Abstract
Background
Fibroblast-to-myofibroblast conversion is a major driver of tissue remodelling in organ fibrosis. Distinct lineages of fibroblasts support homeostatic tissue niche functions, yet their specific activation states and phenotypic trajectories during injury and repair have remained unclear.
Methods
We combined spatial transcriptomics, multiplexed immunostainings, longitudinal single-cell RNA-sequencing and genetic lineage tracing to study fibroblast fates during mouse lung regeneration. Our findings were validated in idiopathic pulmonary fibrosis patient tissues in situ as well as in cell differentiation and invasion assays using patient lung fibroblasts. Cell differentiation and invasion assays established a function of SFRP1 in regulating human lung fibroblast invasion in response to transforming growth factor (TGF)β1.
Measurements and main results
We discovered a transitional fibroblast state characterised by high Sfrp1 expression, derived from both Tcf21-Cre lineage positive and negative cells. Sfrp1+ cells appeared early after injury in peribronchiolar, adventitial and alveolar locations and preceded the emergence of myofibroblasts. We identified lineage-specific paracrine signals and inferred converging transcriptional trajectories towards Sfrp1+ transitional fibroblasts and Cthrc1+ myofibroblasts. TGFβ1 downregulated SFRP1 in noninvasive transitional cells and induced their switch to an invasive CTHRC1+ myofibroblast identity. Finally, using loss-of-function studies we showed that SFRP1 modulates TGFβ1-induced fibroblast invasion and RHOA pathway activity.
Conclusions
Our study reveals the convergence of spatially and transcriptionally distinct fibroblast lineages into transcriptionally uniform myofibroblasts and identifies SFRP1 as a modulator of TGFβ1-driven fibroblast phenotypes in fibrogenesis. These findings are relevant in the context of therapeutic interventions that aim at limiting or reversing fibroblast foci formation.
Shareable abstract
This single-cell study discovered a transitional cell state that appears early after injury and precedes the generation of myofibroblasts. This cell state is characterised by expression of SFRP1, which inhibits fibroblast invasion in fibrogenesis. https://bit.ly/3uifxmJ
Introduction
Extracellular matrix (ECM)-producing myofibroblasts are a key therapeutic target to combat tissue fibrosis, one of the biggest unresolved clinical problems across most major chronic diseases. Several recent single-cell RNA-sequencing (scRNAseq) studies described distinct subsets of collagen-producing stromal cells in mouse and human lungs with distinct spatial locations and different functions in supporting epithelial repair [1–5]. How this heterogeneity of fibroblast identities leads to different cell states and functions in fibrotic disease is unclear.
Genetic lineage tracing in mouse models provides evidence for an alveolar lipofibroblast-to-myofibroblast switch after lung injury that is reversible during the resolution of transient fibrosis upon completed epithelial regeneration [6, 7]. The heterogeneity of cellular sources for myofibroblasts in lung fibrosis remains the subject of current investigations. Apart from lipofibroblasts, alveolar pericytes are also potential sources of myofibroblasts [8]. Proliferation, evasion of apoptosis and invasive capacity of myofibroblasts are key hallmarks of fibrotic disease, and transforming growth factor (TGF)β is a known master regulator of these processes [9, 10]. A vast amount of literature demonstrates the TGFβ-induced induction of α-smooth muscle actin (ACTA)2 in fibroblasts, the most widely used marker of mature myofibroblasts in tissues. However, fate mapping and immunofluorescence analysis of fibrotic tissues and myofibroblast foci also show that a substantial fraction of fibroblasts is ACTA2− [11, 12], suggesting additional molecular complexity and heterogeneity among injury-activated fibroblasts. A recent single-cell analysis of collagen-producing cells in lung fibrosis revealed CTHRC1 as a specific marker of highly invasive ACTA2+ myofibroblasts. These cells occur in both mouse and human lung and occupy fibroblastic foci in idiopathic pulmonary fibrosis (IPF) [1].
The regulation of injury-activated fibroblast states is not well understood and of high clinical relevance. Our study reveals the spatiotemporal evolution of distinct fibroblast states towards Cthrc1+ myofibroblasts, highlighting early events of injury-induced fibroblast activation that preceded myofibroblastic differentiation. We discovered a novel SFRP1+/ACTA2− transitional state that is initially noninvasive and only becomes invasive upon TGFβ-driven differentiation towards the CTHRC1+ myofibroblast state. We show that SFRP1 modulates TGFβ1-induced fibroblast invasion and RHOA pathway activity, which constitutes a novel pathway with potential for targeting fibrotic disease mediated by myofibroblasts.
Material and methods
For details of the materials and methods, please refer to the supplementary material.
Experimental study design
C57B6 (males or females) mice were treated with 3.5 units·kg−1 body mass of bleomycin administered intratracheally. Up to two bleomycin-instilled mice were sacrificed at time points (days 0, 2–14, 21, 28, 35, 56) after instillation and subject to scRNAseq using the Drop-seq platform. Tcf21-lineage labelled (Tcf21m-Crem-R26R-tdTomato) mesenchymal cells were analysed using the 10x scRNAseq platform. Two healthy C57B6 mouse lungs were used for SCRINSHOT. Primary human lung fibroblasts (pHLFs) and micro-computed tomography (CT) staged IPF tissues, and data from an integrated IPF cell atlas were used for human validation.
Human tissue and ethics statement
pHLFs of non-chronic lung disease donors were obtained from the CPC-M bioArchive at the Comprehensive Pneumology Center (Munich, Germany). Participants provided written informed consent to participate in this study, in accordance with approval by the local ethics committee of the Ludwig Maximilians University (Germany) (project 333-10). Micro-CT-staged IPF samples were provided from the KU Leuven lung biobank (ethical approval S52174). Samples were derived from explanted lungs, after written informed consent from all patients. Unused donor lungs were included as controls, following Belgian legislation. Three IPF lungs and three controls were included.
Data availability
RNA-seq data were deposited to the Gene Expression Omnibus (GEO) database. The high temporal mesenchymal enriched Drop-seq data can be found with the accession code GSE207851, and the Tcf21-lineage labelled mesenchymal 10x data with the accession code GSE207687. Microarray data of pHLFs (Sfrp1-siRNA knockdown) can be found with the accession code GSE207561.
Results
Heterogeneity of mesenchymal cells at distinct spatial localisations in the lung
The Tcf21+ lineage constitutes the lung lipofibroblast population [13]. To study heterogeneity within the Tcf21+ and Tcf21− fibroblast lineages, we tamoxifen-labelled Tcf21+ cells in lungs at 11 weeks of age using Tcf21m-Crem-R26R-tdTomato mice (supplementary figure S1a). Tcf21 lineage-positive (lin+) cells were indeed found near alveolar type 2 (AT2) cells and alveolar capillaries (supplementary figure S1b). We flow-sorted Tcf21 lin+ and lin− stromal cells and performed scRNAseq (n=4 mice, k=12.068 cells), identifying six distinct cell types with different Tcf21-lineage proportions (figure 1a and supplementary figure S1c and d). Previously established marker genes [1, 14, 15] were used to annotate clusters (supplementary table S1). All cell type identities expressed type 1 collagen (Col1a2) (supplementary figure S2c) and were consistent with previous work [1].
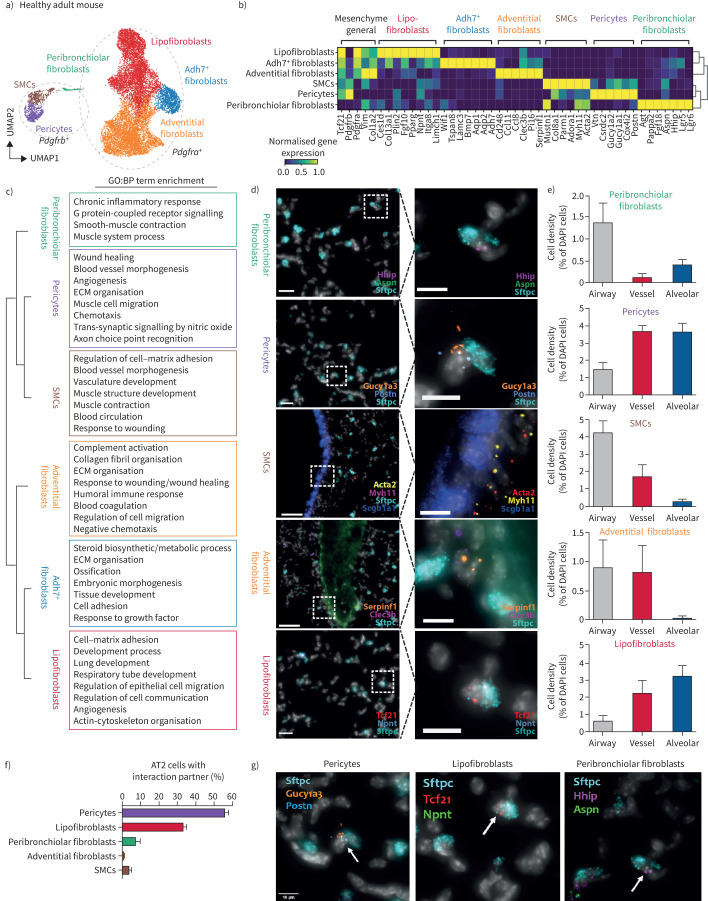
FIGURE 1.
Stromal cell heterogeneity in the adult lung is associated with distinct spatial locations. a) Uniform manifold approximation and projection (UMAP) clustering depicts the seven distinct mesenchymal cell types (12 068 cells) identified in the healthy adult mouse lung. Circles mark Pdgfra- and Pdgfrb-expressing cells. b) The seven mesenchymal cell types are characterised with distinct gene expression profiles, as depicted in the matrixplot. c) Marker genes of the indicated mesenchymal subtypes are enriched for characteristic gene ontology (GO):biological process (BP) terms. False discovery rate <10%. Top 500 marker genes considered. d) Single mRNA multiplexed fluorescence in situ hybridisation (SCRINSHOT) with two cell type specific marker genes per cell type, as well as general markers allowed to identify preferential spatial locations of the mesenchymal cell types in the mouse lung. Scale bars=50 μm (overviews); 10 µm (insets). e) Localisation analysis shows stereotypic localisation of the mesenchymal cell types to peribronchiolar, adventitial and alveolar regions. The spatial location of 976 cells was quantified over five regions with 508 pericytes, 70 peribronchiolar cells, 206 lipofibroblasts, 41 adventitial fibroblasts and 151 smooth muscle cells (SMCs) counted. Graphs show cell density as percentage of cell type specific cells compared to all the cells (4′,6-diamidino-2-phenylindole (DAPI) stain-positive nuclei) in the area. Error bars represent sem. f) Colocalisation analysis of 1269 alveolar type 2 (AT2) cells revealed preferred interaction partners among the mesenchymal cell types. A total of 291 AT2 interactions with mesenchymal cell types in the alveolar space were found: 167 pericytes; 93 lipofibroblasts, 15 peribronchiolar fibroblasts; three adventitial fibroblasts; 13 SMCs. g) SCRINSHOT pictures exemplifying the colocalisation of AT2 cells with distinct mesenchymal subtypes. Scale bars=10 μm. ECM: extracellular matrix.
We identified three main Pdgfra+ populations (lipofibroblasts, adventitial fibroblasts, Adh7+ fibroblasts), two Pdgfrb+ populations (smooth muscle cells and pericytes) with highly distinct marker genes and pathway enrichments (figure 1b and c), and Pdgfra/Pdgfrb double-positive cells, as recently found in the kidney [16]. These cells expressed Lgr5 and Lgr6 (figure 1b), markers for a peribronchiolar fibroblast population [4]. Importantly, a recent study identified similar LGR5+ fibroblast populations in distal human airways [17]. Subclustering of this Hhip+ peribronchiolar fibroblast population revealed additional complexity with Lgr5/Lgr6 single- and double-positive populations (supplementary figure S1e–g). All cell types were lineage-labelled in the Tcf21m-Crem-R26R-tdTomato mouse, with exception of the Hhip+ peribronchiolar fibroblasts (supplementary figure S1c and d). Thus, Tcf21lin− peribronchiolar fibroblasts may constitute a developmentally distinct lineage from Tcf21+ stromal cells.
Next, we used targeted spatial transcriptomics [18] to multiplex the mRNA localisation of 18 cell type marker genes in six representative regions of adult murine lungs (n=2) along the proximal distal axis of the airway tree (figure 1d and supplementary figure S2a–c). Hhip+/Aspn+ peribronchiolar fibroblasts were enriched around airways, with some cells also in alveoli. Myh11+/Acta+ smooth muscle cells and Serpinf1+/Clec3b+ adventitial fibroblasts were enriched around airways and large vessels. Gucy1a3+/Postn+ pericytes localised preferentially to alveolar space and around larger vessels. The Tcf21hi/Npnt+ lipofibroblasts were localised preferentially to alveolar space (figure 1e and supplementary figure S2d and e). Consequently, the number of cells in close physical proximity (direct cell–cell contact) to Sftpc+ AT2 cells was highest for pericytes and lipofibroblasts with some Hhip+/Aspn+ cells also participating in the AT2 cell niche (figure 1f and supplementary figure S2f). Our data highlight the complexity of the AT2 cell niche, which we here demonstrate to contain at least three distinct stromal cell types.
An activated fibroblast state characterised by high Sfrp1 and Col28a1 expression
To follow the fate of fibroblasts during injury and repair we sorted cells from Tcf21m-Crem-R26R-tdTomato mice 14 days after bleomycin-induced lung injury (supplementary figures S1 and S3). Three injury-induced clusters became apparent (figure 2a and b), which were a mixture of Tcf21-lineage-positive and -negative cells. Interestingly, Tcf21-lineage-negative Hhip+ peribronchiolar fibroblasts expanded (supplementary figure S1j, p and q), and some markers (e.g. Lgr5 and Lgr6) were expressed in a myofibroblast subset (supplementary figure S1l), indicating that both Tcf21-lineage negative and positive cells converged into myofibroblasts.
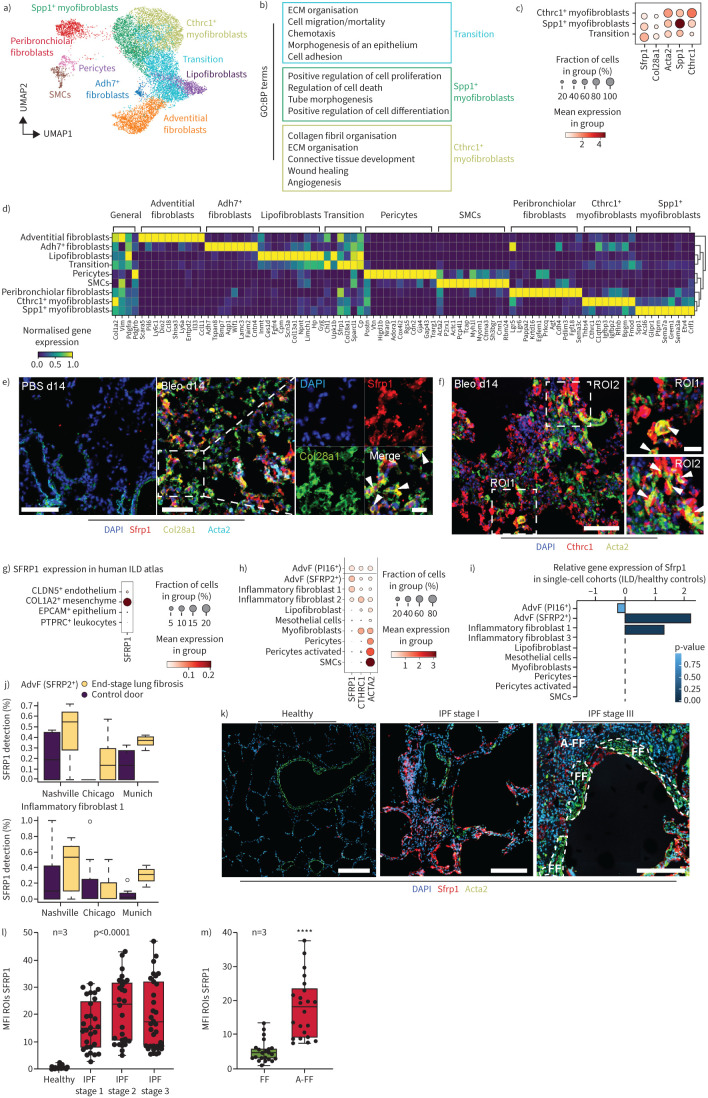
FIGURE 2.
Three distinct activated mesenchymal cell types emerge after bleomycin (bleo)-induced mouse lung injury. a) Uniform manifold approximation and projection (UMAP) depicts mesenchymal cell types (n=12 254 cells) identified at day 14 (d14) after bleomycin injury. b) The marker genes of activated mesenchymal subtypes are enriched for characteristic gene ontology (GO):biological process (BP) terms. False discovery rate <10%. Top 500 marker genes considered. c) The dotplot depicts a gradient of marker gene expression across the activated cell types. d) The mesenchymal cell types are characterised with distinct gene expression profiles, as depicted in the matrixplot. e, f) Immunofluorescence analysis of lung tissue sections from bleomycin-treated mice demonstrates colocalisation of SFRP1 (red) and COL28A1 (green) 14 days after injury. Arrowheads in the magnified insets indicate SFRP1/COL28A1 double-positive cells. SFRP1/COL28A1 double-positive cells did not colocalise with α-smooth muscle actin (ACTA)2 (cyan). No expression of SFRP1 was detected in PBS controls. Scale bars=100 µm (PBS), 100 µm (Bleo d14) and 50 µm (inset). f) At d14 after injury, ACTA2 (green) and CTHRC1 (red) double-positive cells were found (indicated by arrowheads in the magnified insets for ROI1 and ROI2 in the very right panel). Nuclei in blue colour (4′,6-diamidino-2-phenylindole DAPI). Scale bars=100 µm and 20 µm (inset). g, j) Distinct markers from the bleomycin mouse model were analysed in a recently published integrated atlas of human lung fibrosis at single-cell resolution [20]. g) SFRP1 showed specific expression on human COL1A2+ positive mesenchymal cells. h) Marker gene expression across all mesenchymal cell types identified in the human lung. i, j) Differential gene expression analysis between interstitial lung disease (ILD) patient samples and healthy control samples: i) reveals significant regulation of SFRP1 in adventitial fibroblasts (AdvF) and inflammatory fibroblasts cluster 2; j) this regulation is consistent across all three cohorts in the atlas. k) Immunofluorescence analysis of micro-CT staged idiopathic pulmonary fibrosis (IPF)-patient tissues demonstrating an increase of SFRP1 (red) in regions of mild fibrosis (IPF stage I) compared to healthy controls. In regions of severe fibrosis (IPF stage III) and tissue remodelling, fibroblastic foci (white dashed lines, FF) stained positive for α-smooth muscle actin (ACTA2) (green), but mostly not for SFRP1. Adjacent to fibroblastic foci (A-FF) SFRP1 expressing fibroblasts were still present. Scale bars=200 µm. Representative images are shown. l) Quantification of SFRP1's mean fluorescence intensity (MFI) of various regions-of-interest (ROIs) from three different patients (n=3). Statistics: one-way ANOVA. m) Quantification of SFRP1 MFI in late-stage IPF comparing SFRP1 expression within fibroblastic foci (FF) and adjacent to fibroblastic foci (A-FF) from three different patients (n=3). Statistics: unpaired t-test. ****: p<0.0001. SMC: smooth muscle cell; ECM: extracellular matrix.
We identified two Cthrc1+/Acta2+ myofibroblast types, with one subcluster showing enhanced Spp1 expression (figure 2c and d). One cluster coexpressed myofibroblast genes with different fibroblast markers, especially from lipofibroblasts; hence, we named these “transitional fibroblasts” (figure 2b–d). Several genes, including the secreted frizzled-related protein 1 (Sfrp1), showed highest expression in this putative intermediate cell population (figure 2c and d; supplementary table S2). Gene–gene correlation of Sfrp1 with other genes detected a transitional fibroblast core gene set, including Col28a1 (supplementary figure S4d). Tissue proteomics revealed transient induction of COL28A1 and SFRP1 proteins after bleomycin injury (supplementary figure S4c) [19]. Co-staining SFRP1 and COL28A1 confirmed coexpression in ACTA2-low or -negative transitional cells (figure 2e), while CTHRC1+ cells expressed higher levels of ACTA2 (figure 2f).
To address clinical relevance, we compared Col1a2+ cells after bleomycin injury with COL1A2+ cells in ILD patients [20]. SFRP1 was specific to COL1A2+ mesenchymal cells and was weakly expressed in some CLDN5+ endothelial cells (figure 2g). Expression was restricted to adventitial fibroblasts and disease enriched “inflammatory” fibroblast subsets (figure 2g and h). Increased expression of SFRP1 in disease-induced fibroblast states was consistent in all three study cohorts (figure 2i and j). Importantly, in IPF tissues, more SFRP1+ fibroblasts were present in mildly affected (early-stage) regions, while end-stage regions had more ACTA2+ myofibroblasts localised to fibroblast foci (figure 2k–m). These data suggests a similar trajectory of fibroblast states in human lung fibrogenesis as seen in the bleomycin model.
Sfrp1+ transitional fibroblasts precede the appearance of Cthrc1+ myofibroblasts
To follow transcriptional dynamics of Col1a2+ stromal cells during inflammatory, fibrogenic and resolution phases of lung regeneration, we collected Epcam−/Pecam1−/Lyve1−/CD45− stromal cells at 18 time points after bleomycin injury (figure 3a–c). This longitudinal scRNAseq dataset of Col1a2+ cells (69 185 cells) (figure 3b) featured three distinct injury-induced cell states (supplementary figure S4a) similar to the Tcf21-lineage tracing dataset (figure 3c and supplementary table S3). Sfrp1+ transitional fibroblasts peaked at day 3, preceding Spp1+ and Cthrc1+ myofibroblasts at day 9 to day 21 (figure 3d). Proliferation analysis showed a transient increase in proliferation rates, mainly in Spp1+ and Cthrc1+ myofibroblasts, returning to baseline from day 28 onwards (supplementary figure S4b). This suggests the early post-injury increase of Sfrp1+ cells was not from expanding pre-existing cells, but from differentiating baseline fibroblast states. Using immunofluorescence (figure 3e–l), we found that in healthy lungs, signals for SFRP1, CTHRC1 and SPP1 were mostly absent, except for nonspecific signals in the airway epithelium's luminal areas (figure 3e). Filamentous COL28A1 localised around bronchovascular cuffs, whereas ACTA2 was primarily observed in smooth muscle cells near airways and blood vessels (figure 3e). Localisation of adventitial and peribronchial fibroblasts next to bronchovascular cuffs has been described [1] and concurs with our single-molecule fluorescence in situ hybridisation (smFISH) analysis (figure 1).
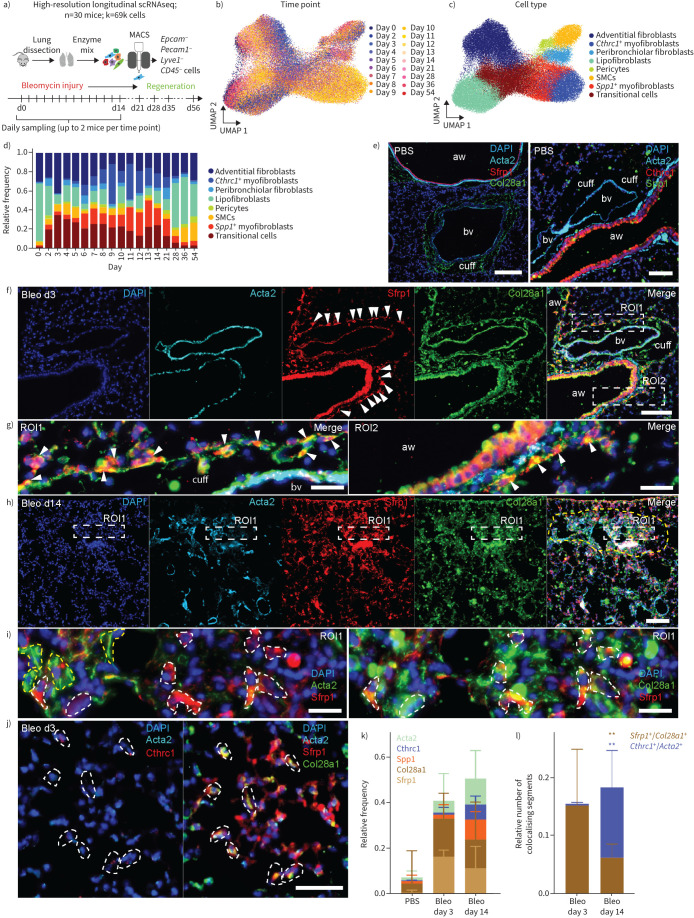
FIGURE 3.
Sfrp1+ transitional fibroblasts emerge in adventitial and alveolar space upon injury preceding the emergence of Cthrc1+ myofibroblasts. a) A high-resolution longitudinal dataset was generated by subjecting magnetic-activated cell-sorting (MACS)-sorted cells from the mesenchymal compartment to single-cell RNA-sequencing (scRNAseq) at the 18 indicated time points. Uniform manifold approximation and projection (UMAP) embedding displays cells coloured by b) time point and c) cell type identity. d) The distribution of cell type frequencies across time points. e) Immunofluorescence analysis of lung tissue sections from PBS control mice. In the left panel, cell nuclei were immunostained with 4′,6-diamidino-2-phenylindole DAPI (blue) and smooth muscle cells (SMCs) with α-smooth muscle actin (ACTA)2 (cyan). SFRP1 (red) staining was absent apart from nonspecific signals in bronchial epithelial cells. COL28A1 (green) was mostly found as a filamentous staining in “cuffs” surrounding blood vessels (bv). In the right panel, cell nuclei were stained with DAPI (blue) and SMCs with ACTA2 (cyan). CTHRC1 (red) staining was absent apart from nonspecific signals in bronchial epithelial cells. SPP1 is depicted in green. Scale bars=100 µm. f) Immunofluorescence analysis of lung tissue sections from bleomycin-treated mice at day 3 after injury (Bleo d3) demonstrating appearance of SFRP1 (red) positive cells surrounding blood vessels and airways (aw) (arrowheads). Scale bar=100 µm. g) Arrowheads in the magnified insets taken from region of interest (ROI)1 and ROI2 of figure 4f indicate SFRP1/COL28A1 double-positive cells surrounding blood vessels in ROI1 reminiscent of adventitial fibroblasts, as well as surrounding airways (aw) in ROI2 reminiscent of peribronchiolar fibroblasts. Scale bars=20 µm. h) Yellow dashed line indicates a fibrotic region of lung tissue after 14 days of bleomycin treatment. ACTA2 staining in cyan exhibits the appearance of myofibroblasts, and concomitant expression of SFRP1 (red) and COL28A1 (green) in this fibrotic region. Cell nuclei are stained with DAPI in blue. Scale bar=100 µm. A magnified view of fibrotic ROI1 is displayed in (i), in the left panel demonstrating mutually exclusive appearance of SFRP1+ (red and dashed white lines) and ACTA2+ (green and dashed yellow lines) cells. Here, for better detection of colocalisation signals between ACTA2 and SFRP1, we switched colours of ACTA2 from cyan as depicted in (h) to green. The right panel denotes SFRP1 (red) and COL28A1 (green) double-positive cells encircled with white dashed lines. Cell nuclei stained with DAPI in blue. Scale bars=20 µm. j) Iterative immunofluorescence staining of parenchymal lung tissue (4i-FFPE) at day 3 after injury indicating COL28A1 (green)/SFRP1 (red) double-positive cells in the left image. The same cells, as indicated by white dashed lines, were found to be CTHRC1- (red) and ACTA2- (cyan) negative. Scale bar=50 µm. A larger overview image can be found in supplementary figure S7a. k) Stacked bar-plot denoting relative frequencies of ACTA2-, CTHRC1-, SPP1-, COL28A1- and SFRP1-positive tissue segments at day 3 and day 14 after bleomycin treatment and compared to PBS controls from software-based segmented images as exemplified in supplementary figure S7c. Three different ROIs (each 1.1 mm2 in size) from two different mice for each condition were analysed. l) Quantification of double-positive SFRP1+/COL28A1+ and CTHRC1+/ACTA2+ tissue segments were analysed by fluorescent-signal colocalisation (supplementary figure S7d), its segmentation and software-based quantification. The number of colocalising tissue segments relative to the total cell count (20 691 cells) is shown. 10–20 different 0.1–0.25-mm2 ROIs (peribronchial/perivascular/parenchymal on day 3 compared to fibrotic on day 14) from two different mice per condition were analysed. Statistics: two-way ANOVA with Tukey's multiple comparison: **: p<0.01.
3 days post-injury, SFRP1/COL28A1 double-positive cells emerged around airways, blood vessels and alveoli, with ACTA2 expressed in bronchovascular smooth muscle cells (figure 3f, g and j). In contrast, at day 14 post-injury, ACTA2-myofibroblasts and SFRP1+/COL28A1+ cells coexisted in fibrotic dense areas (figure 3h and i). Multiplexed immunofluorescence-analysis confirmed the increase of SFRP1+/COL28A1+ transitional cells at day 3, and until day 14 post-injury expanding SPP1+ and CTHRC1+ myofibroblasts (figure 3k and l; total analysis of 20 691 cells, supplementary figure S7c), thus verifying our data in figure 3d on protein level.
Convergence of multiple mesenchymal cell types towards myofibroblast identity
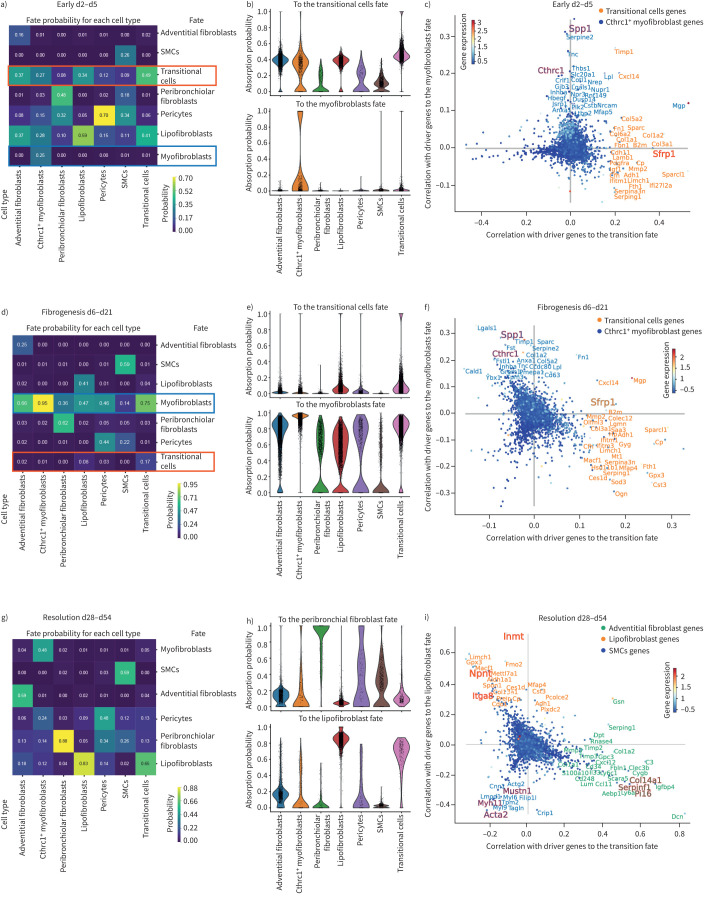
To infer lineage relationships between the different cell types and activation states we used CellRank (figure 4) [21]. We analysed three phases as “early injury” (figure 4a–c), “fibrogenesis” (figure 4d–f) and “resolution” (figure 4g–i). CellRank computed fate probabilities for each cell, showing their specific differentiation potential towards a “terminal state” end-point in the time-course data. Heatmaps demonstrate fate probability (figure 4a, d and g), and violin plots visualise the probability distribution among the cells for selected fates in each cell type (figure 4b, e and h). Key lineage driver genes correlating with different fate trajectories are depicted in scatter plots (figure 4c, f and i). Early injury phase analysis (days 2–5) showed that adventitial fibroblasts and lipofibroblasts had a high fate probability of transitioning to the Sfrp1+ state (figure 4a and b). Probabilities towards myofibroblasts were low, represented only by few Cthrc1+ myofibroblasts (figure 4a and b). Top driver genes towards transitional cells included various collagens, chemokines and notably Sfrp1, while the myofibroblast lineage showed marker genes including Tnc, Spp1 and Thbs1 (figure 4c). In the fibrogenesis phase (day 6 to day 21), fate probabilities predicted Sfrp1+ cells transitioning to myofibroblasts (figure 4d,e). Top driver genes towards myofibroblasts represented classical myofibroblast-associated genes including Lgals1, Sparc and Spp1 as well as Cthrc1 (figure 4f). In the resolution phase, differentiation probabilities suggested a reversion of Cthrc1+ myofibroblasts towards lipofibroblast, peribronchiolar fibroblast and pericyte states (figure 4g and i). This prediction correlates with previous observations of a two-way conversion between lipogenic and myogenic fibroblastic phenotypes [6].
FIGURE 4.
Lineage convergence and two-way conversion of fibroblasts in lung regeneration. a, d, g) CellRank's coarse-grained and directed transition matrix was calculated for the given three phases of the bleomycin lung injury time course, of early phase (day 2 to day 5), fibrogenesis phase (day 6 to day 21) and resolution phase (day 28 to day 54). Terminal macrostates were set manually and annotated according to their overlap with the underlying gene expression clusters. The heatmap represents the mean absorption probabilities of every cell in the given cell types to the terminal fates. b, e, h) Violin plots indicating the absorption probability towards the specified fate for every cell in all the cell types. c, f, i) Scatter plots show top driver genes, predicted to facilitate transition to the given terminal fate, visualised as gene-correlations between two terminal fate lineages. The top genes of a third lineage are visible by the third colour of the gene names. Dots are coloured according to their mean gene expression. SMC: smooth muscle cell.
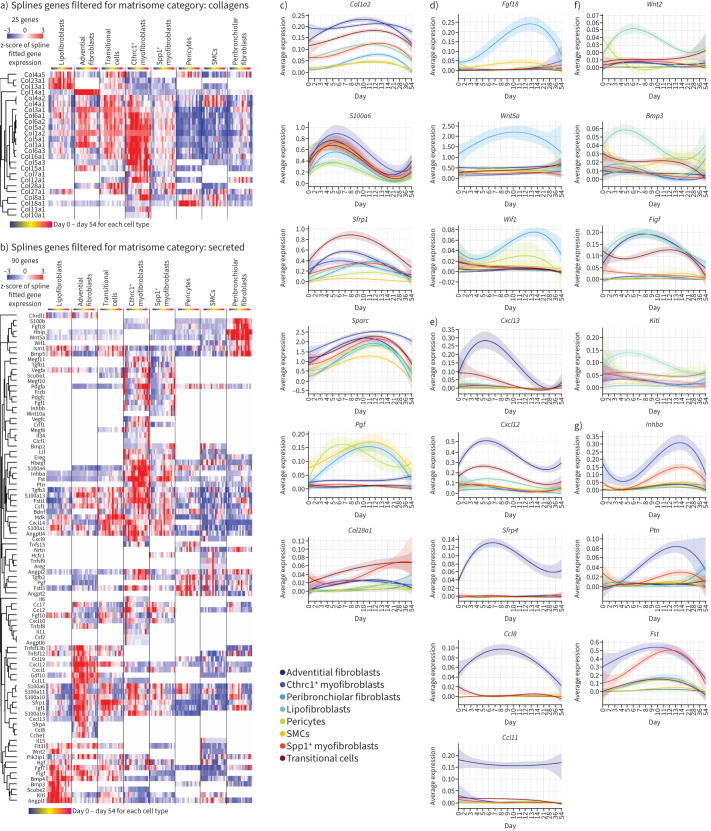
Next, we used a spline regression model revealing genes with differential expression in at least one cell type over time (supplementary table S4). We found 25 different collagens with dynamic expression patterns after injury or between various stromal cells (figure 5a). Additionally, 90 secreted matrisome [22] genes with significant expression changes along the injury time-course were identified (figure 5b). All cell types displayed transiently increased expression of collagen type-I (Col1a2) and the ECM protein Sparc, a marker for myofibroblasts (figure 5c) [23]. We also observed common early fibroblast activation events, like the early post-injury upregulation of S100a6 (figure 5c). Peribronchiolar fibroblasts specifically featured important secreted morphogens like Hhip, Wif1, Fgf18 and Wnt5a (figure 5d). This matters as fibroblast-derived Wnt-signaling, especially Wnt5a, defines a specific AT2 cell niche in normal homeostasis [24], is part of a distinct mesenchymal niche in human distal airways, and is secreted by LGR5+ fibroblasts [17]. Adventitial fibroblasts specifically produced the Wnt modulator Sfrp4, besides injury-induced chemokines like Cxcl13 for B-cell recruitment, and Cxcl12 and Ccl8 to attract T-cells, monocytes and neutrophils. Interestingly, the chemokine eotaxin (Ccl11) specifically marked adventitial fibroblasts, unaffected by injury (figure 5e). Lipofibroblasts specifically expressed the morphogens Wnt2 and Bmp3, plus stem cell factor (SCF) (Kitl) (figure 5f). The SCF-c-Kit pathway is activated in bleomycin-injured lungs, with potential profibrotic effects via recruitment of Kit+ immune cells to the lung [25].
FIGURE 5.
Highly specific expression changes in distinct fibroblast lineages. a, b) Heatmaps display genes that showed differential expression along the time-course in at least one cell type using a spline regression model for a) 25 different collagens and b) 90 secreted matrisome [22] genes. c–g) Line plots show average expression of indicated genes for each cell type along the bleomycin time course, for c) general mesenchymal marker genes, d) genes regulated over time in peribronchial fibroblasts, e) adventitial fibroblast, f) lipofibroblasts and g) Cthrc1+ myofibroblasts. SMC: smooth muscle cell.
Conclusively, our data suggest that early after injury multiple mesenchymal cell types converge towards Sfrp1+ transitional cells, which ultimately give rise to Cthrc1+ myofibroblasts, with a potential to revert towards lipofibroblasts, peribronchiolar fibroblasts and pericytes during the resolution phase.
TGFβ mediates differentiation of Sfrp1+ transitional fibroblasts into myofibroblasts
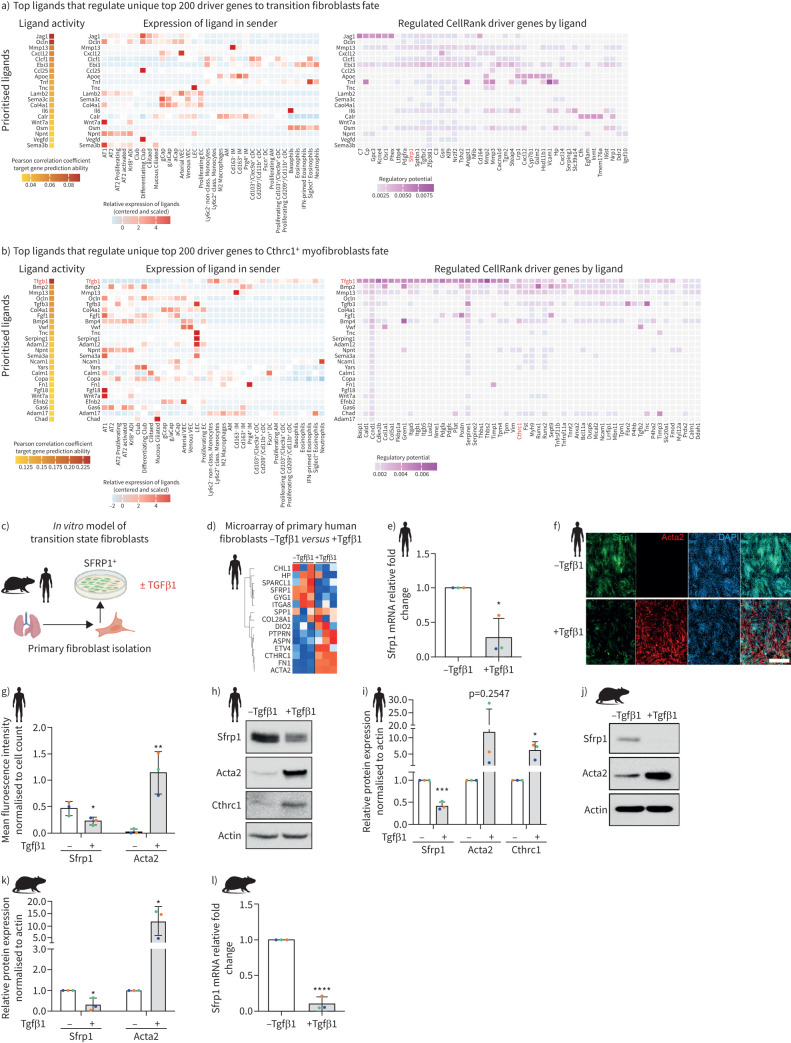
To infer potential regulators of the fibroblast cell state transitions after injury we used NicheNet [26], which predicted the ligands with highest probability to induce expression of the top driver genes from the CellRank outputs (figure 4) for Sfrp1+ transitional cells and Cthrc1+ myofibroblasts (figure 6a and b). The top ligand upstream of Sfrp1+ transitional cells was the Notch ligand Jag1, predominantly expressed in secretory airway epithelial cells, and to a lesser degree in alveolar epithelial and vascular endothelial cells (figure 6a). Consistent with our finding here, Notch deficiency in mesenchymal cells reportedly reduced fibrotic remodelling and myofibroblast differentiation in the bleomycin model [27]. Notably, upstream of Sfrp1+ transitional cell state driver genes the top ligands excluded Tgfβ, which was the top ranked ligand for Cthrc1+ myofibroblast driver genes. Tgfβ1 was the top ligand, primarily expressed by myeloid lineage immune cells, along with Tgfβ3 expressed by activated AT2 cells and lymphatic endothelial cells (figure 6b).
FIGURE 6.
Tgfβ1 drives conversion of Sfrp1+ transitional cells to Cthrc1+ myofibroblasts. a, b) NicheNet analysis based prediction of regulatory ligands upstream of fibroblast states. Heatmaps show top ranked ligands with highest potential of regulating the top 200 driver genes towards the fate of a) Sfrp1+ transitional fibroblasts and b) Cthrc1+ myofibroblasts. The left panel shows scaled expression of those ligands across cell types of the whole lung, while the right panel shows the top downstream target genes of each ligand. c) Experimental design. d) Heatmap shows z-scored gene expression from bulk transcriptomics after transforming growth factor (TGF)β1 treatment of primary human fibroblasts grown in vitro. e) mRNA (quantitative (q)PCR) expression of SFRP1 48 h after TGFβ1-treatment of primary human lung fibroblasts grown in vitro. f) Immunofluorescence staining and g) quantification thereof of primary human lung fibroblasts showing the expression of SFRP1 and ACTA2 with and without 48 h of TGFβ1-treatment. Scale bar=250 µm. h) Protein expression of SFRP1, ACTA2 and CTHRC1 by Western blotting and i) its quantification using human lung fibroblasts cultured in vitro for 48 h with and without TGFβ1. Representative of n=3. j) Protein expression of SFRP1 and ACTA2 by Western blotting and k) its quantification using mouse lung fibroblasts cultured in vitro for 48 h with and without TGFβ1. Representative of n=3. l) mRNA (qPCR) expression of Sfrp1 48 h after TGFβ1-treatment of primary mouse lung fibroblasts grown in vitro. All data are shown as mean±sd with n=3 from different mice or human donors. *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001 by unpaired two-tailed t-tests.
Culturing isolated primary mouse and human lung fibroblasts in vitro, we observed marker gene expression consistent with the in vivo Sfrp1+ transitional state (figure 6c and d). Myofibroblast identifiers like CTHRC1 and SPP1, and fibrosis-relevant ECM marker FN1, were upregulated upon TGFβ1 stimulation, while SFRP1 was downregulated (figure 6d). We validated these observations in human and mouse primary lung fibroblasts using quantitative PCR (figure 6e and l), immunofluorescence (figure 6f and g) and Western blotting (figure 6i–k). Thus, we validated our computational predictions, demonstrating TGFβ1 as a master switch controlling expression of SFRP1 and myofibroblast markers in primary fibroblasts.
SFRP1 modulates TGFβ1-induced invasion and RHOA activity in patient fibroblasts
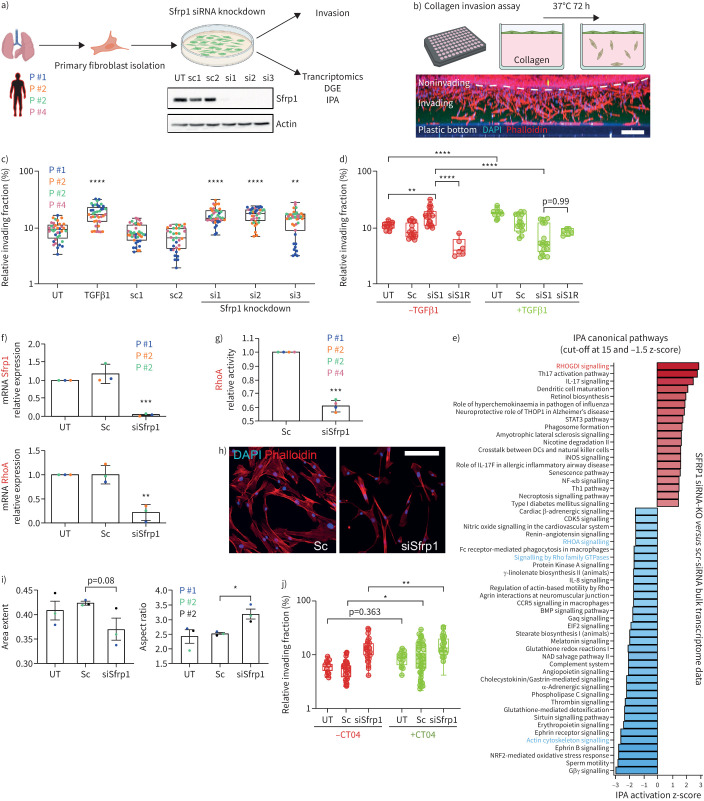
Secreted SFRP1 is an inhibitor of the Wnt signalling pathway [28, 29] and regulating tumour cell invasion [30, 31]. We previously identified a transcriptomic signature of collagen-invading lung fibroblasts characterised by a robust reduction in Sfrp1 expression [32]. In fibrogenesis, activated fibroblasts are thought to migrate into damaged tissue regions to form fibrotic foci. Transplantation experiments in mouse lungs post-bleomycin injury demonstrated superior migratory capacity of Cthrc1+ myofibroblasts compared to other fibroblast states [1]. To examine SFRP1's role in fibroblast invasion, we siRNA-depleted SFRP1 in pHLFs from four donors and performed collagen invasion assays and transcriptome analyses (figure 7a and b). SFRP1-deficient cells considerably increased their invasive capacity, similar to TGFβ1-treatment, compared to controls (p<0.001) (figure 7c). Surprisingly, combined SFRP1-siRNA and TGFβ1 treatments gave an inverted phenotype abrogating the TGFβ1-induced invasion. Notably, both increased invasion in untreated SFRP1 knockdown cells and reduced invasion in TGFβ1-stimulated ones, were at least partially restored by reconstituting SFRP1, indicating that SFRP1-dependent pathways modulate TGFβ1-driven fibroblast phenotypes (figure 7d). Additionally, direct treatment of invading fibroblasts with SFRP1 decreased their invasiveness (supplementary figure S8).
FIGURE 7.
SFRP1 in transition state fibroblasts regulates invasion, RHOA activity and cell shape. a) Primary human fibroblasts were isolated from lungs of human patients and cultured on rigid two-dimensional surfaces. SFRP1 expression was diminished in these cells by application of three different siRNAs targeting various exons. The successful knockdown of SFRP1 protein was confirmed by Western blot analysis. b) Primary human fibroblasts were applied on top of a collagen extracellular matrix (ECM) and left to invade for 72–96 h. Cells were stained for nuclei (4′,6-diamidino-2-phenylindole (DAPI), blue) and filamentous actin (phalloidin, red). A maximum intensity projection of a z-stack from one entire well is depicted as xz-view. Scale bar=200 µm. c) Transforming growth factor (TGF)β1-treatment as well as SFRP1-depletion augmented the ECM invasion capacity of primary human lung fibroblasts. Data are presented as mean±sd. One-way ANOVA with Tukey's multiple comparison test, n=4 (fibroblasts from four different patients displayed as distinct colours). Logarithmic (log10) y-axis. d) Addition of recombinant SFRP1 (R) rescued the increased invasion of SFRP1-depleted human lung fibroblasts (siS1R−TGFβ1) as well as partially the reduced invasion of TGFβ1-treated and SFRP1-depleted fibroblasts (siS1R+TGFβ1). Sc shows pooled data from Sc1-2. SiRNA shows pooled data from siRNA1-3. One-way ANOVA with Tukey's multiple comparison test, n=1–3 (fibroblasts from up to three different patients). Logarithmic (log10) y-axis. e) Ingenuity pathway analysis (IPA) canonical pathway analysis based on bulk transcriptome data analysis of SFRP1-siRNA-depleted compared to SFRP1-expressing primary human lung fibroblasts. IPA identified RHOA-signalling as a highly affected pathway in SFRP1-siRNA-depleted fibroblasts. f) Transcript analysis by quantitative PCR confirmed the successful reduction of Sfrp1 mRNA in Sfrp1-siRNA-treated primary human lung fibroblasts. RHOA mRNA was found to be considerably reduced in SFRP1-depleted fibroblasts. Data are presented as mean±sd. Unpaired two-tailed t-test. n=3 (fibroblasts from three different patients). g) Consistent with IPA analysis shown in (e) as well as reduction of RHOA mRNA displayed in (f), RHOA-GTPase activity was significantly reduced in SFRP1-depleted primary human lung fibroblasts. Data are presented as mean±sd. Unpaired two-tailed t-test. n=4 (fibroblasts from four different patients). h) Immunofluorescence labelling of filamentous actin cytoskeleton (phalloidin in red) indicated a morphological switch towards smaller and elongated cell shapes in SFRP1-depleted primary human lung fibroblasts. Scale bar=100 µm. i) Detailed cell morphological analysis using an automatised workflow in CellProfiler software. The unbiased quantification of 1000 cells from untreated (UT), scrambled-siRNA-treated (Sc) and SFRP1-siRNA-treated (siSfrp1) human lung fibroblasts confirmed a substantial switch towards more elongated (smaller area extent, higher aspect ratio) cell morphologies in SFRP1-depleted primary human lung fibroblasts. In total we analysed up to 15 000 single cells from each condition and patient. Data are presented as mean±sd. Unpaired t-test. p=0.08, n=3 (fibroblasts from three different patients). j) RHOA-GTPase inhibition by CT04 (C3 transferase) triggered an increase in the invasive capacity of primary human lung fibroblasts. Sc shows pooled data from Sc1-2. SiRNA shows pooled data from siRNA1-3. One-way ANOVA with Tukey's multiple comparison test, n=4 (fibroblasts from four different patients). Logarithmic (log10) y-axis. *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001.
Bulk transcriptomic analysis of SFRP1-depleted fibroblasts indicated a downregulation of RHOA-signalling-related pathways possibly through upregulation of RhoGDI inhibitory signals (figure 7e). We confirmed diminished expression of RHOA mRNA (figure 7f) and reduced RHOA-GTPase activity (figure 7g). Additionally, the SFRP1 knockdown induced cell morphology changes towards elongated cell shapes (figure 7h and i). To test how RhoA inhibition affects fibroblast invasion we used the RhoA inhibitor CT04 in SFRP1-depleted and control cells and found that CT04 treatment alone significantly increased cell invasion (figure 7j). However, we did find a small but significant additive effect on cell invasion using CT04 in SFRP1-decificient cells, suggesting that not all of the modulatory effects of SFRP1 on cell invasion strictly depend on the RhoA pathway (figure 7j).
In summary, our loss of function experiments in pHLFs reveal a function of SFRP1 in modulating TGFβ1 induced fibroblast invasion partially via regulation of the RHOA pathway.
Discussion
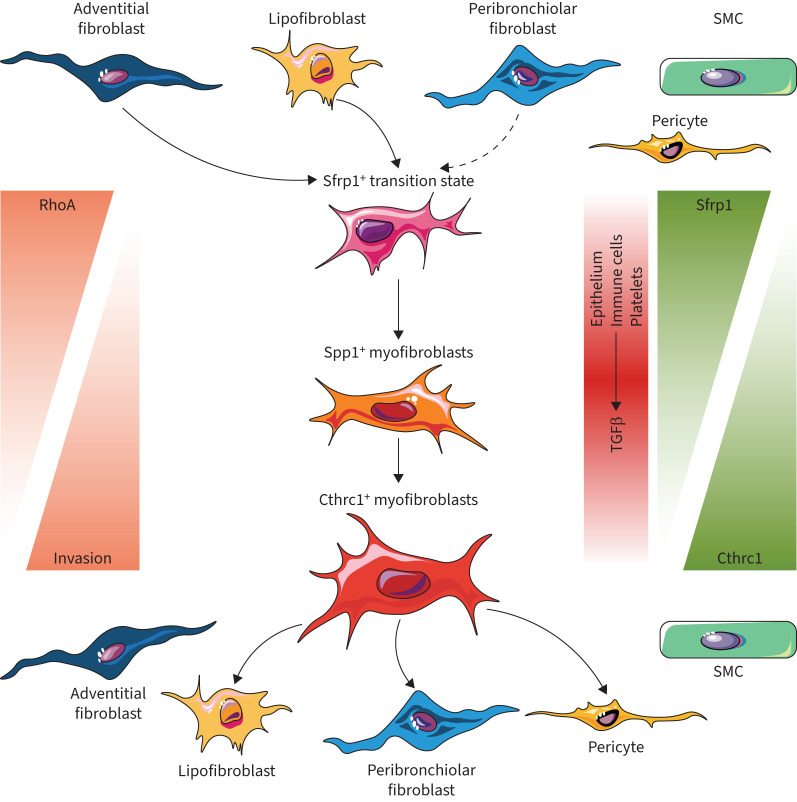
Fibroblasts are master orchestrators of tissue homeostasis, immune reactions and wound healing. We delineated injury-induced activation states of fibroblast lineages with specific niches in peribronchiolar, adventitial and alveolar lung locations. Longitudinal single-cell and lineage-tracing experiments uncovered a novel transitional cell state and highly cell-type- and lineage-specific paracrine signals mediating specialised functions in lung regeneration. Our analysis further reveals SFRP1 as a key modulator of TGFβ1-induced fibroblast invasion during the myofibroblastic transition in pulmonary fibrosis (figure 8).
FIGURE 8.
Model illustrating fibroblast phenotypic trajectories in lung repair and regeneration. Based on our data we propose the following model of the spatiotemporal evolution of distinct fibroblast states after lung injury. We discovered a novel transitional fibroblast state which is characterised by the expression of Sfrp1, as well as its emergence from adventitial, peribronchial and lipofibroblasts early after injury. Expression of Sfrp1 initially prevents transforming growth factor (TGF)β1 induced fibroblast invasion and RhoA activity. Prolonged exposure to TGFβ1 will in turn lead to increased RhoA activity, cell invasion, downregulation of Sfrp1 expression, and finally induction of the myofibroblast programme, including genes such as ACTA2, SPP1 and CTHRC1. Upon completion of epithelial regeneration, the Cthrc1+ myofibroblasts can resolve by differentiation back to a normal homeostatic phenotype. SMC: smooth muscle cell.
The current model of IPF/ILD pathogenesis involves aberrant and/or persistent repair associated with specific (epi-)genetic factors. The bleomycin mouse model likely recapitulates early stages of IPF/ILD pathogenesis more accurately and loses relevance at later stages of disease progression. Indeed, we found interesting similarities of SFRP1+ transitional fibroblasts in both early stages after bleomycin injury and mildly affected regions in IPF explants (scored using micro-CT for disease severity). This advocates a high clinical relevance for fibroblast activation states in the bleomycin mouse model mirroring early stages of ILD/IPF.
Our trajectory inference benefits from the very high time resolution after injury (daily sampling) and suggests that adventitial fibroblasts, peribronchiolar and alveolar fibroblasts transcriptionally converge after injury. To our knowledge, this has not been demonstrated before and will require the development of specific genetic lineage tracing tools for experimentally validating our computational predictions. We used single-cell transcriptomics on lungs from the Tcf21-Cre reporter mice to address the lineage trajectory of lipofibroblasts. However, a limitation was that all other mesenchymal cell types with exception of the Hhip+ peribronchiolar fibroblasts were also lineage-labelled due to low-level Tcf21 expression. Tcf21-lineage-negative Hhip+ cells were interestingly heavily expanded after injury. We show that these cells can reside in direct physical contact with AT2 cells and are the exclusive source of morphogens such as Wnt5a after injury, which probably has important implications for AT2 progenitor cell function [24].
Previous work established that fibroblast invasion contributes to lung fibrosis progression and severity [1, 33–35]. We here show that the early Sfrp1+ transitional state of injury-activated fibroblasts is noninvasive, suggesting that injury-activated fibroblasts serve local niche-functions, like recruiting immune cells by adventitial fibroblasts, or activating epithelial stem cells by lipofibroblasts, pericytes and Hhip+ fibroblasts. Our data indicate that recruited myeloid cells, which heavily accumulate early in the bleomycin model [36], plus activated epithelial and lymphatic endothelial cells, produce Tgfβ to switch the noninvasive Sfrp1+ transitional cells into highly invasive Cthrc1+/Spp1+ myofibroblasts. Indeed, a profibrotic cell circuit between macrophages and fibroblasts has been described, which requires cadherin-11 mediated direct cell–cell interactions to promote latent TGFβ activation [37].
Interestingly, the classical view of sessile actomyosin based contractile myofibroblasts contradicts our observations of highly invasive and mobile CTHRC1+/ACTA2+ myofibroblasts. Our cell invasion data demonstrate that availability of SFRP1 in pHLFs dictates whether TGFβ1 induces pro-invasive or anti-invasive behaviour. This interesting crosstalk of SFRP1 and TGFβ1 signalling may enable precise timing of distinct fibroblast functions (e.g. invasion versus ECM remodelling and contraction) during the highly concerted tissue repair post-injury. In pathologies like IPF, myofibroblasts invade to organise themselves into dense accumulations called fibroblast foci, a process which is also recapitulated in the bleomycin model [34]. One limitation of the current study was the uncertainty regarding the commercial anti-SFRP1 antibodies’ capacity to neutralise SFRP1 activity. Thus, the mechanisms discovered in this study have direct implications for IPF opening ways for new therapeutic interventions to limit or reverse fibroblast foci formation.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary figures and methods. ERJ-01326-2023.Supplement (2MB, pdf)
Supplementary table S1. Marker gene per cell type for 10x Tcf21 PBS subset. ERJ-01326-2023.Table_S1 (899KB, xlsx)
Supplementary table S2. Marker gene per cell type for 10x Tcf21 bleomycin day14 subset. ERJ-01326-2023.Table_S2 (1.8MB, xlsx)
Supplementary table S3. Marker gene per cell type for Dropseq bleomycin time course. ERJ-01326-2023.Table_S3 (191.5KB, xlsx)
Supplementary table S4. Spline table for DGE genes across time points per cell type. ERJ-01326-2023.Table_S4 (13.9MB, xlsx)
Supplementary table S5. Sequences for SCRINSHOT probes. ERJ-01326-2023.Table_S5 (23.3KB, xlsx)
Supplementary table S6. List of antibodies. ERJ-01326-2023.Table_S6 (14KB, xlsx)
Supplementary table S7. Deparaffinisation protocol. ERJ-01326-2023.Table_S7 (13.6KB, xlsx)
Supplementary table S8. Sequence of primers used for qRT-PCR studies. ERJ-01326-2023.Table_S8 (13.4KB, xlsx)
Shareable PDF
Acknowledgements
We gratefully acknowledge the provision of human biomaterial (primary fibroblasts) and clinical data from the CPC-M bioArchive and its partners at the Asklepios Biobank Gauting, the LMU Hospital and the Ludwig-Maximilians-Universität München. We thank the patients and their families for their support. We are grateful to M. Neumann and A. van den Berg (Comprehensive Pneumology Center (CPC), Munich, Germany) for providing superb technical support. We also thank Inti Alberto de la Rosa Velazquez and the team from the genomics core facility of Helmholtz Munich for expert sequencing service.
Footnotes
Author contributions: Conceptualisation and supervision: G. Burgstaller and H.B. Schiller. Methodology and investigation: C.H. Mayr, A. Sengupta, M. Ansari, J.C. Pestoni, P. Ogar, I. Angelidis, I.E. Fernandez, A. Liontos, J.A. Rodriguez-Castillo, N.J. Lang, M. Strunz, S. Asgharpour, D. Porras-Gonzalez, B. Oehrle, V. Viteri-Alvarez, M. Irmler, M. Tallquist, M. Gerckens, R.M. Wasnick and K. Ahlbrecht. Bioinformatic analysis and software: C.H. Mayr, M. Ansari and F.J. Theis. Surgical work and human tissue: G.M. Stoleriu, J. Behr, N. Kneidinger, W.A. Wuyts and L.J. De Sadeleer. Visualisation and writing: H.B. Schiller, G. Burgstaller, C.H. Mayr, A. Sengupta and R.M. Wasnick. Resources and funding: H.B. Schiller, G. Burgstaller, O. Eickelberg, A.Ö. Yildirim, F.J. Theis, R.E. Morty and C. Samakovlis.
This article has an editorial commentary: https://doi.org/10.1183/13993003.02188-2023
Conflict of interest: M. Gerckens reports grants from Stiftung Atemweg e.V. and a patent pending EP21178481 “Novel anti-fibrotic drugs”, outside the submitted work. M. Tallquist reports support for the present manuscript from NIH (5R21HL156112). J. Beckers reports funding for the current manuscript, as well as funding for consumables outside the submitted work, from Helmholtz Zentrum München GmbH. O. Eickelberg reports support for the present manuscript from R01 HL146519; in addition, O. Eickelberg reports consulting fees from Blade Therapeutics, Yap Therapeutics and Pieris Pharmaceuticals, stock or stock options from Blade Therapeutics, outside the submitted work. J. Behr reports a leadership role as Chair of Guideline Committee of the German Respiratory Society (DGP), outside the submitted work. W.A. Wuyts reports grants, consulting fees, lecture honoraria and advisory board participation from Roche, Pliant, Boehringer Ingelheim, Alentis and Galapagos. K. Ahlbrecht reports support for the present manuscript from Max Planck Society, German Center for Lung Research (Deutsches Zentrum für Lungenforschung; DZL), Federal Ministry of Higher Education, Research and the Arts of the State of Hessen LOEWE, Programme through grant UGMLC; in addition, K. Ahlbrecht reports grants from Rhon Klinikum AG (grant FI_71), outside the submitted work. R.E. Morty reports leadership roles as Editor-in-Chief, American Journal of Physiology – Lung Cellular and Molecular Physiology and Group Chair, Group 07.08 Lung and Airway Development, at the European Respiratory Society, outside the submitted work. C. Samakovlis reports grants from Swedish Research Council, Swedish Cancer Society, DFG, Stockholm University, Stockholm, Sweden, Justus-Liebig University, Giessen, Germany, DiscovAir, EU, payment for expert testimony from Swedish Cancer Society and Wallengberg Foundation, and a leadership role with Royal Academy of Science, Sweden, outside the submitted work. F.J. Theis reports support for the present manuscript from the Chan Zuckerburg Foundation (grant number 2019- 002438), as well as consulting fees from Roche, Immunai, Singularity, Omniscope and CytoReason, lecture honoraria from Genentech Research Organisation, AMGEN GmbH, Munich, Roche Germany, Roche, Basel, ETH Zurich, Vizgen, ThirdRockVentures and Pfizer, advisory board participation with Max Planck Institute for Intelligent Systems, Berlin Institute of Health and EMBL, and stock or stock options from Cellarity, outside the submitted work. H.B. Schiller reports support for the present manuscript from Helmholtz Association, Deutsches Zentrum für Lungenforschung (DZL) and CZI/H2020 (discovair). The remaining authors have no potential conflicts of interest to disclose.
Support statement: We acknowledge support by the German Center for Lung Research (DZL), the Helmholtz Association, the European Union's Horizon 2020 research and innovation programme (grant agreement 874656) and the Chan Zuckerberg Initiative (CZF2019-002438). Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Tsukui T, Sun K-H, Wetter JB, et al. . Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Commun 2020; 11: 1920. doi: 10.1038/s41467-020-15647-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zepp JA, Zacharias WJ, Frank DB, et al. . Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell 2017; 170: 1134–1148. doi: 10.1016/j.cell.2017.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie T, Wang Y, Deng N, et al. . Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep 2018; 22: 3625–3640. doi: 10.1016/j.celrep.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J-H, Tammela T, Hofree M, et al. . Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell 2017; 170: 1149–1163. doi: 10.1016/j.cell.2017.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riccetti M, Gokey JJ, Aronow B, et al. . The elephant in the lung: integrating lineage-tracing, molecular markers, and single cell sequencing data to identify distinct fibroblast populations during lung development and regeneration. Matrix Biol 2020; 91–92: 51–74. doi: 10.1016/j.matbio.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Agha E, Moiseenko A, Kheirollahi V, et al. . Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell 2017; 20: 571. doi: 10.1016/j.stem.2017.03.011 [DOI] [PubMed] [Google Scholar]
- 7.Kheirollahi V, Wasnick RM, Biasin V, et al. . Metformin induces lipogenic differentiation in myofibroblasts to reverse lung fibrosis. Nat Commun 2019; 10: 2987. doi: 10.1038/s41467-019-10839-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung C, Linn G, Chow Y-H, et al. . Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 2013; 188: 820–830. doi: 10.1164/rccm.201212-2297OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frangogiannis N. Transforming growth factor-β in tissue fibrosis. J Exp Med 2020; 217: e20190103. doi: 10.1084/jem.20190103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinz B, Lagares D. Evasion of apoptosis by myofibroblasts: a hallmark of fibrotic diseases. Nat Rev Rheumatol 2020; 16: 11–31. doi: 10.1038/s41584-019-0324-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biasin V, Crnkovic S, Sahu-Osen A, et al. . PDGFRα and αSMA mark two distinct mesenchymal cell populations involved in parenchymal and vascular remodeling in pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2020; 318: L684–L697. doi: 10.1152/ajplung.00128.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun K-H, Chang Y, Reed NI, et al. . α-Smooth muscle actin is an inconsistent marker of fibroblasts responsible for force-dependent TGFβ activation or collagen production across multiple models of organ fibrosis. Am J Physiol Lung Cell Mol Physiol 2016; 310: L824–L836. doi: 10.1152/ajplung.00350.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J, Ivey MJ, Deana Y, et al. . The Tcf21 lineage constitutes the lung lipofibroblast population. Am J Physiol Lung Cell Mol Physiol 2019; 316: L872–L885. doi: 10.1152/ajplung.00254.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Travaglini KJ, Nabhan AN, Penland L, et al. . A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020; 587: 619–625. doi: 10.1038/s41586-020-2922-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angelidis I, Simon LM, Fernandez IE, et al. . An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat Commun 2019; 10: 963. doi: 10.1038/s41467-019-08831-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuppe C, Ibrahim MM, Kranz J, et al. . Decoding myofibroblast origins in human kidney fibrosis. Nature 2021; 589: 281–286. doi: 10.1038/s41586-020-2941-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadur Lakshminarasimha Murthy P, Sontake V, Tata A, et al. . Human distal lung maps and lineage hierarchies reveal a bipotent progenitor. Nature 2022; 604: 111–119. doi: 10.1038/s41586-022-04541-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sountoulidis A, Liontos A, Nguyen HP, et al. . SCRINSHOT enables spatial mapping of cell states in tissue sections with single-cell resolution. PLoS Biol 2020; 18: e3000675. doi: 10.1371/journal.pbio.3000675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiller HB, Fernandez IE, Burgstaller G, et al. . Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Mol Syst Biol 2015; 11: 819. doi: 10.15252/msb.20156123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayr CH, Simon LM, Leuschner G, et al. . Integrative analysis of cell state changes in lung fibrosis with peripheral protein biomarkers. EMBO Mol Med 2021; 13: e12871. doi: 10.15252/emmm.202012871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange M, Bergen V, Klein M, et al. . CellRank for directed single-cell fate mapping. Nat Methods 2022; 19: 159–170. doi: 10.1038/s41592-021-01346-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis MN, Horne-Badovinac S, Naba A. In-silico definition of the Drosophila melanogaster matrisome. Matrix Biol Plus 2019; 4: 100015. doi: 10.1016/j.mbplus.2019.100015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klingberg F, Hinz B, White ES. The myofibroblast matrix: implications for tissue repair and fibrosis. J Pathol 2013; 229: 298–309. doi: 10.1002/path.4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nabhan AN, Brownfield DG, Harbury PB, et al. . Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 2018; 359: 1118–1123. doi: 10.1126/science.aam6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding L, Dolgachev V, Wu Z, et al. . Essential role of stem cell factor-c-Kit signalling pathway in bleomycin-induced pulmonary fibrosis. J Pathol 2013; 230: 205–214. doi: 10.1002/path.4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods 2020; 17: 159–162. doi: 10.1038/s41592-019-0667-5 [DOI] [PubMed] [Google Scholar]
- 27.Hu B, Wu Z, Bai D, et al. . Mesenchymal deficiency of Notch1 attenuates bleomycin-induced pulmonary fibrosis. Am J Pathol 2015; 185: 3066–3075. doi: 10.1016/j.ajpath.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, Sheng T, Stelter AA, et al. . Suppressing Wnt signaling by the hedgehog pathway through sFRP-1. J Biol Chem 2006; 281: 35598–35602. doi: 10.1074/jbc.C600200200 [DOI] [PubMed] [Google Scholar]
- 29.Gibb N, Lavery DL, Hoppler S. sfrp1 promotes cardiomyocyte differentiation in Xenopus via negative-feedback regulation of Wnt signalling. Development 2013; 140: 1537–1549. doi: 10.1242/dev.088047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y-L, Wang T-H, Hsu H-C, et al. . Overexpression of CTHRC1 in hepatocellular carcinoma promotes tumor invasion and predicts poor prognosis. PLoS One 2013; 8: e70324. doi: 10.1371/journal.pone.0070324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baharudin R, Tieng FYF, Lee L-H, et al. . Epigenetics of SFRP1: the dual roles in human cancers. Cancers 2020; 12: 445. doi: 10.3390/cancers12020445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oehrle B, Burgstaller G, Irmler M, et al. . Validated prediction of pro-invasive growth factors using a transcriptome-wide invasion signature derived from a complex 3D invasion assay. Sci Rep 2015; 5: 12673. doi: 10.1038/srep12673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahluwalia N, Grasberger PE, Mugo BM, et al. . Fibrogenic lung injury induces non-cell-autonomous fibroblast invasion. Am J Respir Cell Mol Biol 2016; 54: 831–842. doi: 10.1165/rcmb.2015-0040OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukui T, Ueha S, Abe J, et al. . Qualitative rather than quantitative changes are hallmarks of fibroblasts in bleomycin-induced pulmonary fibrosis. Am J Pathol 2013; 183: 758–773. doi: 10.1016/j.ajpath.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Jiang D, Liang J, et al. . Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J Exp Med 2011; 208: 1459–1471. doi: 10.1084/jem.20102510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strunz M, Simon LM, Ansari M, et al. . Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat Commun 2020; 11: 3559. doi: 10.1038/s41467-020-17358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lodyga M, Cambridge E, Karvonen HM, et al. . Cadherin-11-mediated adhesion of macrophages to myofibroblasts establishes a profibrotic niche of active TGF-β. Sci Signal 2019; 12: eaao3469. doi: 10.1126/scisignal.aao3469 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary figures and methods. ERJ-01326-2023.Supplement (2MB, pdf)
Supplementary table S1. Marker gene per cell type for 10x Tcf21 PBS subset. ERJ-01326-2023.Table_S1 (899KB, xlsx)
Supplementary table S2. Marker gene per cell type for 10x Tcf21 bleomycin day14 subset. ERJ-01326-2023.Table_S2 (1.8MB, xlsx)
Supplementary table S3. Marker gene per cell type for Dropseq bleomycin time course. ERJ-01326-2023.Table_S3 (191.5KB, xlsx)
Supplementary table S4. Spline table for DGE genes across time points per cell type. ERJ-01326-2023.Table_S4 (13.9MB, xlsx)
Supplementary table S5. Sequences for SCRINSHOT probes. ERJ-01326-2023.Table_S5 (23.3KB, xlsx)
Supplementary table S6. List of antibodies. ERJ-01326-2023.Table_S6 (14KB, xlsx)
Supplementary table S7. Deparaffinisation protocol. ERJ-01326-2023.Table_S7 (13.6KB, xlsx)
Supplementary table S8. Sequence of primers used for qRT-PCR studies. ERJ-01326-2023.Table_S8 (13.4KB, xlsx)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-01326-2023.Shareable (741.3KB, pdf)
Data Availability Statement
RNA-seq data were deposited to the Gene Expression Omnibus (GEO) database. The high temporal mesenchymal enriched Drop-seq data can be found with the accession code GSE207851, and the Tcf21-lineage labelled mesenchymal 10x data with the accession code GSE207687. Microarray data of pHLFs (Sfrp1-siRNA knockdown) can be found with the accession code GSE207561.