Abstract
Objectives
Antimicrobial-resistant Neisseria gonorrhoeae (NG) is a concern. Little is known about antimicrobial susceptibility profiles and associated genetic resistance mechanisms of NG in Madagascar. We report susceptibility data of NG isolates obtained by the medical laboratory (CBC) of the Institut Pasteur de Madagascar, Antananarivo, Madagascar, during 2014–2020. We present antimicrobial resistance mechanisms data and phenotype profiles of a subset of isolates.
Methods
We retrieved retrospective data (N=395) from patients with NG isolated during 2014−2020 by the CBC. We retested 46 viable isolates including 6 found ceftriaxone and 2 azithromycin resistant, as well as 33 isolated from 2020. We determined minimal inhibitory concentrations for ceftriaxone, ciprofloxacin, azithromycin, penicillin, tetracycline and spectinomycin using Etest. We obtained whole-genome sequences and identified the gene determinants associated with antimicrobial resistance and the sequence types (STs).
Results
Over the study period, ceftriaxone-resistant isolates exceeded the threshold of 5% in 2017 (7.4% (4 of 54)) and 2020 (7.1% (3 of 42)). All retested isolates were found susceptible to ceftriaxone, azithromycin and spectinomycin, and resistant to ciprofloxacin. The majority were resistant to penicillin (83% (38 of 46)) and tetracycline (87% (40 of 46)). We detected chromosomal mutations associated with antibiotic resistance in gyrA, parC, penA, ponA, porB and mtrR genes. None of the retested isolates carried the mosaic penA gene. The high rate of resistance to penicillin and tetracycline is explained by the presence of bla TEM (94.7% (36 of 38)) and tetM (97.5% (39 of 40)). We found a high number of circulating multilocus STs. Almost half of them were new types, and one new type was among the four most predominant.
Conclusions
Our report provides a detailed dataset obtained through phenotypical and genotypical methods which will serve as a baseline for future surveillance of NG. We could not confirm the occurrence of ceftriaxone-resistant isolates. Our results highlight the importance of implementing quality-assured gonococcal antimicrobial resistance surveillance in Madagascar.
Keywords: NEISSERIA GONORRHOEAE; AFRICA; Drug Resistance, Bacterial; MOLECULAR TYPING
WHAT IS ALREADY KNOWN ON THIS TOPIC.
Neisseria gonorrhoeae (NG) is increasingly becoming resistant to antimicrobials.
WHAT THIS STUDY ADDS
There is a paucity of data concerning the antimicrobial resistance (AMR) of NG circulating in Madagascar. We provide data on the phenotypes and associated antimicrobial resistance mechanisms of NG isolated in a medical laboratory in Antananarivo, Madagascar, and have set a baseline of circulating genotypes.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Quality-assured gonococcal AMR surveillance should be implemented in Madagascar. National Reference Laboratories should be assigned to ensure the quality of the AMR data. Whole-genome sequencing to identify resistance mechanisms in NG isolates is feasible and should be considered in the design of AMR surveillance of NG in Madagascar.
Introduction
Since the emergence of antimicrobials to treat gonorrhoea, Neisseria gonorrhoeae (NG) has gained mechanisms to evade their harmful effects. Today, antimicrobial-resistant NG is a global threat1 2 and features on the WHO’s and Centers for Disease Prevention and Control’s priority lists.1 3
The NG antimicrobial-resistance (AMR) mechanisms are chromosomally mediated, acquired through mutations, horizontal gene transfer and homologous recombination, or are introduced by mobile genetic elements such as plasmids.4 Several chromosomal-mediated determinants leading to resistance have been identified.4 The mutations in genes like penA, ponA, mtrR, porB, rpsJ, gyrA and parC alter or modify antimicrobial targets or influx/efflux. NG can also obtain genes through plasmid acquisition.4 Several β-lactamases, hydrolysing β-lactam antimicrobials and β-lactamase-encoding plasmids have been described.4 5 tetM is another plasmid-borne gene-mediating tetracycline resistance by replacing tetracycline from its ribosome target resulting in further protein synthesis.4 The gene is carried by conjugative plasmids.5 Plasmid-mediated resistance results in rapid high resistance levels, while chromosomally mediated resistance is acquired gradually through mutations or recombination in the chromosome.4 5
Whole-genome sequencing (WGS) is useful for identifying genetic determinants and predicting AMR and can guide treatment guidelines in specifying which antibiotics to exclude.6 In addition, molecular typing schemes based on WGS are used to determine the spread of genotypes and describe their relatedness and evolution.6
A national surveillance programme such as the WHO-Enhanced Gonococcal Antimicrobial Surveillance Programme has not been implemented in Madagascar.7 Also, studies conducted on circulating NG and their antimicrobial susceptibility (AMS) profiles are rare. Consequently, data on the NG AMS profiles in Madagascar are scarce.
Overall, laboratory-based diagnosis of sexually transmitted infections does not exist in Madagascar and the syndromic approach is widely applied in all levels of healthcare. The medical laboratory (CBC) of the Institut Pasteur de Madagascar is one of few laboratories performing molecular testing for NG and Chlamydia trachomatis and to our knowledge the only medical laboratory to conduct AMS Etesting for NG.
We aimed to describe the AMS profiles of NG isolated from a patient population attending the CBC from 2014 to 2020. In addition, we performed phenotypical and genomic characterisation of AMR in a subset of isolates including all viable isolates from 2020.
Methods
Study method
We retrieved retrospective patient data collected by the CBC, Antananarivo, Madagascar, from 2014 to 2020. Physicians refer patients to the CBC for medical biological analysis. The CBC’s medical laboratory staff (nurses, midwives) collect the specimens. The final diagnosis is made by the referring physician who will prescribe treatment if needed. The patient (or their employer or health insurance, if applicable) will pay for the costs.
We exported coded patient and specimen data including minimal inhibitory concentrations (MICs) of ceftriaxone, azithromycin and ciprofloxacin, as well as β-lactamase results of the NG isolates from the laboratory information system.
The CBC used Modified Thayer-Martin medium, incubated at 35±2°C in a 5% CO2 atmosphere for 18–48 hours, for NG isolation. NG-suspected colonies were identified by Matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. MICs for ceftriaxone, ciprofloxacin and azithromycin were determined using Etest (bioMérieux, Marcy-l’Etoile, France) on enriched (PolyViteX) chocolate agar incubated at 35±2°C in a 5% CO2 atmosphere for 18–24 hours. β-lactamase production was detected using a cefinase disk (bioMérieux, Marcy-l’Etoile, France). Isolates were stored in skim milk +20% glycerol at −80°C.
Staff members of the experimental bacteriology laboratory (UBEX) retrieved all NG isolates stored at the CBC and transported them (<100 m) under frozen conditions using cool boxes. At UBEX, we revived and retested a subset of isolates using the same method as described above, except that we revived the isolates using enriched (PolyViteX) chocolate agar media. We determined MICs for ceftriaxone, ciprofloxacin, azithromycin, penicillin, tetracycline and spectinomycin using Etest as described above. We used WHO reference gonococcal strains (WHO F, L, O, P, W, V, X, Y, Z) for quality control (QC).8
We interpreted MIC results using the clinical breakpoints recommended by the Comité de l’Antibiogramme de la Société Française de Microbiologie, 2021, V.1.0 April, based on European Committee on AMS Testing recommendations (online supplemental table 1). We defined isolates as reduced susceptible when they were categorised as ‘susceptible, increased exposure, category I’. This definition means that there is a likelihood of therapeutic success with an increased exposure to the antimicrobial.
sextrans-2023-055878supp001.xlsx (38.8KB, xlsx)
Whole-genome sequencing
We extracted genomic DNA from purified cultures using QIAamp DNA Mini Kit on a Qiacube automate (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. We checked DNA quality using nanodrop before shipment to the National Institute for Communicable Diseases, Johannesburg, South Africa, where WGS was performed using Illumina MiSeq next-generation sequencing technology with DNA libraries prepared using Nextera XT DNA Library Preparation Kit (Illumina, San Diego, California, USA), followed by 2×300 paired-end sequencing runs with ~100 times coverage.
Bioinformatic analysis
We used TORMES pipeline V.1.2.19 for whole-genome analysis as briefly described hereafter. We applied Prinseq10 and Trimmomatic11 for QC and filtering of the reads, respectively. We performed de novo assembly using SPAdes12 and assembly statistics using QUAST.13 We used Kraken214 to infer the taxonomy of each isolate, Prodigal15 and Prokka16 for gene prediction and annotation. We screened for AMR genes using ABRicate (https://github.com/tseemann/abricate) with the databases ResFinder,17 CARD18 and ARG-ANNOT.19
We removed contigs sizes <500 bp with a local script and used quality thresholds of assemblies as previously described.20
We obtained multilocus sequence types (STs) and genetic AMR determinants from Pathogenwatch.21
We identified plasmids associated with tetM and bla TEM genes using the Blastn program and a custom plasmid database (online supplemental table 2). We considered a coverage and identity score of >99% and ≥99%, respectively.
We constructed the phylogeny using the core genome single nucleotide polymorphism (SNP) analysis on the Pathogenwatch platform combined with Microreact for visualisation and rooted the tree at the midpoint.22
We completed our study dataset with the reference sequences from WHO, available under BioProject accession no. PRJEB14020 in the European Nucleotide Archive ENA.8
Statistical method
We summarised coded patient and bacteriological data using medians and IQRs for continuous variables and frequencies and proportions for categorical data.
We defined MIC50 and MIC90 as the lowest concentration of the antimicrobial at which 50% and 90% of the isolates were inhibited, respectively.
Results
NG isolates
Over the study period, the CBC identified 395 NG isolates. More than half of them (58%) were obtained from women. Overall, the age of the patients ranged from <1 year to 73 years, women (n=229) being slightly younger (median: 30 years) compared with men (n=166) (median: 34 years).
NG was mainly isolated from urogenital specimens (388 of 395), but also from sperm (n=3), blood (n=1), puncture fluid (n=2) and a not specified wound (n=1).
Antimicrobial susceptibility of NG over time
Of the 389 isolates with ceftriaxone MIC results, 372 (96%) isolates were susceptible and 17 (4%) were resistant. The resistance rate exceeded the 5% threshold in 2017 and 2020. Over the years, the MIC50 remained at 0.002 µg/mL, and the MIC90 varied from 0.004 to 0.02 µg/mL, with lower concentrations during the last 3 years (table 1).
Table 1.
Susceptibility to ceftriaxone and ciprofloxacin of Neisseria gonorrhoeae over the study period 2014–2020
| Year | Total number of isolates (N=395) | Ceftriaxone (n=389) | Ciprofloxacin (n=364) | |||||||||
| No data | R | S | MIC50 (µg/mL) | MIC90 (µg/mL) | No data | R | RS | S | MIC50 (µg/mL) | MIC90 (µg/mL) | ||
| 2014 | 31 | 3 | 0 | 28 | 0.002 | 0.02 | 2 | 29 | 0 | 0 | 2 | 4 |
| 2015 | 67 | 0 | 3 | 64 | 0.002 | 0.006 | 21 | 46 | 0 | 0 | 2 | 3 |
| 2016 | 35 | 0 | 1 | 34 | 0.002 | 0.02 | 4 | 31 | 0 | 0 | 1 | 2 |
| 2017 | 56 | 2 | 4 | 50 | 0.002 | 0.016 | 1 | 50 | 2 | 3 | 1 | 3 |
| 2018 | 82 | 1 | 3 | 78 | 0.002 | 0.004 | 2 | 79 | 0 | 1 | 1.5 | 3 |
| 2019 | 82 | 0 | 3 | 79 | 0.002 | 0.006 | 1 | 81 | 0 | 0 | 2 | 4 |
| 2020 | 42 | 0 | 3 | 39 | 0.002 | 0.004 | 0 | 42 | 0 | 0 | 1.5 | 3 |
Breakpoints used: for ceftriaxone: S: ≤0.125 µg/mL, R: >0.125 µg/mL; for ciprofloxacin: S: ≤0.03 µg/mL, R: >0.06 µg/mL, RS: >0.03–0.06 µg/mL.
MIC50: lowest concentration of the antimicrobial at which 50% of the isolates are inhibited.
MIC90: lowest concentration of the antimicrobial at which 90% of the isolates are inhibited.
MIC, minimum inhibitory concentration; R, resistant; RS, reduced susceptible; S, susceptible.
Among the 364 isolates tested against ciprofloxacin, 358 (98%) were resistant. Susceptible isolates were detected in 2017 (n=3) and 2018 (n=1). Two isolates from 2017 tested reduced susceptible. The MIC50 (range 1–2 µg/mL) and MIC90 (range 2–4 µg/mL) values persisted over the years (table 1).
Azithromycin was tested for 95 isolates: 53 in 2019 and 42 in 2020. Three isolates had MICAZT >0.5 µg/mL and were deemed resistant. One resistant strain from 2019 had a MICAZT of 1.5 µg/mL and the two resistant strains from 2020 had MICAZT of 0.64 and 0.75 µg/mL. The MIC90 evolved from 0.064 µg/mL in 2019 to 0.32 µg/mL in 2020.
Almost all isolates produced β-lactamase. Exceptions were found in 2016, 2017 and 2019 with 6%, 2% and 4% of the NG isolates lacking β-lactamase, respectively.
Subset of NG isolates
We retrieved all isolates from 2020 and those found ceftriaxone and/or azithromycin resistant or reduced susceptible in previous years from the CBC collection. We recovered 68 isolates, of them 46 were identified as NG: 33 isolates from 2020, 6 previously determined resistant to ceftriaxone, 2 previously tested resistant to azithromycin, 5 isolates had no specific characteristics.
Online supplemental tables 3 and 4 show the sequence quality data and MICs of the tested antimicrobials and associated AMR mechanisms of the NG isolates, respectively.
All isolates were susceptible to ceftriaxone using Etest, and all penA alleles coding for the penicillin-binding protein 2 were non-mosaic: the most prevalent was penA.14 (n=20) followed by penA.19 (n=12), penA.2 (n=7), penA.22 (n=6) and penA.9 (n=1) (online supplemental table 4).
All tested susceptible to azithromycin. A 57 adenosine deletion (57Adel) in mtrR promoter was found in three isolates (MICAZT (0.064–0.125 µg/mL)), two were combined with a mutation G45D in mtrR coding region. One isolate (MICAZT=0.064 µg/mL) carried a disrupted mtrR.
In contrast, all isolates were found phenotypically resistant to ciprofloxacin. We detected higher MICCIP when mutations in both gyrA and parC occurred. Conversely, two isolates did not possess any mutation in the parC genes and displayed a low-level resistance to ciprofloxacin (MICCIP≤0.75 µg/mL) (table 2).
Table 2.
Genomic resistance mechanisms and minimal inhibitory concentration of ciprofloxacin (MICCIP)
| parC | gyrA | MICCIP (µg/mL) | |
| S91F, D95A | S91F, D95G | Median (min; max) | |
| WT | 1 | 1 | (0.125; 0.75)* |
| D86N | 2 | – | 1.12 (0.75; 1.5) |
| S87R | 6 | – | 1.5 (1; 4) |
| S87N | 33 | – | 2 (0.75; 4) |
| S87N, E91K | 3 | – | 3 (1.5; 4) |
| Total | 45 | 1 | |
*Median is not presented as only two data points were obtained.
max, maximum; min, minimum; WT, wild-type.
Eight (17%) isolates had reduced susceptibility (MICPEN range: 0.064−1 µg/mL) to penicillin, the remaining 38 (83%) were resistant (MICPEN range: 1.5−256 µg/mL). All isolates had the insertion D345 in penA. The isolates with reduced susceptibility carried SNP L421P in the ponA gene, mutations in the mtrR coding region A39T or G45D, and 57Adel in the mtrR promoter, combined (n=6) or not (only L421P n=2). In addition to these mutations, the SNP A121S in porB1b was detected in two resistant isolates.
The β-lactamase bla TEM gene was detected in 36 penicillin-resistant isolates and 2 reduced susceptible isolates. Table 3 summarises the distribution of the bla TEM genes and plasmid types.
Table 3.
β-lactamase genes, plasmid types and associated minimal inhibitory concentration of penicillin (MICPEN)
| bla TEM allele | β-lactamase plasmid type | Total N Median MICPEN (µg/mL) (min; max) |
|||
| African type (pJD5) n (%) | Johannesburg type (pEM1) n (%) | Asian type (pJD4) n (%) | Not determined n (%) | ||
| TEM-1 | 17 (59) | 8 (27) | – | 4 (14) | 29 32 (0.125; 256) |
| TEM-135 | – | – | 6 (100) | – | 6 72 (16; 256) |
| TEM-206 | 2 (67) | – | 1 (33) | – | 3 3 (2; 12) |
| Total | 19 (50) | 8 (21) | 7 (18) | 4 (11) | 38 |
max, maximum; min, minimum.
Forty of 46 isolates (87%) were phenotypically resistant to tetracycline. All isolates, except one resistant, carried rpsJ gene V57M. The isolate lacking the mutation carried the tetM gene and an mtrR coding sequence with the A39T mutation. Almost all tetracycline-resistant isolates (39 of 40) contained plasmids with a tetM gene, one susceptible isolate carried a disrupted tetM gene. The American plasmid type was the most predominant (n=30, 77%), while the other nine (23%) isolates contained the Dutch plasmid. The only resistant isolate (MICTET=12 µg/mL) lacking the tetM gene had mutations G45D in the mtrR coding sequence, 57Adel in the mtrR promoter and carried a porB wild-type gene. Thirty-three isolates carried both the β-lactamase and the conjugative plasmid.
All isolates were susceptible to spectinomycin, and all 16S rRNA genes were wild-type.
Antimicrobial resistance prediction by Pathogenwatch
The Pathogenwatch AMR predictions for azithromycin, spectinomycin, ceftriaxone and ciprofloxacin concurred with the MIC results. Pathogenwatch predicted reduced susceptibility for penicillin in two isolates with a resistant phenotype and predicted resistance in two other isolates with a reduced susceptibility phenotype. Pathogenwatch predicted tetracycline-resistant all isolates phenotypically defined tetracycline resistant. However, of the six phenotype-susceptible isolates, five were predicted reduced susceptible and one resistant.
Molecular typing and phylogenetic analysis
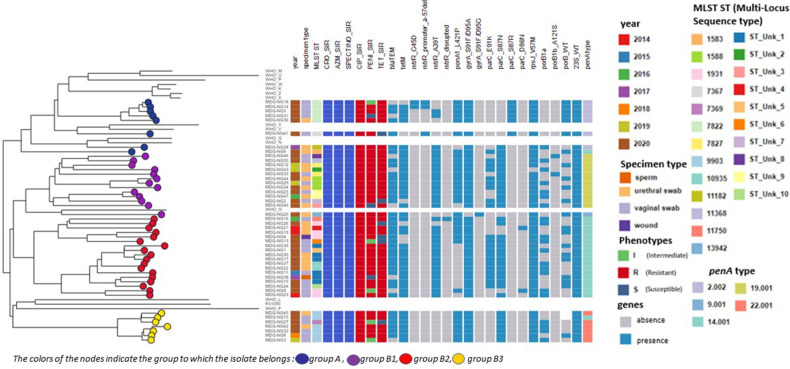
Among the 46 isolates, we identified 23 STs of which 10 were new profiles (figure 1). Eighteen STs were identified among the 33 isolates from 2020, half pertained to four STs: ST7822 (n=5); ST9903 (n=4); STUnk_1 (n=4); ST1931 (n=3) (figure 1).
Figure 1.
Phylogenetic tree using core genome SNP analysis for 46 Neisseria gonorrhoeae isolates from Madagascar, 14 WHO strains and N. gonorrhoeae FA 1090 genome. SNP, single nucleotide polymorphism.
Genomic core analysis resulted in two main lineages (figure 1). The first, group A, clustered nine isolates; and the second contained three groups, B1, B2 and B3, with 12, 18 and 7 isolates, respectively. Each group clustered penA allele types: penA.2, penA.19, penA.14, penA.22 clustered with group A, B1, B2 and B3, respectively. The highly penicillin-resistant isolates carrying bla TEM-135 belonged exclusively to ST7822 clustered in group A and ST9903 clustered in group B3. Group B2 included two predominant STs, STunk_1 and ST1931, while all ST1588 were clustered in B1. All isolates in group B3 carried the tetM gene.
Discussion
To our knowledge, this study is one of the only reports of the last decade that provides information on AMR and WGS of NG isolates from Madagascar.
The CBC reported almost all NG isolates (98%) resistant to ciprofloxacin in agreement with reports from other African countries.23 24 They reported 4.6% and 3.1% resistance to ceftriaxone and azithromycin (only tested in 2019 and 2020), respectively. The MICCRO decreased over the last 3 study years and an increase in MICAZT was observed from 2019 to 2020. β-lactamase production was common (94%).
We aimed to retest and include in the WGS all isolates from 2020, to set a baseline for further surveillance, and all isolates found resistant or reduced susceptible to ceftriaxone and azithromycin over the previous years, to confirm the phenotypical observation and determine the AMR determinants. Unfortunately, only a fraction of isolates obtained over the study period was available. In addition, less than two-thirds of retrieved isolates were viable, highlighting challenges medical laboratories in low-income and middle-income countries (LMICs) may encounter in storing fastidious microorganisms for surveillance purposes.
We could retest 6 of the 17 isolates defined ceftriaxone resistant and found them susceptible with low MICCRO values. Our finding underscores the need to flag and retest isolates with MICCRO exceeding the breakpoints.7 Two isolates considered azithromycin resistant were retested and found susceptible. Both isolates were obtained in 2020 and may explain in part the observed rise of MIC90 in 2020.
We did not detect any specific genomic determinant associated with ceftriaxone, azithromycin or spectinomycin resistance among the sequenced isolates, concordant with the AMS phenotypes obtained after retest. We observed incremental increases in the MICCIP when mutations in gyrA and parC genes accumulated, in agreement with previous reports.4
Almost all penicillin-resistant isolates carried the bla TEM gene conferring high-level resistance. Three bla TEM were identified: TEM-1 was the most prevalent and is usually found,4 TEM-206 has not been described previously, but TEM-135 is the most worrisome. TEM-135 requires only one SNP to evolve into extended-spectrum β-lactamase inducing resistance to extended-spectrum cephalosporins (ESCs).4 All TEM-135 were carried by the Asian plasmid type, in agreement with studies from China25 and the UK26 but in contrast with a global study27 reporting that bla TEM-135 was commonly associated with the Toronto/Rio plasmid type. Moreover, a chromosomally mediated resistance or reduced susceptibility to penicillin through the accumulation of mutations was observed in all isolates. We found a high proportion of isolates carrying the tetM gene acquired through conjugate plasmids and associated with higher MICTET values. Although penicillin and tetracycline are no longer employed to treat gonorrhoea, the majority of the isolates contained plasmids carrying resistance genes (n=33). The high prevalence of plasmids in NG isolates from Africa was reported previously.28–30 In addition, our finding supports the hypothesis that the plasmids present in NG do not impose a fitness cost.28
More than half (n=24) of the isolates carried an accumulation of chromosomal modifications in ponA and mtrR genes, two carried a mutation in porB1b gene. The resulting overexpression of the efflux pump and structural changes in the membrane contribute to the resistance or decreased susceptibility of a wide range of antimicrobials.4
The genome-based Pathogenwatch AMR prediction correlated well with the phenotypical resistance profile. Few discrepancies were observed in case of plasmid-mediated resistance. Pathogenwatch classified all tetracycline-susceptible isolates and those without the tetM gene reduced susceptibility due to the presence of other genetic resistance determinants. It classified two isolates penicillin resistant because they carried a bla TEM gene, but Etesting defined them reduced susceptible. Notwithstanding, isolates containing β-lactamase plasmids and having a MIC below the resistance threshold have been reported previously.31 The opposite was also observed: Pathogenwatch classified two penicillin-resistant isolates but lacking the bla TEM gene as reduced susceptible.
We found a wide diversity of STs (n=23), but two-thirds were singletons. We hypothesise that the lack of clonal transmission within our study population is due to our study design and small number of isolates. However, almost half of the isolates pertained to four major STs: ST9903, ST7822, ST1931 and Unk_1. The identified STs present in the Pathogenwatch database were reported previously in Europe (the UK, Norway, Sweden, Portugal, Greece), Australia, the USA and Vietnam.32 ST7822 has been prevalent in France since 2018,33 and both ST1931 and ST1588 were reported in Africa: Burkina Faso, South Africa and Kenya.34–36 On the other hand, the observation of STUnk_1 as a predominant ST suggests the circulation of a possible new clone in Antananarivo, Madagascar. We did not identify ST9363 or ST1901 associated with high-level resistance to ceftriaxone and azithromycin. However, we detected two ceftriaxone-susceptible isolates belonging to ST7827, an ST associated with ESC resistance.37
The isolates pertained to two lineages with one presenting three clusters. We did not observe a clustering of isolates associated with their phenotypical AMS profiles. This observation may be inconsistent with previous reports,38 and is probably due to the obtained dichotomy: all isolates were fully susceptible to three antimicrobials and almost fully resistant to the other three.
The Malagasy national treatment guidelines recommend ceftriaxone in combination with azithromycin in case of coinfection with C. trachomatis. We could not confirm the data submitted to the GLobal Antimicrobial resistance and use Surveillance System suggesting that ceftriaxone is no longer a valid antimicrobial for first-line treatment in Madagascar.39 Moreover, our findings support the need to implement national NG AMS surveillance and assign reference laboratories for reporting accurate data and providing warnings in case of AMR to the Ministry of Health. The benefit of retesting by reference laboratories was previously concluded, although in a high-resource context, by Harris et al, as they observed that discrepancies between phenotype and genotype data were solved after retesting isolates.40
In conclusion, surveillance of NG is mainly laboratory based requiring culture and AMS testing. Consequently, NG surveillance in many LMICs is limited. We demonstrated that genetic detection of AMR determinants is feasible. However, developing NG DNA capture methods and addressing WGS capabilities in LMICs will be essential for effective NG surveillance and management. Furthermore, with appropriate epidemiological data, a more comprehensive genomic epidemiological surveillance could be considered.
sextrans-2023-055878supp002.pdf (100.3KB, pdf)
Acknowledgments
The authors wish to express their gratitude to Jean-Marc Collard, researcher in Enteric Bacterial Pathogens Unit & French National Reference Center for Escherichia coli, Shigella and Salmonella, Institut Pasteur in Paris, France, for facilitating the contacts with NICD. They extend their thanks to Magnus Unemo, WHO centre for gonorrhoea and other Sexually Transmitted Infections, Örebro University, Örebro, Sweden, and the team of the STI reference laboratory at the Institute of Tropical Medicine, Antwerp, Belgium, for sending them the WHO reference and other quality control strains. A very grateful thanks to Irith De Baetselier for her critical reading of the first draft of the manuscript and to Matthieu Schoenhals for his final proofreading.
Footnotes
Handling editor: Erica L Plummer
Presented at: Part of the results was presented at the 23rd IUSTI World Congress, September 2022, Zimbabwe.
Contributors: LFR, MANR and TC designed the study. ERH and FR were responsible for the routine analysis at the CBC and contributed to the data. LFR performed additional laboratory testing. LFR and MANR conducted the bioinformatic analysis. AS performed whole-genome sequencing and made the data available to the study group. TC was the guarantor. LFR and TC wrote the original draft. All authors contributed to reviewing and editing.
Funding: The sequencing part of this study was made possible by support from the SEQAFRICA Project which is funded by the Department of Health and Social Care’s Fleming Fund using UK aid.
Disclaimer: The views expressed in this publication are those of the authors and not necessarily those of the UK Department of Health and Social Care or its Management Agent, Mott MacDonald.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. This Whole Genome Shotgun Project has been deposited at DDBJ/ENA/GenBank under the Accession BioProject PRJNA929018.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
This study involves human participants. Ethical clearance was waived by the National Ethical Committee of Madagascar for routinely collected and coded data analysed for surveillance purposes. Therefore, ethical clearance was not needed in this study.
References
- 1. WHO . Antibiotic-resistant gonorrhoea on the rise, new drugs needed, . 2017. Available: https://www.who.int/news/item/07-07-2017-antibiotic-resistant-gonorrhoea-on-the-rise-new-drugs-needed [Accessed 7 Dec 2022].
- 2. WHO . Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae, . 2012. Available: http://apps.who.int/iris/bitstream/10665/44863/1/9789241503501_eng.pdf [Accessed 7 Dec 2022].
- 3. Centers for Disease Prevention and Control . U.S. Department of Health and Human Services Atlanta, GA; Antibiotic Resistance Threats in the United States, . 2019. Available: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf [Google Scholar]
- 4. Unemo M, Shafer WM. Antimicrobial resistance in Neisseria Gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 2014;27:587–613. 10.1128/CMR.00010-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cehovin A, Lewis SB. Mobile genetic elements in Neisseria Gonorrhoeae: movement for change. Pathog Dis 2017;75. 10.1093/femspd/ftx071 [DOI] [PubMed] [Google Scholar]
- 6. Abrams AJ, Trees DL. Genomic sequencing of Neisseria Gonorrhoeae to respond to the urgent threat of antimicrobial-resistant Gonorrhea. Pathog Dis 2017;75:ftx041. 10.1093/femspd/ftx041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. WHO . Licence: CC BY-NC-SA 3.0 IGO. ISBN 978-92-4-002134-1. Enhanced Gonococcal Antimicrobial Surveillance Programme (EGASP): general protocol. Geneva: World Health Organization, 2021. [Google Scholar]
- 8. Unemo M, Golparian D, Sánchez-Busó L, et al. The novel 2016 WHO Neisseria Gonorrhoeae reference strains for global quality assurance of laboratory investigations: Phenotypic, genetic and reference genome characterization. J Antimicrob Chemother 2016;71:3096–108. 10.1093/jac/dkw288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Quijada NM, Rodríguez-Lázaro D, Eiros JM, et al. TORMES: an automated pipeline for whole bacterial genome analysis. Bioinformatics 2019;35:4207–12. 10.1093/bioinformatics/btz220 [DOI] [PubMed] [Google Scholar]
- 10. Schmieder R, Edwards R. Quality control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011;27:863–4. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible Trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prjibelski A, Antipov D, Meleshko D, et al. Using spades de novo assembler. Curr Protoc Bioinformatics 2020;70:e102. 10.1002/cpbi.102 [DOI] [PubMed] [Google Scholar]
- 13. Gurevich A, Saveliev V, Vyahhi N, et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013;29:1072–5. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wood DE, Lu J, Langmead B. Improved Metagenomic analysis with Kraken 2. Genome Biol 2019;20:257. 10.1186/s13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hyatt D, Chen G-L, Locascio PF, et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010;11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seemann T. Prokka: rapid Prokaryotic genome annotation. Bioinformatics 2014;30:2068–9. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 17. Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012;67:2640–4. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McArthur AG, Waglechner N, Nizam F, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 2013;57:3348–57. 10.1128/AAC.00419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gupta SK, Padmanabhan BR, Diene SM, et al. ARG-ANNOT, a new Bioinformatic tool to discover antibiotic resistance genes in bacterial Genomes. Antimicrob Agents Chemother 2014;58:212–20. 10.1128/AAC.01310-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sánchez-Busó L, Cole MJ, Spiteri G, et al. Centre for Genomic pathogen surveillance and the Euro-GASP study group. Europe-wide expansion and eradication of multidrug-resistant Neisseria Gonorrhoeae lineages: a Genomic surveillance study. The Lancet Microbe 2022;3:e452–63. [DOI] [PubMed] [Google Scholar]
- 21. Sánchez-Busó L, Yeats CA, Taylor B, et al. A community-driven resource for Genomic epidemiology and antimicrobial resistance prediction of Neisseria Gonorrhoeae at Pathogenwatch. Genome Med 2021;13:61. 10.1186/s13073-021-00858-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Argimón S, Abudahab K, Goater RJE, et al. Microreact: Visualizing and sharing data for Genomic epidemiology and Phylogeography. Microb Genom 2016;2:e000093. 10.1099/mgen.0.000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maduna LD, Kock MM, van der Veer BMJW, et al. Antimicrobial resistance of Neisseria Gonorrhoeae isolates from high-risk men in Johannesburg, South Africa. Antimicrob Agents Chemother 2020;64:e00906-20. 10.1128/AAC.00906-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Crucitti T, Belinga S, Fonkoua MC, et al. Sharp increase in ciprofloxacin resistance of Neisseria Gonorrhoeae in Yaounde, Cameroon: analyses of a laboratory database period 2012–2018. Int J STD AIDS 2020;31:579–86. 10.1177/0956462419897227 [DOI] [PubMed] [Google Scholar]
- 25. Yan J, Zhang J, van der Veen S. High prevalence of TEM-135 expression from the Asian Plasmid in Penicillinase-producing Neisseria Gonorrhoeae from Hangzhou, China. Int J Antimicrob Agents 2019;54:361–6. 10.1016/j.ijantimicag.2019.06.012 [DOI] [PubMed] [Google Scholar]
- 26. Cole MJ, Unemo M, Grigorjev V, et al. Genetic diversity of blaTEM Alleles, antimicrobial susceptibility and molecular Epidemiological characteristics of Penicillinase-producing Neisseria Gonorrhoeae from England and Wales. J Antimicrob Chemother 2015;70:3238–43. 10.1093/jac/dkv260 [DOI] [PubMed] [Google Scholar]
- 27. Muhammad I, Golparian D, Dillon J-AR, et al. Characterisation of blaTEM genes and types of Β-Lactamase Plasmids in Neisseria Gonorrhoeae - the prevalent and conserved blaTEM-135 has not recently evolved and existed in the Toronto Plasmid from the origin. BMC Infect Dis 2014;14:454. 10.1186/1471-2334-14-454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cehovin A, Jolley KA, Maiden MCJ, et al. Association of Neisseria Gonorrhoeae Plasmids with distinct lineages and the economic status of their country of origin. J Infect Dis 2020;222:1826–36. 10.1093/infdis/jiaa003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fayemiwo SA, Müller EE, Gumede L, et al. Plasmid-mediated penicillin and Tetracycline resistance among Neisseria Gonorrhoeae isolates in South Africa: prevalence, detection and typing using a novel molecular assay. Sex Transm Dis 2011;38:329–33. 10.1097/OLQ.0b013e3181fc695a [DOI] [PubMed] [Google Scholar]
- 30. Karim S, Bouchikhi C, Banani A, et al. Molecular antimicrobial resistance of Neisseria Gonorrhoeae in a Moroccan area. Infect Dis Obstet Gynecol 2018;2018:7263849. 10.1155/2018/7263849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harrison OB, Clemence M, Dillard JP, et al. Genomic analyses of Neisseria Gonorrhoeae reveal an Association of the Gonococcal genetic Island with antimicrobial resistance. J Infect 2016;73:578–87. 10.1016/j.jinf.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sánchez-Busó L, Yeats CA, Taylor B, et al. [Pathogenwatch Repository]. A community-driven resource for genomic epidemiology and antimicrobial resistance prediction of Neisseria gonorrhoeae at Pathogenwatch, Available: https://pathogen.watch/genomes/all?genusId=482&speciesId=485 [Accessed 27 Aug 2023]. [DOI] [PMC free article] [PubMed]
- 33. Bébéar C, Berçot B, Dupin N. Centre National de Référence des Infections Sexuellement Transmissibles bactériennes Bilan 2017-2021, Available: https://www.cnr-ist.fr/ressources/editeur/CNR-IST-Bilan-2017-2021.pdf [Accessed 27 Aug 2023].
- 34. Congo-Ouedraogo M, Poncin T, Sangaré L, et al. Genomic and antimicrobial resistance analyses of Neisseria Gonorrhoeae isolates, Burkina Faso, 2018–2019. J Eur Acad Dermatol Venereol 2022;36:e565–8. 10.1111/jdv.18037 [DOI] [PubMed] [Google Scholar]
- 35. Mitchev N, Singh R, Allam M, et al. Antimicrobial resistance mechanisms, Multilocus sequence typing, and NG-STAR sequence types of diverse Neisseria Gonorrhoeae isolates in Kwazulu-natal, South Africa. Antimicrob Agents Chemother 2021;65:e00759-21. 10.1128/AAC.00759-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Juma M, Sankaradoss A, Ndombi R, et al. Antimicrobial resistance profiling and Phylogenetic analysis of Neisseria Gonorrhoeae clinical isolates from Kenya in a resource-limited setting. Front Microbiol 2021;12:647565. 10.3389/fmicb.2021.647565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Korne-Elenbaas J, Bruisten SM, de Vries HJC, et al. Emergence of a Neisseria Gonorrhoeae clone with reduced Cephalosporin susceptibility between 2014 and 2019 in Amsterdam, the Netherlands, revealed by Genomic population analysis. J Antimicrob Chemother 2021;76:1759–68. 10.1093/jac/dkab082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pinto M, Borges V, Isidro J, et al. Neisseria Gonorrhoeae clustering to reveal major European whole-genome-sequencing-based Genogroups in association with antimicrobial resistance. Microb Genom 2021;7:000481. 10.1099/mgen.0.000481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Unemo M, Lahra MM, Escher M, et al. WHO global antimicrobial resistance surveillance for Neisseria Gonorrhoeae 2017-18: a retrospective observational study. Lancet Microbe 2021;2:e627–36. 10.1016/S2666-5247(21)00171-3 [DOI] [PubMed] [Google Scholar]
- 40. Harris SR, Cole MJ, Spiteri G, et al. Public health surveillance of multidrug-resistant clones of Neisseria Gonorrhoeae in Europe: a Genomic survey. Lancet Infect Dis 2018;18:758–68. 10.1016/S1473-3099(18)30225-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sextrans-2023-055878supp001.xlsx (38.8KB, xlsx)
sextrans-2023-055878supp002.pdf (100.3KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. This Whole Genome Shotgun Project has been deposited at DDBJ/ENA/GenBank under the Accession BioProject PRJNA929018.