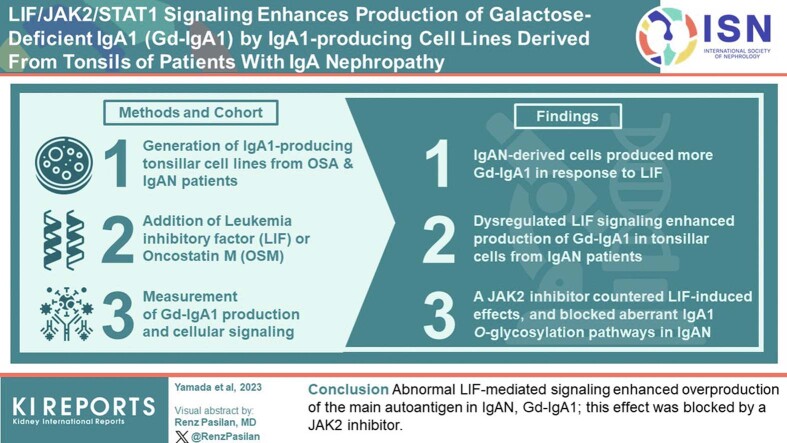
Abstract
Introduction
Galactose-deficient IgA1 (Gd-IgA1) plays a key role in the pathogenesis of IgA nephropathy (IgAN). Tonsillectomy has been beneficial to some patients with IgAN, possibly due to the removal of tonsillar cytokine-activated cells producing Gd-IgA1. To test this hypothesis, we used immortalized IgA1-producing cell lines derived from tonsils of patients with IgAN or obstructive sleep apnea (OSA) and assessed the effect of leukemia inhibitory factor (LIF) or oncostatin M (OSM) on Gd-IgA1 production.
Methods
Gd-IgA1 production was measured by lectin enzyme-linked immunosorbent assay; JAK-STAT signaling in cultured cells was assessed by immunoblotting of cell lysates; and validated by using small interfering RNA (siRNA) knock-down and small-molecule inhibitors.
Results
IgAN-derived cells produced more Gd-IgA1 than the cells from patients with OSA, and exhibited elevated Gd-IgA1 production in response to LIF, but not OSM. This effect was associated with dysregulated STAT1 phosphorylation, as confirmed by STAT1 siRNA knock-down. JAK2 inhibitor, AZD1480 exhibited a dose-dependent inhibition of the LIF-induced Gd-IgA1 overproduction. Unexpectedly, high concentrations of AZD1480, but only in the presence of LIF, reduced Gd-IgA1 production in the cells derived from patients with IgAN to that of the control cells from patients with OSA. Based on modeling LIF-LIFR-gp130-JAK2 receptor complex, we postulate that LIF binding to LIFR may sequester gp130 and/or JAK2 from other pathways; and when combined with JAK2 inhibition, enables full blockade of the aberrant O-glycosylation pathways in IgAN.
Conclusion
In summary, IgAN cells exhibit LIF-mediated overproduction of Gd-IgA1 due to abnormal signaling. JAK2 inhibitors can counter these LIF-induced effects and block Gd-IgA1 synthesis in IgAN.
Keywords: galactose-deficient IgA1, genome-wide association studies, JAK2 inhibitor, leukemia inhibitory factor, oncostatin M, phosphorylated STAT1
Graphical abstract
IgAN is the most common primary glomerulonephritis in the world, with progression to kidney failure in up to 40% of patients. Development of disease-specific therapies is urgently needed. A potential association between the mucosal immune system and IgAN has been proposed because the main production sites of IgA1 are in the mucosal tissues, and a common clinical feature of IgAN is macroscopic hematuria concurrent with upper respiratory tract infection.1,2 In addition, urinary abnormalities often occur after tonsillar irritation.3
The “multi-hit hypothesis” has been suggested for the pathobiology of IgAN,4 wherein Gd-IgA1 glycoforms form immune complexes with Gd-IgA1-specific IgG autoantibodies. Some of these complexes deposit in the glomeruli and induce kidney injury. Serum levels of Gd-IgA1 and IgG autoantibodies are associated with disease progression.5, 6, 7, 8 Although the major sources of Gd-IgA1 production in vivo are still under investigation, the production of Gd-IgA1 by tonsillar B cells has been proposed as one possible source and could explain that tonsillectomy improved clinical symptoms in some patients with IgAN.9,10
It has also been suggested that Gd-IgA1 in the circulating blood may be produced, in part, by palatine-tonsil B cells.11 Previous studies showed that a tumor-necrosis-factor family member, a proliferation-inducing ligand (APRIL) was overexpressed in B cells located in germinal centers of tonsils from patients with IgAN, and its expression levels correlated with the severity of proteinuria. Serum level of Gd-IgA1 in many patients with IgAN decreased after tonsillectomy.12 A nationwide retrospective study in Japan found that tonsillectomy is associated with improved renal survival rates in patients with IgAN.13 As for supporting, a randomized controlled trial reported tonsillectomy with corticosteroid pulse therapy being effective14; specifically, the rate of proteinuria reduction was significant in the tonsillectomy with pulse therapy group compared to the steroid pulse monotherapy group during the observation period of 1 year after the start of treatment.
Genome-wide association studies (GWAS) in IgAN have identified a number of cytokine-related and mucosal-immunity-related loci. The Chr.22q12 locus with HORMAD2/LIF/OSM genes was identified in several studies, and is associated with the regulation of IgA production in the mucosa and serum IgA levels.15, 16, 17 Furthermore, another GWAS study revealed that the same locus was associated with acute tonsilitis and chronic inflammation of tonsils leading to tonsillectomy.18
Previous studies of LIF/OSM cytokines revealed that LIF stimulation of immortalized IgA1-producing cell lines derived from peripheral blood of patients with IgAN increased Gd-IgA1 production.19 Signaling studies implicated abnormal activation of Src family kinase represented by Lyn.20 In addition, clinical studies of peripheral-blood cells suggest that changes in phosphorylation of STAT1 (phospho-STAT1; pSTAT1) may affect the progression of renal lesions.21 Elevated numbers of IgA-positive cells,9,22,23 enhanced expression of B-cell activating factor (BAFF),24 and enhanced expression of APRIL12,25 have been reported in tonsils from patients with IgAN. However, the mechanisms of the signal transduction alteration are still not clear.
In this study, we analyzed the effects of LIF or OSM on IgA1 and Gd-IgA1 production using tonsillar cells from patients with IgAN and disease controls. The purpose of this study was to obtain a better understanding of IgA1-producing tonsillar cells from patients with IgAN and the effect of cytokines encoded in one of the IgAN-associated GWAS loci.
Methods
Patient Cohorts
Palatine tonsils were obtained from patients with biopsy-proven IgAN and patients with OSA (disease controls) who had undergone tonsillectomy at the Department of Otorhinolaryngology of Juntendo University Hospital. Protocols for obtaining the tonsil samples for isolation of cells were approved by the Institutional Review Board for Human Use of the Juntendo University Faculty of Medicine, and the samples were obtained after written informed consent. In Table 1, we provide selected clinical and laboratory data of the volunteers.
Table 1.
Clinical and laboratory data of the volunteers
| Patients profile | Obstructive sleep apnea | IgA nephropathy |
|---|---|---|
| n | 3 | 3 |
| Age, year | 24.67 ± 4.73 | 28.0 ± 5.57 |
| Sex | M = 3 | M = 1, F = 2 |
| sCr, mg/dl | 0.75 ± 0.04 | 0.70 ± 0.18 |
| BUN, mg/dl | 13.00 ± 2.65 | 13.67 ± 2.52 |
| eGFR, ml/min/1.73 m2 | 106.00 ± 6.06 | 93.67 ± 11.95 |
| Protein-to-urine Cr ratio, g/gCr | – | 0.51 ± 0.43 |
| Hematuria (RBCs per HPF) | ||
| <20 | – | 0 |
| 20–49 | – | 2 |
| >50 | – | 1 |
Data expressed as mean ± SD. M, male: F, female; BUN, blood urea nitrogen; sCr, serum creatinine; eGFR, estimated glomerular filtration rate; Cr, creatinine; –, not done.
Hematuria was assessed by assigning scores according to the number of red blood cells (RBCs) per high-power field (HPF).
Generation of Epstein-Barr Virus-Immortalized IgA1-Secreting Cell Lines
Tonsillar tissue specimens were dissected into small pieces that were then mechanically dissociated using a 100-μm cell strainer. Mononuclear cells were isolated using Ficoll-Hypaque density gradient and thereafter immortalized by infection with Epstein-Barr virus (EBV).26
IgA1-secreting cell lines were subcloned by limiting dilution, as described before.26 We then randomly selected IgA1-secreting cell lines from 3 patients with IgAN and 3 patients with OSA (Table 1). The selected cell lines exhibited stable growth, production of IgA, and the degree of galactose deficiency of the secreted IgA1. Cells were cultured in RPMI 1640 supplemented with 20% fetal bovine serum, 100 U/ml of penicillin, and 0.1 mg/ml of streptomycin in a humidified CO2 (5%) incubator at 37 °C. Cell viability was assessed by using trypan blue exclusion.
Treatment of IgA1-Secreting Cells With LIF and JAK2 Inhibitors
IgA1-secreting cells were plated at 1 × 105 cells/well in 24-well plates, stimulated with LIF (40 ng/ml) or OSM (40 ng/ml) in the absence or presence of JAK2 inhibitor AZD1480 (LC Laboratories, Woburn, MA) (0 to 0.3 μM) or AG490. The cells were preincubated with the inhibitor for 60 minutes before the addition of cytokine. The cells were then incubated with LIF or OSM for 30 minutes after preincubation with the inhibitor or a mock control. Samples of culture medium were harvested after 5 days for analyses of total IgA1 and Gd-IgA1.
Determination of Total IgA Concentration
Total IgA was measured by enzyme-linked immunosorbent assay. Ninety-six-well plates were coated with 0.1 μg/well of goat IgG F(ab’)2 specific for human IgA (Jackson ImmunoResearch Inc., West Grove, PA), blocked with 1% BSA in phosphate-buffered saline (PBS) with 0.05% Tween 20 (PBS-T), washed, and incubated with serially diluted samples of cell-culture supernatants from IgA-producing cells. Serially diluted standardized serum (Bio-Rad, Hercules, CA) was used to generate a calibration curve for IgA quantification. Bound IgA was detected after addition of biotinylated goat IgG F(ab’)2 specific for human IgA (Biosource, San Diego, CA), followed by avidin–horseradish peroxidase conjugate (Extravidin; Sigma, St. Louis, MO) and peroxidase substrate o-phenylenediamine-hydrogen peroxide (Sigma). Optical densities (ODs) were measured at 490 nm using an EL808 microplate reader (BioTek, Winooski, VT).
As indicated above, we have cloned IgA-secreting cells and confirmed that the secreted IgA is exclusively IgA1.26 However, because the enzyme-linked immunosorbent assay is not specific for IgA1 because of the use of polyclonal anti-IgA antibody, we refer to the measured analyte as IgA for formal reasons.
Gd-IgA1 Assay
Amount of Gd-IgA1 secreted in culture medium was determined by lectin enzyme-linked immunosorbent assay. Microplates coated with 0.25 μg/well of goat IgG F(ab’)2 specific for human IgA (Jackson ImmunoResearch Inc.) were blocked with 1% BSA in PBS-T, washed, and then cell-culture supernatant with 100 ng of IgA per well was added and incubated overnight at 4 °C. Captured IgA was desialylated using neuraminidase (Arthrobacter ureafaciens; Glyko, Toronto, Canada) at 2 mU/ml for 3 hours at 37 °C. Gd-IgA1 was detected using biotinylated N-acetylgalactosamine (GalNAc)-specific lectin from Helix aspersa (HAA, Sigma; 2 μg/ml in 1% BSA in PBS-T), followed by avidin–horseradish peroxidase conjugate (Sigma) and peroxidase substrate o-phenylenediamine-hydrogen peroxide (Sigma). ODs were measured at 490 nm. The Gd-IgA1 concentration was expressed in Units defined as the ratio of OD determined for the individual sample to the OD for a standard Gd-IgA1 (Ale) myeloma protein. Specifically, 100 Units was defined as the OD of 100 ng of the standard Gd-IgA1 protein.27
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis and Western Blot Analysis
LIF-treated cells were pelleted by centrifugation, washed using ice-cold PBS, and lysed in M-PER lysis buffer containing protease inhibitor cocktail and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA). Cell debris was removed by centrifugation for 10 minutes at 20,000g at 4 °C. Protein concentrations in the supernatants were measured using a protein assay kit (Bio-Rad); aliquots corresponding to 7 μg/lane of total protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane for Western-blot analysis. After the transfer, the membranes were blocked using Superblock (Thermo Fisher Scientific) and incubated with phospho-Y701-STAT1-specific rabbit polyclonal antibody, diluted 1:800 in blocking buffer, or with STAT1-specific mouse monoclonal antibody diluted 1:10,000 (R&D Systems, Minneapolis, MN). Bound antibodies were detected by addition of HRP-conjugated anti-rabbit (1:10,000) or anti-mouse (1:10,000) IgG antibodies (Southern Biotech, Birmingham, AL), respectively, followed by addition of chemiluminescence substrate (Thermo Fisher Scientific). The detected bands were visualized on a Kodak radiography film. Densitometric evaluation with ImageJ software was used to express the amount of phospho-Y701-STAT1 relative to STAT1 protein in cellular lysates. Actin-specific antibody served as an additional control of protein load.
Real-Time Quantitative Polymerase Chain Reaction
RNA was isolated from 2 × 105 cells using RNeasy 96 Mini Kit (Qiagen, Hilden, Germany), converted to complementary DNA by SuperScript III First-Strand Synthesis SuperMix kit (Invitrogen, Carlsbad, CA). Levels of STAT1 and GAPDH transcripts were determined by real-time quantitative polymerase chain reaction using LightCycler 480 DNA SYBR Green I Master chemistry on LightCycler 480 instrument (Roche, Basel, Switzerland), as reported before.20 The primer sets for STAT1 and GAPDH genes are provided in the Supplementary Table S1. Results were expressed as the fold change versus the control groups after the results were normalized with corresponding GAPDH housekeeping gene mRNA values using the 2–ΔΔCT method.28
STAT1 siRNA Knock-Down
IgA1-producing cell lines derived from tonsillar cells from 3 patients with IgAN and 3 patients with OSA were transfected using ON-TARGETplus SMARTpool siRNAs (Thermo Fisher Scientific) specific for human STAT1. ON-TARGETplus Non-targeting SMARTpool siRNAs served as controls. Cells were inoculated at density 5 × 105/ml for 24 hours before siRNAs were added, following our previously published protocol.27 Before transfection, the cells were harvested by centrifugation for 10 minutes at 300g and resuspended at room temperature in Nucleofector Solution C (Lonza, Basel, Switzerland) at density of 2.5 × 106 cells per 100 μl for each transfection. After addition of 1.4 μg of individual siRNA, cells were pulsed in Amaxa nucleofector II (Lonza) using program X-001, immediately transferred to a 24-well panel containing 1.4 ml of cell-culture medium and incubated in humidified CO2 incubator at 37 °C. Twenty-four hours after transfection, the knock-down efficiency was determined by real-time quantitative polymerase chain reaction. The knock-down effect was expressed as complementary DNA level of the individual gene normalized to the level of GAPDH, after respective siRNA treatment, divided by respective value obtained after treatment by nontargeting siRNA.
Statistical Analyses
Results were expressed as mean ± SD values. Data were analyzed using 2-tailed Student’s t test (unpaired or paired, as applicable). For analyzing multiple groups, 1-way analysis of variance was used, and values of P < 0.05 were regarded significant.
Results
IgA1-Producing Cell Lines From Tonsils of Patients With IgAN and Those With OSA
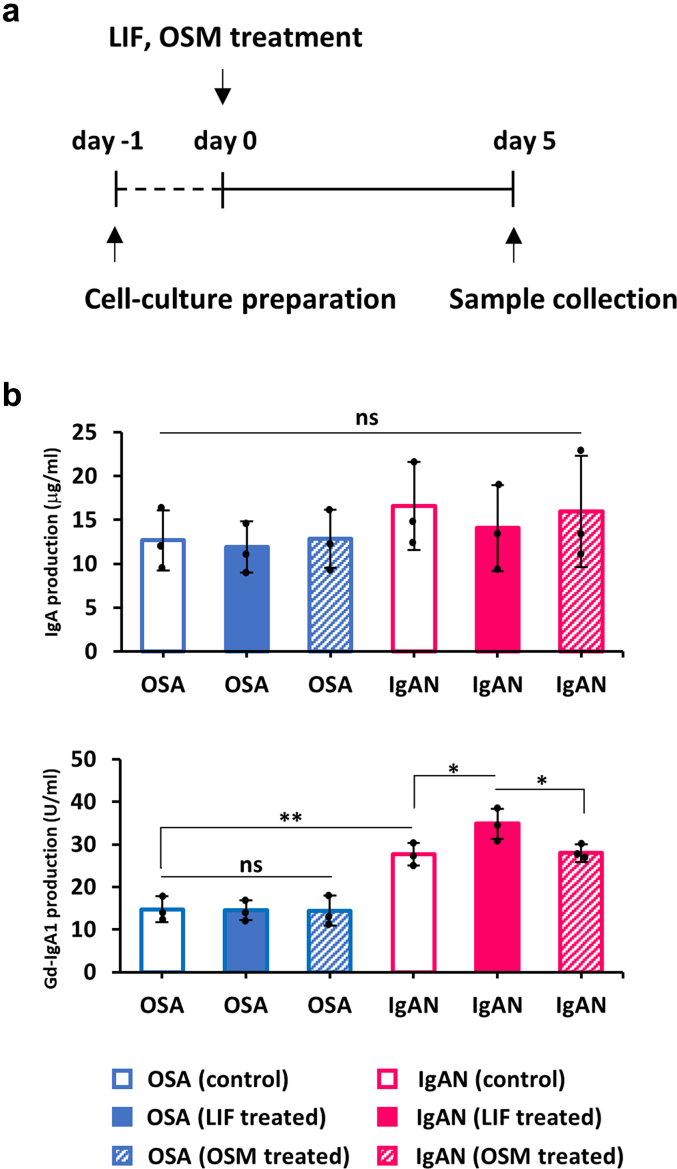
EBV-immortalized IgA1-producing cell lines derived from tonsils of patients with IgAN and those with OSA secreted comparable amounts of IgA1 into the culture medium under baseline conditions. LIF supplementation did not alter total IgA1 production in any group (Figure 1b, top). Tonsillar IgA1-secreting cell lines from patients with IgAN produced more Gd-IgA1 than those from OSA (P < 0.01) (Figure 1b, bottom). However, LIF but not OSM increased Gd-IgA1 production by tonsillar IgA1-secreting cell lines from patients with IgAN but not from patients with OSA (P < 0.05) (Figure 1b, bottom).
Figure 1.
IgA1 and Gd-IgA1 production by IgA1-producing cell lines derived from tonsils and the effect of LIF or OSM stimulation. (a) Experimental protocol for treatment of IgA1-secreting cells. Cells were stimulated with LIF (40 ng/ml), OSM (40 ng/ml), or control on day 0. Cell-culture medium was harvested on day 5 for determination of IgA and Gd-IgA1. (b) There were no differences in the concentrations of IgA in the cell-culture medium between cells from OSA versus patients with IgAN (Figure 1b, top). Treatment with LIF or OSM did not alter IgA concentration in the medium. For Gd-IgA1, baseline production was higher in the cells from patients with IgAN than from OSA group. LIF increased Gd-IgA1 production in the cells from patients with IgAN but not from OSA group. Treatment with OSM did not cause significant changes in OSA or IgAN group. One hundred Units of Gd-IgA1 was defined as 100 ng of the standard Gd-IgA1 (Figure 1b bottom). All data are presented as mean ± SD values and mean values for individual cell lines are shown by black circles. ∗P < 0.05; ∗∗P < 0.01. IgAN, IgA nephropathy; ns, not statistically significant; LIF, leukemia inhibitory factor; OSA, obstructive sleep apnea; OSM, oncostatin M.
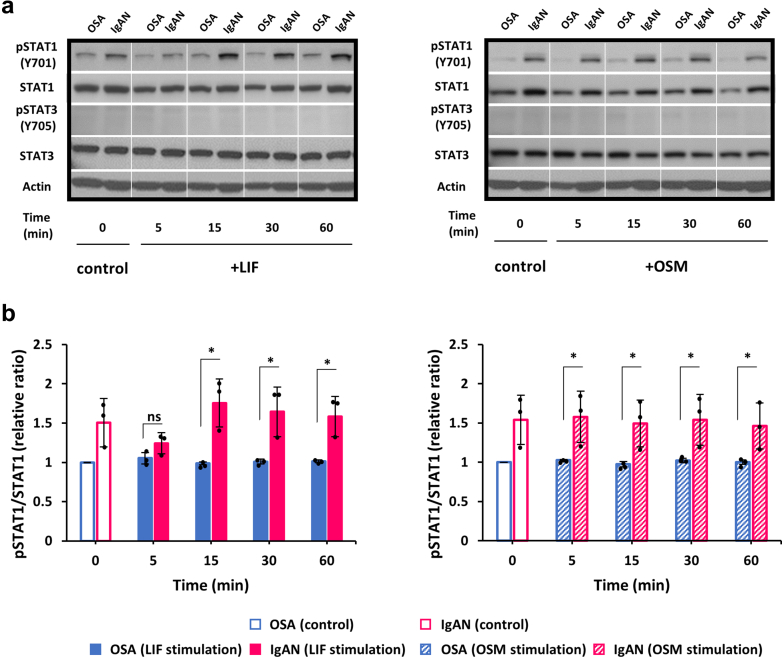
LIF Stimulation Induced STAT1 Phosphorylation in IgA1-Secreting Cells From Tonsils of Patients With IgAN to a Greater Extent Than in Cells From OSA
Baseline pSTAT1 was elevated in the cell lines from patients with IgAN compared to those from OSA group (Figure 2). LIF stimulation initially down-regulated pSTAT1 in IgAN-derived cells at 5 minutes (Figure 2a and b, left panels, Supplementary Figure S1) and, after this initial downregulation, increased at 15 minutes and remained elevated up to 60 minutes. Treatment with OSM did not increase pSTAT1 amounts in the cells from OSA group, nor did it enhance the upregulated pSTAT1 in cells from patients with IgAN (Figure 2a and b, right panels). Because STAT3 is activated by some IL-6-family cytokines, we tested whether LIF or OSM can induce STAT3 phosphorylation. We found that LIF or OSM stimulation did not alter pSTAT3 at Y705 in the cells from IgAN and OSA groups (Figure 2a, both panels).
Figure 2.
Activation of STAT1 and STAT3 in IgA1-secreting tonsillar cell lines by LIF or OSM. (a) Representative blots show phosphorylation of STAT1 (pSTAT1) (Y701) after 5-, 15-, 30-, and 60-minute stimulation with LIF (left panels) or OSM (right panels). Total STAT1 protein served as a loading control. Baseline pSTAT1 was enhanced in cells from IgAN group compared to those from OSA group. Treatment with LIF further enhanced pSTAT1 at different time points, reaching peak at 15 minutes. Treatment with OSM did not induce significant changes of pSTAT1 at different time points. STAT3 was not phosphorylated in any of the tested groups at baseline or after LIF or OSM stimulation. (b) Densitometric analyses of pSTAT1 normalized to total STAT1 (n = 3 in each group). pSTAT1 relative to total STAT1 level in OSA samples (control) was set to 1 for the comparisons. Actin blot is shown as an additional control for protein load. All data are presented as mean ± SD values and mean values for individual cell lines are shown by black circles. ∗P < 0.05. IgAN, IgA nephropathy; ns, not statistically significant; LIF, leukemia inhibitory factor; OSA, obstructive sleep apnea; OSM, oncostatin M.
JAK2 Mediates LIF-Induced Overproduction of Gd-IgA1
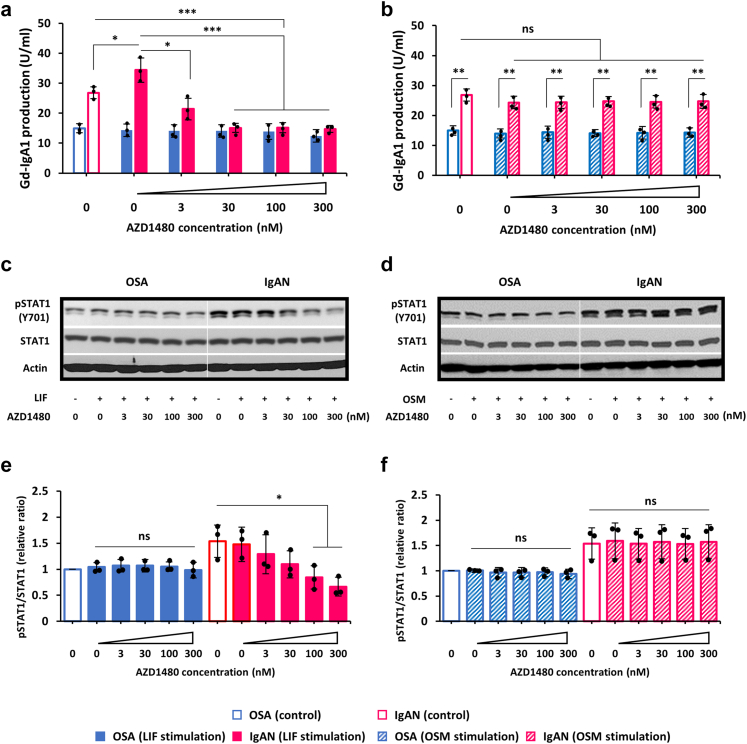
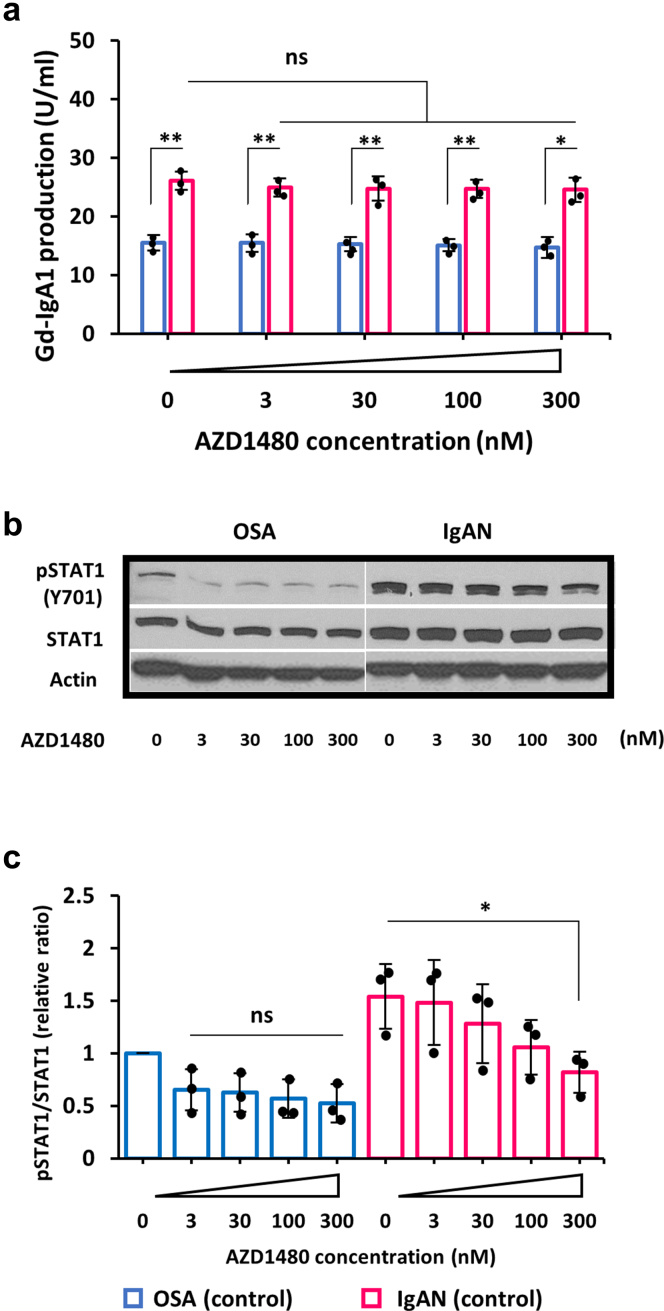
To assess if JAK2 pathway was involved in LIF-mediated overproduction of Gd-IgA1, a JAK2 inhibitor, AZD1480, was used in the LIF stimulation study. AZD1480 inhibited in a dose-dependent fashion the LIF-enhanced production of Gd-IgA1 in IgA1-producing cell lines from patients with IgAN. Notably, AZD1480 at 30 nM and higher concentrations reduced production of Gd-IgA1 by IgAN cells to the level observed for OSA cells (Figure 3a), but only in the presence of LIF (Figure 3a) and not OSM (Figure 3b). Similar effect was observed for another JAK2 inhibitor, AG490, that also reduced production of Gd-IgA1 in IgAN cells stimulated with LIF to the level observed for OSA cells (Supplementary Figure S2). The enhanced pSTAT1 induced by LIF was significantly reduced by AZD1480 (Figure 3c and e). These results indicated that LIF signaling was mediated by JAK2-STAT1 pathway. However, AZD1480 treatment without LIF did not inhibit production of Gd-IgA1 in IgAN (Figure 4a). AZD1480 inhibited pSTAT1 in a dose-dependent manner (Figure 4b and c).
Figure 3.
Impact of JAK2 inhibitor AZD1480 on Gd-IgA1 production and activation of STAT1 by LIF versus OSM in IgA1-secreting tonsillar cell lines. (a) LIF-induced overproduction of Gd-IgA1 in tonsillar IgA1-producing cells from patients with IgAN was inhibited by AZD1480 in a dose-dependent manner. (b) Conversely, OSM alone or OSM with AZD1480 did not alter Gd-IgA1 production. The cells were preincubated with the inhibitor for 60 minutes before addition of a cytokine. One-hundred Units of Gd-IgA1 was defined as 100 ng of the standard Gd-IgA1. (c) LIF enhanced phosphorylation of STAT1 (pSTAT1) and this effect was downregulated by AZD1480 in a dose-dependent manner. (d) Conversely, OSM or OSM with AZD1480 did not alter pSTAT1. In panels c and d, the cells were incubated with LIF or OSM for 30 minutes. Actin blots are shown as additional controls for protein load. (e and f) Densitometric analysis of pSTAT1 of data from b and d was performed for each sample (n = 3). pSTAT1 relative to total STAT1 level in OSA control cells (control) was set to 1. Statistical analysis for (a, b, e, and f) was performed by 1-way analysis of variance followed by Tukey’s multiple comparison test. All data are presented as mean ± SD values and mean values for individual cell lines are shown by black circles. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.005. IgAN, IgA nephropathy; ns, not statistically significant; LIF, leukemia inhibitory factor; OSA, obstructive sleep apnea; OSM, oncostatin M.
Figure 4.
JAK2 inhibitor AZD1480 alone does not affect production of Gd-IgA1 in IgA1-secreting tonsillar cell lines. (a) Gd-IgA1 production by tonsillar IgA1-producing cell lines from patients with OSA and IgAN was evaluated in the absence and presence of AZD1480 (3–300 nM). AZD 1480 did not change baseline levels of Gd-IgA1 in the tested cell lines. (b) Representative western blot of phosphorylation of STAT1 (pSTAT1) and total STAT1 in the absence or presence of AZD1480 (3–300 nM) in mock-stimulated tonsillar IgA1-producing cells. Baseline phosphorylation of STAT1 was inhibited by treatment of AZD1480 in a dose-dependent manner. Actin blot is shown as an additional control for protein load. (c) Densitometric analysis of pSTAT1 immunoblot data from panel b was performed in all samples (n = 3 in each group). pSTAT1 relative to total STAT1 level in OSA (control) was set to 1. All data are presented as mean ± SD values and mean values for individual cell lines are shown by black circles. ∗P < 0.05, ∗∗P < 0.01. IgAN, IgA nephropathy; ns, not statistically significant; OSA, obstructive sleep apnea.
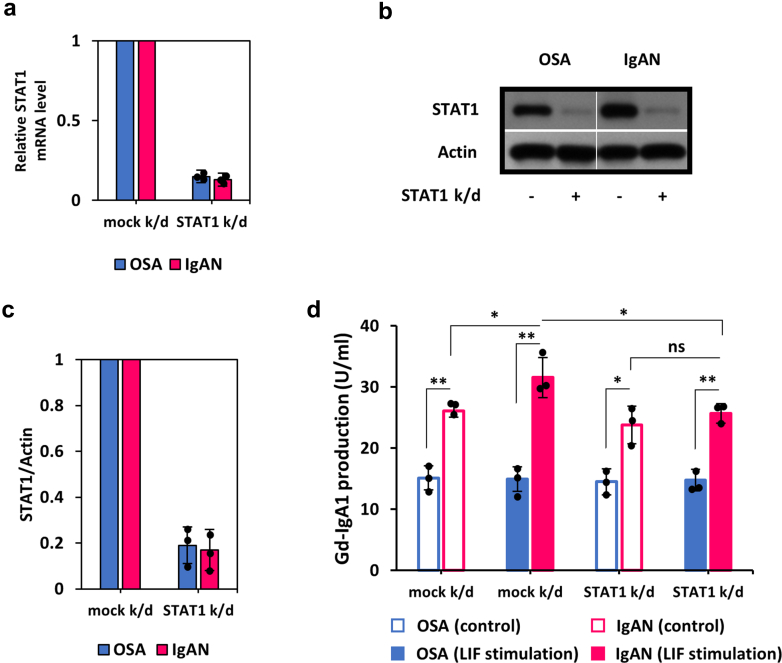
STAT1 siRNA Knock-Down Reduced LIF-Mediated Overproduction of Gd-IgA1
To confirm that STAT1 mediated LIF-induced Gd-IgA1 overproduction in IgA1-producing cell lines from patients with IgAN, we used a gene-specific siRNA knock-down. STAT1 siRNA knock-down reduced STAT1 expression in cell lines from patients with IgAN as well as OSA by approximately 90% (Figure 5a). Corresponding reduction of STAT1 protein (>80%) was confirmed by immunodetection (Figure 5b and c). Furthermore, pSTAT1 was also reduced (data not shown). Analysis of secreted IgA1 revealed that STAT1 siRNA knock-down significantly reduced the LIF-induced overproduction of Gd-IgA1 in IgAN-derived cell lines (Figure 5d), without impacting total IgA1 production (Supplementary Figure S3).
Figure 5.
STAT1 siRNA knock-down reduced LIF-induced Gd-IgA1 overproduction. (a–c) Quantitative polymerase chain reaction analysis of STAT1 gene expression (a) and SDS-PAGE/Western blotting of STAT1 protein (b and c) showed that siRNA knock-down of STAT1 decreased the expression of STAT1 in the cells from OSA and patients with IgAN. Actin blot is shown as an additional control for protein load. (d) siRNA knock-down of STAT1 reduced LIF-mediated overproduction of Gd-IgA1 in IgAN-derived cells. siRNA knock-down of STAT1 did not affect production of Gd-IgA1 after being stimulated with LIF in OSA-derived cells. One hundred Units of Gd-IgA1 was defined as 100 ng of the standard Gd-IgA1. Statistical analysis for (d) was performed by 1-way analysis of variance followed by Tukey’s multiple comparison test. All data are presented as mean ± SD values and mean values for individual cell lines are shown by black circles. ∗P < 0.05; ∗∗P < 0.01. IgAN, IgA nephropathy; k/d, knock-down; ns, not statistically significant; OSA, obstructive sleep apnea.
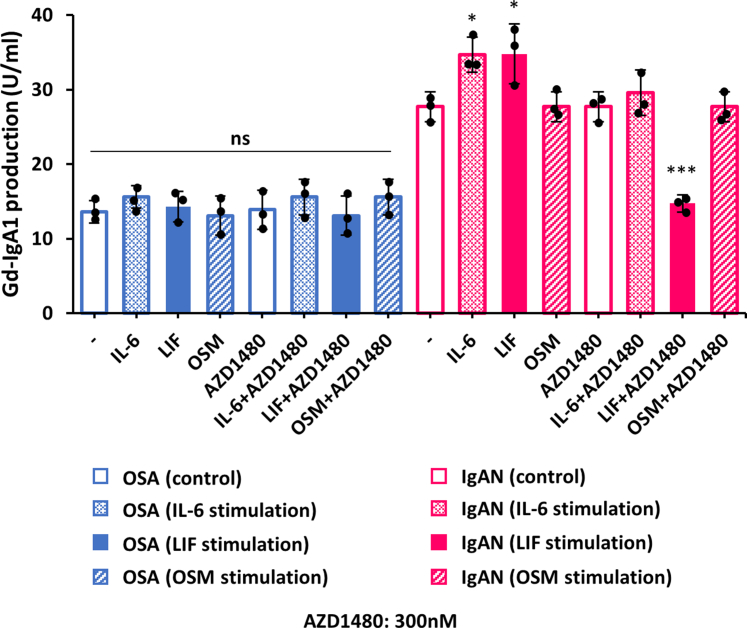
JAK2 Inhibitor AZD1480 Inhibited Gd-IgA1 Overproduction Induced by IL-6-Family Cytokines
The effects of JAK2 inhibitors on IL-6, LIF, or OSM-mediated effects were assessed for IgA1 cell lines from patients with IgAN and those with OSA (Figure 6). LIF and IL-6, but not OSM, increased Gd-IgA1 production in the cell lines from patients with IgAN. AZD1480 inhibitor blocked this increase. Notably, LIF-induced Gd-IgA1 production was reduced by the JAK2 inhibitor to the levels of control cells. Production of Gd-IgA1 by the cell lines from patients with OSA was not affected by IL-6, LIF, OSM, or by JAK2 inhibitor, AZD1480.
Figure 6.
Effect of JAK2 inhibitor AZD1480 on Gd-IgA1 production in the presence or absence of IL-6, LIF, or OSM. Cell lines derived from OSA (blue) and IgAN (red) tonsils were stimulated with IL-6, LIF, OSM alone or in the presence of JAK2 inhibitor AZD1480 (300nM). None of these treatments altered Gd-IgA1 production in OSA group. In IgAN group, LIF and IL-6 increased Gd-IgA1 productions. Furthermore, AZD1480 with LIF stimulation reduced Gd-IgA1 production. All data are presented as mean ± SD values and mean values for individual cell lines are shown by black circles. ∗P < 0.05, ∗∗∗P < 0.005. ns, not statistically significant. IgAN, IgA nephropathy; ns, not statistically significant; LIF, leukemia inhibitory factor; OSA, obstructive sleep apnea; OSM, oncostatin M.
Discussion
Macroscopic hematuria and other signs of disease exacerbation often occur along with or after an upper respiratory tract infection. In Japan, tonsillectomy combined with steroid-pulse therapy has shown favorable clinical outcomes, and this treatment regime is performed frequently in some patients with IgAN.13,14,29,30 Based on these pieces of evidence, Kidney Disease: Improving Global Outcomes (KDIGO) guidelines have been revised, and in Japan, it is indicated that tonsillectomy should be considered, depending on the individual case.31
Recently, GWAS research on IgAN involving Asian and Western patients has progressed and revealed multiple candidate genes involved in mucosal immunity.15,32,33 Genes related to mucosa-related lymphoid tissues involved in IgA production, such as ITGAM and TNFSF13, have risk alleles. A subset of patients with IgAN had elevated serum levels of APRIL, a cytokine related to BAFF.34 Factors such as Toll-like receptor 9 (TLR9) may be involved in the pathogenesis of IgAN via APRIL and BAFF pathways.12,24 Other risk-associated loci include HORMAD2 locus that encodes multiple genes including LIF and OSM. Moreover, a recent study showed that the involvement of the MTMR3/HORMAD2/LIF/OSM locus in IgAN pathogenesis is likely mediated by TLR9 pathways.35 Interestingly, LIF was shown as a drug target gene with a high score by the GWAS additional functional criteria evaluation.17 In this study, we examined the signaling in the LIF/JAK/STAT system related to elevated Gd-IgA1 production.
We previously generated EBV-immortalized IgA1-producing cell lines derived from peripheral blood of healthy individuals and patients with IgAN and used those cells as resource to examine the mechanisms of aberrant IgA1 glycosylation and cytokine-mediated intracellular signaling leading to enhanced galactose deficiency of IgA1. The production of Gd-IgA1 in IgAN is related to altered expression and/or activity of 2 glycosyltransferases. Specifically, reduced expression of core 1 β1,3-galactosyltransferase, which adds galactose to GalNAc and elevated expression of α-N-acetylgalactosaminide α-2,6-sialyltransferase 2, which adds sialic acid to GalNAc are related to Gd-IgA1 production.36 In the cells from patients with IgAN, STAT3 and STAT1 signaling abnormalities were associated with Gd-IgA1 overproduction due to IL-6 family cytokines.20,27 Abnormal STAT phosphorylation of immune cells obtained from patients with IgAN also affects renal tissue lesions, and in particular, changes in pSTAT1 are associated with disease severity (proteinuria, renal function).21
Tonsils are located mainly at the gateway of the respiratory tract and are reportedly one of the secondary lymphatic organs of the immune system. The palatine tonsil is composed of B cell-dominant lymphocytes and a small number of myeloid cells. In our study, pSTAT1 was highly expressed in tonsillar cells from patients with IgAN indicating its possible roles in Gd-IgA1 production.
LIF is an IL-6 related cytokine that uses gp130 for signal transduction and has been previously implicated in mucosal immunity.37 LIF stimulation induced pSTAT1 (Y701) production in IgA1-producing cells from tonsils of patients with IgAN to a greater degree compared to those from OSA. LIF signaling did not involve STAT3 activation (Figure 2). JAK2 inhibitor (AZD1480) inhibited LIF-induced pSTAT1 in the cells from patients with IgAN, and inhibited Gd-IgA1 overproduction induced by LIF (Figure 3). These results suggest that the LIF-LIFR/gp130-JAK2-STAT1 pathway in the tonsillar cells of patients with IgAN plays a key role in Gd-IgA1 production compared to disease controls.
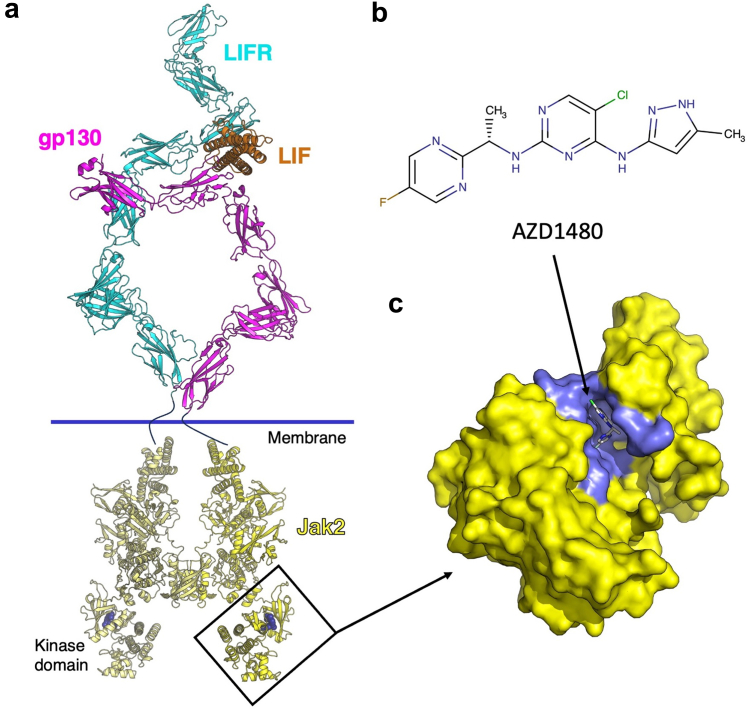
Furthermore, these results offered 2 possible hypotheses that can explain why the inhibitor together with LIF, but not the inhibitor alone, reduced Gd-IgA1 production in IgAN-derived cells to the level of Gd-IgA1 production by OSA-derived cells. In Figure 7, we provide a model of the LIF signaling complex. LIF drives association of gp130 and LIFR. In addition to LIFR, gp130 forms extracellular receptor complexes with other cytokine receptor chains, including IL-6R,48, 49, 50, 51, 52 IL-11R,53 OSMR,54 IL-27R,55 IL-12Rβ2.56 Thus, gp130 enables signal transduction for multiple cytokine receptors. One possible mechanism to explain our findings is based on the fact that LIF drives formation of the receptor complex, which causes sequestration of intracellular gp130 from other receptor complexes. The dimeric receptor then “scavenges” the intracellular homodimeric JAK2 through interaction with the cytoplasmic tails of LIFR and gp130. Decreased Gd-IgA1 production could be due to the combined effect of reducing available gp130 and inhibiting pSTAT1 production by AZD1480 at the level of JAK2. Notably, IL-6-mediated Gd-IgA1 overproduction was also blocked by JAK2 inhibitor. However, this signaling pathway was mediated by STAT3, and JAK2 inhibitor did not reduce Gd-IgA1 production to that by OSA-derived cells (Supplementary Figure S4a). Therefore, the JAK2 inhibitor effects seem to be distinct for LIF-LIFR-STAT1 pathway and we have confirmed that by using another JAK2 inhibitor, AG490 (Supplementary Figure S2).
Figure 7.
LIFR, gp130, and LIF ternary complex and inhibition of the JAK/STAT signaling pathways. Upon binding to the LIFR/gp130 heterodimeric receptor complex, cytokine LIF can activate the JAK/STAT pathway, leading to the altered downstream gene expression. (a) The extracellular ternary complex of gp130/LIFR/LIF complex and a composite model of the intracellular protein JAK2 are shown in ribbon model with each protein shaded magenta (gp130), cyan (LIFR), orange (LIF), or yellow (JAK2). The inhibitor AZD1480 is shown in spheres representation (dark blue). (b) shows the Kekulé diagram of AZD1480, an inhibitor of JAK2 kinase activity used in these studies. (c) The kinase domain of JAK2 is shown in yellow surface representation with AZD-1480 bound. Amino acid residues in proximity to the inhibitor are shaded in slate color. The following protein coordinates were used to generate models in this figure: LIFR [PDB ID:2Q7N],38 and LIF/gp130 [PDB ID:1PVH].39 The initial core composite gp130/LIFR/LIF structure was generated according to methods in recent papers.19,38 Additional domains for LIFR (domains 6–8) were generated by AlphaFold,40 and domains 4–6 of gp130 were taken from PDB ID: 3L5I.41 All domains were rigidly fit to the cryo-reconstruction in (see, Zhou Y, et al.).42 To generate the homology model of JAK2, structures of JAK2 domains FERM-SH2 [PDB ID: 6E2Q],43 pseudokinase [PDB ID: 4FVQ],44 and kinase with AZD1480 [PDB ID: 2XA4]45 were aligned on the JAK1 dimeric complex [PDB ID: 7T6F].46 A composite of the JAK2 domains was submitted to SWISS-MODEL47 to fill in missing gaps between structures with the dimeric JAK1 structure as a guide. Two individual JAK2 structures were aligned to the 2 copies of JAK1 to yield the dimeric JAK2. LIF, leukemia inhibitory factor; LIFR, leukemia inhibitory factor receptor.
JAK/STAT is a major pathway that responds to and transduces inflammatory signals from extracellular ligands such as cytokines and chemokines.57 GWAS revealed a strong association of the genomic locus that encodes LIF with the risk of IgAN.15,58 The abnormal LIF/LIFR-gp130/JAK2/STAT1 signaling in tonsillar cells in patients with IgAN may play a key role for disease progression.
The Helix aspersa lectin used in this study is a lectin specific for GalNAc. Therefore, an alternative way to explain the effect of JAK2 inhibitor with LIF stimulation might be based on a reduced content of GalNAc in the secreted IgA1. Future studies should include assessment of GalNAc-transferases that add GalNAc to Ser/Thr residues of IgA1. Furthermore, LIF increased Gd-IgA1 production, but did not increase pSTAT1/STAT1 ratio compared to the unstimulated cells. Given the AZD1480 and STAT1 siRNA knock-down results, this would imply the necessary but not sufficient role for STAT1 in Gd-IgA1 production. There might be as yet unknown transcription factors/other regulatory components involved. In IgA1-producing cells from peripheral blood from patients with IgAN, LIF-induced Gd-IgA1 production by increased pSTAT1 with Src family kinase activation, not via JAK2.20 To determine whether these differences are universal for all patients with IgAN, we plan in our future studies to compare peripheral-blood-derived and tonsil-derived IgA1-secreting cells obtained from the same individuals. In the future, it is necessary to identify and analyze the causative cells in Gd-IgA1 production based on the differences in signal transduction involved in the pathogenesis of IgAN, which may help to identify possible therapeutic regimes.
Disclosure
HS, JN, and YS are coinventors on US patent application 14/318,082 (assigned to UAB Research Foundation). JN is a co-founder and co-owner of and consultants for Reliant Glycosciences, LLC. All the other authors declared no competing interests.
Acknowledgments
The authors thank all patients involved in this study and the clinicians at Juntendo University Faculty of Medicine, the staff at the University of Alabama at Birmingham for assistance and advice on EBV immortalization and generation of IgA1-producing cell lines. This study was supported in part by Grant-in-Aid for Scientific Research (C) KAKENHI 22K08362, grants DK078244, DK082753, DK134489, and AI149431 from the National Institutes of Health, and a gift from the IGA Nephropathy Foundation.
Author Contributions
KY, Z-QH, JN, and YS designed the study. KY, CR, and HS performed the experiments and/or data analyses. TJG performed structural modelling of LIF-LIF receptor complex. KY wrote the original draft and JN edited the initial manuscript, with all authors providing feedback. All authors read and approved the final version of the manuscript.
Footnotes
Figure S1. Temporal trends of STAT1 activation on LIF or OSM stimulation.
Figure S2. Effect of JAK2 inhibitor AG490 on LIF-mediated Gd-IgA1 overproduction in tonsillar cell lines.
Figure S3. IgA1 production was not altered by STAT1 siRNA knock-down.
Figure S4. Effect of AZD1480 on IL-6-mediated STAT3 activation and overproduction of Gd-IgA1 by IgA1-secreting tonsillar cell lines.
Table S1. The primer sets for qPCR of STAT1 and GAPDH genes.
Supplementary Material
Figure S1. Temporal trends of STAT1 activation on LIF or OSM stimulation.
Figure S2. Effect of JAK2 inhibitor AG490 on LIF-mediated Gd-IgA1 overproduction in tonsillar cell lines.
Figure S3. IgA1 production was not altered by STAT1 siRNA knock down.
Figure S4. Effect of AZD1480 on IL-6-mediated STAT3 activation and overproduction of Gd-IgA1 by IgA1-secreting tonsillar cell lines.
Table S1. The primer sets for qPCR of STAT1 and GAPDH genes.
References
- 1.Wyatt R.J., Julian B.A. IgA nephropathy. N Engl J Med. 2013;368:2402–2414. doi: 10.1056/NEJMra1206793. [DOI] [PubMed] [Google Scholar]
- 2.Kiryluk K., Novak J. The genetics and immunobiology of IgA nephropathy. J Clin Invest. 2014;124:2325–2332. doi: 10.1172/JCI74475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Y., Chen X., Nishi S., Narita I., Gejyo F. Relationship between tonsils and IgA nephropathy as well as indications of tonsillectomy. Kidney Int. 2004;65:1135–1144. doi: 10.1111/j.1523-1755.2004.00486.x. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki H., Kiryluk K., Novak J., et al. The pathophysiology of IgA nephropathy. J Am Soc Nephrol. 2011;22:1795–1803. doi: 10.1681/ASN.2011050464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki H., Fan R., Zhang Z., et al. Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest. 2009;119:1668–1677. doi: 10.1172/JCI38468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao N., Hou P., Lv J., et al. The level of galactose-deficient IgA1 in the sera of patients with IgA nephropathy is associated with disease progression. Kidney Int. 2012;82:790–796. doi: 10.1038/ki.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berthoux F., Suzuki H., Thibaudin L., et al. Autoantibodies targeting galactose-deficient IgA1 associate with progression of IgA nephropathy. J Am Soc Nephrol. 2012;23:1579–1587. doi: 10.1681/ASN.2012010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maixnerova D., Ling C., Hall S., et al. Galactose-deficient IgA1 and the corresponding IgG autoantibodies predict IgA nephropathy progression. PLoS One. 2019;14 doi: 10.1371/journal.pone.0212254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horie A., Hiki Y., Odani H., et al. IgA1 molecules produced by tonsillar lymphocytes are under-O-glycosylated in IgA nephropathy. Am J Kidney Dis. 2003;42:486–496. doi: 10.1016/s0272-6386(03)00743-1. [DOI] [PubMed] [Google Scholar]
- 10.Inoue T., Sugiyama H., Hiki Y., et al. Differential expression of glycogenes in tonsillar B lymphocytes in association with proteinuria and renal dysfunction in IgA nephropathy. Clin Immunol. 2010;136:447–455. doi: 10.1016/j.clim.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Nakata J., Suzuki Y., Suzuki H., et al. Changes in nephritogenic serum galactose-deficient IgA1 in IgA nephropathy following tonsillectomy and steroid therapy. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muto M., Manfroi B., Suzuki H., et al. Toll-like receptor 9 stimulation induces aberrant expression of a proliferation-inducing ligand by tonsillar germinal center B cells in IgA nephropathy. J Am Soc Nephrol. 2017;28:1227–1238. doi: 10.1681/ASN.2016050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano K., Matsuzaki K., Yasuda T., et al. Association between tonsillectomy and outcomes in patients with immunoglobulin A nephropathy. JAMA Netw Open. 2019;2 doi: 10.1001/jamanetworkopen.2019.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komatsu H., Fujimoto S., Hara S., Sato Y., Yamada K., Kitamura K. Effect of tonsillectomy plus steroid pulse therapy on clinical remission of IgA nephropathy: a controlled study. Clin J Am Soc Nephrol. 2008;3:1301–1307. doi: 10.2215/CJN.00310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gharavi A.G., Kiryluk K., Choi M., et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet. 2011;43:321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L., Khan A., Sanchez-Rodriguez E., et al. Genetic regulation of serum IgA levels and susceptibility to common immune, infectious, kidney, and cardio-metabolic traits. Nat Commun. 2022;13:6859. doi: 10.1038/s41467-022-34456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiryluk K., Sanchez-Rodriguez E., Zhou X.J., et al. Genome-wide association analyses define pathogenic signaling pathways and prioritize drug targets for IgA nephropathy. Nat Genet. 2023;55:1091–1105. doi: 10.1038/s41588-023-01422-x. [DOI] [PubMed] [Google Scholar]
- 18.Feenstra B., Bager P., Liu X., et al. Genome-wide association study identifies variants in HORMAD2 associated with tonsillectomy. J Med Genet. 2017;54:358–364. doi: 10.1136/jmedgenet-2016-104304. [DOI] [PubMed] [Google Scholar]
- 19.Person T., King R.G., Rizk D.V., Novak J., Green T.J., Reily C. Cytokines and production of aberrantly O-glycosylated IgA1, the main autoantigen in IgA nephropathy. J Interferon Cytokine Res. 2022;42:301–315. doi: 10.1089/jir.2022.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada K., Huang Z.Q., Raska M., et al. Leukemia inhibitory factor signaling enhances production of galactose-deficient IgA1 in IgA nephropathy. Kidney Dis (Basel) 2020;6:168–180. doi: 10.1159/000505748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao J., Mariani L., Eddy S., et al. JAK-STAT Activity in peripheral blood cells and kidney tissue in IgA nephropathy. Clin J Am Soc Nephrol. 2020;15:973–982. doi: 10.2215/CJN.11010919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bene M.C., Faure G., Hurault de Ligny B., Kessler M., Duheille J. Immunoglobulin A nephropathy. Quantitative immunohistomorphometry of the tonsillar plasma cells evidences an inversion of the immunoglobulin A versus immunoglobulin G secreting cell balance. J Clin Invest. 1983;71:1342–1347. doi: 10.1172/jci110886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy J., Brandtzaeg P. Tonsillar distribution of IgA and IgG immunocytes and production of IgA subclasses and J chain in tonsillitis vary with the presence or absence of IgA nephropathy. Scand J Immunol. 1988;27:393–399. doi: 10.1111/j.1365-3083.1988.tb02362.x. [DOI] [PubMed] [Google Scholar]
- 24.Goto T., Bandoh N., Yoshizaki T., et al. Increase in B-cell-activation factor (BAFF) and IFN-γ productions by tonsillar mononuclear cells stimulated with deoxycytidyl-deoxyguanosine oligodeoxynucleotides (CpG-ODN) in patients with IgA nephropathy. Clin Immunol. 2008;126:260–269. doi: 10.1016/j.clim.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Takahara M., Nagato T., Nozaki Y., et al. A proliferation-inducing ligand (APRIL) induced hyper-production of IgA from tonsillar mononuclear cells in patients with IgA nephropathy. Cell Immunol. 2019;341 doi: 10.1016/j.cellimm.2019.103925. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki H., Moldoveanu Z., Hall S., et al. IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J Clin Invest. 2008;118:629–639. doi: 10.1172/JCI33189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada K., Huang Z.Q., Raska M., et al. Inhibition of STAT3 signaling reduces IgA1 autoantigen production in IgA nephropathy. Kidney Int Rep. 2017;2:1194–1207. doi: 10.1016/j.ekir.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Hotta O., Miyazaki M., Furuta T., et al. Tonsillectomy and steroid pulse therapy significantly impact on clinical remission in patients with IgA nephropathy. Am J Kidney Dis. 2001;38:736–743. doi: 10.1053/ajkd.2001.27690. [DOI] [PubMed] [Google Scholar]
- 30.Kawamura T., Yoshimura M., Miyazaki Y., et al. A multicenter randomized controlled trial of tonsillectomy combined with steroid pulse therapy in patients with immunoglobulin A nephropathy. Nephrol Dial Transplant. 2014;29:1546–1553. doi: 10.1093/ndt/gfu020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kidney Disease: Improving Global Outcomes (KDIGO) Glomerular Diseases Work Group KDIGO 2021 Clinical practice guideline for the management of glomerular diseases. Kidney Int. 2021;100:S1–S276. doi: 10.1016/j.kint.2021.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Feehally J., Farrall M., Boland A., et al. HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol. 2010;21:1791–1797. doi: 10.1681/ASN.2010010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X.Q., Li M., Zhang H., et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet. 2011;44:178–182. doi: 10.1038/ng.1047. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy D.D., Kujawa J., Wilson C., et al. Mice overexpressing BAFF develop a commensal flora-dependent, IgA-associated nephropathy. J Clin Invest. 2011;121:3991–4002. doi: 10.1172/JCI45563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y.N., Gan T., Qu S., et al. MTMR3 risk alleles enhance Toll like receptor 9-induced IgA immunity in IgA nephropathy. Kidney Int. 2023;104:562–576. doi: 10.1016/j.kint.2023.06.018. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki H., Raska M., Yamada K., et al. Cytokines alter IgA1 O-glycosylation by dysregulating C1GalT1 and ST6GalNAc-II enzymes. J Biol Chem. 2014;289:5330–5339. doi: 10.1074/jbc.M113.512277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cella M., Fuchs A., Vermi W., et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huyton T., Zhang J.G., Luo C.S., et al. An unusual cytokine: Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor. Proc Natl Acad Sci U S A. 2007;104:12737–12742. doi: 10.1073/pnas.0705577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boulanger M.J., Bankovich A.J., Kortemme T., Baker D., Garcia K.C. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol Cell. 2003;12:577–589. doi: 10.1016/s1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- 40.Jumper J., Evans R., Pritzel A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y., Kershaw N.J., Luo C.S., et al. Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes. J Biol Chem. 2010;285:21214–21218. doi: 10.1074/jbc.C110.129502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y., Stevis P.E., Cao J., et al. Structural insights into the assembly of gp130 family cytokine signaling complexes. Sci Adv. 2023;9 doi: 10.1126/sciadv.ade4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrao R.D., Wallweber H.J., Lupardus P.J. Receptor-mediated dimerization of JAK2 FERM domains is required for JAK2 activation. eLife. 2018;7 doi: 10.7554/eLife.38089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bandaranayake R.M., Ungureanu D., Shan Y., Shaw D.E., Silvennoinen O., Hubbard S.R. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat Struct Mol Biol. 2012;19:754–759. doi: 10.1038/nsmb.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ioannidis S., Lamb M.L., Wang T., et al. Discovery of 5-chloro-N2−[(1S)-1-(5-fluoropyrimidin-2-yl)ethyl]-N4-(5-methyl-1H-pyrazol-3-yl)pyrimidine-2,4-diamine (AZD1480) as a novel inhibitor of the Jak/Stat pathway. J Med Chem. 2011;54:262–276. doi: 10.1021/jm1011319. [DOI] [PubMed] [Google Scholar]
- 46.Glassman C.R., Tsutsumi N., Saxton R.A., Lupardus P.J., Jude K.M., Garcia K.C. Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation. Science. 2022;376:163–169. doi: 10.1126/science.abn8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waterhouse A., Bertoni M., Bienert S., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirano T., Yasukawa K., Harada H., et al. Complementary DNA for a novel human interleukin (BSF-2) that induces B lymphocytes to produce immunoglobulin. Nature. 1986;324:73–76. doi: 10.1038/324073a0. [DOI] [PubMed] [Google Scholar]
- 49.Kaleeba J.A., Bergquam E.P., Wong S.W. A rhesus macaque Rhadinovirus related to Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 encodes a functional homologue of interleukin-6. J Virol. 1999;73:6177–6181. doi: 10.1128/JVI.73.7.6177-6181.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neipel F., Albrecht J.C., Ensser A., et al. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/JVI.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholas J., Ruvolo V.R., Burns W.H., et al. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 52.Boulanger M.J., Chow D.C., Brevnova E.E., Garcia K.C. Hexameric structure and assembly of the interleukin-6/IL-6 α-receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 53.Paul S.R., Bennett F., Calvetti J.A., et al. Molecular cloning of a cDNA encoding interleukin 11, a stromal cell-derived lymphopoietic and hematopoietic cytokine. Proc Natl Acad Sci U S A. 1990;87:7512–7516. doi: 10.1073/pnas.87.19.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malik N., Kallestad J.C., Gunderson N.L., et al. Molecular cloning, sequence analysis, and functional expression of a novel growth regulator, oncostatin M. Mol Cell Biol. 1989;9:2847–2853. doi: 10.1128/mcb.9.7.2847-2853.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pflanz S., Hibbert L., Mattson J., et al. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J Immunol. 2004;172:2225–2231. doi: 10.4049/jimmunol.172.4.2225. [DOI] [PubMed] [Google Scholar]
- 56.Collison L.W., Delgoffe G.M., Guy C.S., et al. The composition and signaling of the IL-35 receptor are unconventional. Nat Immunol. 2012;13:290–299. doi: 10.1038/ni.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Shea J.J., Plenge R., Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36:542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiryluk K., Li Y., Scolari F., et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet. 2014;46:1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.