Abstract
Introduction
C3 glomerulopathy (C3G) is an ultrarare renal disease characterized by deposition of complement component C3 in the glomerular basement membrane (GBM). Rare and novel genetic variation in complement genes and autoantibodies to complement proteins are commonly identified in the C3G population and thought to drive the underlying complement dysregulation that results in renal damage. However, disease heterogeneity and rarity make accurately defining characteristics of the C3G population difficult.
Methods
Here, we present a retrospective analysis of the Molecular Otolaryngology and Renal Research Laboratories C3G cohort. This study integrated complement biomarker testing and in vitro tests of autoantibody function to achieve the following 3 primary goals: (i) define disease profiles of C3G based on disease drivers, complement biomarkers, and age; (ii) determine the relationship between in vitro autoantibody tests and in vivo complement dysregulation; and (iii) evaluate the association between autoantibody function and disease progression.
Results
The largest disease profiles of C3G included patients with autoantibodies to complement proteins (48%) and patients for whom no genetic and/or acquired drivers of disease could be identified (43%). The correlation between the stabilization of convertases by complement autoantibodies as measured by in vitro modified hemolytic assays and systemic biomarkers that reflect in vivo complement dysregulation was remarkably strong. In patients positive for autoantibodies, the degree of stabilization capacity predicted worse renal function.
Conclusion
This study implicates complement autoantibodies as robust drivers of systemic complement dysregulation in approximately 50% of C3G but also highlights the need for continued discovery-based research to identify novel drivers of disease.
Keywords: complement dysregulation, c3 glomerulopathy, nephritic factors
In 2013, C3G was defined by consensus as a glomerular disease in which deposition of the third complement component, C3, is present in the GBM with an intensity that is at least 2-fold higher than any other immune complexes.1 Disease-specific treatments are lacking and as a result, end-stage renal disease develops in approximately 50% of patients within 10 years of diagnosis. Prognosticating disease outcome, however, in individual patients is not yet possible.2,3 Dysregulation of the alternative pathway of complement drives the disease and with multiple anticomplement therapies now in clinical trials, it has become imperative to define as accurately as possible the underlying causes of complement dysregulation. This added clarity may inform patient inclusion criteria and/or a more optimal selection of a trial, ultimately enabling precisely targeted anti-complement therapy.
A renal biopsy with immunofluorescence is required to diagnosis C3G.1 Disease subclassification into either C3 glomerulonephritis (C3GN) or dense deposit disease (DDD) is based on electron microscopy. The former is characterized by amorphous, subepithelial deposits, whereas the latter is distinguished by thick, intramembranous deposits.1 Low serum C3 levels are present in approximately 60% of patients and indicate ongoing systemic complement dysregulation.4 To characterize and quantify potential causes of in vivo complement dysregulation, patients should be screened for genetic variation in complement genes, autoantibodies to complement proteins, and serum levels of complement proteins and their cleavage products.5
Options for managing C3G include blood pressure and proteinuria control, general immunosuppression,6,7 kidney transplant,8,9 and complement blockade therapies10,11; however, given the rarity and heterogeneity of C3G, data supporting the efficacy of specific treatment regimens are relatively sparse. The optimal treatment to prevent progression to end-stage renal disease for those with more aggressive disease is yet to be determined.
The complement system is a network of proteins that bridges innate and adaptive immunity by identifying pathogenic material, activating proinflammatory pathways, and inducing opsonization and lysis of identified pathogens. Its activity is driven primarily by the alternative pathway, which is continuously activated by the spontaneous hydrolysis of C3 in a process known as tick-over.12,13 Once activated, the complement response is amplified by the C3 convertase, a molecular serine protease complex comprised of C3b and Bb that cleaves C3, leading to the generation of C5 convertase and terminal pathway activation.14 Proper regulation of the complement response to mitigate self-damage is provided by both soluble and cell-surface regulators.15
Patients with C3G often carry factors—immunologic and/or genetic—that are believed to be permissive to dysregulation of the alternative pathway. Most common among the immunologic factors are autoantibodies to C3 and C5 convertases, known as C3 and C5 nephritic factors (C3Nefs and C5Nefs), respectively, which are identified in 40% to 60% of patients.16, 17, 18, 19, 20 Given the labile nature of convertases, the identification of C3Nefs and C5Nefs is not trivial, and consequently their in vivo impact has been debated. Several assays18,21, 22, 23, 24 are available to quantify convertase activity and characterize the mechanism of action of Nefs; however, the interpretation of these tests and their relationship to in vivo complement biomarkers has remained unclear. This limitation reflects the small sample size of most studies and/or the inclusion of patients with a variety of complement-mediated diseases in the same cohort.25, 26, 27, 28, 29 As a result, data describing Nefs as primary drivers of systemic complement dysregulation in C3G remain conflicting.30, 31, 32, 33, 34
In addition to C3Nefs and C5Nefs, other autoantibodies reported at much lower rates in C3G include those to factor B (FB) (FBAAs; 3% of patients),35, 36, 37 factor H (FH) (FHAAs; 3% of patients),38, 39, 40, 41 and C4 (C4Nefs; 5%–15% of patients).37,42, 43, 44 The C3G population is also enriched for persons with rare genetic variations (∼20%) in complement proteins, which can contribute to abnormal complement activity.45,46 In aggregate, these assorted immunologic and genetic factors create a complicated picture.
Here, we present a large-scale, retrospective analysis of our C3G cohort that supports a primary role for C3Nefs, C4Nefs, and C5Nefs as drivers of in vivo systemic complement dysregulation. Our data highlight the diversity of Nef function across patients with C3G and demonstrate that in the patient subset positive for Nefs, Nef stabilization capacity is predictive of renal function. These data also emphasize the importance of following Nef activity over time and support the need for targeted discovery-based studies for novel drivers of local rather than systemic complement dysregulation. To that end, we propose a framework for evaluating the different patient cohorts within the C3G population.
Methods
C3G Cohort
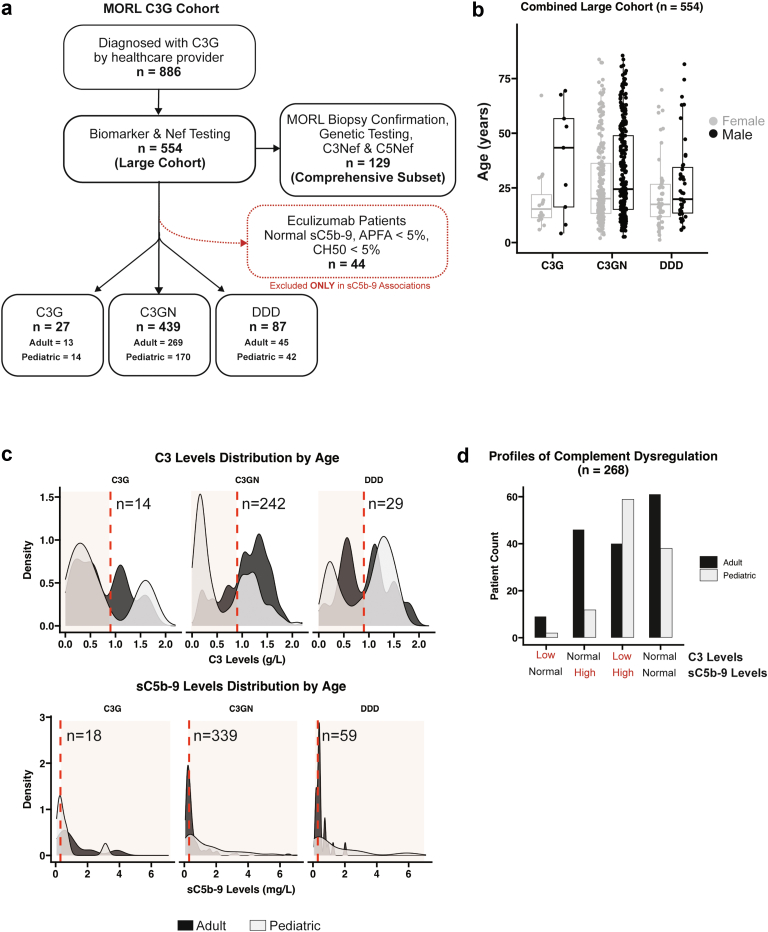
Patients for this study were selected based on a C3G diagnosis as provided from the referring health care provider. Of 886 patients in our C3G cohort, 554 were included in this study based on concurrent testing (with 60 days) for Nefs and complement biomarkers. A smaller subset (n = 129) included patients with comprehensive genetic testing, Nef and biomarker analysis, and manual review of biopsy images by scientists at the Molecular Otolaryngology and Renal Research Laboratories, with consensus classification of each patient as either C3GN or DDD. Pediatric patients were defined as patients younger than 18 years old at the time of testing. Forty-four patients on eculizumab were excluded from any analyses that include sC5b-9 as a variable (Figure 1a). The diagnosis, sex (male or female, as reported in medical records), and age at clinical testing is shown in Figure 1b. The Institutional Review Board of the University of Iowa approved this study, and the described research was performed in accordance with the Declaration of Helsinki.
Figure 1.
Data from 886 patients in the Molecular Otolaryngology and Renal Research Laboratories C3 glomerulopathy database. (a) Of these, 554 patients with C3 glomerulopathy had complement biomarker, functional, and autoantibody testing performed within a 60-day window of diagnosis (defining the biopsy diagnosis as time zero). In 129 patients (comprehensive subset), biopsy images were manually reviewed to confirm a diagnosis of dense deposit disease or C3 glomerulonephritis and genetic screening was performed. (b) In the large cohort, in 27 individuals only a diagnosis of C3 glomerulopathy was made. In 1 patient, age was unknown. (c) C3 and sC5b-9 testing was available in 285 and 416 patients, respectively (shaded red, abnormal levels). (d) A total of 268 patients had both C3 and sC5b-9 testing; 25% of patients had only 1 convertase dysregulated (low C3 or high sC5b-9), 37% had both convertases dysregulated (low C3 and high sC5b-9), and 37% had normal biomarkers (normal C3 and sC5b-9). C3G, C3 glomerulopathy; DDD, dense deposit disease.
Only the comprehensively tested and biopsy-reviewed subset of patients (n = 129) was used to draw conclusions regarding what percentage of the C3G population carries a certain disease profile. There were no significant differences between age, sex, Nef occurrence, or abnormal biomarker occurrence between the large Molecular Otolaryngology and Renal Research Laboratories C3G cohort and the smaller comprehensive patient subset (Supplementary Table S1).
Sample Preparation
Serum and plasma were prepared by centrifugation of whole blood samples, aliquoted, and stored at −80 ⁰C. Patient-specific IgG was isolated using the Melon Gel IgG Purification Kit (Thermo Scientific, Rockford, IL).
DNA Extraction and Genetic Testing
Genomic DNA was extracted from peripheral blood and evaluated on 1% agarose gel and by NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) to assess integrity and purity, respectively. Samples tested before January 2014 were sequenced using Sanger Sequencing47,48; samples tested after January 2014 were sequenced on a targeted, high throughput, next generation sequencing panel.46 C3G-associated complement genes included CFH, CFI, CD46 (MCP), CFB, CFHR5, and C3.
C3Nef and C5Nef Detection
Convertase stabilization by Nefs was detected using hemolytic-based assays (C3 convertase stabilizing assay, and C3 convertase stabilizing assay with properdin to detect C3Nefs and C5Nefs, respectively) as previously described.49 In brief, to detect C3Nefs, C3 convertase is built on the surface of C3b-decorated sheep erythrocytes using purified FB and factor D. The amount of FB is titrated to yield approximately 1 convertase per erythrocyte at 30 ⁰C after 5 minutes, at which time patient-purified IgG is added. Twenty minutes are allowed to lapse for natural decay of the convertase and then rat serum in ethylenediamine tetraacetic acid is added to supply terminal complement components. The addition of rat serum leads to cell lysis in the presence of Nef-stabilized C3 convertase. C3 convertase stabilizing assay with properdin includes properdin to facilitate C5 convertase formation on the erythrocyte, with a longer incubation period to accommodate the longer C5 convertase half-life. Nef stabilization capacity is measured as a percentage of hemolysis relative to a negative control. Hemolysis is binned as <20%, 20% to 40%, 41% to 60%, 61% to 80% and >80%, corresponding to negative, 1+, 2+, 3+, and 4+ hemolysis, respectively.
C4Nef Detection
C4Nefs were detected as described using a hemolytic assay to quantitate antibody-mediated stabilization of the classical pathway C3 convertase.43 In brief, patient-purified IgG and complement component C2 are incubated with C4b-decorated sheep erythrocytes to allow for convertase formation and subsequent decay, and rat serum in the presence of ethylenediamine tetraacetic acid is added to trigger activity of the terminal pathway of complement and cell lysis in the presence of C4Nef-stabilized convertase.43,50 As described above, Nef stabilization capacity is measured as the percentage of hemolysis relative to a negative control, binning percent hemolysis in a similar manner.
FB and FH autoantibody detection
Enzyme-linked immunosorbent assays were used for FB and FH autoantibody detection, as described.38,49,51 In short, microtiter plates were coated with FB or FH. Horseradish peroxidase-tagged, antihuman IgG is used to identify binding of patient-purified IgG to the protein of interest after incubation.
Tests of Fluid-Phase C3 Convertase Activity
A modified immunofixation electrophoresis (IFE) was performed to assess fluid-phase C3 convertase activity by mixing patient serum and pooled normal human serum. Following an incubation period at 37 ⁰C and immunoprecipitation by an anti-C3 antibodies, C3 convertase activity is measured by quantifying the generation of C3 activation products by agarose gel electrophoresis.49
Biomarker Quantification
Complement component C3 was detected in patient plasma using radial immunodiffusion (The Binding Site, Birmingham, United Kingdom), and soluble C5b-9 by an enzyme-linked immunosorbent assay-based method (sC5b-9, Quidel Corporation, San Diego, CA).
Proteinuria Measurement
The urine protein-to-creatinine ratios (UPCRs) measured within 1 year of Nef testing were used to quantify proteinuria. Thirty-six patients had both UPCR and Nef testing available; 23 also had C3 biomarker data.
Statistics and Data Visualization
Statistics and data visualization were completed using R,52 RStudio,53 and visualization packages.54,55 Statistical significance was evaluated using a t-test, 1-way analysis of variance or χ2 test, as indicated.56,57
Results
Defining the Disease Profiles in C3G
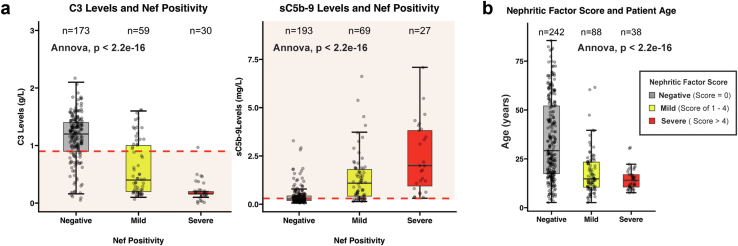
Biomarkers used to quantify C3 convertase activity (i.e., C3) and C5 convertase activity (i.e., sC5b-9) were measured in patient plasma and represent an in vivo snapshot of systemic complement activity. In the large cohort, C3 levels distributed in a bimodal pattern, with a large portion of patients having normal C3 levels and a large portion having almost entirely depleted levels. Soluble C5b-9 biomarkers were also normal in a large portion of patients. When biomarker distribution was stratified by age, 69 of 121 (57%) pediatric patients and 51 of 163 (31%) adult patients had low C3 levels (P < 0.0001). High sC5b-9 was identified in 109 of 175 (62%) pediatric patients and 129 of 240 (54%) adult patients (P < 0.0883) (Figure 1c). In 268 patients with both C3 and sC5b-9 levels, 69 patients (26%) showed dysregulation of only 1 convertase (marked by either low C3 or high sC5b-9, but not both), 99 patients (37%) had dysregulation of both convertases, and 100 patients (37%) had biomarkers indicating no active systemic complement dysregulation (Figure 1d). Patients with a biomarker profile of both convertases dysregulated were more frequently pediatric patients (Figure 1d).
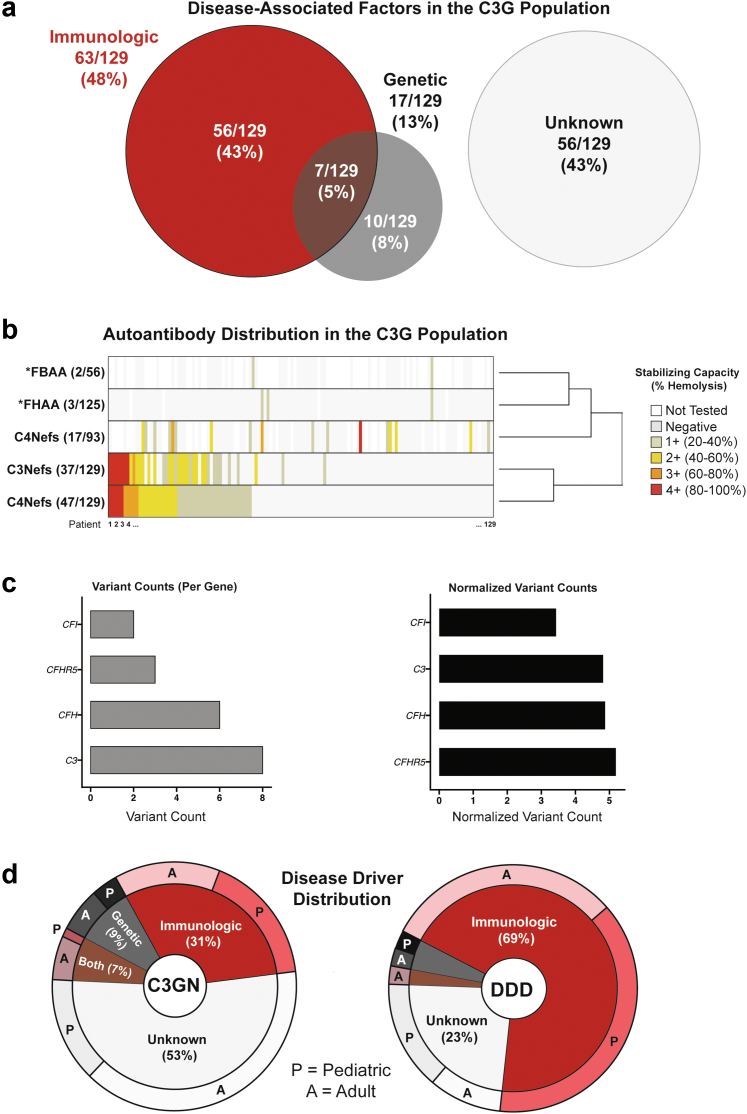
The comprehensive clinical subset (n = 129) was evaluated to determine disease profiles in C3G and how frequently each profile occurs. The general disease profiles were as follows: (i) 56 patients (43%) with immunologic driver(s) only, (ii) 56 patients (43%) with no identified disease-associated factors, (iii) 10 patients (8%) with genetic driver(s) only, and (iv) 7 patients (5%) with both immunologic and genetic driver(s) (Figure 2a). Of the known immunologic drivers, patient positivity for C5Nefs was most common (47/129, 36.4%), followed by positivity for C3Nefs (37/129, 29%), C4Nefs (17/93 patients tested; 18%), FBAAs (2/56 patients tested; 4%), and FHAAs (3/125 patients tested; 2%). Copositivity for C3Nefs and C5Nefs occurred most frequently. Of patients positive for C4Nefs, a plurality occurred in concert with C5Nefs (47%), whereas a smaller portion were copositive with C3Nefs (29%). FBAAs and FHAAs did not occur in the presence of C3Nefs or C5Nefs. Of the 3 patients positive for FHAAs, 1 patient was copositive for C4Nefs, and 1 was copositive for FBAAs. The second patient testing positive for FBAA had no other immunologic factors (Figure 2b).
Figure 2.
Disease drivers in C3 glomerulopathy. (a) Of 129 patients, 63 and 17 carried an immunologic and/or genetic driver, respectively. (b) C5Nefs were the most common driver, followed closely by C3Nefs. Both were frequently found together with similar stabilization capacities. An asterisk (∗) indicates that autoantibodies were detected using enzyme-linked immunosorbent assay-based assays as described in methods. In these cases, all positive findings were considered 1+. (c) Likely pathogenic genetic drivers were found in 17 patients. A variant was considered to be a likely pathogenic driver if it had a minor allele frequency <0.01 and a combined annotation dependent depletion score >15 or if it was predicted to disrupt canonical splicing. (d) Distribution of these drivers was uneven; patients with dense deposit disease were more likely to carry an immunologic driver of disease, whereas patients with C3 glomerulonephritis were most likely to have no known driver of disease. C3G, C3 glomerulopathy; DDD, dense deposit disease; Nefs, nephrotic factors.
Rare genetic variants in disease-associated genes were identified in 13% (17 of 129) of patients screened. Variants were included if they occurred at <1% in any population in the gnomAD database and either carried a combined annotation dependent depletion score58 of >15 or were predicted by Human Splice Finder59 to disrupt canonical splicing (Table 1). After normalizing genes by base pair length, C3, CFH, and CFHR5 all had a similar genetic burden followed closely by CFI (Figure 2c). When subgrouped into C3GN and DDD cohorts, immunologic drivers were identified in approximately 70% of patients with DDD as compared to 30% of patients with C3GN; however, patients with C3GN were almost 3 times more likely to carry genetic variants as compared to patients with DDD. Over 50% of patients with C3GN and 24% of patients with DDD had no identifiable driver of disease; the majority of these patients were adults (Figure 2d).
Table 1.
Genetic variants in complement 3 glomerulopathy cohort
| Patient | Diagnosis | Gene | Genomic coordinate | Protein change | MAF (gnomAD) | Maximum MAF (gnomAD) | CADD score | Splice prediction | Adapted ACMG interpretation | Plasma C3 levels (g/L) | Plasma sC5b-9 levels (mg/L) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | DDD | C3 | chr19:6718304:G>C | p.Tyr129Stop | 0 | 0 | 35 | None | Pathogenic | <0.16 | 3.84 |
| B | C3GN | C3 | chr19:6718166:C>T | p.Arg148Gln | 7.072E-06 | 0.00001549 | 27.3 | None | Likely Pathogenic | 0.30 | 0.32 |
| C | C3GN | C3 | chr19:6714178:G>A | p.Tyr227Tyr | 0.0001026 | 0.002029 | 12.15 | ESE altered | VUS | 1.00 | 0.20 |
| D | C3GN | C3 | chr19:6709748:C>T | p.Val598Met | 3.187E-05 | 0.0001149 | 27.2 | None | VUS | NT | 0.17 |
| E | C3GN | C3 | chr19:6707877:C>G | p.Gly637Arg | 0.000223 | 0.0005082 | 21.9 | None | VUS | 0.30 | 3.60 |
| F | C3GN | C3 | chr19:6707251:C>A | p.Cys694Phe | 0 | 0 | 25.8 | None | VUS | 0.60 | 0.74 |
| G | C3GN | C3 | chr19:6679429:C>T | p.Arg1512His | 0.0001025 | 0.000178 | 23.4 | None | VUS | 0.70 | 0.24 |
| H | DDD | CFH | chr1:196642262:G>A | p.Trp71Stop | 4.003E-06 | 8.897E-06 | 32 | None | Pathogenic | 1.20 | 0.29 |
| I | C3GN | CFH | chr1:196646746:A>G | p.Met190Val | 0 | 0 | 15.69 | None | VUS | NT | 1.24 |
| J | C3GN | CFH | chr1:196648872:G>- | p.Gly247GluFSa34 | 0 | 0 | 36 | None | Likely Pathogenic | NT | 0.33 |
| K | C3GN | CFH | chr1:196709833:C>T | p.Thr956Met | 0.001294 | 0.003364 | 14.99 | None | Benign | 0.70 | 0.26 |
| L | DDD | CFH | chr1:196709833:C>T | p.Thr956Met | 0.001294 | 0.003364 | 14.99 | None | Benign | NT | 1.51 |
| M | C3GN | CFHR5 | NA | Complex Rearrangement | 0 | 0 | N/A | aSplicing altered | Likely pathogenic | 1.3 | 0.33 |
| N | C3GN | CFHR5 | NA | Complex Rearrangement | 0 | 0 | N/A | aSplicing altered | Likely pathogenic | 1.2 | 0.23 |
| O | C3GN | CFHR5 | chr1:196973872:G>A | p.Gly471Glu | 0.0006083 | 0.001386 | 15.42 | None | VUS | 1.00 | 0.42 |
| P | C3GN | CFI | chr4:110681720:A>C | p.Ile244Ser | 0.0000358 | 0.0003806 | 23 | None | VUS | NT | 0.21 |
| Q | C3GN | CFI | chr4:110670411:C>T | p.Gly371Ser | 0 | 0 | 31 | None | VUS | NT | 0.23 |
ACMG, American College of Medical Genetics; CADD, combined annotation dependent depletion; C3GN, complement 3 glomerulonephritis; DDD, dense deposit disease; ESE, exonic splice enhancer; MAF, minor allele frequency; NT, not tested; VUS, variants of uncertain significance.
Splicing effect determined in-house.
Characterizing C3Nefs , C4Nefs , and C5Nefs
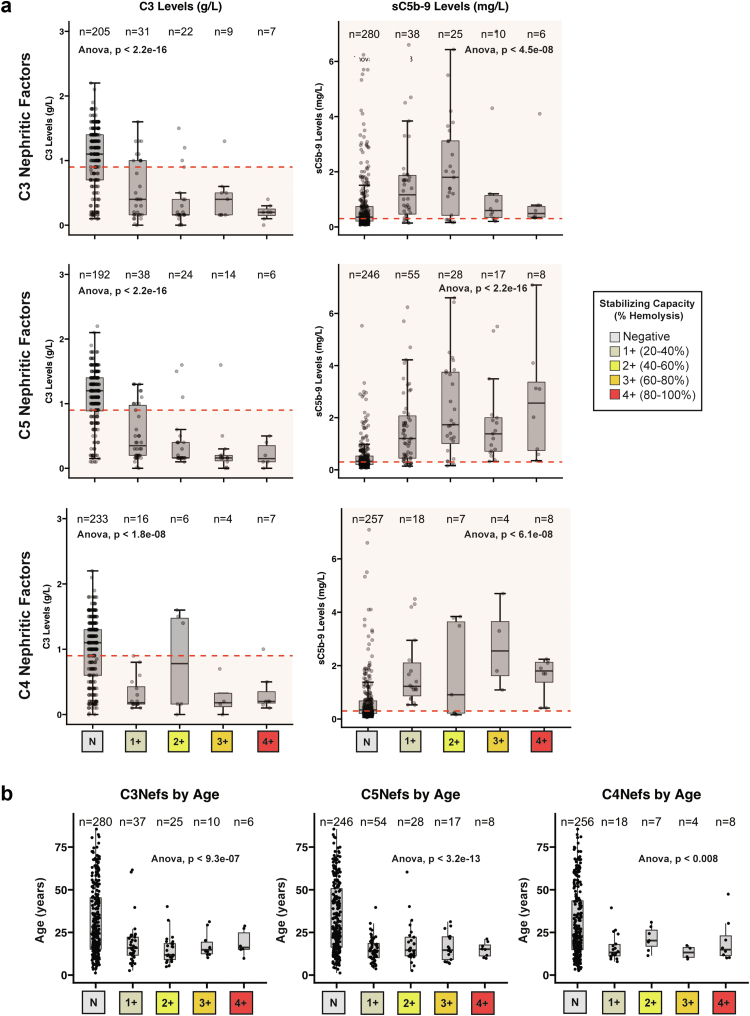
Because autoantibodies to convertases (including C3Nefs, C4Nefs, and C5Nefs) are quantified in the absence of complement regulatory proteins, Nef stabilization capacity as measured in vitro was compared to complement biomarkers measured in vivo to determine whether there was any correlation. Two hundred seventy-four patients had both serum C3 measured and C3Nef and C5Nef activity quantitated. Patients positive for C3Nefs had lower C3 levels as compared to patients negative for C3Nefs (P < 2.2 × 10−16). Of patients tested for C3Nefs and C5Nefs, 359 and 354 patients, respectively had sC5b-9 biomarkers tested (and were not on eculizumab). Soluble C5b-9 was increased in patients with mild (1–2+) C3Nef stabilization capacity but not significantly elevated in patients with 3–4+ C3Nef stabilization capacity (P < 4.5 × 10−08) (Figure 3a, Supplementary Figure S1A). However, patients positive for C5Nefs had significantly lower C3 (P < 2.2 × 10−16) and significantly higher sC5b-9 (P < 2.2 × 10−16) compared to patients negative for C5Nefs (Figure 3a, Supplementary Figure S1B), as did patients positive for C4Nefs (266 and 307 patients with C4Nef testing had C3 levels and sC5b-9 levels available, respectively) (Figure 3a, Supplementary Figure S1C). When patients were stratified as either Nef-positive, or Nef-negative, these differences remained (Supplementary Figure S2). When patients were broken down by Nef type and Nef stabilization category, Nef stabilization capacity also correlated with younger age (Figure 3b, Supplementary Figure S1).
Figure 3.
In vivo complement dysregulation. (a) In patients tested for C3Nefs, C5Nefs, and C4Nefs, serum C3 levels were lower (indicating C3 convertase overactivity), and sC5b-9 was higher (indicating C5 convertase overactivity) in most patients positive for the nephritic factor as compared to the patients who did not carry a nephritic factor. (b) Patients positive for nephritic factors of any kind tended to be younger than patients testing negative. Nefs, nephrotic factors.
Fluid-Phase C3 Convertase Activity in C3G
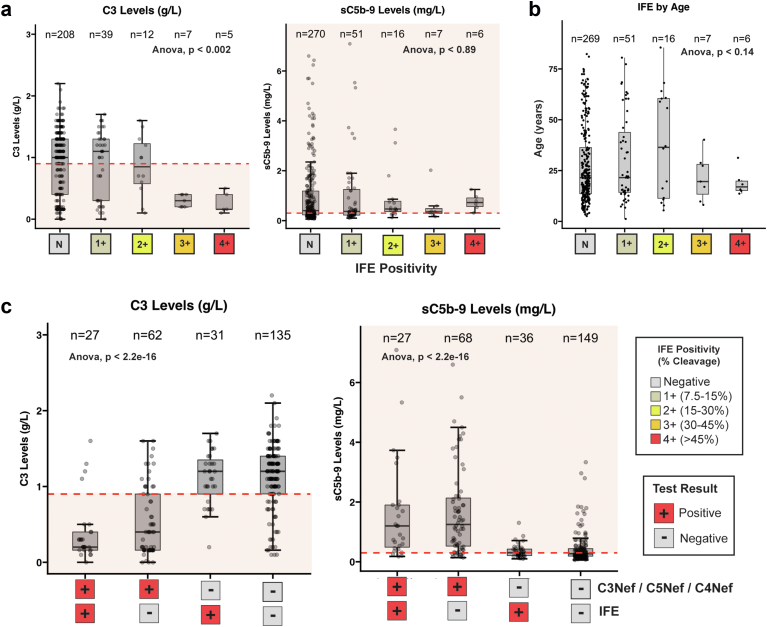
Fluid phase C3 convertase activity as measured by IFE was evaluated in 271 and 350 patients in whom C3 and sC5b-9 levels were available, respectively (Figure 4a). IFE positivity only correlated with in vivo C3 levels when IFE was robust (3+ and 4+) (P < 0.002) and did not correlate with sC5b-9 levels at all (P < 0.89) (Figure 4a, Supplementary Figure S3A). IFE positivity did not correlate with age (Supplementary Figure S3B). Patients positive for C3Nefs, C4Nefs, and/or C5Nefs were significantly enriched for decreased C3 and increased sC5b-9 levels regardless of IFE status (P < 2.2 × 10−16) (Figure 4c).
Figure 4.
The IFE test. (a) IFE positivity does not correlate well with C3 levels unless IFE is strongly positive (3+ and 4+). (b) Severe IFE scores are found in the younger age group. (c) Nephritic factors are much stronger predictors of complement dysregulation than is IFE regardless of the IFE score. IFE, immunofixation electrophoresis
Nefs and Proteinuria
Considering that copositivity for C3Nefs, C4Nefs, and C5Nefs was common, an aggregate score of C3Nef, C4Nef, and C5Nef positivity was calculated on a scale of 0 to 12. Patients with mild positivity (sum of Nef stabilization capacity ≤4) had significantly higher C3 levels and lower sC5b-9 levels as compared to patients with severe positivity (≥5) (P < 2.2 × 10−16) (Figure 5a, Supplementary Figure S4A). As expected, Nef score also correlated with age (Figure 5b, Supplementary Figure S4B). In 36 patients, UPCR and Nef activity were evaluated. When patients were divided into Nef-positive and Nef-negative groups, no significant differences in UPCR distribution were observed (Supplementary Figure S4C); however, when patients were stratified by Nef score, patients with a higher Nef score had worse UPCR as compared to patients with mild or negative Nef scores (Supplementary Figure S4D). In 23 patients where UPCR, C3 levels and Nef testing were available, UPCR was compared to C3 levels. No major trends were identified when all patients were included regardless of Nef status (R2 < 0.01). However, when only patients positive for Nefs were included, a slight negative correlation between UPCR and C3 levels was detected (R2 = 0.10) (Supplementary Figure S4E).
Figure 5.
Summed Nef scores. (a) Because Nefs often occur in combination, a sum score of C3Nef positivity (1–4+), C5Nef positivity (1–4+), and C4Nef positivity (1–4+) was calculated (mild Nef score, 1–4; severe Nef score >4). The relationship between Nef score and (b) age was evaluated. Nef, nephrotic factor.
Biomarker Profiles by Immunologic Status
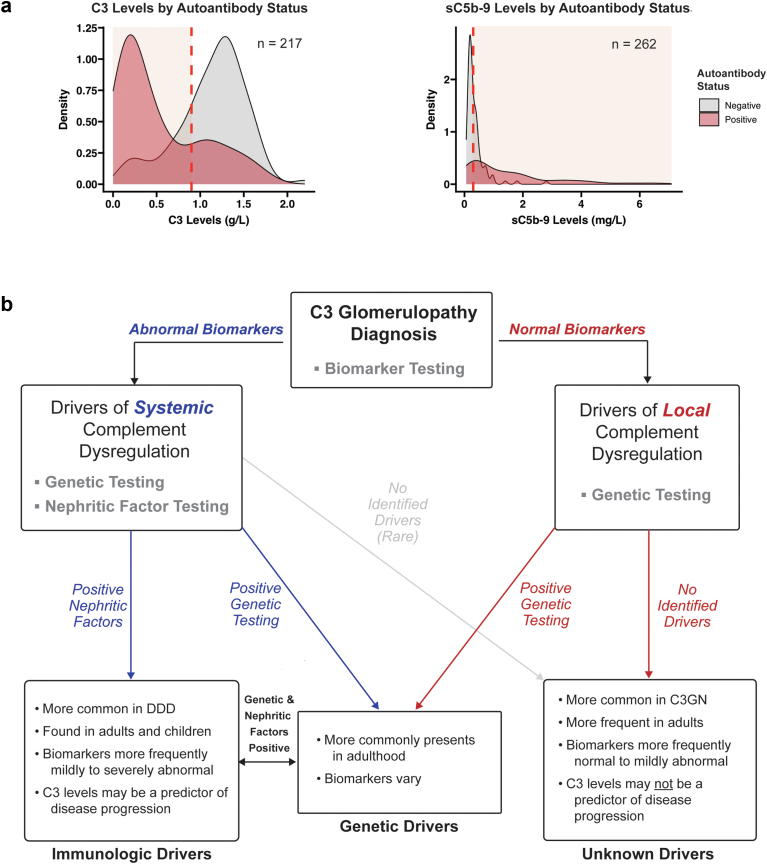
When biomarker profiles were stratified by either the presence or absence of any autoantibodies (C3Nefs, C5Nefs, C4Nefs, FBAAs, and/or FHAAs), 79 of 107 (74%) of patients with normal C3 levels and 60 of 80 (75%) of patients with normal sC5b-9 levels were negative for these drivers of systemic complement dysregulation (Figure 6a).
Figure 6.
Stratifying patients with C3 glomerulopathy. (a) Patients testing positive for 1 or more autoantibodies most often have a complement biomarker profile consistent with systemic complement dysregulation. Conversely, patients negative for autoantibodies most often have normal complement biomarkers. (b) Predictors of disease can be identified by subgrouping patients based on local or systemic drivers of disease. C3G, C3 glomerulopathy; DDD, dense deposit disease.
Discussion
Numerous studies have explored the role of Nefs as drivers of complement dysregulation in C3G, but our understanding of this association remains conflicting and incomplete.17,29,32,33,49,60 In this large retrospective analysis of patients with C3G, we provide a refined view of C3Nefs, C4Nefs, and C5Nefs and their strong association with systemic in vivo complement dysregulation in C3G. Our data show the following: (i) the in vitro stabilization capacity of C3Nefs, C4Nefs, and C5Nefs correlates with in vivo systemic complement dysregulation; (ii) Nefs are diverse and cause varying degrees of in vivo complement dysregulation; (iii) a high Nef score correlates with proteinuria; and (iv) patients who lack biomarker evidence of systemic complement dysregulation are most frequently negative for Nefs.
Defining the role of Nefs as potential drivers of systemic complement dysregulation in C3G has been challenging. Early research focused on their characterization primarily in studies of hypocomplementemic patients, thus limiting ability to define the extent to which Nefs may or may not associate specifically with complement dysregulation.17,34,61 More recent studies have demonstrated an association between C3Nefs and C5Nefs and decreased C3 and increased sC5b-9 levels, respectively.20,23,28 However, Nefs have also been reported in persons with renal disease and normal complement biomarkers, and in healthy persons.29,32,33,60,62,63 This spectrum of presentations has fueled a debate as to whether Nefs play a pathogenic role in driving complement dysregulation or are an epiphenomenon triggered by some other process.64 Clarifying their role is important because Nefs are the most commonly identified abnormalities in patients with C3G (Figure 2a and b).
Difficulty in determining the relationship between complement dysregulation and Nefs can be attributed in part to the difficulty in identifying these autoantibodies themselves. They target the labile and transient C3 and C5 convertases and as such technical expertise is required to isolate and functionally characterize these autoantibodies. We used a modified hemolytic assay49 and built C3 or C5 convertases on the surface of sheep erythrocytes using the relevant complement proteins. Patient-purified IgG was then added to determine whether convertase-stabilizing IgG was present. Using this assay, we identified a strong correlation between in vitro stabilization capacity of C3Nefs, C4Nefs, and C5Nefs and in vivo complement dysregulation as measured by C3 and sC5b-9 serum levels (Figure 3a). The correlation between increased complement dysregulation and increased Nef stabilization capacity was strongest with in vivo C3 levels in agreement with data reported by Marinozzi et al.20 in 2017.
A second challenge in characterizing Nefs is their potential diversity. Many studies suggest that Nefs show structural diversity and have various mechanisms of action, including varied ability to stabilize convertases in the presence of complement regulatory proteins and/or properdin.20,22,65, 66, 67 In this study, Nef diversity is reflected in the association between stabilization capacity and serum C3 dysregulation (Figure 5a) and in the association between disease severity (estimated by UPCR) and summed Nef scores (Supplementary Figure S4D). In aggregate, our data show that Nefs almost universally lead to C3 consumption; however, the degree of C3 consumption varies across individuals. Whether this variation reflects differences in specific antigenic epitopes, Nef concentrations, or genetic variation predisposing to complement overactivation is not known. When Nef positivity is interpreted with IFE results, patients positive for C3Nefs, C4Nefs, and/or C5Nefs and IFE trend toward lower C3 serum levels as compared to patients positive only for Nefs (Figure 4c). These intricate and complex correlations show that Nefs drive complement dysregulation and are not nonfunctional antibodies generated randomly as an epiphenomenon of complement dysregulation.
To frame these findings in the context of the C3G population as a whole, we evaluated the frequency with which immunologic drivers (C3Nefs, C5Nefs, C4Nefs, FHAAs, and FBAAs) were detected in patients with abnormal systemic complement biomarkers as compared to patients with normal systemic complement biomarkers and found that approximately 40% of patients with C3G were positive for autoantibodies; of this subset, approximately 90% had abnormal complement biomarkers. In patients with a normal biomarker profile, approximately 75% were negative for Nefs (Figure 6a). This association supports the classification of Nefs as drivers of systemic complement dysregulation but equally importantly, focuses our attention on 2 other patient subgroups: those with genetic drivers of disease (13%) and those with no identified drivers of disease (43%). The last subgroup is particularly intriguing. The majority of these patients have C3GN and may have complement dysregulation localized to the glomerular microenvironment, thus explaining the absence of abnormal systemic complement biomarkers, a hypothesis that warrants investigation (Figure 2d, Figure 5a, and Figure 6a).
Distinguishing between systemic and local drivers of disease is necessary to select biomarkers that accurately predict disease progression. When we divided our cohort into Nef-positive and Nef-negative groups, there was no statistically significant difference in the UPCR (Supplementary Figure S4C). However, when stratified by Nef scores, patients with severe Nef scores had higher UPCRs than patients with mild or negative Nef scores (Supplementary Figure S4D). This association shows that though the presence or absence of Nefs may not predict disease progression, if present, their stabilization capacity may do that. Similarly, when we evaluated the relationship between UPCR and C3 levels, an inverse relationship was detected only when patients positive for Nefs were evaluated alone. When all patients—regardless of their Nef status—were evaluated, this relationship was masked. Therefore, though C3 levels may be a good predictor of disease progression in patients with systemic drivers of complement dysregulation, they fail to predict outcome in patients lacking a systemic driver of disease (Supplementary Figure S4E, Figure 6). These associations are based on small sample sizes and emphasize the need for studies focusing on the natural history of disease course, which we are currently completing.
The possibility that C3G can be driven by local complement dysregulation mediated by properties of the glomerular microenvironment is an important hypothesis to consider in patients lacking systemic complement dysregulation. The glomerular microenvironment includes fenestrated endothelial cells, the GBM, and epithelial podocytes. Blanketing the endothelial cells and covering the GBM is the glycocalyx, a mesh of proteoglycans, glycoproteins, and glycolipids that extends into the lumen of glomerular capillaries. This highly specialized extracellular matrix is an important protector of the GBM and has many components such as heparan sulfate and sialic acid that are important for FH-mediated regulation of C3. Disrupting this environment could result in a permissive extracellular matrix that renders the GBM susceptible to local overactivation of C3. A key factor in this patient subset would be that though their systemic complement biomarker profile may look normal, disease is still driven by chronic (though localized) complement dysregulation. Stratification of patients with C3G based on local versus systemic drivers of complement dysregulation may be necessary to define robust predictors of disease progression and select or develop effective therapeutic strategies. Patients with local complement dysregulation may benefit from complement therapeutics or other therapies that target complement dysregulation at the GBM. For example, heparan sulfate mimetics are being developed for use in diseases where heparan sulfate-binding proteins are driving the underlying disease process.68 Possibly, these agents may be beneficial in some patients with C3G by sequestering FH-related proteins that perpetuate C3 activation.69
The major limitation of this study is the dearth of longitudinal data to define the natural history of C3G. Nefs are known to change over time, and these changes remain poorly understood. For example, it remains possible that some patients with no current identifiable driver of disease may have been Nef-positive at some point in time. Collection of consistent, longitudinal data will resolve this weakness and aid in defining with greater resolution the disease course across different subsets of patients with C3G over time. A similar study in patients with immune complex membranoproliferative glomerulonephritis would also enhance our understanding of Nefs in complement-mediated renal disease, because this patient population has many overlapping features with Nef-driven C3G.
Conclusions
These data show that Nefs are robust drivers of in vivo complement dysregulation. Differences in stabilization capacity of Nefs can be quantitated and impact renal outcome. As a clinical correlate, patients with C3G should be stratified by disease driver type in order to identify biomarkers relevant to disease progression.
In addition to the characterization of C3Nefs, C4Nefs, and C5Nefs as drivers of systemic complement dysregulation, this study highlights the need for novel disease driver discovery. A large subset of patients lacks any identifiable driver of disease. The correlation between the absence of a disease driver and normal complement biomarkers in these patients points to disease mechanisms that drive local complement dysregulation in the glomerular microenvironment in the absence of systemic complement dysregulation.
Disclosure
CMN is the site principal investigator for NCT03369236 (A proof-of-concept study for 6 month treatment in patients with C3 glomerulopathy), Achillion Pharmaceuticals, Inc; is the data and safety monitoring board chair, NCT02949128 (single arm study of ALXN1210 in complement inhibitor treatment-naïve adult and adolescent patients with atypical hemolytic uremic syndrome) and NCT03131219 (Study of ALXN1210 in children and adolescents with atypical hemolytic uremic syndrome), Alexion Pharmaceuticals, Inc. RJHS is the director of the Molecular Otolaryngology and Renal Research Laboratories, which offers genetic and functional complement testing for patients with complement-mediated renal diseases and is an advisor to Novartis. All the other authors report no conflicting interests.
Acknowledgments
This work was supported in part by NIDDK R01 110023 to CMN and RJHS and a National Science Foundation Graduate Research Fellowship 000390183 to JJH. We also thank the health care providers and patients who have allowed us to participate in their care.
Footnotes
Figure S1. Post hoc test of significance for nephritic factors.
Figure S2. Positive versus negative nephritic factor biomarkers.
Figure S3. Post hoc test of significance for immunofixation electrophoresis.
Figure S4. Post hoc test of significance for nephritic factor scores and proteinuria across nephritic factor groups.
Table S1. Comparison of large and comprehensive cohorts.
Supplementary Material
Figure S1. Post hoc test of significance for nephritic factors.
Figure S2. Positive versus negative nephritic factor biomarkers.
Figure S3. Post hoc test of significance for immunofixation electrophoresis.
Figure S4. Post hoc test of significance for nephritic factor scores and proteinuria across nephritic factor groups.
Table S1. Comparison of large and comprehensive cohorts.
References
- 1.Pickering M.C., D’Agati V.D., Nester C.M., et al. C3 glomerulopathy: consensus report. Kidney Int. 2013;84:1079–1089. doi: 10.1038/ki.2013.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao X., Pickering M.C., Smith R.J.H. C3 glomerulopathy: the genetic and clinical findings in dense deposit disease and c3 glomerulonephritis. Semin Thromb Hemost. 2014;40:465–471. doi: 10.1055/s-0034-1376334. [DOI] [PubMed] [Google Scholar]
- 3.Lu D.-F., Moon M., Lanning L.D., McCarthy A.M., Smith R.J.H. Clinical features and outcomes of 98 children and adults with dense deposit disease. Pediatr Nephrol. 2012;27:773–781. doi: 10.1007/s00467-011-2059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medjeral-Thomas N.R., O’Shaughnessy M.M., O’Regan J.A., et al. C3 glomerulopathy: clinicopathologic features and predictors of outcome. Clin J Am Soc Nephrol. 2014;9:46–53. doi: 10.2215/CJN.04700513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidney Disease: Improving Global Outcomes (KDIGO) Glomerular Diseases Work Group KDIGO 2021 Clinical Practice Guideline for the Management of Glomerular Diseases. Kidney Int. 2021;100:S1–S276. doi: 10.1016/j.kint.2021.05.021. [DOI] [PubMed] [Google Scholar]
- 6.Sethi S., Fervenza F.C., Zhang Y., et al. C3 glomerulonephritis: clinicopathological findings, complement abnormalities, glomerular proteomic profile, treatment, and follow-up. Kidney Int. 2012;82:465–473. doi: 10.1038/ki.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabasco C., Cavero T., Román E., et al. Effectiveness of mycophenolate mofetil in C3 glomerulonephritis. Kidney Int. 2015;88:1153–1160. doi: 10.1038/ki.2015.227. [DOI] [PubMed] [Google Scholar]
- 8.McCaughan J.A., O’Rourke D.M., Courtney A.E. Recurrent dense deposit disease after renal transplantation: an emerging role for complementary therapies. Am J Transplant. 2012;12:1046–1051. doi: 10.1111/j.1600-6143.2011.03923.x. [DOI] [PubMed] [Google Scholar]
- 9.Zand L., Lorenz E.C., Cosio F.G., et al. Clinical findings, pathology, and outcomes of C3GN after kidney transplantation. J Am Soc Nephrol. 2014;25:1110–1117. doi: 10.1681/ASN.2013070715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruseva M.M., Peng T., Lasaro M.A., et al. Efficacy of targeted complement inhibition in experimental C3 glomerulopathy. J Am Soc Nephrol. 2016;27:405–416. doi: 10.1681/ASN.2014121195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nester C.M., Smith R.J.H. Complement inhibition in C3 glomerulopathy. Semin Immunol. 2016;28:241–249. doi: 10.1016/j.smim.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Bexborn F., Andersson P.O., Chen H., Nilsson B., Ekdahl K.N. The tick-over theory revisited: formation and regulation of the soluble alternative complement C3 convertase (C3(H(2)O)Bb. Mol Immunol. 2008;45:2370–2379. doi: 10.1016/j.molimm.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isenman D.E., Isenman D.E. the role of the thioester bond in C3 and C4 in the determination of the conformational and functional states of the molecule. Ann N Y Acad Sci. 1983;421:277–290. doi: 10.1111/j.1749-6632.1983.tb18115.x. [DOI] [PubMed] [Google Scholar]
- 14.Sarma J.V., Ward P.A. The complement system. Cell Tissue Res. 2011;343:227–235. doi: 10.1007/s00441-010-1034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James K. Complement: activation, consequences, and control. Am J Med Technol. 1982;48:735–742. [PubMed] [Google Scholar]
- 16.Zhang Y., Nester C.M., Martin B., et al. Defining the complement biomarker profile of C3 glomerulopathy. Clin J Am Soc Nephrol. 2014;9:1876–1882. doi: 10.2215/CJN.01820214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spitzer R.E., Vallota E.H., Forristal J., et al. Serum C’3 lytic system in patients with glomerulonephritis. Science. 1969;1979:436–437. doi: 10.1126/science.164.3878.436. [DOI] [PubMed] [Google Scholar]
- 18.Daha M.R., Fearon D.T., Austen K.F. C3 nephritic factor (C3NeF): stabilization of fluid phase and cell-bound alternative pathway convertase. J Immunol. 1976;116:1–7. doi: 10.4049/jimmunol.116.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Schreiber R.D., Medicus R.G., Gïtze O., Müller-Eberhard H.J. Properdin- and nephritic factor-dependent C3 convertases: requirement of native C3 for enzyme formation and the function of bound C3b as properdin receptor. J Exp Med. 1975;142:760–772. doi: 10.1084/jem.142.3.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marinozzi M.C., Chauvet S., Le Quintrec M., et al. C5 nephritic factors drive the biological phenotype of C3 glomerulopathies. Kidney Int. 2017;92:1232–1241. doi: 10.1016/j.kint.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 21.Koch F.J., Jenis E.H., Valeski J.E. Test for C3 nephritic factor activity by immunofixation electrophoresis. Am J Clin Pathol. 1981;76:63–67. doi: 10.1093/ajcp/76.1.63. [DOI] [PubMed] [Google Scholar]
- 22.Paixão-Cavalcante D., López-Trascasa M., Skattum L., et al. Sensitive and specific assays for C3 nephritic factors clarify mechanisms underlying complement dysregulation. Kidney Int. 2012;82:1084–1092. doi: 10.1038/ki.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donadelli R., Pulieri P., Piras R., et al. Unraveling the molecular mechanisms underlying complement dysregulation by nephritic factors in C3G and IC-MPGN. Front Immunol. 2018;9:2329. doi: 10.3389/fimmu.2018.02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michels M.A.H.M., van de Kar N.C.A.J., van den Bos R.M., et al. Novel assays to distinguish between properdin-dependent and properdin-independent C3 nephritic factors provide insight into properdin-inhibiting therapy. Front Immunol. 2019;10:1–16. doi: 10.3389/fimmu.2019.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mollnes T.E., Ng Y.C., Peters D.K., Lea T., Tschopp J., Harboe M. Effect of nephritic factor on C3 and on the terminal pathway of complement in vivo and in vitro. Clin Exp Immunol. 1986;65:73–79. [PMC free article] [PubMed] [Google Scholar]
- 26.Jelezarova E., Schlumberger M., Sadallah S., Späth P.J., Schifferli J.A., Lutz H.U. A C3 convertase assay for nephritic factor functional activity. J Immunol Methods. 2001;251:45–52. doi: 10.1016/s0022-1759(01)00295-2. [DOI] [PubMed] [Google Scholar]
- 27.West C.D., Bissler J.J. Nephritic factor and recurrence in the renal transplant of membranoproliferative glomerulonephritis type II. Pediatr Nephrol. 2008;23:1867–1876. doi: 10.1007/s00467-008-0887-x. [DOI] [PubMed] [Google Scholar]
- 28.Iatropoulos P., Daina E., Curreri M., et al. Cluster analysis identifies distinct pathogenetic patterns in c3 glomerulopathies/immune complex–Mediated membranoproliferative GN. J Am Soc Nephrol. 2018;29:283–294. doi: 10.1681/ASN.2017030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwertz R., Rother U., Anders D., Gretz N., Schärer K., Kirschfink M. Complement analysis in children with idiopathic membranoproliferative glomerulonephritis: a long-term follow-up. Pediatr Allergy Immunol. 2001;12:166–172. doi: 10.1034/j.1399-3038.2001.012003166.x. [DOI] [PubMed] [Google Scholar]
- 30.Ravindran A., Fervenza F.C., Smith R.J.H., De Vriese A.S., Sethi S. C3 glomerulopathy: 10-years experience at the Mayo Clinic HHS public access. Mayo Clin Proc. 2018;93:991–1008. doi: 10.1016/j.mayocp.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiramatsu M., Tsokos G.C. Epstein-Barr virus transformed B cell lines derived from patients with systemic lupus erythematosus produce a nephritic factor of the classical complement pathway. Clin Immunol Immunopathol. 1988;99:91–99. doi: 10.1016/0090-1229(88)90009-8. [DOI] [PubMed] [Google Scholar]
- 32.Spitzer R.E., Stitzel A.E., Tsokos G.C. Evidence that production of autoantibody to the alternative pathway C3 convertase is a normal physiologic event. J Pediatr. 1990;116:3–8. doi: 10.1016/s0022-3476(05)82711-8. [DOI] [PubMed] [Google Scholar]
- 33.Ohi H., Watanabe S., Fujita T., Yasugi T. Significance of C3 nephritic factor (C3NeF) in non-hypocomplementaemic serum with membranoproliferative glomerulonephritis (MPGN) Clin Exp Immunol. 1992;89:479–484. doi: 10.1111/j.1365-2249.1992.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohi H., Yasugi T. Occurrence of C3 nephritic factor and C4 nephritic factor in membranoproliferative glomerulonephritis (MPGN) Clin Exp Immunol. 1994;95:316–321. doi: 10.1111/j.1365-2249.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strobel S., Zimmering M., Papp K., Prechl J., Jozsi M. Anti-factor B autoantibody in dense deposit disease. Mol Immunol. 2010;47:1476–1483. doi: 10.1016/j.molimm.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Marinozzi M.C., Roumenina L.T., Chauvet S., et al. Anti-factor B and anti-C3b autoantibodies in C3 glomerulopathy and Ig-associated membranoproliferative GN. J Am Soc Nephrol. 2017;28:1603–1613. doi: 10.1681/ASN.2016030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauer J.J., Shao D., Zhang Y., Nester C.M., Smith R.J.H. Factor B and C4B2a autoantibodies in C3 glomerulopathy. Front Immunol. 2019;10:2–7. doi: 10.3389/fimmu.2019.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanc C., Togarsimalemath S.K., Chauvet S., et al. Anti-factor H autoantibodies in C3 glomerulopathies and in atypical hemolytic uremic syndrome: one target, two diseases. J Immunol. 2015;194:5129–5138. doi: 10.4049/jimmunol.1402770. [DOI] [PubMed] [Google Scholar]
- 39.Goodship T.H.J., Pappworth I.Y., Toth T., et al. Factor H autoantibodies in membranoproliferative glomerulonephritis. Mol Immunol. 2012;52:200–206. doi: 10.1016/j.molimm.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Victor K.D., Pascual V., Stitzel A.E., Tsokos G.C., Capra J.D., Spitzer R.E. Nucleotide sequence of a human autoantibody to the alternative pathway C3/C5 convertase (C3NeF) Hybridoma. 1993;12:231–237. doi: 10.1089/hyb.1993.12.231. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Ghiringhelli Borsa N., Shao D., et al. Factor H autoantibodies and complement-mediated diseases. Front Immunol. 2020;11:1–10. doi: 10.3389/fimmu.2020.607211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLean R.H., Nilson S.H. C3 nephritic factor stabilization of the classic C3 convertase: a role for C2 in C3 nephritic factor activity. Proc Soc Exp Biol Med. 1979;161:358–363. doi: 10.3181/00379727-161-40553. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., Meyer N.C., Fervenza F.C., et al. C4 nephritic factors in C3 glomerulopathy: a case series. Am J Kidney Dis. 2017;70:834–843. doi: 10.1053/j.ajkd.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garam N., Prohászka Z., Szilágyi Á., et al. C4 nephritic factor in patients with immune-complex-mediated membranoproliferative glomerulonephritis and C3-glomerulopathy. Orphanet J Rare Dis. 2019;14:247. doi: 10.1186/s13023-019-1237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osborne A.J., Breno M., Borsa N.G., et al. Statistical validation of rare complement variants provides insights into the molecular basis of atypical hemolytic uremic syndrome and C3 glomerulopathy. J Immunol. 2018;200:2464–2478. doi: 10.4049/jimmunol.1701695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bu F., Borsa N.G., Jones M.B., et al. High-throughput genetic testing for thrombotic microangiopathies and C3 glomerulopathies. J Am Soc Nephrol. 2016;27:1245–1253. doi: 10.1681/ASN.2015040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maga T.K., Nishimura C.J., Weaver A.E., Frees K.L., Smith R.J.H. Mutations in alternative pathway complement proteins in American patients with atypical hemolytic uremic syndrome. Hum Mutat. 2010;31:1445–1460. doi: 10.1002/humu.21256. [DOI] [PubMed] [Google Scholar]
- 48.Roumenina L.T., Loirat C., Dragon-Durey M.A., Halbwachs-Mecarelli L., Sautes-Fridman C., Fremeaux-Bacchi V. Alternative complement pathway assessment in patients with atypical HUS. J Immunol Methods. 2011;365:8–26. doi: 10.1016/j.jim.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., Meyer N.C., Wang K., et al. Causes of alternative pathway dysregulation in dense deposit disease. Clin J Am Soc Nephrol. 2012;7:265–274. doi: 10.2215/CJN.07900811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blom A.M., Corvillo F., Magda M., et al. Testing the activity of complement convertases in serum/plasma for diagnosis of C4NeF-mediated C3 glomerulonephritis. J Clin Immunol. 2016;36:517–527. doi: 10.1007/s10875-016-0290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chauvet S., Berthaud R., Devriese M., et al. Anti-factor B antibodies and acute postinfectious GN in children. J Am Soc Nephrol. 2020;31:829–840. doi: 10.1681/ASN.2019080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R Core Team R: a language and environment for statistical computing. https://www.scirp.org/reference/ReferencesPapers?ReferenceID=2342186 Published 2018.
- 53.RStudio Team RStudio: integrated development environment for R. https://www.r-project.org/conferences/useR-2011/abstracts/180111-allairejj.pdf Published 2020.
- 54.Wickham H. Springer-Verlag; New York: 2016. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 55.Larsson J., Gustafsson P. A case study in fitting area-proportional euler diagrams with ellipses using eulerr. CEUR Workshop Proceedings. https://portal.research.lu.se/en/publications/a-case-study-in-fitting-area-proportional-euler-diagrams-with-ell
- 56.Kassambara A. Ggpubr: ‘ggplot2’ Based Publication Ready Plots. https://github.com/kassambara/ggpubr Published 2020.
- 57.Larsson J. “eulerr”: area-proportional Euler and Venn diagrams with ellipses. https://cran.r-project.org/web/packages/eulerr/eulerr.pdf Published 2021.
- 58.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:1–14. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schena F.P., Pertosa G., Stanziale P., Vox E., Pecoraro C., Andreucci V.E. Biological significance of the C3 nephritic factor in membranoproliferative glomerulonephritis. Clin Nephrol. 1982;18:240–246. [PubMed] [Google Scholar]
- 61.Skattum L., Mårtensson U., Sjöholm A.G. Hypocomplementaemia caused by C3 nephritic factors (C3 NeF): clinical findings and the coincidence of C3 NeF type II with anti-C1q autoantibodies. J Intern Med. 1997;242:455–464. doi: 10.1111/j.1365-2796.1997.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 62.Spitzer R.E., Stitzel A.E., Tsokos G.C. Autoantibody to the alternative pathway C3/C5 convertase and its anti-idiotypic response. A study in affinity. J Immunol. 1992;148:137–141. doi: 10.4049/jimmunol.148.1.137. [DOI] [PubMed] [Google Scholar]
- 63.Paz Z., Tsokos G.C. Autoantibodies. 3rd ed. Elsevier; 2014. Nephritic factor autoantibodies; pp. 561–565. [DOI] [Google Scholar]
- 64.West C.D. Nephritic factors predispose to chronic glomerulonephritis. Am J Kidney Dis. 1994;24:956–963. doi: 10.1016/s0272-6386(12)81068-7. [DOI] [PubMed] [Google Scholar]
- 65.Clardy C.W., Judith F., Strife C.F., West C.D. A properdin dependent nephritic factor slowly activating C3, C5, and C9 in membranoproliferative glomerulonephritis, types I and III. Clin Immunol Immunopathol. 1989;50:333–347. doi: 10.1016/0090-1229(89)90141-4. [DOI] [PubMed] [Google Scholar]
- 66.Tanuma Y., Ohi H., Hatano M. Two types of C3 nephritic factor: properdin-dependent C3NeF and properdin-independent C3NeF. Clin Immunol Immunopathol. 1990;56:226–238. doi: 10.1016/0090-1229(90)90144-f. [DOI] [PubMed] [Google Scholar]
- 67.Zhao F., Afonso S., Lindner S., et al. C3-glomerulopathy autoantibodies mediate distinct effects on complement C3- and C5-convertases. Front Immunol. 2019;10:1030. doi: 10.3389/fimmu.2019.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morla S. Glycosaminoglycans and glycosaminoglycan mimetics in cancer and inflammation. Int J Mol Sci. 2019;20:1963. doi: 10.3390/ijms20081963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loeven M.A., Maciej-hulme M.L., Yanginlar C., Hubers M.C. Selective binding of heparin / heparan sulfate oligosaccharides to factor H and factor H-related proteins : therapeutic potential for C3 glomerulopathies. 2021;12 doi: 10.3389/fimmu.2021.676662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.