and some known TRP53 target genes (Fig. 2G, blue arrows), including Dusp1 [24], Perp [25], Dusp5 [26], Fos [27], Rgs2 [28], Spry1 [29], Plk2 [30] and Zfp36L2 [31]. Additionally, gene-set enrichment analysis (GSEA) comparing wt with Puma−/−p21−/−Zmat3−/− thymocytes showed a significant enrichment for TRP53 regulated genes (Fig. 2G). Thus, these data indicate that ZMAT3 may function in tumour suppression by regulating TRP53 function. To assess the contribution of PUMA and p21 to the observed transcriptomic changes, we compared gene expression profiles between Puma−/−p21−/−Zmat3−/− and Zmat3−/− thymocytes. This revealed only 9 significantly DE genes (Fig. S3C), with Rnps1 and Rps2 being the only genes overlapping between the two sets of comparisons (Fig. S3B, C). This demonstrates that the absence of PUMA and p21 have only minor impact on the transcriptome changes seen in Puma−/−p21−/−Zmat3−/− cells. Collectively, our findings support the model in which ZMAT3 regulates the expression of a range of genes involved in TRP53 regulation of various cellular processes.
Consequences of combined loss of ZMAT3 and p21 on γ-radiation induced thymic lymphoma development
Exposure of mice from a young age (starting at ~4 weeks) to repeated low-dose γ-radiation drives thymic lymphoma development through a mechanism that is suppressed by TRP53 [32, 33]. To investigate whether TRP53-mediated induction of ZMAT3 could restrain γ-radiation induced thymic lymphoma development, we examined the consequences of individual or combined loss of ZMAT3 and p21 in this model. Mice lacking PUMA alone or in combination with ZMAT3 and p21 were not included as it has been previously shown that mice defective in TRP53-induced apoptosis due to loss of PUMA are profoundly resistant to γ-radiation-induced thymic lymphoma development [33, 34]. We found that p21−/−Zmat3−/− as well as Zmat3−/− and p21−/− mice developed thymic lymphoma at a similar rate to wt mice (Fig. 3A). Immunophenotyping of the tumours from sick p21−/−Zmat3−/−, Zmat3−/− and p21−/− mice confirmed that these lymphomas were all of T lymphoid origin expressing THY1 and variable expression of CD4 and CD8 (Figs. 3B, S4A). Lymphoma burden, as determined by thymus weight, was smaller in p21−/−Zmat3−/− mice compared to those detected in Zmat3−/−, p21−/− and wt mice (Fig. 3C). However, no differences between these genotypes were seen in spleen weights or white blood cell counts (Fig. S4B, C), consistent with no difference in tumour burden. Interestingly, high levels of p19ARF which are indicative of loss of TRP53 pathway function, were apparent in 2 out of 4 wt, 2 out of 5 Zmat3−/−, 2 out of 3 p21−/− but none out of 8 p21−/−Zmat3−/− lymphomas tested by Western blotting (Fig. 3D). We therefore sequenced Trp53 exons in all available tumours and found only 1/11 p21−/−Zmat3−/− tumours to harbour a Trp53 mutation, with considerable higher frequencies seen in lymphomas from the other tested genotypes, p21−/− or Zmat3−/− (Fig. 3E, F, Table S4). These findings demonstrate that in this mouse model of lymphomagenesis tumour suppression by TRP53 does not depend on ZMAT3 and/or p21 function. However, the reduction of readily detectable defects in TRP53 pathway function in the lymphomas from the p21−/−Zmat3−/− mice indicates that the absence of these two TRP53 target genes may obviate the selection for mutations in Trp53 in this malignant disease.
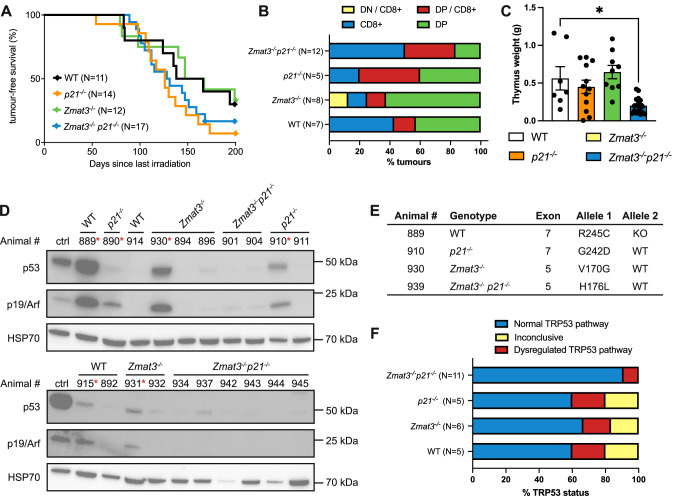
Fig. 3. The combined absence of ZMAT3 and p21 does not accelerate γ-radiation induced thymic lymphoma development.
A Kaplan-Meier curves showing percentages of tumour-free mice of the indicated genotypes after exposure to four weekly doses of γ-radiation (1.5 Gy for each dose). Differences in thymic lymphoma incidence between wt and p21−/−Zmat3−/− were not statistically significant. P value determined by log-rank (Mantel-Cox) test p = 0.5. B Immunophenotyping of γ-radiation-induced thymic lymphomas arising in mice of the indicated genotypes, as assessed by cell surface marker staining and flow cytometric analysis of tumour cells from the thymus. Data are presented as the frequency of the indicated phenotypes for each genotype. Double negative CD4-CD8- (DN) or double positive CD4+CD8+ (DP) lymphoma cells found in the thymus that show a trend towards the CD8 T cell lineage (DP/CD8+ or DN CD8+, respectively). N number of mice. C Thymus weights from sick mice of the indicated genotypes. Significant differences were observed in spleen weights between the sick p21−/−Zmat3−/− and sick wt γ-irradiated mice. p21−/−Zmat3−/− (N = 12), p21−/− (N = 7), Zmat3−/− (N = 7) and wt (N = 5). Mean ± SEM, Unpaired Students t-test **p = 0.0065. D Western blot analysis of p19ARF, TRP53 and HSP70 (loading control) in thymic lymphomas of the indicated genotypes. Red asterisk indicates samples with dysregulated TRP53 pathway. The Trp53 mutant Eµ-Myc lymphoma cell line EMRK1172 [51] was used as a control for p19ARF and mutant TRP53 protein overexpression. Protein size standards in kilodaltons (kDa) are indicated. The numbers represent the identification of individual mice in the colony. E Trp53 exons 4-11 were sequenced from tumours (N = 26, Table S4). Table summary of mutations found, listed by amino acid change, Trp53 knock out (KO) or Trp53 wild-type (WT) alleles. F Summary of the TRP53 status in γ-radiation-induced thymic lymphomas arising in mice of the indicated genotypes as assessed by Western blot analysis and/or Trp53 exon sequencing (see also Table S4). Inconclusive refers to lymphomas where TRP53 pathway dysregulation was seen by only Western blot analysis or exon sequencing but not in both assays.
Zmat3−/−Puma−/−p21−/− mice are prone to spontaneous tumour development
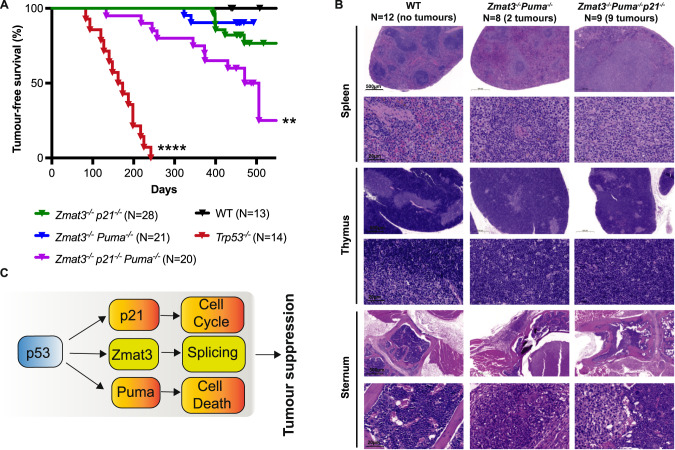
To examine the impact of combined loss of ZMAT3, PUMA and p21 in all tissues on the development of cancer, we aged Puma−/−p21−/−Zmat3−/− TKO as well as Puma−/−Zmat3−/−, p21−/−Zmat3−/− and as controls Trp53−/− as well as wt mice for 450–500 days. These animals were monitored for tumour development and major tissues were collected either at the time of sickness or at 450–500 days (study endpoint) regardless of health status. Interestingly, TKO as well as Puma−/−Zmat3−/− and p21−/−Zmat3−/− DKO mice were significantly more prone to spontaneous tumour development compared to wt controls, with TKO mice showing a cancer incidence of nearly 50% by 500 days (Fig. 4A). The majority of sick Puma−/−Zmat3−/− and Puma−/−p21−/−Zmat3−/− mice presented with lymphoma (Figs. 4B, S5, Table 1) as determined by histopathological analysis, while most sick Zmat3−/−p21−/− mice presented with solid tumours (Table 1). Consistent with previous reports [35], Trp53−/− mice all develop cancer, mostly thymic lymphomas, within 250 days of age [35] (Fig. 4A, Table 1). None of the control wt mice developed tumours during the 500 day observation period (Fig. 4A, B). Collectively, these results show that the combined absence of the three TRP53 target genes Zmat3, Puma and p21 causes a high incidence of spontaneous tumour development. It also shows that most malignancies arising in the compound mutant mice lacking PUMA are lymphomas whereas mice lacking ZMAT3 and p21 develop solid cancers.
Fig. 4. The combined absence of ZMAT3, PUMA and p21 causes spontaneous tumour development in mice.
A Kaplan-Meier curves showing tumour-free survival of Puma−/−p21−/−Zmat3−/−, Puma−/−Zmat3−/− and p21−/−Zmat3−/− mice. Data from wt and Trp53−/− mice are included as controls. Log-rank (Mantel-Cox) test P values comparing to wt mice; Puma−/−p21−/−Zmat3−/− p = 0.003**; Puma−/−p21−/− p = 0.06; Puma−/−Zmat3−/− p = 0.26; ****p < 0.0001. B Representative H&E-stained sections of spleen, bone marrow (sternum) and thymus of sick Puma−/−p21−/−Zmat3−/− (#454,615) and Puma−/−Zmat3−/− (#471,624) mice, and an age-matched healthy wt (#953) mouse for comparison. Scale bar denotes 500 µm or 20 µm, respectively. N number of mice of a particular genotype analysed. C Model: ZMAT3 loss drives spontaneous tumour development in collaboration with additional loss of PUMA and p21.
Table 1.
Tumour types observed in Zmat3, Puma, p21 compound mutant mice.
| Animal # | Sex | Age (days) | Tumour type(s) observed |
|---|---|---|---|
| Zmat3−/−;Puma−/− | |||
| 624 | M | 340 | Early B- (Spl) and T-cell (Thy) lymphoma, infiltration (Liv, Kid, SC) |
| 471 | M | 323 | B-cell (Spl) and T-cell (Thy) lymphoma, metastasis (Liv, Kid) |
| 6 M/F | 450–493 | No tumours observed | |
| 13 M/F | 450–469 | N.D. No tumours observed | |
| Zmat3−/−;p21−/− | |||
| 222 | M | 572 | Hepatocellular carcinoma |
| 276 | M | 399 | Hepatocellular carcinoma |
| 283 | F | 470 | Sarcoma |
| 285 | F | 423 | Sarcoma |
| 297 | F | 394 | Sarcoma |
| 293 | M | 400 | Subcutaneous tumour resembling amelanotic melanoma |
| 831 | M | 400 | Lymphoma (Spl), metastasis (Liv, Kid) |
| 832 | M | 549 | N.D. Enlarged spleen (0.3 g), mottled liver (0.26 g) observed |
| 2 M/F | 461–517 | No tumours observed | |
| 18 M/F | 459–517 | N.D. No tumours observed | |
| Zmat3−/−;p21−/−;Puma−/− | |||
| 615 | M | 245 | Early B-cell (Spl) and T-cell (Thy, MLN) lymphoma |
| 616 | M | 257 | Early B-cell (Spl) and T-cell (Thy) lymphoma |
| 696 | F | 133 | Early B-cell (Spl) and T-cell (Thy) lymphoma |
| 454 | F | 430 | B-cell lymphoma (Spl), myeloid tumour (SC/BM) |
| 473 | F | 345 | B-cell lymphoma (Liv) |
| 507 | M | 374 | B-cell (Spl) and T-cell (MLN) lymphoma, metastasis (L, Spl) |
| 773 | M | 218 | B-cell (Thy, LN) and T-cell (Spl) lymphoma |
| 187 | F | 506 | N.D. Splenomegaly (2.9 g) observed |
| 651 | F | 453 | Follicular hyperplasia (Spl) |
| 562 | F | 373 | Myelogenous leukaemia |
| 10 M/F | 450–590 | N.D, No tumours observed | |
| WT | |||
| 12 M/F | 438–508 | No tumours observed | |
| Trp53−/− | |||
| 14 M/F | 83–424 | N.D. 5x thymic lymphoma, 8x lymphoma, 1x solid tumour | |
Aged mice from Zmat3−/−;p21−/− Puma−/− inter-crosses were euthanised at ethical endpoint (sick, hind leg paralysis, tumour volume, anaemia). Wild-type (WT) and Trp53−/− mice housed in the same facility are included as comparative controls. Tumour types were determined by pathological analysis of H&E-stained organs and tumour sections. Organs involved noted in brackets.
Spl spleen, Thy thymus, Liv liver, Kid kidney, SC spinal cord, BM bone marrow, MLN mesenteric lymph node, L lung, LN lymph node, N.D. histology not done, noted observations made at autopsy.
Discussion
TP53 is a critical suppressor of cancer development and progression [1–3, 36]. TP53 functions as a transcription factor that can directly regulate the expression of ~500 target genes, and indirectly many more [4, 36, 37]. Direct transcriptional targets of TP53/TRP53 that are critical for the induction of apoptotic cell death, cell cycle arrest and senescence have been considered as essential effectors of TP53/TRP53-mediated tumour suppression [4]. Unexpectedly, however, genetic inactivation of these three TRP53-activated cellular processes, for example due to the combined absence of PUMA, NOXA and p21, does not render mice tumour prone [10, 38, 39]. In contrast, 100% of TRP53-deficient mice spontaneously develop tumours, on a C57BL/6 background mostly thymic T cell lymphoma, before 270 days of age [40, 41]. Genetic screens and genetically modified mouse strains have provided useful tools to examine the importance and biological functions of a range of TRP53 target genes in TRP53-mediated tumour suppression. Utilising in vivo CRISPR and shRNA screening in oncogene expressing mouse embryonic fibroblast in vitro [6] and haematopoietic stem/progenitor cells in vivo [5], respectively, identified the zinc-finger RNA-binding protein ZMAT3 as a critical factor contributing to TRP53-mediated tumour suppression.
Here we show that the combined absence of ZMAT3, PUMA and p21, the latter two critical for TRP53-mediated induction of apoptosis [12, 13] and cell cycle arrest/cell senescence [14], respectively, resulted in nearly 50% of spontaneous tumour development by 500 days of age. Given that PUMA is a key factor in TRP53-mediated apoptosis in haematopoietic cells [1, 12, 13, 15], it is perhaps not surprising that the majority of sick Puma−/−Zmat3−/− and Puma−/−p21−/−Zmat3−/− mice presented with lymphoma. Interestingly, some Zmat3−/−p21−/− sick mice developed a range of solid cancers at a relatively old age. Their incidence of such malignant disease was significantly higher compared to wt mice. This indicates that loss of apoptotic function, cell cycle arrest/cell senescence and ZMAT3-regulated alternative splicing or not yet identified functions of ZMAT3 contribute to the tumour suppressive function of TRP53 in both haematological and solid cancers, with the combination of the genes lost determining cancer type. Notably, Puma−/−Zmat3−/− and Puma−/−p21−/−Zmat3−/− mice presented with lymphoma, and Zmat3−/−p21-/ mice mainly developed solid tumours (6/8). This indicates that the loss of PUMA is particularly critical for spontaneous development of haematopoietic malignancies, consistent with the profound impact of its absence on apoptotic death of lymphoid and myeloid cells [12, 13, 15].
Examination of the lymphoid tissues from young Puma−/−Zmat3−/−, Zmat3−/−p21−/−Puma−/−and p21−/−Zmat3−/− mice revealed that they were largely normal, although we did observe a significant, albeit minor, increase in LSK HSPCs in Puma−/−p21−/−Zmat3−/− mice compared to wt controls. This is consistent with earlier reports that the absence of TRP53 itself does not cause detectable abnormalities in the haematopoietic system of young healthy mice although it predisposes them to spontaneous lymphoma development [15, 40–42].
Several direct TRP53 target genes have been implicated in DNA damage-induced apoptosis signalling, but only the BH3-only proteins, PUMA and to lesser extent NOXA, have been shown to be essential for TRP53-induced apoptosis [12, 13, 15, 43]. As expected, thymocytes from Puma−/−p21−/−Zmat3−/− and Puma−/−Zmat3−/− mice displayed significant resistance to etoposide (comparable to thymocytes from Puma−/− mice) to etoposide, a cytotoxic agent that kills these cells via a TRP53-dependent pathway [42, 44, 45]. This indicates that the additional loss of ZMAT3, alone or with further removal of p21, does not increase the protection against TRP53-dependent or TRP53-independent apoptosis that is caused by the absence of PUMA. Consistent with previous studies [5], this demonstrates that ZMAT3 does not have a prominent role in apoptosis, at least in lymphoid cells.
ZMAT3 is a double-stranded RNA binding protein and known TP53/TRP53 target gene, that has been shown to modulate splicing of genes involved in TP53/TRP53 regulation, various cellular processes, and splicing itself [46]. Similarly, we found here that ZMAT3 regulates expression of transcripts encoding proteins involved in the regulation of TP53/TRP53 function. The targets identified include, RPS2 is a ribosomal protein that was reported to interact with MDM2 to control the levels of TP53/TRP53 [16], SIK1 serine/threonine kinase that regulates TP53 function in apoptosis triggered by lack of adhesion to suppress breast cancer metastasis [21]. Additional genes included JUN, transcription factor and apoptosis repressor with caspase recruitment domain (ARC) that are commonly induced in cancers [22, 23]. Notably, CEBPB and KLF4 transcription factors were described to suppress the expression of TP53/TRP53 [20, 47] and frequently dysregulated in acute myeloid leukaemias [17]. In addition, we found that proteins encoded by known TP53/TRP53 target genes, including dual-specific threonine and tyrosine phosphatases DUSP1 [24] and DUSP5 [26], the FOS oncoprotein [27], RGS2 [28], SPRY1 [29], PLK2 [30] and ZFP36L2 [31] are regulated by ZMAT3. Collectively, these data indicate that ZMAT3 may suppress tumorigenesis by modulating the expression of the genes that control and maintain TP53/TRP53 activity.
Previous reports have shown that thymic T cell lymphoma development in mice induced by γ-radiation can be significantly accelerated by loss of Trp53 (even loss of one allele of Trp53) [32, 34, 48]. This is likely due to impairment of DNA damage-induced apoptosis, cell cycle arrest/cell senescence and coordination of DNA repair, which constitute critical processes for TRP53-mediated tumour suppression [1–3, 36, 49, 50]. Whole body γ-irradiation activates TRP53 and causes induction of its downstream target genes p21, Puma and Zmat3 in thymocytes as well as fibroblasts [5, 11] and probably many (if not all) other cell types. However, our investigations show that the absence of ZMAT3, either alone or in combination with loss of p21, did not accelerate (or slow) γ-radiation induced thymic T cell lymphoma development, in contrast to the loss of even a single allele of Trp53. Of note, while many Zmat3−/− and p21−/− thymic T cell lymphomas had acquired defects in TRP53 function, as revealed by high levels of p19ARF and DNA sequencing analysis, most p21−/−Zmat3−/− lymphomas tested had retained wt TRP53 function. These findings demonstrate that ZMAT3 loss is not essential for the suppression of γ-radiation induced thymic T cell lymphoma development, probably because TRP53 can activate many at least partially redundant tumour suppressive processes in response to DNA damage.
In conclusion, our results confirm and extend the notion that ZMAT3 is a direct TRP53 target gene that contributes to TRP53-mediated tumour suppression. It probably does this by regulating a large number of mRNAs that function in the TP53/TRP53 network, thereby impacting, perhaps in a feed-forward activation loop, several TP53/TRP53-activated cellular responses [8, 46]. Of note, on its own loss of ZMAT3 does not cause cancer in mice but its tumour suppressive function becomes apparent when TRP53-mediated apoptosis (loss of PUMA) and/or cell cycle arrest/cell senescence (loss of p21) are concomitantly disabled. Similarly, the impact of loss of ZMAT3 in mouse models of lung and liver cancer was less pronounced than that caused by the absence of TRP53 [6, 11]. Additional support for the idea that in addition to the absence of ZMAT3, additional TP53/TRP53-regulated processes must also be disabled for spontaneous tumour development comes from the observation that mutations in ZMAT3 are not prevalent in human cancers compared to the very high frequency of TP53 mutations [6, 46]. Since the combined loss of ZMAT3, PUMA and p21 does not result in the same tumour spectrum and duration of tumour-free survival as loss of TRP53 alone. Hence other TP53/TRP53 regulated processes for tumour prevention such as coordination of DNA repair to maintain genome stability [5] are required. Thus, our findings further cement the importance of the coordinated action of multiple TP53/TRP53-activated cellular responses for tumour suppression. This may have implications for developing novel strategies for treating cancer as simultaneous activation of several of these responses might be needed for effective therapy.
Materials and methods
Mice
Zmat3−/−5, Puma−/−p21−/−21 and Trp53−/−41mice have been previously described. All mice were maintained on a C57BL/6-WEHI background. To produce Puma−/−p21−/−Zmat3−/−, Puma−/−Zmat3−/− and p21−/−Zmat3−/− mice, we serially inter-crossed Zmat3−/− and Puma−/−p21−/− mice. Genotyping was performed by polymerase chain reaction (PCR) to confirm the absence of Zmat3, Puma and/or p21 genes (for PCR primers used, see Supplementary Table 5).
Histology
Organs from mice were fixed in 10% buffered formalin solution and subsequently embedded in paraffin. Slides were prepared and stained with haematoxylin and eosin (H&E). Histological examination of the organs was performed by Phenomics Australia Histopathology and Slide Scanning Service, University of Melbourne.
Thymic T cell lymphoma induction
p21−/−Zmat3−/−, p21−/−, Zmat3−/− and wt mice (starting at 27 ± 6 days of age) were γ-irradiated weekly for 4 weeks with 1.5 Gy from a 60 Co source (Theratron Phoenix, Theratronics). Treated mice were then monitored for 200 days for signs of illness, and tumour onset was calculated from the last (4th) dose of γ-irradiation.
Trp53 exon sequencing
Genomic DNA was isolated from 5 × 106 thymic lymphoma cells using the DNeasy Blood & Tissue Kit (Qiagen). A total of 9 PCR primers pairs spanning exons 4–11 of Trp53 [51] were used to amplify coding regions. Subsequently, PCR products were subjected to a second PCR using indexing primers, purified using Ampure XP beads (Beckman Coulter), then pooled and sequenced using the MiSeq platform (Illumina).
Immunostaining and flow cytometric analysis
Thymus, spleen, and bone marrow were harvested, and single cell suspensions were prepared in PBS (Gibco) with 5 mM EDTA (Merck), supplemented with 5% foetal calf serum (FCS; Sigma-Aldrich) for staining. Fluorochrome-conjugated monoclonal antibodies used are as follows; B220 (RA3-6B2), IgM (RMM-1), IgD (11.26C), TCRβ (H57.597), CD4 (GK1.5), CD8 (53-6.7), MAC1 (M1/70) SCA1 (E13-161.7), c-KIT (2B8), NK1.1(PK136), Ter119 (TER119), Ly6G (1A8-4-10-9), F4/80 (BM8), CD2 (RM2.1), and were obtained from eBioscience, BioLegend, or generated in-house. Staining with propidium iodide (PI Sigma-Aldrich; 1 μg/mL) was used to exclude dead cells. Whole-organ cell counts were determined by mixing a known concentration of APC Calibrite beads (Becton Dickinson) with each sample. Data were collected using LSRFortessa X-20 or FACSymphony analysers and examined using FlowJo 10 (Becton Dickson).
Cell viability and activated (cleaved) caspase-3 assays
Thymi were harvested from mice of the indicated genotypes and single cell suspensions were prepared by gentle mashing through a 100 µm nylon strainer (Falcon). Cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal calf serum, 50 μM 2-mercaptoethanol, 100 mM asparagine, 100 U/mL penicillin, and 100 mg/mL streptomycin. For viability assay, 5 × 104 cells were plated into 96-well flat-bottom plates in triplicate and treated, as described [35], with DMSO (control), 1 nM dexamethasone (Sigma-Aldrich), 1 µg/mL etoposide (Sigma-Aldrich), 1 µg/mL ionomycin (Sigma-Aldrich) or subjected to serum starvation by culturing in medium containing only 1% foetal calf serum. Cell viability was assessed at 24 h by resuspending cells in Annexin-V binding buffer (0.1 M Hepes (pH 7.4)), 1.4 M NaCl, 25 mM CaCl2 containing fluorescein isothiocyanate (FITC)-conjugated Annexin-V and 1 µg/mL PI. For activated (i.e. cleaved) caspase-3 staining, 5 × 104 cells were plated into 96-well flat-bottom plates in triplicate and treated with 5 µg/mL etoposide for 6 h, fixed (FIX & PERM, Invitrogen), and stained with an antibody to detect cleaved caspase-3 conjugated to FITC (#9661, Cell Signalling). Cells were examined by flow cytometric analysis using a BD-Biosciences LSR-II analyser. Ten thousand events were recorded per sample and data were analysed using FlowJo 10 analysis software.
Western blot analysis
For Western blot analysis, cell lysates were prepared in radio-immunoprecipitation assay (RIPA) buffer supplemented with PhosSTOP and complete protease inhibitor cocktail (Roche). Protein concentration was determined by Bradford assay using the Protein Assay Dye Reagent Concentrate (Bio-Rad, Hercules, CA). Samples of 10 μg of protein were prepared in Laemmli buffer, boiled for 5 min and size-fractionated by gel electrophoresis on NuPAGE 10% Bis-Tris 1.5-mm gels (Life Technologies) in 2-(N-morpholino) ethanesulfonic acid (MES) buffer. Proteins were then transferred onto nitrocellulose membranes (Life Technologies) using the iBlot membrane transfer system (Bio-Rad). Antibody dilution and blocking were performed in 5% skim milk, 0.1% Tween 20 in phosphate-buffered saline (PBS). The following antibodies were used for probing; mouse TRP53 (clone CM5, Novocastra), p19/ARF (clone 5.C3.1, Rockland) and HSP70 (clone N6, gift from Dr. R Anderson, Olivia Newton-John Cancer Research Institute, Melbourne, VIC, Australia), the latter used as a control for protein loading. Secondary antibodies used include goat anti-mouse IgG and goat anti rabbit-IgG, both conjugated to horseradish peroxidase (HRP) (Southern Biotech). Forte Western HRP substrate (Millipore, Billerica, MA) was used for developing the signal, and membranes were imaged and analysed using the ChemiDoc XRS1 machine with ImageLab software (Bio-Rad).
RNA sequencing
RNA was extracted from thymocytes using the miRNeasy Micro Kit (Qiagen) according to the manufacturer’s protocol, including a DNase digestion step. An input of 100 ng total RNA for each sample was indexed separately using the TruSeq RNA Prep Kit v2 (Illumina). Each library was quantified using the Agilent Tapestation. The indexed libraries were pooled and diluted to 750 pM for paired end sequencing (2 × 116 cycles) on the NextSeq 2000 instrument using the P3 200 cycle High Output Kit (Illumina) according to the manufacturer’s instructions. The base calling and quality scoring were determined using Real-Time Analysis on board software v2.4.6, while the FASTQ file generation and de-multiplexing utilised bcl2fastq conversion software v2.15.0.4. Raw FASTQ files were processed, and quality control performed using the nf-core/rnaseq v3.10.1 [52, 53]. Reads were pseudo-aligned and quantified using the Salmon option [54]. The GRCm38.p6 version of the mouse reference genome and the GENCODE vM25 GTF annotation files were used. We filtered out genes: (i) with obsolete Entrez Gene IDs, (ii) ribosomal RNA (rRNA), (iii) non-protein coding immunoglobulin genes (iv) the Xist gene and (v) genes that are on chromosome Y. Next, we filtered out lowly expressed genes by discarding those that did not show a minimum reliable level of expression of 20 counts per million reads of the smallest library size, in at least all the samples of the smallest group [55, 56]. The DESeq2 package (v1.40.0) [55, 57] was used for the differential expression analysis. We adjusted for the sex as a batch effect correction and surrogate variables were calculated with SVA [58]. Genes with adjusted p < 0.05 (5% FDR) and absolute log2FC > 1.5 were considered as statistically significant. Functional enrichment analysis of the biological processes was conducted with the Gene Ontology (GO) database using the clusterProfiler package [59] GSEA [60] was carried out in R using the fgsea package v1.24.0 [61] using the hallmark and curated gene sets from the mouse MsigDb [60, 62–64] GSEA was used to test for enrichment of specific gene sets within a ranked list based on p-value and log2FC. Gene sets with a p < 0.05 were considered as statistically significant.
Supplementary information
Acknowledgements
We thank all members of the Blood Cells and Blood Cancer Division at The Walter and Eliza Hall Institute (WEHI) for support and advice; G. Siciliano, D. Fayle, H. Johnson, C. Gatt and their team for animal husbandry; Dr. S. Monard and his team at the WEHI Flow Cytometry Unit for help with FACS analysis; Thomas Nikolaou and Kevin Weston for haematology support. This study utilised the Phenomics Australia Histopathology and Slide Scanning Service, University of Melbourne.
Author contributions
MSB, MJH, AJ and AS conceptualised and planned the study and wrote the manuscript. MSB, AJ, KB, GH, LGibson, LGangoda and MAP performed experiments and analysed data. EL provided Trp53−/− mouse survival data. MSB and SW performed RNA sequencing, with data analysed by GRS.
Funding
This work was supported by grants and fellowships from the Australian Phenomics Network (APN), the Australian National Health and Medical Research Council (NHMRC) to MJH and AS (1143105), Programme Grant to AS (1016701), Investigator grant to AS (2007887), Investigator Grant to MJH (2017971); the Leukaemia and Lymphoma Society of America to AS and MJH (LLS SCOR 7015-18); the Cancer Council of Victoria Project grant to AS (1052309) and Venture Grant to MJH and AS; support to AS from the estate of Anthony (Toni) Redstone OAM; support to AJ from Spanish Ministry of Economy and Development Grant (PID2021-127710OB-I00) and Programa Captació de Talent Investigador ‘La Caixa’ foundation (51110009). AJ is supported by Ramon y Cajal Research Fellowship (RYC2018-025244-I), AS and MJH are supported by NHMRC Fellowships (1020363 and 1156095), MSB is supported by Cancer Council Victoria Postdoctoral Fellowship and Swedish Cancer Society (21 0355 PT). This work was made possible through the Victorian Government Operational Infrastructure Support and Australian Government, the ‘Unidad de Excelencia María de Maeztu’ funded by the Spanish Government.
Data availability
RNA-seq data have been deposited in the Gene Expression Omnibus (ncbi.nlm.nih.gov/geo) under accession number GSE248747. The code used for the RNA-Seq analysis is available at https://github.com/Geriroso/ZMAT3_analysis. All knockout mice will be made available to the scientific community upon request.
Competing interests
The authors declare no competing interests.
Ethics approval
All experiments with mice were performed with the approval of The Walter and Eliza Hall Institute Animal Ethics Committee and according to the Australian Code of Practice for the care and use of animals for scientific purposes.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Andreas Strasser, Marco J. Herold, Ana Janic.
Contributor Information
Andreas Strasser, Email: strasser@wehi.edu.au.
Marco J. Herold, Email: herold@wehi.edu.au
Ana Janic, Email: ana.janic@upf.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01250-w.
References
- 1.Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25:104–13. doi: 10.1038/cdd.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–78. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 4.Boutelle AM, Attardi LD. p53 and tumor suppression: it takes a network. Trends Cell Biol. 2021;31:298–310. doi: 10.1016/j.tcb.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janic A, Valente LJ, Wakefield MJ, Di Stefano L, Milla L, Wilcox S, et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat Med. 2018;24:947–53. doi: 10.1038/s41591-018-0043-5. [DOI] [PubMed] [Google Scholar]
- 6.Bieging-Rolett KT, Kaiser AM, Morgens DW, Boutelle AM, Seoane JA, Van Nostrand EL, et al. Zmat3 is a key splicing regulator in the p53 tumor suppression program. Mol Cell. 2020;80:452–469.e9. doi: 10.1016/j.molcel.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bersani C, Xu L-D, Vilborg A, Lui W-O, Wiman KG. Wig-1 regulates cell cycle arrest and cell death through the p53 targets FAS and 14-3-3σ. Oncogene. 2014;33:4407–17. doi: 10.1038/onc.2013.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilborg A, Bersani C, Wilhelm MT, Wiman KG. The p53 target Wig-1: a regulator of mRNA stability and stem cell fate? Cell Death Differ. 2011;18:1434–40. doi: 10.1038/cdd.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muys BR, Anastasakis DG, Claypool D, Pongor L, Li XL, Grammatikakis I, et al. The p53-induced RNA-binding protein ZMAT3 is a splicing regulator that inhibits the splicing of oncogenic CD44 variants in colorectal carcinoma. Genes Dev. 2020;35:102–16. doi: 10.1101/gad.342634.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–83. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Best SA, Vandenberg CJ, Abad E, Whitehead L, Guiu L, Ding S, et al. Consequences of Zmat3 loss in c-MYC- and mutant KRAS-driven tumorigenesis. Cell Death Dis. 2020;11. 10.1038/s41419-020-03066-9. [DOI] [PMC free article] [PubMed]
- 12.Villunger A, Michalak EM, Coultas L, Müllauer F, Böck G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins Puma and Noxa. Science. 2003;302:1036–8. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 13.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–8. doi: 10.1016/S1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 14.Deng C, Zhang P, Wade Harper J, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development but are defective in G1 checkpoint control. Cell. 1995;82:675–84. doi: 10.1016/0092-8674(95)90039-X. [DOI] [PubMed] [Google Scholar]
- 15.Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008;15:1019–29. doi: 10.1038/cdd.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho J, Park J, Shin SC, Kim JH, Kim EEK, Song EJ. Ribosomal protein S2 interplays with MDM2 to induce p53. Biochem Biophys Res Commun. 2020;523. 10.1016/j.bbrc.2020.01.038. [DOI] [PubMed]
- 17.Seipel K, Marques MT, Bozzini MA, Meinken C, Mueller BU, Pabst T. Inactivation of the p53-KLF4-CEBPA axis in acute myeloid leukemia. Clin Cancer Res. 2016;22. 10.1158/1078-0432.CCR-15-1054. [DOI] [PubMed]
- 18.Miyajima C, Inoue Y, Hayashi H. Pseudokinase tribbles 1 (TRB1) negatively regulates tumor-suppressor activity of p53 through p53 deacetylation. Biol Pharm Bull. 2015;38. 10.1248/bpb.b15-00003. [DOI] [PubMed]
- 19.Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, et al. P53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22. 10.1038/nm.4135. [DOI] [PubMed]
- 20.Ewing SJ, Zhu S, Zhu F, House JS, Smart RC. C/EBPβ represses p53 to promote cell survival downstream of DNA damage independent of oncogenic Ras and p19Arf. Cell Death Differ. 2008;15. 10.1038/cdd.2008.105. [DOI] [PMC free article] [PubMed]
- 21.Cheng H, Liu P, Wang ZC, Zou L, Santiago S, Garbitt V, et al. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Sci Signal. 2009;2. 10.1126/scisignal.2000369. [DOI] [PMC free article] [PubMed]
- 22.Schreiber M, Kolbus A, Piu F, Szabowski A, Möhle-Steinlein U, Tian J, et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13. 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed]
- 23.Foo RSY, Nam YJ, Ostreicher MJ, Metzl MD, Whelan RS, Peng CF, et al. Regulation of p53 tetramerization and nuclear export by ARC. Proc Natl Acad Sci USA. 2007;104. 10.1073/pnas.0710017104. [DOI] [PMC free article] [PubMed]
- 24.Liu YX, Wang J, Guo J, Wu J, Lieberman HB, Yin Y. DUSP1 is controlled by p53 during the cellular response to oxidative stress. Mol Cancer Res. 2008;6. 10.1158/1541-7786.MCR-07-2019. [DOI] [PubMed]
- 25.Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, et al. PERP, an apoptosis-associated target of p53, in a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14. 10.1101/gad.14.6.704. [PMC free article] [PubMed]
- 26.Ueda K, Arakawa H, Nakamura Y. Dual-specificity phospha tase 5 (DUSP5) as a direct transcriptional target of tumor suppressor p53. Oncogene. 2003;22. 10.1038/sj.onc.1206845. [DOI] [PubMed]
- 27.Elkeles A, Juven-Gershon T, Israeli D, Wilder S, Zalcenstein A, Oren M. The c- fos proto-oncogene is a target for transactivation by the p53 tumor suppressor. Mol Cell Biol. 1999;19. 10.1128/mcb.19.4.2594. [DOI] [PMC free article] [PubMed]
- 28.Buckbinder L, Velasco-Miguel S, Chen Y, Xu N, Talbott R, Gelbert L, et al. The p53 tumor suppressor targets a novel regulator of G protein signaling. Proc Natl Acad Sci USA. 1997;94. 10.1073/pnas.94.15.7868. [DOI] [PMC free article] [PubMed]
- 29.Łasut-Szyszka B, Rusin M The Wheel of p53 Helps to Drive the Immune System. Int J Mol Sci. 2023;24. 10.3390/ijms24087645. [DOI] [PMC free article] [PubMed]
- 30.Burns TF, Fei P, Scata KA, Dicker DT, El-Deiry WS. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol Cell Biol. 2003;23. 10.1128/mcb.23.16.5556-5571.2003. [DOI] [PMC free article] [PubMed]
- 31.Kouzu H, Tatekoshi Y, Chang HC, Shapiro JS, McGee WA, De Jesus A, et al. ZFP36L2 suppresses mTORc1 through a P53-dependent pathway to prevent peripartum cardiomyopathy in mice. J Clin Investig. 2022;132. 10.1172/JCI154491. [DOI] [PMC free article] [PubMed]
- 32.Kemp CJ, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet. 1994;8:66–9. doi: 10.1038/ng0994-66. [DOI] [PubMed] [Google Scholar]
- 33.Michalak EM, Vandenberg CJ, Delbridge ARD, Wu L, Scott CL, Adams JM, et al. Apoptosis-promoted tumorigenesis: γ-irradiation-induced thymic lymphomagenesis requires Puma-driven leukocyte death. Genes Dev. 2010;24:1608–13. doi: 10.1101/gad.1940110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labi V, Erlacher M, Krumschnabel G, Manzl C, Tzankov A, Pinon J, et al. Apoptosis of leukocytes triggered by acute DNA damage promotes lymphoma formation. Genes Dev. 2010;24:1602–7. doi: 10.1101/gad.1940210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dudgeon C, Chan C, Kang W, Sun Y, Emerson R, Robins H, et al. The evolution of thymic lymphomas in p53 knockout mice. Genes Dev. 2014;28:2613–20. doi: 10.1101/gad.252148.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas AF, Kelly GL, Strasser A. Of the many cellular responses activated by TP53, which ones are critical for tumour suppression? Cell Death Differ. 2022. 10.1038/S41418-022-00996-Z. [DOI] [PMC free article] [PubMed]
- 37.Mello SS, Attardi LD. Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol. 2018;51:65–72. doi: 10.1016/j.ceb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valente LJ, Gray DHD, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep. 2013;3:1339–45. doi: 10.1016/j.celrep.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 39.Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–83. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 41.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/S0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 42.Strasser A, Harris AW, Jacks T, Cory S. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 1994;79:329–39. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 43.Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK, et al. Puma and to a lesser extent, Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 2009;16:684–96. doi: 10.1038/cdd.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–9. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 45.Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;; 362:849–52. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 46.Bieging-Rolett KT, Attardi LD. Zmat3 splices together p53-dependent tumor suppression. Mol Cell Oncol. 2021;8. 10.1080/23723556.2021.1898523. [DOI] [PMC free article] [PubMed]
- 47.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7. 10.1038/ncb1314. [DOI] [PubMed]
- 48.Kaplan HS, Brown MB. A quantitative dose-response study of lymphoid-tumor development in irradiated C57 black mice. J Natl Cancer Inst. 1952;13:185–208. [PubMed] [Google Scholar]
- 49.Wang Z, Strasser A, Kelly GL. Should mutant TP53 be targeted for cancer therapy? Cell Death Differ. 2022;29:911–20. doi: 10.1038/s41418-022-00962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aubrey BJ, Janic A, Chen Y, Chang C, Lieschke EC, Diepstraten ST, et al. Mutant TRP53 exerts a target gene-selective dominant-negative effect to drive tumor development. Genes Dev. 2018;32. 10.1101/gad.314286.118. [DOI] [PMC free article] [PubMed]
- 51.Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O’Connor L, Milla L, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–32. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 52.Patel H et al. nf-core/rnaseq: nf-core/rnaseq v3.0—Silver Shark. 2020.
- 53.Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38. 10.1038/s41587-020-0439-x. [DOI] [PubMed]
- 54.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed]
- 55.Love MI, Anders S, Kim V, Huber W. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000Res 2015;4. 10.12688/f1000research.7035.1. [DOI] [PMC free article] [PubMed]
- 56.Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12. 10.1101/gad.12.19.2973. [DOI] [PubMed]
- 57.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed]
- 58.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3. 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed]
- 59.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2. 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed]
- 60.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed]
- 61.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. 10.1101/060012.
- 62.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011; 27. 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed]
- 63.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular signatures database hallmark gene set collection. Cell Syst. 2015;1. 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed]
- 64.Castanza AS, Recla JM, Eby D, Thorvaldsdóttir H, Bult CJ, Mesirov JP. Extending support for mouse data in the Molecular Signatures Database (MSigDB). Nat Methods. 2023. 10.1038/s41592-023-02014-7. [DOI] [PMC free article] [PubMed]
- 65.Martínez-Cruz AB, Santos M, Lara MF, Segrelles C, Ruiz S, Moral M, et al. Spontaneous squamous cell carcinoma induced by the somatic inactivation of retinoblastoma and Trp53 tumor suppressors. Cancer Res 2008;68. 10.1158/0008-5472.CAN-07-3049. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the Gene Expression Omnibus (ncbi.nlm.nih.gov/geo) under accession number GSE248747. The code used for the RNA-Seq analysis is available at https://github.com/Geriroso/ZMAT3_analysis. All knockout mice will be made available to the scientific community upon request.