Abstract
Differential diagnosis between constitutional mismatch repair deficiency (CMMRD) and neurofibromatosis type 1 (NF1) is crucial as treatment and surveillance differ. We report the case of a girl with a clinical diagnosis of sporadic NF1 who developed a glioblastoma. Immunohistochemistry for MMR proteins identified PMS2 loss in tumour and normal cells and WES showed the tumour had an ultra-hypermutated phenotype, supporting the diagnosis of CMMRD. Germline analyses identified two variants (one pathogenic variant and one classified as variant(s) of unknown significance) in the PMS2 gene and subsequent functional assays on blood lymphocytes confirmed the diagnosis of CMMRD. The large plexiform neurofibroma of the thigh and the freckling were however more compatible with NF1. Indeed, a NF1 PV (variant allele frequencies of 20%, 3% and 9% and in blood, skin and saliva samples, respectively) was identified confirming a mosaicism for NF1. Retrospective analysis of a French cohort identified NF1 mosaicism in blood DNA in 2 out of 22 patients with CMMRD, underlining the existence of early postzygotic PV of NF1 gene in patients with CMMRD whose tumours have been frequently reported to exhibit somatic NF1 mutations. It highlights the potential role of this pathway in the pathogenesis of CMMRD-associated gliomas and argues in favour of testing MEK inhibitors in this context.
Keywords: DNA Repair, Genetic Counselling, Genetic Predisposition to Disease, Neoplasms, Paediatrics
Introduction
Neurocutaneous syndromes are associated with the risk of brain tumour, the most frequent being neurofibromatosis type 1 (NF1)1 for which café-au-lait macule (CALM) is often the most obvious clinical sign in young patients. Other conditions such as constitutional mismatch repair deficiency (CMMRD) syndrome2–4 can also be associated with brain tumours and CALMs. Pilocytic astrocytoma5 (WHO grade I), often located in the optic pathway (optic pathway glioma), is the typical brain tumour observed a child or adolescent with NF1, whereas patients with CMMRD of this age develop malignant tumours (mainly high-grade gliomas and medulloblastomas).
In CMMRD, patient’s prognosis is worse, the risk of successive tumours is considerably higher and therapeutic strategies clearly differ. Unfortunately, the phenotypic overlap between these two cancer predisposing syndromes may lead to a delayed diagnosis of CMMRD in some patients.6–8 We describe here the case of a patient, initially diagnosed with NF1, in whom CMMRD syndrome was suspected after the occurrence of a glioblastoma. Genetic testing identified germline pathogenic variants (PVs) of both PMS2 alleles. Nevertheless, a PV in NF1 was detected in a mosaicism form in DNAs from blood, skin and saliva. Our observation raises important issues with respect to diagnosis and pathogenesis of CMMRD-associated glial tumours.
Patients and methods
Patients
After the identification of a mosaic NF1 in a patient with CMMRD, we questioned the frequency of NF1 mosaicism in a series of 22 French patients diagnosed with CMMRD and for whom blood samples were available for DNA analysis. For all the patients, consents for genetic testing have been obtained from the parents/patients, according to research ethics requirements during a genetic counselling. CMMRD was confirmed in case of the identification of two variants in any of the four MMR genes, classified as (likely) PV and confirmed to be in trans by genetic testing. For patients with monoallelic or biallelic variant(s) of unknown significance (VUS), confirmation of constitutional microsatellite instability (MSI) by a functional test (see below) was performed to confirm the diagnosis of CMMRD. Clinical and molecular data of patients with CMMRD were collected in the ‘Observatory of Genetic Cancer Predisposition Syndromes in Children and Adolescents’ French database (Observatoire des syndromes de prédisposition génétique au cancer des enfants et des adolescents, PREDCAP, IRB00003888).
Nucleic acid extractions and next-generation sequencing
DNA extraction and experiments were performed at the next-generation sequencing (NGS) panel facility of Gustave Roussy, Villejuif and Cochin Hospital, Paris (Assistance Publique- Hôpitaux de Paris, AP-HP). NF1 was sequenced in the patient and in a large series of 22 patients with CMMRD for whom blood samples were available, using a targeted NGS panel, as previously described.9 10 Sequencing libraries were prepared from 10 ng of DNA per sample according to the TruSeq Custom Amplicon Library Preparation Guide (Illumina, San Diego, California, USA). The custom primer panel targets the entire NF1-coding exons, intron boundaries (25 bp), and the 5′ and 3′ untranslated regions with amplicons with an average size of 150 bp. The pooled libraries were paired-end (2 × 150) and sequenced with NextSeq 500 Mid Output Kit v2 on a NextSeq500 instrument (Illumina). The number of samples tested per run was adapted to allow an increase in sequencing depth (mean sequencing depth per sample: 1500X). After demultiplexing and generation of FASTQ files, the sequence analysis was performed according to the Genome Analysis Tool Kit (GATK) guidelines. Sequence alignment, variant calling and variant annotation were performed using the MOABI pipeline and Polyweb pipeline (Paris Cité University). The VAF threshold used for the analysis was set at 3% to detect low mosaic variations. Variants were named at the coding DNA, RNA and protein levels according to the Human Genome Variation Society recommendations. An assessment of variants’ pathogenicity was performed according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology guidelines. Assessment of variants implication was mainly performed based on population databases (gnomAD), variant databases (ClinVar, HGMD, LOVD and COSMIC) and prediction software. In silico predictions of the effect of the variant were performed with CADD, SPiP, dbscSNV, Human Splice Finder and PROVEAN. For patients in whom a mosaic was identified in NF1, the PV was also searched by sequencing in samples of different origin (ie, saliva and skin).
Additional and functional tests for CMMRD diagnosis
An immunohistochemical (IHC) analysis of PMS2, MLH1, MSH2 and MSH6 protein expression in non-neoplastic cells was performed using formalin-fixed paraffin-embedded tumour sections available to assess loss of the affected MMR protein.
Due to the presence of a VUS of the PMS2 gene, functional tests, that is, methylation tolerance (MT) and ex vivo microsatellite instability (evMSI) assays,11 were performed on immortalised lymphocytes. Studies were completed with NGS-based analysis of the large panel of selected markers, recently published by Gallon et al.12
Results
Case presentation (patient 1)
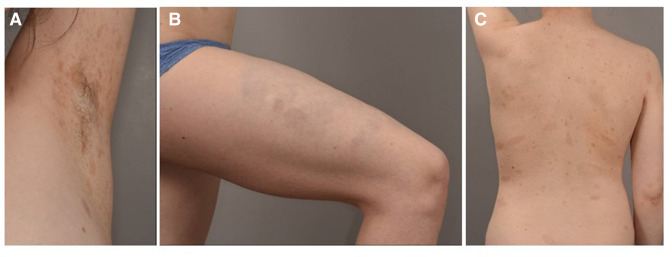
We describe the case of a girl who presented numerous CALMs, bilateral axillar freckling and a plexiform neurofibroma of the right thigh (figure 1A–C), suggesting NF1,13 but no molecular screening for NF1 was performed at the time of diagnosis. No history of tumour or NF1 phenotype in relatives has been reported. She had been treated for scoliosis since the age of 4. She had an ophthalmological screening at the age of 6 which did not find neither Lisch nodules nor visual alteration, but no brain MRI was performed during childhood.
Figure 1.
Skin features of patient 1: (A) axillar frecklings, (B) plexiform neurofibroma of the right thigh, (C) numerous café-au-lait macules.
At the age of 16, she was diagnosed with a right posterior parieto-occipital brain tumour revealed by headaches accompanied by hypoesthesia of the left upper limb. Brain MRI showed two synchronous contiguous lesions with a contrast enhancement, peripheral oedema and infiltration towards the corpus callosum. Multiple developmental venous brain anomalies were also reported but no focal area of signal intensity on T2/FLAIR was observed. A partial excision of the occipital part of the tumour was performed allowing for pathological diagnosis and somatic molecular analyses. The histopathology examination of the tissue biopsy revealed a glioblastoma with a few giant or multinucleated cells. IHC staining demonstrated expression of OLIG2, ATRX and P53 in the tumour cells. The occurrence of a high-grade glioma, a typical CMMRD-associated paediatric tumour, raised doubts about the NF1 diagnosis. Thus, immunostaining for MMR protein expression was performed and showed PMS2 loss in normal and tumour cells. Whole-tumour-exome sequencing revealed a typical pattern for CMMRD associated tumour14 with an ultra-high tumour burden (229 mut/Mb; above the threshold of 100 mut/Mb defined for ultra-hypermutator phenotype), a somatic POLE driver PV (c.857C>G p.Pro286Arg), multiple PVs in genes such as NF1 (two variants: c.2033dup p.(Ile679Aspfs*21) and c.532G>T p.(Glu178*)), PMS2 (two variants: c.695G>T p.(Gly232Val) and c.2275+1G>A), TP53, ATR and FANCA (table 1). Neither IDH1 p.Arg132His somatic mutation nor histone H3K27M or H3G34R/V somatic mutation was found. No microsatellite instability (MSI) (2.4%, ie, 202/7940) was reported according to MSI sensor (https://github.com/ding-lab/msisensor).
Table 1.
Molecular analysis of patient 1: germline and somatic alterations identified by RNA sequencing and whole exome sequencing of blood and tumour (glioblastoma, percentage of tumour cells: 80%)
| Gene name | Variant classification | HGVSc | HGVSp | Consequence | Constitutional DNA VAF (%) | Tumour DNA VAF (%) | Tumour RNA VAF (%) |
| PMS2 | PV/SNV |
NM_000088.4: c.695G>T |
NP_000526.2: p.(Gly232Val) | Missense variant | 55 | 50 | 68 |
| NF1 | PV/insertion |
NM_000267.3: c.2033dup |
NP_000258.1: p.(Ile679Aspfs*21) | Frameshift variant | 20 | 47 | 0 |
| PMS2 | PV/SNV |
NM_000535.7: c.2275+1G>A |
. | Splice donor variant | 45 | 29 | 0 |
| TP53 | PV/SNV |
NM_000546.6: c.817C>T |
NP_000537.3: p.(Arg273Cys) |
Missense variant | . | 48 | 77 |
| FANCA | PV/SNV |
NM_000135.4: c.3624C>T |
NP_000126.2: p.(Ser1208=) | Splice region and synonymous variant | . | 47 | 0 |
| TGFBR2 | PV/SNV |
NM_003242.6: c.1336G>A |
NP_001020018.1: p.(Asp446Asn) | Missense variant | . | 46 | 15 |
| SETD2 | PV/insertion |
NM_001349370.3: c.4087dup |
NP_001336299.1: p.(Arg1363Lysfs*8) | Frameshift variant | . | 45 | 22 |
| SMO | PV/SNV |
NM_005631.5: c.1965G>A |
NP_005622.1: p.(Trp655*) |
Nonsense | . | 44 | 42 |
| KDM6A | PV/SNV |
NM_001291415.2: c.4207C>T |
NP_001278344.1: p.(Arg1403*) | Nonsense | . | 41 | 0 |
| POLE | PV/SNV |
NM_006231.4 c.857C>G |
NP_006222.2: p.(Pro286Arg) | Missense variant | . | 41 | 58 |
| ATR | PV/SNV |
NM_001184.4: c.7597C>T |
NP_001175.2: p.(Arg2533*) | Nonsense | . | 38 | 31 |
| NF1 | PV/SNV |
NM_000267.3: c.532G>T |
NP_000258.1: p.(Glu178*) |
Nonsense | . | 37 | 50 |
| APC | PV/SNV |
NM_000038.6: c.2626C>T |
NP_000029.2: p.(Arg876*) |
Nonsense | . | 34 | 50 |
| TP53 | PV/SNV |
NM_001126112.3: c.586C>T |
NP_001119584.1: p.(Arg196*) | Nonsense | . | 30 | 0 |
| PPP2R1A | PV/SNV |
NM_001363656.2: c.7C>T |
NP_001350585.1: p.(Arg3Trp) | Missense variant | . | 25 | 28 |
| ARID1B | PV /SNV |
NM_001346813.1: c.2455C>T |
NP_001333742.1: p.(Gln819*) | Nonsense | . | 14 | 16 |
PV, pathogenic variant; SVN, small nucleotide variant; VAF, variant allele frequency.
In view of the patient’s phenotypic and tumour characteristics, constitutional genetic analyses were performed after genetic counselling. Germline genetic analysis revealed two different variants (compound heterozygous) in the PMS2 gene identified by NGS on two independent blood samples: one (c.2275+1G>A) classified as pathogenic inherited from the mother and the second (c.695G>T,p.(Gly232Val)), initially classified as VUS, inherited from the father. In addition, the germline genetic analysis also identified a PV in exon 18 of the NF1 gene (NM_001042492.3): c.2033dup, p.(Ile679Aspfs*21) (variant allele frequency (VAF): ~20% in a blood sample, 3% in a skin sample and 9% in a saliva sample) confirming the diagnosis of a mosaic NF1 associated with CMMRD in the patient (online supplemental figure S1). Cultured fibroblasts were enriched with cells with the NF1 PV (VAF=50% by Sanger sequencing).
jmg-2023-109235supp001.pdf (105.5KB, pdf)
Since one PMS2 variant was classified as VUS, we performed additional ancillary testing in order to confirm the diagnosis of CMMRD. Functional assays demonstrated that the cells of the patient displayed methylation tolerance and ex vivo MSI,11 confirming the diagnosis of CMMRD, although analyses showed clear MSI only after prolonged culture. The patient was included in the study of Gallon et al,12 her cMSI score confirmed the diagnosis of CMMRD in this patient, although she has one of the lowest cMSI score among the PMS2-associated CMMRD samples, probably due to the PMS2 missense variant causing their CMMRD syndrome. Based on these additional analyses, the c.695G>T p.(Gly232Val) PMS2 variant was finally reclassified as likely pathogenic (class 4), according to the ACMG-AMP criteria.15
After the partial surgery, the patient received adjuvant treatment combining radiotherapy to the right occipital lobe (54 Gy at the rate of 1.8 Gy per session) and immune checkpoint inhibition therapy with anti-PD1 as per the NIVOGLIO protocol (NCT04267146), without temozolomide. She had a good clinical and radiological response with a significant decrease of the residual brain tumour and, as per protocol, immunotherapy was discontinued after 1 year of treatment. She is in continuous remission 1.5 years after immunotherapy withdrawal and 2.5 years after the initial diagnosis. The identification of the CMMRD tumour predisposition syndrome enabled appropriate oncological surveillance to be proposed to this patient and her parents. Upper gastrointestinal endoscopy was normal. Colonoscopy at the age of 17 revealed a single 3 mm sessile polyp from the middle rectum, which has been excised. The pathological examination was in favour of a low-grade adenoma.
Cohort analysis
Among a series of 22 patients, we identified 2 additional patients with a NF1 mosaicism in blood DNA: c.4751dup, p.(Lys1585Glnfs*37) with VAF ~4% (100/2560 reads) for patient 2 and c.184_185dup, p.(Leu62Phefs*2) with VAF ~16% (191/1167 reads) for patient 3 (online supplemental figure S1). Patient 2 has previously been published in the C4CMMRD report on brain tumours.3 He was diagnosed with CMMRD after occurrence of T lymphoblastic lymphoma, with identification of PMS2 biallelic germline PV (c.2007–2A>G, p.?). Subsequently, during childhood, he developed a brain anaplastic astrocytoma grade III (IDH wild type with angiocentric features) with a loss of PMS2 expression by immunostaining and a high mutation load (rate=184 coding SNV/Mb). He is still alive with stable disease after a recurrence of his brain tumour treated with a further focal radiation therapy 2.5 years ago. Clinically, he presents multiple CALMs with hypopigmented skin lesions but no neurofibroma or other NF1-associated features.
Patient 3 is the brother of a boy who developed an acute leukaemia in the first years of life and a subsequent colonic adenocarcinoma, from which he died. Both brothers are carriers of compound heterozygous PMS2 PVs (c.1020_1021del p.(Arg341Alafs*23) and exons 13–15 deletion). CMMRD was identified in patient 3 prior to the occurrence of an acute B-lymphoblastic leukaemia (as young teenager) treated according to the French CAALL protocol. He is in complete remission 2 years after the end of the treatment. He presents with thoracic CALMs with segmental distribution exclusively on the right thoracic side and disseminated hypopigmented skin lesions. Like his brother, he also has multiple pilomatricomas, some of which have been surgically removed.
Discussion
We report three patients who carry a combination of a CMMRD syndrome caused by biallelic germline PMS2 PVs and a postzygotic NF1 PV. For patient 1, initially diagnosed with NF1, CMMRD was suspected after the occurrence of a glioblastoma. The tumour characteristics (ie, IHC and high mutation burden) and additional functional assays confirm CMMRD in this patient. Accordingly, both parents had a Lynch syndrome. Identification of the CMMRD and Lynch underlying diseases enables appropriate oncological surveillance for the patient and her parents, to be proposed according to published recommendations.4 16 17
Since the first descriptions, the occurrence of CALM, especially with a segmental distribution, in patients with CMMRD is intriguing.18 19 To date, only one patient carrying both a biallelic MMR PV and a de novo germline NF1 PV (c.3721C>T p.(Arg1241*)) has been reported,6 20 while no NF1 PV was reported in any of the other CMMRD cases in whom NF1 analysis was performed.6 19 Our report highlights that NF1 postzygotic PVs exist in patients with CMMRD, and 3 out of 23 (13%) patients have this combined genotype in the CMMRD cohort studied. Although tumour spectrum of CMMRD and NF1 may partly overlaps, tumour histologies are different between these two conditions. This observation underlines the fact that the identification of a mosaic NF1 PV does not rule out the diagnosis of CMMRD. Considering the rarity of such a situation associating CMMRD and a mosaic NF1 mutation, additional explorations to confirm or exclude CMMRD in patients with mosaic NF1 should however be proposed for patients with one or more additional features strongly suggestive of CMMRD such as a typical CMMRD-associated tumour, digestive polyps or a diagnosis of Lynch syndrome in one of the parents.2 7
The large size of the NF1 gene and its high mutation rate reflected in the fact that at least 50% of all NF1 cases are sporadic, may render the NF1 gene highly susceptible to postzygotic mutations.18 In addition, the c.2033dup variant identified in patient 1 is a hotspot variant in a 7 cytosine stretch which is the longest mononucleotide repeat in the NF1 coding regions. We can speculate that in patients with CMMRD, postzygotic mutations of NF1 may occur more frequently than in the normal population due to a non-functional MMR system. It is worth noting that the three NF1 mosaic variants identified in our study are all duplications (c.2033dup, c.4751dup and c.184_185dup), suggesting a mechanism of replicative slippage mutagenesis favoured by the MMR system deficiency. The very high frequency of NF1 somatic mutations in MMR-deficient tumours21 22 and the lack of recurrent mutations reported in other large genes, also support the hypothesis that NF1 is a target of MMR deficiency. NF1 haploinsufficiency has been shown to increase astrocyte proliferation23 and is associated with increased angiogenesis24 and perturbations of cell cycle and DNA repair pathways.25 It is conceivable that NF1 heterozygous mutations may confer a growth advantage and a positive selection of NF1-mutated cells in a CMMRD context. We suggest that CMMRD may alter NF1 at different developmental times and heightens the pressure to acquire NF1 postzygotic mutations early in development. For patient 1, the mutation probably appeared very early before the individualisation of the neuro-ectodermal and mesenchymal leaflets.
These three cases support the hypothesis that NF1-associated phenotypic features in patients with CMMRD are a consequence of early NF1 mutations. In two out of the three patients described, CALMs were segmental, which is consistent with the clinical features of usual mosaic NF1. In patient 1, molecular analysis of the glioblastoma showed two hits in NF1 (including the mosaic PV), indicating the contribution of NF1 inactivation to the development of this cancer. The actual impact of such genetic conditions on oncological risks for patients who carry both MMR and NF1 PVs remains to be determined. The risk of neurofibroma transformation in the context of NF1 is not negligible.26 Even though no malignant peripheral nerve sheath tumours (MPNST) have been reported as far as we know in patients with CMMRD, it could be speculated that the risk of transformation could be higher in the context of MMR deficiency. There is currently no evidence of the need for specific surveillance guidelines for plexiform neurofibroma in the context of CMMRD. Future study will have to analyse whether the current guidelines for patients with NF1-associated plexiform neurofibroma27 are adapted for patients with CMMRD.
This report demonstrates that the phenotypic overlap between CMMRD and NF1 syndromes can sometimes be explained by mosaic NF1 and suggests the involvement of the RAS-MAPK pathway in the pathogenesis of CMMRD-associated glial neoplasms. These observations support the previous suggestion22 to explore the combination of MEK inhibitors with immune checkpoint inhibitors for the treatment of these tumours.
jmg-2023-109235supp002.pdf (198.8KB, pdf)
Footnotes
Contributors: Study concept and design, collection and assembly of data: LG-R, LB, JG. Molecular somatic and germline data analyses: EP, MM, TA-D-B, OC, SC, EG, ER, SZ, DV. Neuropathological examinations: PV. Medical care to patients: LG-R, SA, LB, CC, PD, CD, JG. Data interpretation: LG-R, EP, MM, LB, DV, JG. Manuscript writing: LG-R, EP, LB, JG. Manuscript review and approval: All authors.
Funding: The authors declare that they have been supported by the Pediatric Campaign of the Fondation Gustave Roussy “Guérir les Cancers des Enfants au XXI siècle” and a grant from the Société Française des Cancers de l’Enfant (SFCE). SZ is supported by a grant from the Ecole Normale Supérieure.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s)
References
- 1. Kratz CP, Jongmans MC, Cavé H, et al. Predisposition to cancer in children and adolescents. Lancet Child Adolesc Health 2021;5:142–54. 10.1016/S2352-4642(20)30275-3 [DOI] [PubMed] [Google Scholar]
- 2. Wimmer K, Kratz CP, Vasen HFA, et al. Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium ‘care for CMMRD’ (C4CMMRD). J Med Genet 2014;51:355–65. 10.1136/jmedgenet-2014-102284 [DOI] [PubMed] [Google Scholar]
- 3. Guerrini-Rousseau L, Varlet P, Colas C, et al. Constitutional mismatch repair deficiency-associated brain tumors: report from the European C4CMMRD consortium. Neurooncol Adv 2019;1:vdz033. 10.1093/noajnl/vdz033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aronson M, Colas C, Shuen A, et al. Diagnostic criteria for constitutional mismatch repair deficiency (CMMRD): recommendations from the International consensus working group. J Med Genet 2022;59:318–27. 10.1136/jmedgenet-2020-107627 [DOI] [PubMed] [Google Scholar]
- 5. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 2021;23:1231–51. 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wimmer K, Rosenbaum T, Messiaen L. Connections between constitutional mismatch repair deficiency syndrome and neurofibromatosis type 1. Clin Genet 2017;91:507–19. 10.1111/cge.12904 [DOI] [PubMed] [Google Scholar]
- 7. Suerink M, Ripperger T, Messiaen L, et al. Constitutional mismatch repair deficiency as a differential diagnosis of neurofibromatosis type 1: consensus guidelines for testing a child without malignancy. J Med Genet 2019;56:53–62. 10.1136/jmedgenet-2018-105664 [DOI] [PubMed] [Google Scholar]
- 8. Guerrini-Rousseau L, Suerink M, Grill J, et al. Patients with high-grade gliomas and café-au-lait macules: is neurofibromatosis type 1 the only diagnosis AJNR Am J Neuroradiol 2019;40:E30–1. 10.3174/ajnr.A6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pasmant E, Parfait B, Luscan A, et al. Neurofibromatosis type 1 molecular diagnosis: what can NGS do for you when you have a large gene with loss of function mutations Eur J Hum Genet 2015;23:596–601. 10.1038/ejhg.2014.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lobón-Iglesias MJ, Laurendeau I, Guerrini-Rousseau L, et al. NF1-like optic pathway gliomas in children: clinical and molecular characterization of this specific presentation. Neurooncol Adv 2020;2:i98–106. 10.1093/noajnl/vdz054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bodo S, Colas C, Buhard O, et al. Diagnosis of constitutional mismatch repair-deficiency syndrome based on microsatellite instability and lymphocyte tolerance to methylating agents. Gastroenterology 2015;149:1017–29. 10.1053/j.gastro.2015.06.013 [DOI] [PubMed] [Google Scholar]
- 12. Gallon R, Phelps R, Hayes C, et al. Constitutional microsatellite instability, genotype, and phenotype correlations in constitutional mismatch repair deficiency. Gastroenterology 2023;164:579–92. 10.1053/j.gastro.2022.12.017 [DOI] [PubMed] [Google Scholar]
- 13. Legius E, Messiaen L, Wolkenstein P, et al. Revised diagnostic criteria for neurofibromatosis type 1 and Legius syndrome: an international consensus recommendation. Genet Med 2021;23:1506–13. 10.1038/s41436-021-01170-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Campbell BB, Light N, Fabrizio D, et al. Comprehensive analysis of hypermutation in human cancer. Cell 2017;171:1042–56. 10.1016/j.cell.2017.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–24. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vasen HFA, Ghorbanoghli Z, Bourdeaut F, et al. Guidelines for surveillance of individuals with constitutional mismatch repair-deficiency proposed by the European consortium ‘care for CMMR-D’ (C4CMMR-D). J Med Genet 2014;51:283–93. 10.1136/jmedgenet-2013-102238 [DOI] [PubMed] [Google Scholar]
- 17. Seppälä TT, Latchford A, Negoi I, et al. European guidelines from the EHTG and ESCP for Lynch syndrome: an updated third edition of the Mallorca guidelines based on gene and gender. Br J Surg 2021;108:484–98. 10.1002/bjs.11902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Q, Lasset C, Desseigne F, et al. Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res 1999;59:294–7. [PubMed] [Google Scholar]
- 19. Auclair J, Leroux D, Desseigne F, et al. Novel biallelic mutations in MSH6 and PMS2 genes: gene conversion as a likely cause of PMS2 gene inactivation. Hum Mutat 2007;28:1084–90. 10.1002/humu.20569 [DOI] [PubMed] [Google Scholar]
- 20. Alotaibi H, Ricciardone MD, Ozturk M. Homozygosity at variant MLH1 can lead to secondary mutation in NF1, neurofibromatosis type I and early onset leukemia. Mutat Res 2008;637:209–14. 10.1016/j.mrfmmm.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 21. Wang Q, Montmain G, Ruano E, et al. Neurofibromatosis type 1 gene as a mutational target in a mismatch repair-deficient cell type. Hum Genet 2003;112:117–23. 10.1007/s00439-002-0858-4 [DOI] [PubMed] [Google Scholar]
- 22. Campbell BB, Galati MA, Stone SC, et al. Mutations in the RAS/MAPK pathway drive replication repair-deficient hypermutated tumors and confer sensitivity to MEK inhibition. Cancer Discov 2021;11:1454–67. 10.1158/2159-8290.CD-20-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gutmann DH, Loehr A, Zhang Y, et al. Haploinsufficiency for the neurofibromatosis 1 (NF1) tumor suppressor results in increased astrocyte proliferation. Oncogene 1999;18:4450–9. 10.1038/sj.onc.1202829 [DOI] [PubMed] [Google Scholar]
- 24. Wu M, Wallace MR, Muir D. NF1 haploinsufficiency augments angiogenesis. Oncogene 2006;25:2297–303. 10.1038/sj.onc.1209264 [DOI] [PubMed] [Google Scholar]
- 25. Pemov A, Park C, Reilly KM, et al. Evidence of perturbations of cell cycle and DNA repair pathways as a consequence of human and murine NF1-haploinsufficiency. BMC Genomics 2010;11:194. 10.1186/1471-2164-11-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brems H, Beert E, de Ravel T, et al. Mechanisms in the pathogenesis of malignant tumours in neurofibromatosis type 1. Lancet Oncol 2009;10:508–15. 10.1016/S1470-2045(09)70033-6 [DOI] [PubMed] [Google Scholar]
- 27. Carton C, Evans DG, Blanco I, et al. ERN GENTURIS tumour surveillance guidelines for individuals with neurofibromatosis type 1. EClinicalMedicine 2023;56:101818. 10.1016/j.eclinm.2022.101818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmg-2023-109235supp001.pdf (105.5KB, pdf)
jmg-2023-109235supp002.pdf (198.8KB, pdf)