Abstract
Introduction
Sodium-glucose cotransporter 2 inhibitors (SGLT2i) have emerged as novel therapeutics to treat diabetic kidney disease (DKD). Although the beneficial effects of SGLT2i have been demonstrated, their target mechanisms on kidney function are unknown. The current study aimed to elucidate these mechanisms by studying SGLT2i-induced changes in the urinary proteome of persons with type 2 diabetes (T2D) and DKD.
Methods
A total of 40 participants with T2D were enrolled in a double-blinded randomized cross-over trial at the Steno Diabetes Center Copenhagen, Denmark. They were treated with 10 mg of dapagliflozin for 12 weeks. Thirty-two participants with complete urinary proteomics measures before and after the trial were included. All participants received renin-angiotensin system blockade and had albuminuria, (urine albumin-to-creatinine ratio [UACR] ≥30 mg/g). A type 1 diabetes (T1D) cohort consisting of healthy controls and persons with DKD was included for validation. Urinary proteome changes were analyzed using Wilcoxon signed-rank test. Functional enrichment analysis was conducted to discover affected biological processes.
Results
Dapagliflozin treatment significantly (Padjusted < 0.05) affected 36 urinary peptide fragments derived from 19 proteins. Eighteen proteins were correspondingly reflected in the validation cohort. A multifold change in peptide abundance was observed in many proteins (A1BG, urinary albumin [ALB], Caldesmon 1, COLCRNN, heat shock protein 90-β [HSP90AB1], IGLL5, peptidase inhibitor 16 [PI16], prostaglandin-H2-D-isomerase [PTGDS], SERPINA1). These also included urinary biomarkers of kidney fibrosis and function (type I and III collagens and albumin). Biological processes relating to inflammation, wound healing, and kidney fibrosis were enriched.
Conclusion
The current study discovers the urinary proteome impacted by the SGLT2i, thereby providing new potential target sites and pathways, especially relating to wound healing and inflammation.
Keywords: dapagliflozin, diabetic kidney disease, randomized controlled trial, SGLT2, sodium-glucose cotransporter 2 inhibitors, urinary proteomics
Graphical abstract
DKD is among the most common diabetic complications affecting approximately 40% of persons with T2D.1 Traditionally, DKD is treated with renin-angiotensin-system blockade.2 However, more recently, SGLT2 inhibition has been added to the standard-of-care because multiple studies have reported beneficial effects of SGLT2i on the risk of cardiovascular events, progression to end-stage kidney disease, and death from chronic kidney disease (CKD).3, 4, 5, 6, 7 SGLT2i blocks the reabsorption of sodium and glucose in the proximal renal tubules, which according to the current understanding leads to changes in the tubuloglomerular feedback system.8,9 However, molecular mechanisms underlying SGLT2i treatment effects in DKD are currently incompletely understood but presumed to be associated with multiple pathophysiological disturbances in DKD.10
Urinary proteomics has emerged as a method to explore patho-mechanisms of different kidney-related diseases and to develop disease risk classifiers such as CKD273,11, 12, 13 because 70% of the urinary proteome is derived from the kidney.14 Several studies have shown renoprotective effects of SGLT2i, including ours demonstrating that SGLT2i has a significant impact on the kidney disease risk classifier, CKD273.15 These data argued for an in-depth investigation of SGLT2i on the urinary proteome to explore potential molecular mechanisms of the drug. The current study aims to analyze the effects of SGLT2i on the urinary proteome of patients with DKD and to link the observed changes in the urinary proteins to biological processes with central roles in the development of DKD. This is done by investigating the effect of the SGLT2i, dapagliflozin on urinary proteomics in a previously published placebo-controlled, double-blinded, randomized crossover trial.16
The objectives of the current study are as follows: to (i) identify individual urinary peptides significantly affected by SGLT2i treatment, (ii) compare the SGLT2i-induced changes in urinary proteome to the differences observed between the urinary proteome from healthy controls and DKD cases in an independent T1D cohort, and (iii) map the observed changes in the urinary proteome to biological processes and molecular functions.
Methods
The study is a randomized, placebo-controlled, double-blinded crossover trial carried out to investigate the changes dapagliflozin causes on the urinary peptidome (NCT02914691) as the primary outcome. The full study population has been previously described.15,16 A total of 40 participants with T2D with DKD recruited at Steno Diabetes Center Copenhagen, Denmark, were randomized in a 1:1 ratio. All participants had albuminuria (UACR >30 mg/g in at least 2 out of 3 morning spot urine samples). Participants received 10 mg of dapagliflozin orally for 12 weeks during the active medication period. No washout period was used in the study when the 2 sequence groups crossed over. All participants received renin-angiotensin-aldosterone system blockade treatment throughout the study. In Supplementary Figures S1 and S2, we show the study design and CONSORT reporting guideline flow. The clinical factors and urinary proteomics were obtained at baseline (study start), and at the end of both placebo and dapagliflozin treatment periods, producing 3 data points for each participant, although measured glomerular filtration rate (GFR) was not obtained at baseline. GFR was estimated using the creatinine CKD-Epidemiology Collaboration 2009 equation.17 Four participants were excluded from the study due to various reasons (death, administrative error, and side effects: skin rash and myocardial infarction). Four additional participants lacked urinary proteomics data from some visit and were excluded. Thirty-two participants were included in the final analysis as before.15 More details are presented in Supplementary Methods. All participants gave a written consent, and the study was approved by the Regional Ethics Committee and the Danish Medicine Agency. The study was conducted in accordance with Good Clinical Practice and the declaration of Helsinki.
Validation Cohort 1 (PROTON): Type 1 DKD Versus Healthy Controls
To confirm whether the beneficial effects of dapagliflozin observed through changes in the urinary proteome (discovery study) also reflects differences in urinary proteome of individuals with and without DKD, an independent cohort (DKD vs. healthy controls) from Denmark was used (NCT03509454). The cohort consisted of 50 healthy nondiabetic controls and 110 participants with T1D and DKD, of whom 60 had macroalbuminuria (UACR ≥300 mg/g) and 50 had microalbuminuria (UACR 30–300 mg/g) as described previously.18,19 See Supplementary References.
Validation Cohort 2 (PROVALID): T2D Persons Treated With SGLT2i
We further investigated urinary proteome changes in 88 participants from the PROVALID study, an observational study of persons with T2D treated at the primary level of healthcare.20 Complete urinary peptidome data was available for 88 persons before and after SGLT2i treatment.21
Urinary Proteomics
The urinary proteomics comprised sample preparation and capillary electrophoresis coupled with mass spectrometry (CE-MS/MS or LC-MS/MS), data processing, and peptide sequencing as described previously.15 Details are in the Supplementary Methods. See Supplementary References.
Statistical Analyses
We examined dapagliflozin treatment induced urinary peptide changes as primary post hoc analyses, and changes in clinical factors as secondary post hoc analyses, between the treatment and placebo arms. The end point versus end point comparisons (visits 2 and 3 in Supplementary Figure S1) were modeled using linear mixed-effects models with treatment, sequence, and period as fixed effects and using patient-specific random intercepts packages lmerTest (v. 3.1.3), lme4 (v. 1.1.25), and nlme (v. 3.1.140). Only the end points from placebo and dapagliflozin treatment periods were considered in the models. Restricted maximum likelihood was employed in the maximum likelihood estimation. The 95% confidence intervals were obtained using Kenward-Rogers approximation of degrees of freedom. Nonnormally distributed variables were log-transformed. Lastly, the models with significant results were subjected to a 3-step adjustment procedure, first for age and gender, second for glycated hemoglobin and 24-hour ambulatory systolic blood pressure, and third for UACR and estimated GFR, as described previously.16 The baseline values from the first visit in Supplementary Figure S1 were used in the 3-step adjustment.
Wilcoxon signed-rank test with Benjamini-Hochberg (BH)-adjustment as the multiple testing correction method, was employed to analyze changes in the urinary peptide fragment abundance, and its applicability to nonnormal data.22 End points, only from placebo and dapagliflozin treatment periods were used. Mann-Whitney U test followed by BH-adjustment was used to examine whether the affected peptides differed significantly between healthy controls and DKD cases in the independent cohort (PROTON). The directionality of dapagliflozin-induced changes in the discovery study was compared to the difference in the urinary abundances of these peptide fragments in the independent cohort, that is, if the urinary abundance of a peptide fragment decreased because of dapagliflozin treatment and healthy controls had significantly lower urinary abundances of the same peptide fragment compared to the DKD cases, it is likely that such change represents a beneficial effect on the urinary proteome. All statistical analyses were conducted using Python 3,23 and statistical program R (v. 3.6.1)15,24 with RStudio, (RStudio, PBC, Boston, MA, USA).
Functional Gene and Pathway Enrichment
Functional gene enrichment analyses were performed using the Search Tool for the Retrieval of Interacting Genes/Proteins (v. 11, Cytoscape plug-in stringApp v. 1.6.0) and ClueGO (v. 2.5.7.) together with CluePedia (v. 1.5.7) plug-ins on Cytoscape (v. 3.8.2.).25, 26, 27, 28 For the functional enrichment analyses, lower confidence level was used, because all peptides affected by dapagliflozin at P < 0.05 using the raw, BH-unadjusted P-values were included. The Search Tool for the Retrieval of Interacting Genes/Proteins was used to visualize the physical and functional interactions of the proteins affected by dapagliflozin. Detailed description is in the Supplementary Methods. See Supplementary References.
Results
Study Characteristics and Changes in Clinical Factors
The discovery study was a randomized, double blind, placebo-controlled, cross-over study comprising a total of 32 participants with T2D where 10 mg of dapagliflozin were compared to matching placebo16 with complete information on urinary proteomics profiling (before and after treatment). The study participants had a mean (±SD) age of 63.1 (8.3) years, diabetes duration of 15.9 (4.7) years, and glycated hemoglobin 72.8 (14.1) mmol/mol (8.8 [1.2] %). Of the participants, 34.4% had microalbuminuria and 87.5% were men. Participant baseline clinical characteristics are available in Table 1. The detailed study design and the changes in some clinical factors have been published.15,16 In brief, 7 of the 10 investigated clinical factors were significantly affected by dapagliflozin treatment with decreases in blood pressure, weight, measured GFR, glycated hemoglobin, and UACR (Supplementary Tables S1, S2, and S3). Adverse events are presented in Supplementary Table S4.
Table 1.
Characteristics of participating studies at baseline
| Variable | Discovery study (n = 32) | PROTON: Independent cohort |
|
|---|---|---|---|
| Healthy controls (n = 50) | DKD cases (n = 110) | ||
| Age | 63.1 (8.3) | 58.6 (12.5) | 61.0 (9.8) |
| Gender (male, %) | 87.5 | 56.0 | 59.1 |
| Diabetes duration (years) | 15.9 (4.7) | 0 | 45.2 (13.1) |
| Weight (kg) | 105.4 (20.2) | 74.0 (13.0) | 78.6 (17.6) |
| Body mass index (kg/m2) | 33.7 (5.4) | 24.4 (3.2) | 26.4 (4.5) |
| HbA1c (mmol/mol) | 72.8 (14.1) | 35.7 (2.7) | 62.3 (10.7) |
| HbA1c (%) | 8.8 (1.2) | 5.4 (0.2) | 7.8 (1.0) |
| UACR (mg/g) | 153.8 (94.2–328.9) | 3.6 (2.6–4.6) | 42.9 (11.0–195.8) |
| Office systolic BP (mm Hg) | 141.1 (15.3) | 125.5 (15.2) | 135.5 (19.6) |
| Office diastolic BP (mm Hg) | 82.9 (10.2) | 76.7 (9.7) | 72.7 (9.3) |
| Ambulatory systolic BP (mm Hg) | 146.5 (11.7) | 133.1 (12.4) | 139.2 (13.0) |
| Ambulatory diastolic BP (mm Hg) | 82.9 (7.9) | 79.7 (6.6) | 76.3 (6.3) |
| Serum creatinine (μmol/l) | 80.0 (22.3) | 75.0 (15.1) | 105.1 (49.3) |
| eGFR (ml/min per 1.73 m2, CKD-EPI) | 85.5 (19.1) | 88.7 (13.7) | 68.5 (25.6) |
| LDL cholesterol (mmol/l) | 1.6 (0.7) | 3.2 (0.8) | 2.1 (0.7) |
| ALAT (U/l) | 40.3 (15.6) | 31.1 (6.7) | 35.6 (9.9) |
| Macroalbuminuria (%) | 34.4 | 0 | 54.5 |
ALAT, alanine aminotransferase; BMI, body mass index; BP, blood pressure; CKD-EPI, chronic kidney disease-epidemiology collaboration; eGFR, estimated glomerular filtration rate; HbA1c, glycated hemoglobin; LDL, low-density lipoprotein; UACR, urine albumin-to-creatinine ratio.
Characteristics from the DapKid study and the PROTON study, which was used as the independent cohort in the current study. Continuous, normally distributed variables are reported as mean and SD, whereas nonnormally distributed variables are reported as median and interquartile range. The full study population of the DapKid study (n = 36) has been described previously.14 Geometric mean of 3 morning visits.
Changes in Urinary Proteome (Discovery Study)
A total of 626 peptide fragments from 138 proteins were left for analysis when fragments with at most 50% missingness were included. The Wilcoxon signed-rank test yielded significant changes in 36 individual peptides originating from 19 proteins (Table 2 and Supplementary Table S5). Dapagliflozin significantly decreased the urinary abundance of 23 peptides, whereas the remaining 13 peptides increased (PBH-adjusted < 0.05; Table 2 and Supplementary Table S5). Especially, collagen-derived (type I and III) fragments significantly increased, whereas peptides derived from albumin, α-1-antitrypsin (SERPINA1), and α-1B-glycoprotein decreased. In Supplementary Table S6, we show sequences for the significantly affected peptides.
Table 2.
The table shows the mean values of individual urinary peptide fragment abundance after placebo (n = 32) and dapagliflozin (n = 32) treatment periods, and the mean urinary levels of these peptide fragments in the independent cohort from healthy controls (n = 50) and DKD cases (n = 110). In addition, mass (in Daltons) and migration time (minutes) are given for each peptide fragment, as well as the log-2 transformed fold change induced by dapagliflozin and fold difference calculated between healthy controls and DKD cases in the validation cohort. The reported P-values were adjusted for multiple testing using the Benjamini-Hochberg method
| Human UniProt ID | Peptide sequence ID | Mass (Da) | CE-time (min) | Gene Symbol | Protein | Placebo (mean) | Dapagliflozin (mean) | Dapagliflozin ind. change | BH-adj. P-value | DKD (mean) | Healthy (mean) | DKD ind. change |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P04217 | 11163 | 2181.205 | 20.616 | A1BG | α-1B-glycoprotein | 257.834 | 19.502 | −3.725 | 0.019 | 46.823 | 0 | + |
| 12127 | 2423.331 | 21.122 | A1BG | α-1B-glycoprotein | 3617.333 | 758.044 | −2.254 | 0.027 | 2137.423 | 2.469 | 9.758 | |
| 12950 | 2310.242 | 20.925 | A1BG | α-1B-glycoprotein | 1196.521 | 306.267 | −1.966 | 0.031 | 1781.37 | 4.207 | 8.726 | |
| 13296 | 2493.302 | 19.608 | A1BG | α-1B-glycoprotein | 1052.219 | 33.214 | −4.984 | 0.031 | 379.197 | 0 | + | |
| P02765 | 8180 | 1792.936 | 21.267 | AHSG | α-2-HS-glycoprotein | 355.884 | 1001.749 | 1.493 | 0.042 | 213.442 | 0 | + |
| P02768 | 12518 | 1715.99 | 21.054 | ALB | Serum albumin | 54307.868 | 803.218 | −6.078 | 0.012 | 3134.85 | 28.44 | 6.784 |
| 12970 | 1829.053 | 21.335 | ALB | Serum albumin | 91459.779 | 959.985 | −6.573 | 0.012 | 2663.851 | 25.193 | 6.724 | |
| 13562 | 2540.269 | 19.687 | ALB | Serum albumin | 108201.088 | 236.708 | −8.828 | 0.012 | 7876.335 | 24.974 | 8.301 | |
| 14191 | 2752.422 | 19.881 | ALB | Serum albumin | 326527.011 | 1566.885 | −7.703 | 0.014 | 92739.29 | 423.45 | 7.775 | |
| 14790 | 1460.802 | 22.911 | ALB | Serum albumin | 14059.243 | 507.84 | −4.792 | 0.015 | 2015.093 | 7.35 | 8.099 | |
| 13726 | 2427.187 | 19.577 | ALB | Serum albumin | 10952.877 | 144.342 | −6.243 | 0.016 | 2015.244 | 21.418 | 6.556 | |
| 8491 | 2356.166 | 19.568 | ALB | Serum albumin | 3482.532 | 39.077 | −6.48 | 0.019 | 925.61 | 162.918 | 2.506 | |
| 8062 | 2639.32 | 19.823 | ALB | Serum albumin | 2556.829 | 19.993 | −7.002 | 0.027 | 6433.007 | 12.321 | 9.028 | |
| 7463 | 2566.365 | 19.53 | ALB | Serum albumin | 9486.713 | 19.01 | −8.966 | 0.038 | 2867.477 | 4.667 | 9.263 | |
| 5188 | 1777.952 | 21.01 | ALB | Serum albumin | 1971.827 | 52.188 | −5.238 | 0.045 | 35.267 | 0 | + | |
| P02656 | 19780 | 4113.847 | 24.548 | APOC3 | Apolipoprotein C-III | 178.772 | 476.177 | 1.413 | 0.016 | 354.975 | 506.002 | −0.515 |
| Q05682 | 7837 | 1753.934 | 21.086 | CALD1 | Caldesmon | 2558.923 | 58.12 | −5.461 | 0.035 | 1042.804 | 0 | + |
| P02452 | 17890 | 3416.589 | 31.958 | COL1A1 | Collagen α-1(I) chain | 590.236 | 1123.486 | 0.929 | 0.019 | 960.66 | 1241.408 | −0.377 |
| P08123 | 19745 | 4097.863 | 24.655 | COL1A2 | Collagen α-2(I) chain | 558.697 | 1274.198 | 1.189 | 0.02 | 1804.4 | 2939.349 | −0.713 |
| 13816 | 2825.275 | 24.449 | COL3A1 | Collagen α-1(III) chain | 9461.028 | 16079.857 | 0.765 | 0.011 | 15478.674 | 16458.099 | −0.089 | |
| 15237 | 2809.204 | 24.378 | COL3A1 | Collagen α-1(III) chain | 418.168 | 1003.36 | 1.263 | 0.012 | 608.266 | 679.408 | −0.152 | |
| P02461 | 15129 | 2583.168 | 23.633 | COL3A1 | Collagen α-1(III) chain | 90.217 | 210.593 | 1.223 | 0.02 | 347.915 | 286.865 | 0.275 |
| 8530 | 1834.836 | 24.214 | COL3A1 | Collagen α-1(III) chain | 490.291 | 824.621 | 0.75 | 0.027 | 1033.442 | 634.648 | 0.705 | |
| 11668 | 2248.99 | 26.156 | COL3A1 | Collagen α-1(III) chain | 10751.2 | 14269.938 | 0.408 | 0.031 | 13681.574 | 15627.938 | −0.184 | |
| Q9UBG3 | 883 | 974.524 | 20.641 | CRNN | Cornulin | 566.412 | 92.82 | −2.609 | 0.027 | 219.596 | 0 | + |
| Q8IY81 | 8534 | 1835.712 | 19.874 | FTSJ3 | pre-rRNA processing protein | 207.434 | 549.144 | 1.405 | 0.016 | 837.446 | 2049.9 | −1.286 |
| P08238 | 3285 | 1261.6 | 19.82 | HSP90AB1 | Heat shock protein HSP 90-β | 231.893 | 15.187 | −3.932 | 0.019 | 543.9 | 3.052 | 7.478 |
| B9A064 | 15015 | 2790.385 | 20.225 | IGLL5 | Immunoglobulin λ-like polypeptide 5 | 13096.651 | 106.472 | −6.948 | 0.037 | 22574.673 | 122.896 | 7.521 |
| P04264 | 9412 | 1934.792 | 19.905 | KRT1 | Keratin; type II cytoskeletal 1 | 185.944 | 384.677 | 1.049 | 0.027 | 564.75 | 1126.169 | −1 |
| Q6UXB8 | 8564 | 1838.932 | 20.941 | PI16 | Peptidase inhibitor 16 | 366.042 | 66.723 | −2.456 | 0.018 | 724.295 | 0 | + |
| P01833 | 18732 | 3669.665 | 24.075 | PIGR | Polymeric immunoglobulin receptor | 665.858 | 1579.226 | 1.246 | 0.014 | 1451.439 | 1045.853 | 0.475 |
| P41222 | 8589 | 1842.844 | 24.22 | PTGDS | Prostaglandin-H2 D-isomerase | 2263.559 | 148.626 | −3.928 | 0.045 | 586.969 | 700.124 | −0.252 |
| Q8WVN6 | 12387 | 2340.175 | 20.63 | SECTM1 | Secreted and transmembrane protein 1 | 436.162 | 55.053 | −2.986 | 0.037 | 234.333 | 17.235 | 3.766 |
| P01009 | 9486 | 1943.003 | 24.951 | SERPINA1 | α-1-antitrypsin | 9072.127 | 1854.2 | −2.291 | 0.031 | 9843.728 | 77.306 | 6.992 |
| 11408 | 2215.132 | 32.94 | SERPINA1 | α-1-antitrypsin | 4091.671 | 396.914 | −3.366 | 0.038 | 1826.854 | 0 | + | |
| Q9UPN9 | 10424 | 2067.824 | 20.597 | TRIM33 | E3 ubiquitin-protein ligase TRIM33 | 451.688 | 998.249 | 1.144 | 0.019 | 1329.369 | 2701.35 | −1.029 |
BH, Benjamini-Hochberg; CE-time, capillary electrophoresis migration time; DKD, diabetic kidney disease.
Sensitivity Analysis (Albuminuria Lowering Independent Proteins)
Fourteen out of 19 identified proteins remained significant (P < 0.05) in the sensitivity analysis where individuals with >30% decline in albuminuria (n = 18, post-SGLT2i treatment), were excluded (Supplementary Table S7).
Validation Study 1 (T1D With DKD vs. Healthy Controls)
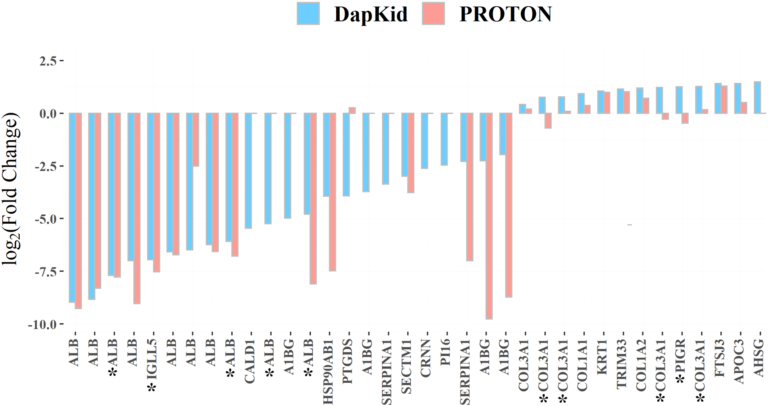
The SGLT2i-induced changes in urinary proteome were compared to the differences in urinary proteome between healthy controls (n = 50) and DKD cases (n = 110) with T1D in an independent cohort (PROTON).18 The comparison against the independent cohort is visualized in Figure 1 showing the base 2 log-transformed fold change induced by dapagliflozin along the base 2 log-transformed fold difference between healthy controls and DKD cases in PROTON. The fold difference was calculated in the independent cohort by dividing the mean urinary abundance of the peptide fragments from the healthy controls by the mean abundance of the same peptide fragments from the DKD cases. When the peptide fragments were analyzed in PROTON, 26 of the 36 peptides were found significantly different between the healthy controls and DKD cases (Supplementary Table S8).
Figure 1.
Changes induced by dapagliflozin in the type 2 diabetes discovery (DapKid) compared to the observed differences in the independent cohort (PROTON). The figure shows the base 2 log-transformed fold change in the urinary abundance of each significantly affected peptide fragment, compared to the base 2 log-difference observed between the healthy cases and DKD controls in the independent cohort (coined PROTON). The urinary abundance of 31 of the 36 significantly affected peptides changed toward healthier values. The peptides marked with an asterisk show the fragments which were not significantly different between the healthy controls and DKD cases in the independent cohort.
Of the peptide fragments, 31 of the 36 changed in the expected direction, suggesting a beneficial effect of SGLT2i; and 24 of these 31 peptide fragments were found to be significantly different between healthy controls and DKD cases in the independent cohort PROTON. The remaining 5 peptide fragments which changed in the opposite direction were derived from polymeric immunoglobulin receptor, prostaglandin-H2 D-isomerase, α-2-HS-glycoprotein, and α1 chain of type III collagen (2 fragments).
Validation Study 2 (T2D Treated With SGLT2i)
PROVALID participants with T2D who received SGLT2i treatment were used as an additional validation cohort. Peptides representing 7 of 19 proteins were available for validation in this study. Most peptides (≥75%) from each protein confirmed similar directionality of effects as in the discovery study. At least 1 peptide from 6 of 7 proteins (ALB, SERPINA1, COL1A1, COL3A1, COL1A2, and PTGDS) differed significantly (P < 0.05) after SGLT2i treatment supporting the discovery results (Table 3 and Supplementary Table S7). Overall, peptides for ALB, PTGDS, and SERPINA1 decreased, whereas those for collagens (COL1A1, COL3A1, COL1A2) and polymeric immunoglobulin receptor increased after SGLT2i treatment in the PROVALID T2D cohort reconfirming our primary findings (Supplementary Table S9).
Table 3.
Summarized potential functions of the differentially excreted proteins. The 19 unique proteins of which the SGLT2i-affected urinary peptide fragments were derived from together with the directionality of dapagliflozin-induced change and their molecular weights (in Daltons).29 The potential roles of the proteins were obtained from the referenced studies and the Human Protein Atlas30
| UniProt ID | Gene Symbol | Protein | Change | Molecular weight (Da) | Potential Role in DKD |
|---|---|---|---|---|---|
| P04217 | A1BG | α-1B-glycoprotein | - | 54,254 | Proteinuria biomarker31 |
| P02768 | ALB | Serum albumin | - | 69,367 | Proteinuria biomarker31 |
| P02765 | AHSG | α-2-HS-glycoprotein | + | 39,341 | Tissue development, endocytosis, inflammation,32 proteinuria biomarker31 |
| P02656 | APOC3 | Apolipoprotein C-III | + | 10,852 | Lipoprotein metabolism & inhibitor of lipolysis33 |
| Q05682 | CALD1 | Caldesmon | - | 93,231 | Actin cytoskeleton organization,34,35 regulation of mesangial cell activation36 |
| P02452 | COL1A1 | Collagen α-1(I) chain | + | 138,941 | Extracellular matrix organization31,32 |
| P02461 | COL3A1 | Collagen α-1(III) chain | + | 138,564 | Extracellular matrix organization31 |
| P08123 | COL1A2 | Collagen α-2(I) chain | + | 129,314 | Extracellular matrix organization31,32 |
| Q9UBG3 | CRNN | Cornulin | - | 53,533 | Repair,31 positive regulation of NF-κB transcription factor activity37 |
| Q8IY81 | FTSJ3 | pre-rRNA processing protein FTSJ3 | + | 96,558 | Unknown |
| P08238 | HSP90AB1 | Heat shock protein HSP 90-β | - | 83,264 | Regulation of TGF-β signaling,38 immunity & inflammation,39 stress response in wound healing40 |
| B9A064 | IGLL5 | Immunoglobulin λ-like polypeptide 5 | - | 23,063 | Inflammation41 |
| P04264 | KRT1 | Keratin; type II cytoskeletal 1 | + | 66,039 | Inflammation31, coagulation,30 extracellular matrix,30 and wound healing30 |
| Q6UXB8 | PI16 | Peptidase inhibitor 16 | - | 49,471 | Extracellular matrix organisation31 |
| P01833 | PIGR | Polymeric immunoglobulin receptor | + | 83,284 | Tubular response to injury42 |
| P41222 | PTGDS | Prostaglandin-H2 D-isomerase | - | 21,029 | Inhibition of TGF-β induced EMT,43,44 GFR biomarker31 |
| Q8WVN6 | SECTM1 | Secreted and transmembrane protein 1 | - | 27,039 | Inflammation,45 NF-κB pathway activation30 |
| P01009 | SERPINA1 | α-1-antitrypsin | - | 46,737 | Inflammation, coagulation and proteinuria biomarker,31,32 kidney fibrosis46 |
| Q9UPN9 | TRIM33 | E3 ubiquitin-protein ligase TRIM33 | + | 122,533 | Regulation of TGF-β pathway,47,48 inflammation & fibrosis47 |
DKD, diabetic kidney disease; EMT, epithelial–mesenchymal transformation; GFR, glomerular filtration rate.
Functional Enrichment Analysis
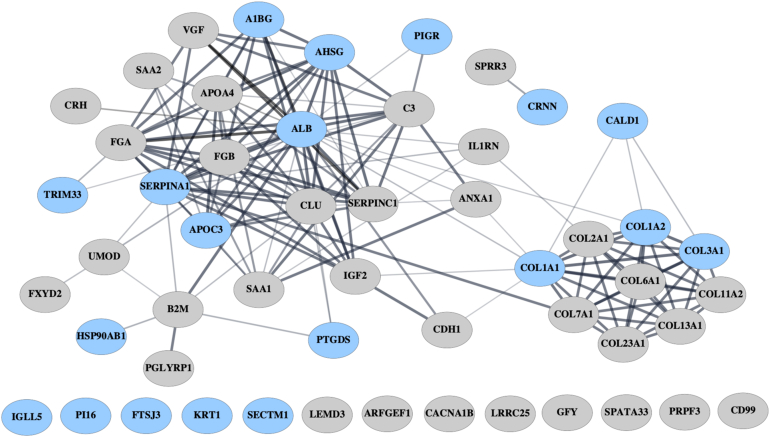
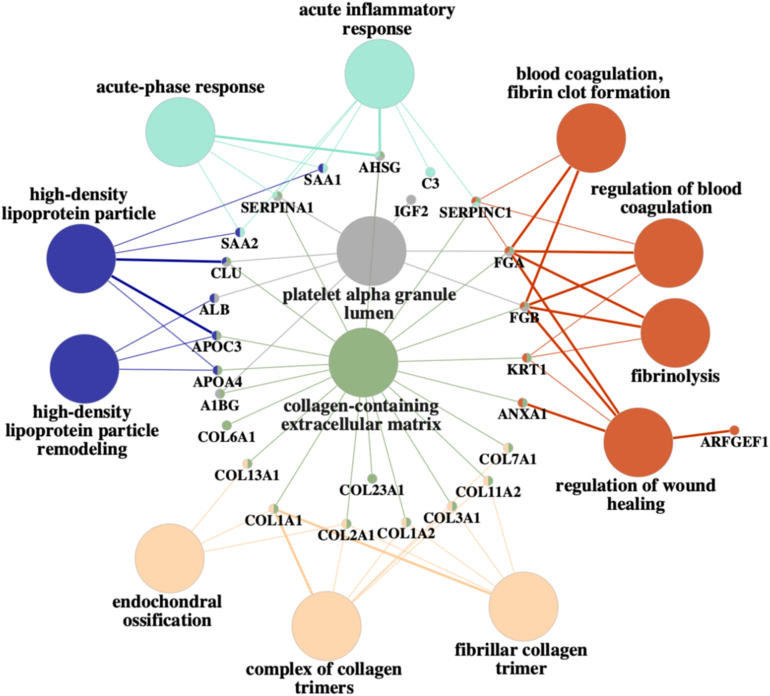
The results from the functional enrichment analyses are visualized in Figures 2 and 3. One hundred twenty peptide fragments derived from 53 proteins were considered (Supplementary Table S10).
Figure 2.
STRING protein-protein interaction network. The proteins colored in light blue denote those proteins that were found to be significantly affected by dapagliflozin using the BH-adjusted P-values and alpha level of 0.05, whereas the remaining proteins were only significant without the BH-adjustment for FDR.
BH, Benjamini-Hochberg; FDR, False Discovery Rate; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins.
Figure 3.
ClueGO Functional enrichment analysis. The results show that, especially coagulation, fibrosis, inflammatory response, and wound healing processes linked to inflammation are significantly enriched. Multiple collagen fragments were associated with extracellular matrix.
The protein-protein interaction network created using the Search Tool for the Retrieval of Interacting Genes/Proteins database is visualized in Figure 2. The network shows high interactivity between 9 unique collagen-related proteins, whereas many of the dapagliflozin-affected proteins seem to have either functional or physical interactions. Especially, apolipoprotein C-III, α-2HS-glycoprotein, albumin, SERPINA1, and α-1B-glycoprotein have high interactivity with several other proteins affected by dapagliflozin.
The results from functional enrichment analysis conducted using ClueGO are shown in Figure 3, which shows the significantly enriched biological processes and cellular components and the proteins that associated to these terms. The produced network shows that the affected urinary proteins are involved in biological processes such as inflammatory response, coagulation, wound healing, and fibrinolysis, and serve as cellular components in for example high-density lipoprotein particles and collagen-containing extracellular matrix (ECM). The most significant group of enriched biological processes was related to wound healing (Supplementary Figures S3 and S4). The urinary collagen proteins were connected to ECM, whereas proteins such as SERPINA1 and alpha-2HS-glycoprotein were linked to inflammatory response related processes. High-density lipoprotein-related processes and components were associated with 2 different apolipoproteins (APOA4 and APOC3) as well as serum amyloid 1 and 2 (SAA1 and 2). Wound healing-related processes (e.g., coagulation and fibrinolysis) were especially linked to fibrinogen alpha and beta chains, KRT1, and annexin A1.
Discussion
We identified 19 target proteins and enriched molecular pathways of SGLT2 inhibition effects investigating urinary proteomic changes occurring before and after dapagliflozin treatment in individuals with T2D with DKD. We verified most of the SGLT2i-induced changes in an independent cohort consisting of DKD cases with T1D and healthy controls. We further reproduced the findings in another study comprising individuals with T2D treated with SGLT2 inhibitors.
The functional enrichment analysis connected the identified proteins to additional biological processes previously linked to DKD development, namely, wound healing, inflammation, and changes in ECM.32
Urine is an optimal biofluid for studying kidney diseases because the changes in the urinary proteome can inform on both functional and structural changes taking place in the kidney.2,14,49 It is suggested that collagen, caldesmon, E3 ubiquitin-protein ligase [TRIM33], and PIR-derived fragments are likely to originate from the nephron, whereas smaller proteins, such as albumin, glycoproteins, apolipoprotein C-III, and alpha-1-antrypsin, are potentially derived from the plasma. This is supported by the fact that the glomerular basement membrane filtration capacity is dependent on protein molecular weight threshold of 70 kDa.50 The differential excretion of small plasma proteins is linked to functional changes of the kidney such as dysfunctional tubular reabsorption51,52 or increasing leakage in glomerular filtration, whereas the changes in excretion rates of larger kidney-derived proteins are associated with structural changes occurring in the kidney; for example, kidney fibrosis.53
SGLT2 inhibition might affect some urinary peptide levels derived from abundant plasma proteins (e.g., albumin) through improved glomerular filtration (and albuminuria reduction) However, many of the significantly changed peptides (including collagens) identified in the current study were in fact increased after SGLT2 treatment, most likely reflecting post glomerular changes. Notably, a sensitivity analysis in the current study controlling for albuminuria changes, showed that most (>70%, Supplementary Table S7) urinary proteins continued to differ significantly post SGLT2 inhibition, suggesting albuminuria independent effects. Our recent report also demonstrates that changes in proteomics classifier of renal function (CKD273) is independent of albuminuria changes, post SGLT2 inhibition.15 These findings might also support the SGLT2 inhibition-based estimated GFR lowering effects observed in large clinical trials, independent of albuminuria,54 further suggesting validation in larger studies.
Albuminuria is a common marker of DKD, and our results showed that dapagliflozin significantly decreased the abundance of 10 urinary peptide fragments derived from ALB. These reflect changes in ALB excretion further linked to changes in glomerular filtration and dysfunctional tubular reabsorption. Unlike ALB, increased levels of urinary collagens have been linked to kidney fibrosis in DKD.11,32,55 Seven individual peptide fragments derived from type I and III collagen, were significantly increased by dapagliflozin treatment. Type I collagen has been suggested to play a role in tubulointerstitial fibrosis with accumulation in arterial walls under pathological conditions whereas type III collagen is overexpressed in the tubulointerstitial space in fibrosis.56 Consequently, the reported increases in urinary collagens might reflect increased turnover of collagen derived peptides in the kidney, suggesting a beneficial effect of SGLT2i on kidney fibrosis. The functional enrichment analysis yielded similar results, because it linked the differentially excreted urinary collagens to ECM, which is known to be a target of kidney fibrosis.56 The protein-protein interaction network in the current study shows high interactivity of 9 unique collagen fragments, suggesting their involvement in response to SGLT2i treatment.
Changes in urinary abundance of other peptides derived from proteins linked to kidney fibrosis were observed. Caldesmon 1 has been connected to actin cytoskeleton organization and regulation of mesangial cell activation, with upregulation particularly in T1D.34, 35, 36 Actin cytoskeleton has been suggested to represent an important target for hyperglycemia-induced damage, which in mesangial cells has been suggested to lead to increased glomerular pressure state and vasodilatation of afferent arteriole, contributing to hyperfiltration.34 Caldesmon 1 protein was also linked to 3 individual type I and III collagens in the protein-protein interaction network. The results showed a significant decrease in urinary levels of 1 fragment from this protein, and this change was confirmed in the PROTON cohort, indicating that this protein may play a role in kidney fibrosis, together with the SGLT2i affected collagens.
Dapagliflozin induced significant changes in urinary abundance of multiple peptides linked to TGF-β signaling, which is overactivated in fibrosis.56, 57, 58 The urinary abundance of fragments from PI16, HSP90AB1, TRIM33, and PTGDS changed significantly. PI16 has been shown to be involved in profibrotic TGF-β signaling, whereas HSP90B1 and TRIM33 have been linked to the regulation of TGF-β signaling.38,43,47,48 HSP90AB1 also plays a role in tissue repair and has important stress response functions in wound healing, especially in hypoxic conditions.40 Similarly, PTGDS has been argued to play a role in TGF-β-induced epithelial-to-mesenchymal transition.44 PTGDS-based peptides decreased after SGLT2i treatment in both T2D cohorts in the current study, although the levels were largely indifferent between patients with DKD and healthy controls within PROTON. The potentially beneficial nature of the changes in TRIM33, HSP90AB1, and PI16-related urinary peptide fragment abundance was suggested by the differences observed in PROTON, because healthy controls had significantly higher levels of urinary TRIM33 fragments and lower levels of urinary HSP90AB1 and PI16 fragments than the DKD cases. Chronic hyperglycemia has been argued to stimulate multiple pathogenic pathways via increased reactive oxygen species generation, leading to TGF-β overexpression2 that decreases ECM degradation and stimulates production of ECM proteins (e.g., type I and IV collagens).59 The significant changes observed in proteins linked to TGF-β signaling thus suggest a beneficial effect of SGLT2i on this pathway.
Hyperglycemia-induced overproduction of mitochondrial reactive oxygen species and the overactivation of pathogenic pathways has also been suggested to underlie inflammation in DKD.60 Kidney inflammation has been linked to elevated levels of NF-κB, which increases proinflammatory gene expression.61 Dapagliflozin impacted the urinary abundance of several proteins suggested to play a role in inflammatory signaling, such as cornulin (CRNN), secreted and transmembrane protein 1 (SECTM1), SERPINA1, and immunoglobulin λ-like polypeptide 5 (IGLL5). The results from the functional enrichment analysis also associated alpha-2HS-glycoprotein and SERPINA1 with inflammatory response-related processes. Generally, these proteins have been linked to NF-κB pathway.37,45,62,63 SECTM has 5 potential binding sites for NF-κB, whereas CRNN has been connected to positive regulation of NF-κB activity.37,45,63 Multiple studies have shown urinary levels of SERPINA1 to be elevated in CKD,11,31,32,64 which is in concordance with our findings in individuals with DKD. Our study confirms this, whereby healthy controls had lower urinary levels of the same SERPINA1 fragments whereas dapagliflozin reduced SERPINA1 levels in both T2D cohorts.
Lastly, functional enrichment analysis showed significant enrichment of high-density lipoprotein-related terms, which has a known role in CKD.65,66 Dapagliflozin significantly increased urinary abundance of 1 apolipoprotein C-III (APOC3)-derived fragment, and similar distribution was observed between healthy cases and DKD controls in PROTON. Serum APOC3 levels are elevated in DKD, hypertriglyceridemia, and coronary heart disease.33,67, 68, 69 Moreover, APOC3 may stimulate inflammatory responses, such as PKC-α, leading to increased expression of NF-κB.69 Assuming increased urinary abundance of APOC3 reflects increased turnover of the same protein, the observed increase might reflect a beneficial effect of SGLT2i on lipoprotein metabolism.
Similar studies investigating the effect of SGLT2i on the full urinary proteome in patients with DKD with T2D have not been conducted previously. Cherney et al.70 studied the effect of the SGLT2i, empagliflozin on the urinary proteome of 40 participants with uncomplicated T1D, focusing only on the subset of urinary peptides used to calculate the CKD273 score.70 The authors reported changes in urinary peptide fragments from SERPINA1, neurosecretory protein VGF, KRT1, TRIM33, and various collagen proteins. Current study results showed that dapagliflozin similarly induced changes on urinary collagens, KRT1, and TRIM33 fragments. Cherney et al.70 reported that empagliflozin significantly increased the urinary abundance of a short peptide fragment of SERPINA1, whereas we found that dapagliflozin significantly decreased urinary level of 2 peptide fragments from SERPINA1.70 The small overlap in the results of the 2 studies might be caused by multiple reasons. Cherney et al.70 did not study the effect of SGLT2i on the full urinary proteome and used participants with uncomplicated T1D. However, because both studies found similar effects of SGLT2i on KRT1 and TRIM33, the role of these proteins in DKD development and SGLT2i action should be further investigated. Higher levels of urinary SERPINA1 and SERPING1 peptides with inflammatory activity have been reported in individuals with DKD,71 where the former targets elastase and other proteases whereas the latter inhibits the complement cascade proteins with known association with circulating IGFBP172 levels that further affect DKD pathogenesis.73
Few studies link the observed changes in urinary peptidome to affected biological processes. For example, Van et al.32 investigated the potential roles of differentially excreted proteins by dividing DKD development into 3 stages basis estimated GFR and albuminuria measurements and running functional enrichment analysis using DKD stage specific proteins. The enriched biological processes in all investigated stages of DKD were related to immune system regulation, coagulation, ECM metabolism, tissue and blood vessel development, and endothelial cell proliferation, that is, processes typically associated with wound healing.32 Interestingly, the dapagliflozin-affected proteins in the current study were linked to similar biological processes, thereby playing a central role in DKD development.10 The current study highlights the benefits of a network-based approach in studying the urinary proteome, because studying the changes in individual proteins neglects the involvement of a network of proteins in disease development.32 However, further studies using multiple data-sources (e.g., gene expression, genetics, and metabolomics) are needed to uncover the molecular mechanisms underlying the reported changes, especially inflammatory and fibrotic processes. Although the current study shows that the SGLT2 inhibition effect-associated urinary proteins are significantly differing in individuals with DKD compared to healthy controls, further studies examining the potential of these urinary proteins to distinguish interindividual drug response (albuminuria lowering vs. no albuminuria lowering) prior to SGLT2i treatment within the diabetic population would be interesting and require larger studies.
There are various limitations and strengths to this study. The discovery study consisted mostly of male participants and was relatively small. It did not include a washout period when the 2 sequence groups crossed over; however, the sequence effect was statistically insignificant in all models looking at changes in clinical factors because of treatment (Supplementary Table S3). Further, the UACR already normalizes within 2 to 4 weeks with no carryover effect as discussed previously.15 The DKD cases in PROTON had T1D whereas the discovery study included participants with T2D. The differences in T1D and T2D should be kept in mind when analyzing the results because persons with T2D typically have more confounding factors contributing to DKD (e.g., hypertension, obesity, and smoking) and are diagnosed with diabetes at a later age.2 However, because most of the observed changes in the urinary proteome of discovery study were confirmed by the T1D independent cohort, in addition to the T2D PROVALID study, it can be argued that the SGLT2i therapy targeted biological processes relevant in both T1D and T2D. The functional enrichment analysis used BH-unadjusted P-values, and these results should be carefully interpreted. The lower confidence level was justified by the nature of enrichment analysis, which is not typically informative when using a low number of proteins or genes. The current study design is unique because it leverages the randomized trial coupled urine proteomics-based discovery and observational validation studies to identify molecular effect pathways of SGLT2i therapy in DKD. Future studies should examine the long-term effect of SGLT2i on identified urinary proteins with more participants, potentially stratified by DKD progression. The newly mapped SGLT2i effect proteins are a key step toward developing improved and tailored treatment options as per the precision medicine in diabetes initiative.74
Disclosure
HM is the cofounder and coowner of Mosaiques Diagnostics. JS is employed by Mosaiques Diagnostics. MKE has served as an educator for Astra Zeneca (all honoraria to institution). PR has served as consultant on advisory boards or as an educator for Astra Zeneca, Astellas, AbbVie, Novo Nordisk, Boehringer Ingelheim, Eli Lilly, Merck, and Bayer (all honoraria to institution); has shares in Novo Nordisk; and has received research grants to institution from Novo Nordisk and Astra Zeneca. FP has served as a consultant on advisory boards or as an educator for Astra Zeneca, Novo Nordisk, Sanofi, Mundipharma, MSD, Boehringer Ingelheim, Novartis, and Amgen; and has received research grants to institution from Novo Nordisk, Amgen, and Astra Zeneca. SD has received a congress support (coverage of the registration fee for the ERA-EDTA Congress 2021) from Astra Zeneca. Astra Zeneca provided the study medication for both studies but had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. All the other authors declared no competing interests.
Acknowledgments
The DapKid study medication was provided by AstraZeneca and the study trial number is NCT02914691. Steno Diabetes Center Copenhagen received a grant from AstraZeneca to carry out the discovery study. Additional tests were funded by the Novo Nordisk Foundation grant NNF 14OC0013659 (FP). TSA was supported by NNF18OC0052457 and Steno Diabetes Center Copenhagen. This research activity is part of the project DC-ren that has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 848011. We thank the participants and research staff of both the DapKid and the PROTON studies.
Author Contributions
Conceptualization was by TSA, TKER, PR, FR, and MKE. Methodology was by TSA and TKER. The investigation was done by MKE, HM, TKER, and TSA. Visualization was done by TKER. Funding acquisition was by MKE, TKA, and PR. Project administration was by PR and FP. Supervision was done by TSA, PR, and FP. Writing of the original draft was done by TSA and TKER. Writing, including review and editing was done by TSA, VRC, PR, and FP. All authors approved the final draft.
Footnotes
Supplementary Methods.
Figure S1. Study design.
Figure S2. Consort flow diagram.
Figure S3. Alternative output from ClueGO functional enrichment analysis.
Figure S4. ClueGO output represented as a pie chart.
Table S1. Comparison of clinical factors at baseline.
Table S2. Changes in clinical factors after dapagliflozin treatment.
Table S3. Detailed model statistics for changes in clinical factors.
Table S4. Events during intervention.
Table S5. Detailed changes in individual peptide fragment abundance after SGLT2i intervention on the Randomized study DapKid.
Table S6. Sequences for the urinary peptide fragments affected by dapagliflozin.
Table S7. Sensitivity analysis: Detailed changes in individual peptide fragment abundance after SGLT2i intervention, independent of albuminuria lowering.
Table S8. Mann-Whitney U test on the urinary proteomics from the independent cohort: validation 1.
Table S9. Mann-Whitney U test on the urinary proteomics from the T2D PROVALID cohort: validation 2.
Table S10. Annotation table for proteins used in functional enrichment analyses.
Supplementary log file from ClueGO.
CONSORT checklist for randomized trial.
Supplementary Material
Supplementary Methods.
Figure S1. Study design.
Figure S2. Consort flow diagram.
Figure S3. Alternative output from ClueGO functional enrichment analysis.
Figure S4. ClueGO output represented as a pie chart.
Table S1. Comparison of clinical factors at baseline.
Table S2. Changes in clinical factors after dapagliflozin treatment.
Table S3. Detailed model statistics for changes in clinical factors.
Table S4. Events during intervention.
Table S5. Detailed changes in individual peptide fragment abundance after SGLT2i intervention on the Randomized study DapKid.
Table S6. Sequences for the urinary peptide fragments affected by dapagliflozin.
Table S7. Sensitivity analysis: Detailed changes in individual peptide fragment abundance after SGLT2i intervention, independent of albuminuria lowering.
Table S8. Mann-Whitney U test on the urinary proteomics from the independent cohort: validation 1.
Table S9. Mann-Whitney U test on the urinary proteomics from the T2D PROVALID cohort: validation 2.
Table S10. Annotation table for proteins used in functional enrichment analyses.
Supplementary log file from ClueGO.
CONSORT checklist for randomized trial.
References
- 1.Alicic R.Z., Rooney M.T., Tuttle K.R. Diabetic kidney disease: challenges, progress, and possibilities. Clin J Am Soc Nephrol. 2017;12:2032–2045. doi: 10.2215/CJN.11491116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas M.C., Brownlee M., Susztak K., et al. Diabetic kidney disease. Nat Rev Dis Primers. 2015;1 doi: 10.1038/nrdp.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heerspink H.J.L., Kosiborod M., Inzucchi S.E., Cherney D.Z.I. Renoprotective effects of sodium-glucose cotransporter-2 inhibitors. Kidney Int. 2018;94:26–39. doi: 10.1016/j.kint.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 4.Zinman B., Wanner C., Lachin J.M., et al. Empagliflozin, cardiovascular outcomes, and mortality in Type 2 diabetes. N Engl J Med. 2015;373:2117–2128. doi: 10.1056/NEJMoa1504720. [DOI] [PubMed] [Google Scholar]
- 5.Neal B., Perkovic V., Mahaffey K.W., et al. Canagliflozin and cardiovascular and renal events in Type 2 diabetes. N Engl J Med. 2017;377:644–657. doi: 10.1056/NEJMoa1611925. [DOI] [PubMed] [Google Scholar]
- 6.Henry R.R., Thakkar P., Tong C., Polidori D., Alba M. Efficacy and safety of canagliflozin, a sodium-glucose cotransporter 2 inhibitor, as add-on to insulin in patients with Type 1 diabetes. Diabetes Care. 2015;38:2258–2265. doi: 10.2337/dc15-1730. [DOI] [PubMed] [Google Scholar]
- 7.Heerspink H.J.L., Stefansson B.V., Correa-Rotter R., et al. Dapagliflozin in patients with chronic kidney disease. N Engl J Med. 2020;383:1436–1446. doi: 10.1056/NEJMoa2024816. [DOI] [PubMed] [Google Scholar]
- 8.Thomas M.C., Cherney D.Z.I. The actions of SGLT2 inhibitors on metabolism, renal function and blood pressure. Diabetologia. 2018;61:2098–2107. doi: 10.1007/s00125-018-4669-0. [DOI] [PubMed] [Google Scholar]
- 9.Anders H.J., Davis J.M., Thurau K. Nephron protection in diabetic kidney disease. N Engl J Med. 2016;375:2096–2098. doi: 10.1056/NEJMcibr1608564. [DOI] [PubMed] [Google Scholar]
- 10.DeFronzo R.A., Reeves W.B., Awad A.S. Pathophysiology of diabetic kidney disease: impact of SGLT2 inhibitors. Nat Rev Nephrol. 2021;17:319–334. doi: 10.1038/s41581-021-00393-8. [DOI] [PubMed] [Google Scholar]
- 11.Good D.M., Zurbig P., Argiles A., et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteomics. 2010;9:2424–2437. doi: 10.1074/mcp.M110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roscioni S.S., de Zeeuw D., Hellemons M.E., et al. A urinary peptide biomarker set predicts worsening of albuminuria in type 2 diabetes mellitus. Diabetologia. 2013;56:259–267. doi: 10.1007/s00125-012-2755-2. [DOI] [PubMed] [Google Scholar]
- 13.Argiles A., Siwy J., Duranton F., et al. CKD273, a new proteomics classifier assessing CKD and its prognosis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decramer S., Gonzalez de Peredo A., Breuil B., et al. Urine in clinical proteomics. Mol Cell Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Curovic V.R., Eickhoff M.K., Ronkko T., et al. Dapagliflozin improves the urinary proteomic kidney-risk classifier CKD273 in Type 2 diabetes with albuminuria: a randomized clinical trial. Diabetes Care. 2022;45:2662–2668. doi: 10.2337/dc22-1157. [DOI] [PubMed] [Google Scholar]
- 16.Eickhoff M.K., Olsen F.J., Frimodt-Moller M., et al. Effect of dapagliflozin on cardiac function in people with type 2 diabetes and albuminuria-a double blind randomized placebo-controlled crossover trial. J Diabetes Complications. 2020;34 doi: 10.1016/j.jdiacomp.2020.107590. [DOI] [PubMed] [Google Scholar]
- 17.Levey A.S., Stevens L.A., Schmid C.H., et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clos-Garcia M., Ahluwalia T.S., Winther S.A., et al. Multiomics signatures of type 1 diabetes with and without albuminuria. Front Endocrinol (Lausanne) 2022;13 doi: 10.3389/fendo.2022.1015557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winther S.A., Henriksen P., Vogt J.K., et al. Gut microbiota profile and selected plasma metabolites in type 1 diabetes without and with stratification by albuminuria. Diabetologia. 2020;63:2713–2724. doi: 10.1007/s00125-020-05260-y. [DOI] [PubMed] [Google Scholar]
- 20.Eder S., Leierer J., Kerschbaum J., et al. A prospective cohort study in patients with type 2 diabetes mellitus for validation of biomarkers (PROVALID) - study design and baseline characteristics. Kidney Blood Press Res. 2018;43:181–190. doi: 10.1159/000487500. [DOI] [PubMed] [Google Scholar]
- 21.Martens D.S., Thijs L., Latosinska A., et al. Urinary peptidomic profiles to address age-related disabilities: a prospective population study. Lancet Healthy Longev. 2021;2:e690–e703. doi: 10.1016/S2666-7568(21)00226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodol) 1995;57:289–300. doi: 10.2307/2346101. [DOI] [Google Scholar]
- 23.van Rossum G., Drake F.L. The Python Language Reference, Release 3.2.3. Python Software Foundation. http://marvin.cs.uidaho.edu/Teaching/CS515/pythonReference.pdf
- 24.R Core Team, R: a language and environment for statistical computing, 2020, R Foundation for Statistical Computing https://scirp.org/reference/referencespapers.aspx?referenceid=3064798 (Accessed 3 January 2024).
- 25.Szklarczyk D., Gable A.L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bindea G., Galon J., Mlecnik B. CluePedia cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bindea G., Mlecnik B., Hackl H., et al. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stelzer G., Rosen N., Plaschkes I., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1 30 31–31:30 33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 30.Sjostedt E., Zhong W., Fagerberg L., et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020;367 doi: 10.1126/science.aay5947. [DOI] [PubMed] [Google Scholar]
- 31.Schanstra J.P., Zurbig P., Alkhalaf A., et al. Diagnosis and prediction of CKD progression by assessment of urinary peptides. J Am Soc Nephrol. 2015;26:1999–2010. doi: 10.1681/ASN.2014050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van J.A., Scholey J.W., Konvalinka A. Insights into diabetic kidney disease using urinary proteomics and bioinformatics. J Am Soc Nephrol. 2017;28:1050–1061. doi: 10.1681/ASN.2016091018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirano T. Abnormal lipoprotein metabolism in diabetic nephropathy. Clin Exp Nephrol. 2014;18:206–209. doi: 10.1007/s10157-013-0880-y. [DOI] [PubMed] [Google Scholar]
- 34.Millioni R., Iori E., Lenzini L., et al. Caldesmon over-expression in type 1 diabetic nephropathy. J Diabetes Complications. 2011;25:114–121. doi: 10.1016/j.jdiacomp.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Conway B.R., Maxwell A.P., Savage D.A., et al. Association between variation in the actin-binding gene caldesmon and diabetic nephropathy in type 1 diabetes. Diabetes. 2004;53:1162–1165. doi: 10.2337/diabetes.53.4.1162. [DOI] [PubMed] [Google Scholar]
- 36.Murphy M., Godson C., Cannon S., et al. Suppression subtractive hybridization identifies high glucose levels as a stimulus for expression of connective tissue growth factor and other genes in human mesangial cells. J Biol Chem. 1999;274:5830–5834. doi: 10.1074/jbc.274.9.5830. [DOI] [PubMed] [Google Scholar]
- 37.Li C., Xiao L., Jia J., et al. Cornulin is induced in psoriasis lesions and promotes keratinocyte proliferation via phosphoinositide 3-kinase/Akt pathways. J Investig Dermatol. 2019;139:71–80. doi: 10.1016/j.jid.2018.06.184. [DOI] [PubMed] [Google Scholar]
- 38.Noh H., Kim H.J., Yu M.R., et al. Heat shock protein 90 inhibitor attenuates renal fibrosis through degradation of transforming growth factor-beta type II receptor. Lab Investig. 2012;92:1583–1596. doi: 10.1038/labinvest.2012.127. [DOI] [PubMed] [Google Scholar]
- 39.Lazaro I., Oguiza A., Recio C., et al. Targeting HSP90 ameliorates nephropathy and atherosclerosis through suppression of NF-kappaB and STAT signaling pathways in diabetic mice. Diabetes. 2015;64:3600–3613. doi: 10.2337/db14-1926. [DOI] [PubMed] [Google Scholar]
- 40.Jayaprakash P., Dong H., Zou M., et al. Hsp90alpha and Hsp90beta together operate a hypoxia and nutrient paucity stress-response mechanism during wound healing. J Cell Sci. 2015;128:1475–1480. doi: 10.1242/jcs.166363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutchison C.A., Cockwell P., Harding S., Mead G.P., Bradwell A.R., Barnett A.H. Quantitative assessment of serum and urinary polyclonal free light chains in patients with type II diabetes: an early marker of diabetic kidney disease? Expert Opin Ther Targets. 2008;12:667–676. doi: 10.1517/14728222.12.6.667. [DOI] [PubMed] [Google Scholar]
- 42.He T., Siwy J., Metzger J., et al. Associations of urinary polymeric immunoglobulin receptor peptides in the context of cardio-renal syndrome. Sci Rep. 2020;10:8291. doi: 10.1038/s41598-020-65154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brennan E.P., Morine M.J., Walsh D.W., et al. Next-generation sequencing identifies TGF-beta1-associated gene expression profiles in renal epithelial cells reiterated in human diabetic nephropathy. Biochim Biophys Acta. 2012;1822:589–599. doi: 10.1016/j.bbadis.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang A., Dong Z., Yang T. Prostaglandin D2 inhibits TGF-beta1-induced epithelial-to-mesenchymal transition in MDCK cells. Am J Physiol Ren Physiol. 2006;291:F1332–F1342. doi: 10.1152/ajprenal.00131.2006. [DOI] [PubMed] [Google Scholar]
- 45.Huyton T., Gottmann W., Bade-Doding C., Paine A., Blasczyk R. The T/NK cell co-stimulatory molecule SECTM1 is an IFN “early response gene” that is negatively regulated by LPS in human monocytic cells. Biochim Biophys Acta. 2011;12:1294–1301. doi: 10.1016/j.bbagen.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 46.Cho J.H., Ryu H.M., Oh E.J., et al. Alpha1-antitrypsin attenuates renal fibrosis by inhibiting TGF-beta1-Induced epithelial mesenchymal transition. PLoS One. 2016;11 doi: 10.1371/journal.pone.0162186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Wang S., Liu S., Li C., Wang J. Role of Smad signaling in kidney disease. Int Urol Nephrol. 2015;47:1965–1975. doi: 10.1007/s11255-015-1115-9. [DOI] [PubMed] [Google Scholar]
- 48.Quere R., Saint-Paul L., Carmignac V., et al. Tif1gamma regulates the TGF-beta1 receptor and promotes physiological aging of hematopoietic stem cells. Proc Natl Acad Sci U S A. 2014;111:10592–10597. doi: 10.1073/pnas.1405546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hijmans R.S., Rasmussen D.G., Yazdani S., et al. Urinary collagen degradation products as early markers of progressive renal fibrosis. J Transl Med. 2017;15:63. doi: 10.1186/s12967-017-1163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lote C.J. Principles of Renal Physiology. Springer; 2012. Book chapter: glomerular filtration; pp. 33–44. [Google Scholar]
- 51.Dickson L.E., Wagner M.C., Sandoval R.M., Molitoris B.A. The proximal tubule and albuminuria: really. J Am Soc Nephrol. 2014;25:443–453. doi: 10.1681/ASN.2013090950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallon V., Thomson S.C. The tubular hypothesis of nephron filtration and diabetic kidney disease. Nat Rev Nephrol. 2020;16:317–336. doi: 10.1038/s41581-020-0256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullen W., Delles C., Mischak H. Euro KUPCa. Urinary proteomics in the assessment of chronic kidney disease. Curr Opin Nephrol Hypertens. 2011;20:654–661. doi: 10.1097/MNH.0b013e32834b7ffa. [DOI] [PubMed] [Google Scholar]
- 54.Heerspink H.J.L., Jongs N., Chertow G.M., et al. Effect of dapagliflozin on the rate of decline in kidney function in patients with chronic kidney disease with and without type 2 diabetes: a prespecified analysis from the DAPA-CKD trial. Lancet Diabetes Endocrinol. 2021;9:743–754. doi: 10.1016/S2213-8587(21)00242-4. [DOI] [PubMed] [Google Scholar]
- 55.Pontillo C., Mischak H. Urinary peptide-based classifier CKD273: towards clinical application in chronic kidney disease. Clin Kidney J. 2017;10:192–201. doi: 10.1093/ckj/sfx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Genovese F., Manresa A.A., Leeming D.J., Karsdal M.A., Boor P. The extracellular matrix in the kidney: a source of novel non-invasive biomarkers of kidney fibrosis? Fibrogenesis Tissue Repair. 2014;7:4. doi: 10.1186/1755-1536-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biernacka A., Dobaczewski M., Frangogiannis N.G. TGF-beta signaling in fibrosis. Growth Factors. 2011;29:196–202. doi: 10.3109/08977194.2011.595714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hills C.E., Squires P.E. The role of TGF-beta and epithelial-to mesenchymal transition in diabetic nephropathy. Cytokine Growth Factor Rev. 2011;22:131–139. doi: 10.1016/j.cytogfr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Ziyadeh F.N. Mediators of diabetic renal disease: the case for TGF-beta as the major mediator. J Am Soc Nephrol. 2004;15(suppl 1):S55–S57. doi: 10.1097/01.asn.0000093460.24823.5b. [DOI] [PubMed] [Google Scholar]
- 60.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 61.Sanz A.B., Sanchez-Nino M.D., Ramos A.M., et al. NF-kappaB in renal inflammation. J Am Soc Nephrol. 2010;21:1254–1262. doi: 10.1681/ASN.2010020218. [DOI] [PubMed] [Google Scholar]
- 62.Pastore N., Ballabio A., Brunetti-Pierri N. Autophagy master regulator TFEB induces clearance of toxic SERPINA1/alpha-1-antitrypsin polymers. Autophagy. 2013;9:1094–1096. doi: 10.4161/auto.24469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsuda A., Suzuki Y., Honda G., et al. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22:3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- 64.Petra E., Siwy J., Vlahou A., Jankowski J. Urine peptidome in combination with transcriptomics analysis highlights MMP7, MMP14 and PCSK5 for further investigation in chronic kidney disease. PLoS One. 2022;17 doi: 10.1371/journal.pone.0262667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vaziri N.D. Lipotoxicity and impaired high density lipoprotein-mediated reverse cholesterol transport in chronic kidney disease. J Ren Nutr. 2010;20(5 suppl):S35–S43. doi: 10.1053/j.jrn.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 66.Rysz J., Gluba-Brzozka A., Rysz-Gorzynska M., Franczyk B. The role and function of HDL in patients with chronic kidney disease and the risk of cardiovascular disease. Int J Mol Sci. 2020;21:601. doi: 10.3390/ijms21020601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rutledge J.C., Ng K.F., Aung H.H., Wilson D.W. Role of triglyceride-rich lipoproteins in diabetic nephropathy. Nat Rev Nephrol. 2010;6:361–370. doi: 10.1038/nrneph.2010.59. [DOI] [PubMed] [Google Scholar]
- 68.Sacks F.M., Alaupovic P., Moye L.A., et al. VLDL, apolipoproteins B, CIII, and E, and risk of recurrent coronary events in the Cholesterol and Recurrent Events (CARE) trial. Circulation. 2000;102:1886–1892. doi: 10.1161/01.cir.102.16.1886. [DOI] [PubMed] [Google Scholar]
- 69.Kawakami A., Aikawa M., Nitta N., Yoshida M., Libby P., Sacks F.M. Apolipoprotein CIII-induced THP-1 cell adhesion to endothelial cells involves pertussis toxin-sensitive G protein- and protein kinase C alpha-mediated nuclear factor-kappaB activation. Arterioscler Thromb Vasc Biol. 2007;27:219–225. doi: 10.1161/01.ATV.0000249620.68705.0d. [DOI] [PubMed] [Google Scholar]
- 70.Cherney D., Perkins B.A., Lytvyn Y., Heerspink H., Rodriguez-Ortiz M.E., Mischak H. The effect of sodium/glucose cotransporter 2 (SGLT2) inhibition on the urinary proteome. PLoS One. 2017;12 doi: 10.1371/journal.pone.0186910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brondani L.A., Soares A.A., Recamonde-Mendoza M., et al. Urinary peptidomics and bioinformatics for the detection of diabetic kidney disease. Sci Rep. 2020;10:1242. doi: 10.1038/s41598-020-58067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahluwalia T.S., Allin K.H., Sandholt C.H., et al. Discovery of coding genetic variants influencing diabetes-related serum biomarkers and their impact on risk of type 2 diabetes. J Clin Endocrinol Metab. 2015;100:E664–E671. doi: 10.1210/jc.2014-3677. [DOI] [PubMed] [Google Scholar]
- 73.Lay A.C., Hale L.J., Stowell-Connolly H., et al. IGFBP-1 expression is reduced in human type 2 diabetic glomeruli and modulates beta1-integrin/FAK signalling in human podocytes. Diabetologia. 2021;64:1690–1702. doi: 10.1007/s00125-021-05427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung W.K., Erion K., Florez J.C., et al. Precision medicine in diabetes: a consensus report from the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD) Diabetes Care. 2020;43:1617–1635. doi: 10.2337/dci20-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.