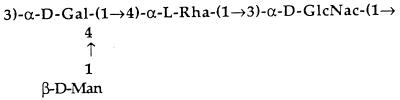Abstract
In order to determine the importance of the O75 O antigen versus the K5 capsular antigen and the bimodal distribution of lipopolysaccharides (LPSs) in protection from complement-mediated lysis, mutants were made by insertion of a cat or an aphA gene in or in place of genes necessary for the synthesis of LPS and/or the K antigen of an O75+ K5+ uropathogenic Escherichia coli strain, GR-12. Mutations were made in the following genes: the rfbD gene (required for the synthesis of TDP-rhamnose), the rfbKM genes (necessary for the synthesis of GDP-mannose), the rol gene (regulating O-antigen length), the kfiC gene (encoding a putative glycosyltransferase), and the kfiC-rfbD genes. The resulting phenotypes were rough (O75−), core plus one partial O-antigen subunit, random distribution of O-antigen chain lengths, acapsular (K5−), and O75− K5−, respectively. All five mutants and GR-12 were analyzed for survival in 80% serum. The GR-12 parent was resistant, exhibiting a 500% increase in numbers. The rol, rfbKM, rfbD, and kfiC-rfbD mutants were sensitive, experiencing 99%, 99.9%, 99.9%, and at least 99.999% killing, respectively, in the first hour. The kfiC mutant, however, increased in numbers in the first hour but experienced delayed sensitivity, decreasing in viability by 80% in the third hour. Single mutants were complemented with the wild-type gene in trans, showing restoration of the wild-type phenotype and serum resistance. Therefore, the O75 antigen is more important for survival in serum than the K5 antigen, and regulation of the O75 O-antigen chain length is crucial for protection of the bacteria from complement-mediated lysis.
Escherichia coli is a major cause of urinary tract infections, septicemia, and neonatal meningitis. E. coli possesses several virulence factors, such as outer membrane proteins, capsules, and lipopolysaccharide (LPS), which allow the bacteria to evade the host defense system and promote infection. LPS and capsules are thought to provide resistance to the bactericidal effects of serum (10, 28–30, 46, 59, 61).
The complex carbohydrate LPS is a major constituent of the outer membrane of gram-negative bacteria. LPS serves as a barrier and shields the bacteria from detergents, free fatty acids, hydrophobic antibiotics, complement, and phagocytosis (40, 56, 63). LPS consists of three parts: lipid A (the toxic portion of the molecule), core oligosaccharide, and O antigen or O polysaccharide. The O antigen is made of sugar residue subunits linked to form a chain. The structure of the O-antigen subunit determines the serogroup of E. coli. The O75 subunit is composed of four sugars: N-acetylglucosamine, rhamnose, galactose, and the side sugar mannose (15).
Specific enzymes construct the oligosaccharide units, polymerize the units together, and ligate the growing O-antigen chain to the core oligosaccharide. Many of the enzymes necessary for the synthesis of the O antigen are encoded by genes in the rfb cluster. The rfb locus maps at 44 min on the E. coli chromosome and encodes enzymes necessary for the synthesis of sugar residues as well as proteins involved in the biosynthesis of the O polysaccharide (for a review, see reference 50). The rfbBDAC genes and the rfbKM genes encode the synthesis of TDP-rhamnose (26) and GDP-mannose (26, 63), respectively. Both of these sugar precursors are necessary to complete the O75 O-antigen subunit. The length of the O-antigen chain is regulated by a protein encoded by the rol (regulator of O-antigen length) gene (3). Normally, a wild-type strain has a bimodal distribution of O-antigen chains, where there exists a higher percentage of long chains (15 to 20 O-antigen lengths) than of short chains (1 to 5 O-antigen lengths). Mutation of the rol gene destroys the ability to regulate O-antigen length, producing a random length of O-antigen chains (3). Other genes, such as rfc and rfaL, are necessary to complete the assembly of the O antigen by polymerizing the O-antigen subunits and ligating the O antigen to the core, respectively (50).
In addition to LPS, many pathogenic bacteria also possess a capsule. The capsular antigens (K antigens) are acidic polysaccharides made of linear polymers of repeating carbohydrate subunits. The K5 antigen consists of repeating units of β-glucuronic acid and N-acetylglucosamine and is identical to an intermediate in the synthesis of heparin (39). The K antigens can be divided into two groups (groups I and II) on the basis of biochemical and physical characteristics (25). The K5 antigen is a group II K antigen. The genes encoding the group II capsules have a common organization. Three gene clusters encode group II capsular antigen synthesis. Regions 1 and 3 are common to all group II capsular antigens, while region 2 is unique for each capsular antigen (6). K5 region 2, consisting of the kfiABCD genes, has been analyzed in the most detail (44).
Of the more than 160 different O antigens, only a small number are associated with disease (42). For example, serotypes O1, O2, O4, O6, O7, O18, and O75 are primarily isolated from urinary tract infections (41). The ability of bacteria to cause disease correlates with the sensitivity of the bacteria to serum. Bacteria able to resist the effects of complement are known as serum resistant; those that are not are considered sensitive to serum. E. coli strains lacking O antigens (rough) are more sensitive to serum (42, 58, 61), while strains which cause severe infections, such as bacteremia and urinary tract infections, have a high percentage of serum resistance (reviewed in reference 60). However, studies by Russo and colleagues using an O4 strain indicated that the loss of the O antigen has only minor effects on serum sensitivity (48).
Conflicting evidence has been presented as to whether capsular polysaccharides actually provide serum resistance. Of the more than 80 types of K antigens, only a few appear to be responsible for serum resistance (2, 9, 16, 34, 47, 48, 64). Evidence suggests that the capsule does not impart resistance by impeding complement components or antibody from binding to the bacterial surface because macromolecules can easily permeate the polysaccharide capsule (54). However, capsular antigens K1 and K54 seem to be protective against the bactericidal effects of serum (10, 34, 47, 64). K5, on the other hand, does not seem to impart serum resistance (8, 55). Thus, only certain O and K antigens may provide resistance to serum.
Many previous studies failed to utilize proven isogenic mutants of wild-type pathogenic strains. Genetically defined mutants are superior for the evaluation of putative virulence factors. We report five new chromosomally defined mutants of an otherwise wild-type O75 pathogenic strain to address the importance of the O75 and K5 antigens in protection from complement-mediated lysis. The genes chosen for mutation were the rfbD, rfbKM, rol, kfiC, and kfiC-rfbD genes, producing distinctly different phenotypes: rough (O75−), core plus one partial O-antigen subunit, random distribution of O-antigen chain lengths, acapsular (K5−), and O75− K5−, respectively. The five mutants were analyzed by LPS analysis, sensitivity to K5-specific phage, outer membrane profiles, and serum assays and compared to the wild-type strain. All mutants were complemented in trans, showing restoration of the wild-type phenotype and serum resistance.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
E. coli GR-12 was originally isolated from a patient with pyelonephritis (57). This strain is O75:K5 and is serum resistant. E. coli XL1-Blue, used for cloning with pBluescript II KS vectors, was obtained from Stratagene (La Jolla, Calif.). E. coli DH1 was used as a host with the pBR322 vectors. For cloning with the suicide vectors pGP704 and pGP704Km, SY327 λ pir (38) was used for initial cloning and SM10 λ pir (53) was used for conjugation. All strains are described in Table 1. Details of the construction of all plasmids used in this study are given in Table 2.
TABLE 1.
Strains used in this study
| Strain | Descriptiona | Reference or source |
|---|---|---|
| GR-12 | Wild-type O75+ K5+ strain | 57 |
| XL1-Blue | F′::Tn10 proA+B+ lacIq Δ(lacZ)M15/recA1 endA1 gyrA96 (Nalr) thi hsdR17 (rK− mK+) supE44 relA1 lac | Stratagene |
| SY327 (λ pir) | F−araD Δ(lac pro) argE(Am) recA56 rpoB gyrA λ pir | 38 |
| SM10 (λ pir) | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr λ pir | 53 |
| DH1 | F−supE44 recA1 endA1 gyrA96 (Nalr) thi-1 hsdR17 (rK− mK+) relA1 spoT1? rfbD1? | 31 |
| SMB20 | GR-12 rfbD::cat Cmr O75− K5+ | This study |
| SMB43 | GR-12 rfbK′-′M::cat Cmr O75P K5+ | This study |
| SMB122 | GR-12 Δrol::cat Cmr O75R K5+ | This study |
| SMB213 | GR-12 kfiC′::cat Cmr O75+ K5− | This study |
| SMB316 | GR-12 kfiC′::aphA rfbD::cat Cmr Kmr O75− K5− | This study |
Superscript P, partial O-antigen subunit; superscript R, random distribution of O-antigen chain lengths.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Purpose | Antibiotic resistance | Reference or source |
|---|---|---|---|---|
| pBluescript II KS | High-copy-number cloning vector | Initial subcloning | Apr | Stratagene |
| pBluescript II KS-Km | HindIII/SmaI-digested wild-type Tn5 treated with the Klenow fragment and ligated to EcoRV-digested pBluescript II KS | Source of aphA gene | Apr Kmr | This study |
| pGP704 | oriR6K mobRP4 | Suicide vector | Apr | 37 |
| pGP704Km | aphA gene inserted into the bla gene of pGP704 | Suicide vector | Kmr | This study |
| pACYC184 | Low-copy-number cloning vector | Complementation | Tcr Cmr | 7 |
| pUC18CML | High-copy-number cloning vector | Source of cat gene | Apr Cmr | 51 |
| pBR322 | Low-copy-number cloning vector | Complementation | Tcr | |
| pRAB2401 | Subclone of O75 rfb gene cluster | Subcloning of rfb genes | Apr | 4 |
| pSMB102 | Intramolecular ligation of pRAB2401 cut with PstI | Cloning of rfbD gene | Apr | This study |
| pSMB103 | cat gene (SmaI/HindII) inserted into SnaBI of pSMB102 | Mutation of rfbD gene | Apr Cmr | This study |
| pSMB104 | EcoRV fragment of pSMB103 ligated to pGP704Km | Subcloning into suicide vector for conjugation | Kmr Cmr | This study |
| pSMB105 | Intramolecular ligation of pRAB2401 cut with ClaI | Cloning of rfbKM genes | Apr | This study |
| pSMB106 | 6.5-kb ClaI/KpnI fragment of pSMB105 inserted into pBluescript II KS | Subcloning of rfbKM genes | Apr | This study |
| pSMB107 | Deletion of 1.1-kb NruI fragment from rfbKM and insertion of cat gene (SmaI/HindII) | Mutation of rfbKM genes | Apr Cmr | This study |
| pSMB108 | EcoRV/KpnI fragment of pSMB107 ligated to pGP704 | Subcloning into suicide vector for conjugation | Apr Cmr | This study |
| pSMB109 | MunI fragment of pSMB106 ligated to pBR322 cut with EcoRI/ScaI | Cloning of rfbKM genes for complementation | Tcr | This study |
| pSMB110 | ugd PCR product (EcoRV/BamHI) ligated to pBluescript II KS | Cloning of sequence upstream of rol gene | Apr | This study |
| pSMB111 | his PCR product (BamHI/XbaI) ligated to pSMB110 | Cloning of upstream and downstream sequences of rol gene | Apr | This study |
| pSMB112 | Insertion of cat gene (BamHI ends) into BamHI site of pSMB111 | cat gene inserted between ugd and his | Apr Cmr | This study |
| pSMB114 | EcoRV/SalI fragment of pSMB112 ligated to pGP704 | Subcloning into suicide vector for conjugation | Apr Cmr | This study |
| pGH1610 | galF rfbBDA′04 from pGH161 (20) cut with PstI/HpaI and ligated to pBR322 cut with PstI/ScaI | Cloning of rfbD gene for complementation | Tcr | This study |
| pRAB10 | rol gene in pACYC184 | Subcloning of rol gene for complementation | Tcr | 3 |
| pSMB250 | kfiCD PCR product (SalI ends) ligated to pBluescript II KS | Cloning of kfiCD genes | Apr | This study |
| pSMB251-Cm | Insertion of cat gene (SmaI/HindII) into NdeI-filled ends of pSMB250 | Mutation of kfiC gene | Apr Cmr | This study |
| pSMB251-Km | Insertion of aphA gene (filled ends) into NdeI-filled ends of pSMB250 | Mutation of kfiC gene | Apr Kmr | This study |
| pSMB252-Cm | SalI fragment of pSMB251-Cm ligated to pGP704 | Subcloning into suicide vector for conjugation | Apr Cmr | This study |
| pSMB252-Km | SalI fragment of pSMB251-Km ligated to pGP704 | Subcloning into suicide vector for conjugation | Apr Kmr | This study |
For standard growth of strains, Luria broth and Luria agar (1.6% [wt/vol] agar) (Difco Laboratories, Detroit, Mich.) were used. In some cases, minimal medium was used for the selection of mutants as described by Davis and Mingioli (13). Minimal medium was supplemented with cysteine (Sigma Chemical Co., St. Louis, Mo.) at a final concentration of 20 μg/ml as required for the growth of GR-12 in minimal medium.
Antibiotics were added to media as needed in final concentrations as follows: 10 to 20 μg/ml for chloramphenicol and tetracycline, 50 μg/ml for kanamycin, and 100 μg/ml for ampicillin (Sigma). Buffered saline-gelatin (BSG), used as a diluent in serum assays, was made as described previously but at a pH of 7.35 for the optimization of complement activity (12). EGTA and MgCl2 were each added to a concentration of 100 mM to BSG to inactivate the classical pathway of complement but to allow the alternative pathway of complement to function.
Mating procedure.
GR-12 (or SMB20) and SM10 with specific plasmids were grown to the mid-log phase and combined in a 1:4 ratio of donors to recipients. The mixture was filtered, and the filter was placed on a fresh Luria-Bertani (LB) plate. The plate was incubated at 37°C for approximately 2 h. The bacteria on the filter were resuspended in 1 ml of BSG, and 50- to 200-μl samples were plated on minimal medium containing cysteine and the appropriate antibiotics. To optimize mating, a nonshaking culture of SM10 (with a plasmid) was mixed with a well-vortexed culture of GR-12 grown to the late stationary phase. After mating, the bacteria were gently resuspended in 1 ml of SOC (36) and incubated at 37°C for 45 min (11). Cells were washed twice with BSG, and samples were plated as before.
Construction of an rfbD mutation in GR-12.
The chloramphenicol acetyltransferase gene (cat) obtained by gel purification from pUC18CML (51) digested with HindII and SmaI was inserted in the rfbD gene of plasmid pSMB102, a subclone of plasmid pRAB2401 (4). Isolates which had the cat gene in the same orientation as the rfbD gene were chosen (22) to avoid disruption of the transcription of upstream genes. The rfbD::cat construct from the new plasmid, pSMB103, was ligated to pGP704Km to make pSMB104. SM10 electroporated with pSMB104 was conjugated with GR-12. The vector pGP704 and its derivative pGP704Km cannot replicate in GR-12 and thus act as suicide vectors. Transconjugants were selected on minimal medium supplemented with cysteine and chloramphenicol to eliminate SM10 (unable to grow on minimal medium). Transconjugants were further screened for kanamycin sensitivity to distinguish single recombinants (in which the entire plasmid was integrated into the chromosome) from double recombinants.
Transconjugants which were Cmr Kms were analyzed by PCR with the rfbD F and rfbD R primers (Table 3). The wild-type amplified product was 2.5 kb, and products resulting from the insertion of the rfbD gene were 1.6 kb larger (4.1 kb). Any transconjugants which had both products were eliminated. Southern analysis was performed to further confirm the correct structure of the transconjugants. Chromosomal DNAs isolated from GR-12, SM10, and transconjugants were digested with EcoRV and probed with a labeled probe for the rfbD gene made with the rfbD F and rfbD R primers.
TABLE 3.
Oligonucleotide primers used in this study
| Gene ampli- fied | Product size (kb) | Primer set | Sequencea |
|---|---|---|---|
| rfbD | 2.5 | rfbD F | 5′-GTGGAATGGTATCTGTC-3′ |
| rfbD R | 5′-TTGTGTTATAGATGTGC-3′ | ||
| rol | 0.7 | rol F | 5′-TGGCAAGATGACAATT-3′ |
| rol R | 5′-AAGTTTATCTCAACGTCAAG-3′ | ||
| ugd | 1.1 | ugd F | 5′-CCGTCACGCGTTGCTATGCTG-3′ |
| ugd R | 5′-TTGGATCCCCGTGAATGCTATAGG-3′b | ||
| his | 0.8 | his F | 5′-TAGGATCCCCGAAGAAAGCAATTCCTTG-3′b |
| his R | 5′-TATCTAGACGGGCAATTTCCTCAATG-3′c | ||
| kfiC | 3.2 | kfiC F | 5′-GGGAAGCTCTATTCTCAGTCGACG-3′d |
| kfiD R | 5′-CGCGACTATAGACTTTGTCGACC-3′d |
Sequences complementary to gene amplified are in boldface.
BamHI sites are underlined.
XbaI sites are underlined.
SalI sites are underlined.
Construction of an rfbKM mutation in GR-12.
A subclone of pRAB2401, pSMB105, was recloned to eliminate the NruI sites, creating pSMB106. A portion of the sequence between the rfbK and rfbM genes was deleted, and a cat gene was inserted. As before, the orientation of the cat gene which corresponded to the predicted direction of transcription of the rfbKM genes was chosen (22). The new plasmid, pSMB107, was subcloned into pGP704. After ligation, the mixture was digested with KspI (found in the pBluescript II KS vector only) before electroporation into SY327 to eliminate self-ligation of pSMB107. The new plasmid, pSMB108, was transferred into SM10 and conjugated with GR-12. Transconjugants were selected as described above.
Chromosomal DNAs were prepared from GR-12, SM10, and selected transconjugants and digested with PstI. A DNA-labeled probe was made from a purified 3.5-kb PstI fragment from pSMB106, and mutants were confirmed by Southern analysis.
Construction of a rol deletion in GR-12.
In order to delete the rol gene, upstream and downstream sequences were amplified with primers with engineered restriction sites so as to clone the products side by side, deleting the rol gene (0.82 kb of sequence deleted). Primers ugd F and ugd R and primers his F and his R (Table 3) were used to amplify by PCR the upstream ugd gene and the downstream his gene, respectively. The ugd PCR product was first cloned into pBluescript II KS (pSMB110), and then the his PCR product was cloned (pSMB111). The cat gene with BamHI ends was inserted between the two cloned genes, creating pSMB112. The ugd′-′his::cat fragment of DNA was cloned into pGP704. The new plasmid, pSMB114, was transferred into SM10 and conjugated with GR-12. Transconjugants were selected as described above.
Transconjugants which were Cmr Kms were amplified by PCR with the flanking primers ugd F and his R. The flanking primers produced a wild-type product of ∼2.7 kb and a rol deletion product of 3.5 kb, since the cat gene was 0.8 kb larger than the portion deleted. Internal primers, rol F and rol R, of the rol gene (Table 3) amplified a 0.7-kb wild-type product and no product for the rol deletion mutants. GR-12 yielded the wild-type product for both PCR amplifications. Several transconjugants had only the rol deletion PCR product, while others had both the wild-type and the deletion products. Isolates with both products resulted from a single recombination event in which the entire plasmid was integrated. Transconjugants with the rol deletion product only and no product with internal primers were chosen for Southern analysis. Some isolates with both products were included as a comparison.
Chromosomal DNAs isolated from GR-12, SM10, and transconjugants were digested with EcoRV and probed with a label made from the PCR product amplified with the ugd F and his R primers. Southern analysis confirmed the PCR results.
Construction of a kfiC mutation in GR-12 and the rfbD mutant.
The kfiCD gene from region 2 of the K5 kps capsular cluster was cloned by PCR amplification with the kfiC F and kfiD R primers (Table 3). The kfiCD PCR product was digested with SalI and ligated to pBluescript II KS, creating pSMB250. The cat gene was inserted into the kfiC gene in the same orientation, resulting in plasmid pSMB251-Cm, for construction of the K5 capsular mutant. The aphA gene from pBluescript II KS-Km was inserted into the kfiC gene in the same orientation, resulting in plasmid pSMB251-Km, for construction of the double mutant, since the rfbD mutant was already Cmr. The new construct was removed from each plasmid (pSMB251-Cm and pSMB251-Km) and ligated to pGP704, making pSMB252-Cm and pSMB252-Km, respectively. The new plasmids were transferred into SM10. SM10 pSMB252-Cm was mated with GR-12 to create a K5− mutant. SM10 pSMB252-Km was mated with SMB20 (rfbD mutant) to create an O75− K5− double mutant.
For the acapsular single mutant, transconjugants which were Cmr Kms were selected and PCR amplified with the kfiC F and kfiD R primers (Table 3). Transconjugants with a PCR product of 4.7 kb (the wild-type product was 3.2 kb) were confirmed by Southern analysis. Chromosomal DNAs isolated from GR-12, SM10, SM10 pSMB252-Cm, and selected transconjugants were digested with EcoRV and probed with a kfiCD-labeled PCR product.
For the double mutant, transconjugants which were Cmr Kmr on minimal medium were selected and amplified by PCR with the kfiC F and kfiD R primers. Transconjugants with a PCR product of 4.5 kb (the wild-type product was 3.2 kb) were confirmed by Southern analysis. Chromosomal DNAs isolated from GR-12, SM10 pSMB252-Km, and selected transconjugants were digested with EcoRV and probed with a kfiCD-labeled PCR product or an aphA-labeled probe.
PCR amplification.
A small amount of a fresh colony was resuspended in 50 μl of PCR mixture (1× PCR buffer, 2 mM MgCl2, 0.5 U of Amplitaq [Perkin-Elmer Cetus, Norwalk, Conn.], 200 μM each deoxynucleoside triphosphate [Boehringer Mannheim Biochemicals, Indianapolis, Ind.]). A DNA concentration of 1 ng was used for plasmid amplification. The mixture was sealed with approximately 60 μl of paraffin oil. The PCR consisted of 27 cycles: 1 min of denaturation at 94°C, 2 min of annealing at 50°C, and 3 min of extension at 72°C. The PCR concluded with 7 min at 72°C and 10 min at 4°C. A modified PCR was used for the kfiC F and kfiC R primers by increasing the annealing temperature to 60°C. In order to amplify products larger than 4.5 kb, it was necessary to use Taq Extender PCR Buffer and Taq Extender PCR Additive from Stratagene. These reactions were done with a 5-min extension time.
DNA manipulations.
In order to purify a small amount of plasmid to screen isolates, the cleared-lysate method of preparation was used as described by Maniatis et al. (36). Larger-scale preparations of plasmids for cloning and storage were made with Qiagen tips (Qiagen, Inc., Chatsworth, Calif.). Chromosomal DNA preparations were made with 0.5-ml overnight cultures in accordance with the protocol of Puregene, Research Triangle Park, N.C. Digestion with restriction endonucleases, ligations, transformations, electrophoresis on agarose gels, and DNA blotting by capillary action were done as described by Maniatis et al. (36). T4 DNA ligase was used for ligations (New England BioLabs, Beverly, Mass.). Restriction endonucleases used for cloning procedures were from New England BioLabs and Boehringer. DNA fragments were cut from high-strength analytical-grade ultra-pure DNA-grade agarose (Bio-Rad, Hercules, Calif.) and purified with Jetsorb according to the manufacturer’s instructions (Genomed, Research Triangle Park, N.C.).
For Southern hybridization, chromosomal DNA was transferred to nylon membranes (Zeta Probe Blotting; Bio-Rad, Richmond, Calif.). DNA was fixed by heating the membranes to 60°C for 2 h under vacuum. Digoxigenin-labeled probes were made by use of the random-primer DNA labeling protocol (Genius kit; Boehringer). DNA for labeling was either PCR amplified in the case of the rfbD, ugd-his, and kfiCD probes or gel purified in the case of the rfbKM probe. Hybridization and detection were done according to manufacturer’s instructions (Genius kit).
LPS analysis.
LPS was isolated from a 1-cm square patch of bacteria on an agar plate by the procedure of Hitchcock and Brown (21). The preparations were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on a 6% stacking–15% separating gel as described by Laemmli (32) with Tris-glycine buffer for GR-12 and mutant strains. For further analysis of the rfbKM mutant, Tricine-SDS buffer was used (49). The samples were electrophoresed at a constant 30 A on a minigel apparatus (Mighty Small II SE 250; Hoefer Scientific, San Francisco, Calif.). The gel was fixed overnight in a solution of 5% acetic acid and 40% ethanol. The gel was silver stained by the procedure of Tsai and Frasch (62).
Sensitivity to K5-specific bacteriophage.
GR-12, mutants, and complemented mutants were tested for sensitivity to K5-specific bacteriophage (G. Schmidt, Borstel, Germany) by cross-streaking of bacteria against a phage suspension on Luria agar.
Outer membrane protein analysis.
Outer membrane proteins were isolated by detergent solubilization as described previously, except that cells were broken by passage through a French press (1). After pelleting of insoluble outer membrane proteins, proteins were washed in saline, centrifuged in Sorvall (SS34; 20,000 rpm, 90 min, 4°C), and resuspended in 25 μl of 2× Laemmli buffer without dye. Proteins were separated by SDS-PAGE on a 4% stacking–12% separating gel at 100 V through the stacking gel and 200 to 250 V through the separating gel on a minigel apparatus (Mighty Small II SE 250). A low-molecular-weight protein ladder was used as a standard (Pharmacia Fine Chemicals, Piscataway, N.J.). Proteins were visualized by staining with 0.25% Coomassie brilliant blue R-250 for 20 min and then were destained as described previously (36).
Serum assays.
Blood was collected from five healthy individuals who gave informed consent and then was allowed to clot. Serum was collected, pooled, and stored at −70°C in aliquots. Serum assays were performed with normal human serum, serum plus EGTA, and heat-inactivated serum. BSG with 100 mM EGTA and MgCl2 was added to serum to chelate calcium, inactivating the classical pathway of complement. Heated serum was serum heated to 56°C for 30 min to inactivate complement components. Assays were performed by a previously described procedure with slight modifications (60). The concentration of serum in the assays was 80%. Bacteria were inoculated into a culture of LB medium from a fresh LB plate and grown to the mid-log phase (optical density at 600 nm, 0.5). Antibiotics were added for the growth of complemented mutants. Cells were added directly or were diluted twofold in saline and then added to normal human serum, serum plus EGTA, or heat-inactivated serum to obtain approximately 3 × 107 or 3 × 105 CFU per ml, respectively, in a serum concentration of 80%. BSG or BSG plus EGTA was used as a diluent for the serum reactions and viable counts. The reaction mixture was incubated at 37°C for 3 h. Viable counts were made by taking samples at 0, 1, 2, and 3 h. At least two samples were taken at each time point. Dilutions from samples were plated onto LB plates and incubated overnight at 37°C, and counts were determined. Percentages for each time point were obtained from the surviving viable count versus the initial viable count. Each strain was assayed at least two different times in 80% serum and 80% heat-inactivated serum.
Complementation of mutants.
Single mutants were complemented with the wild-type gene on a plasmid. The plasmids were electroporated into the mutants, the complemented mutants were selected by resistance to tetracycline, and the presence of the plasmids was confirmed. SMB20 was complemented with the rfbD gene on plasmid pGH1610 (Table 2). This plasmid contains the 3′ end of the galF gene and the entire rfbB and rfbD genes, as well as the 5′ end of the rfbA gene, inserted into the bla gene of pBR322. SMB122 was complemented with the rol gene on plasmid pRAB10 (3). For complementation of SMB43, the rfbKM genes were cloned into plasmid pBR322, creating pSMB109 (Table 2), and electroporated into SMB43. The kfiCD genes were removed from pSMB250 and cloned into pBR322, creating pSMB255 (Table 2). pSMB255 was electroporated into SMB213. The vector plasmid in each case was also electroporated into the mutants to confirm that the vector was not responsible for serum resistance.
RESULTS
In order to study the importance of the O75 O antigen and the K5 capsular antigen in conferring resistance to serum, chromosomal mutations in LPS synthesis and K5 synthesis genes were made in serum-resistant, pathogenic strain GR-12. Several genes determined to be important in LPS synthesis on the basis of preliminary O75 sequence analysis were mutated: the rol gene, the putative rfbD gene, and the putative rfbKM genes (5, 22). The kfiC gene, required for the synthesis of the K5 capsule, was chosen for mutation to make a K5− mutant.
Rationale for mutations in GR-12.
We sought to evaluate and compare the effects of LPS and capsular mutations on serum resistance. A rough mutant, having no O polysaccharide, and a capsule-deficient mutant should establish the role of the O75 and K5 antigens in resistance to complement-mediated lysis. In order to create a rough mutant, the rfbD gene was targeted for mutation. The putative rfbD gene encodes the synthesis of TDP-rhamnose, which is necessary to make the O75 subunit. A diagram of the O75 subunit is shown in Fig. 1. The rfbD gene was inactivated because the sugar rhamnose is unique to the O75 subunit and is not required for the synthesis of other polymers, such as the core oligosaccharide, lipid A, and peptidoglycan (50). Without TDP-rhamnose, the mutant strain is predicted to lack O-polysaccharide units attached to the core. In order to create an acapsular mutant, the kfiC gene, encoding a putative glycosyltransferase necessary for the synthesis of the K5 capsular antigen, was targeted (45, 52). A double mutant was also made with mutations in the kfiC and rfbD genes to determine the effect on serum resistance of the loss of both the O75 and the K5 antigens.
FIG. 1.
Structure of the O75 subunit (15). The O75 subunit is made of four sugars: GlcNAc, N-acetylglucosamine; Rha, rhamnose; Gal, galactose; and Man, mannose (side-chain sugar).
Other LPS mutants were also created to evaluate the resistance provided by O antigens in serum. The rfbK and rfbM genes are necessary for the synthesis of GDP-mannose (26, 63). The sugar mannose is the side-chain sugar of the O75 subunit (Fig. 1). Mutation of either rfbK or rfbM prevents the synthesis of the sugar mannose. Only a partial O75 subunit lacking the side-chain sugar can be synthesized. Bacteria also normally regulate the length of LPS in a bimodal distribution, having a high percentage of long chains. Since long-chain LPS was found to be important in resistance to complement, we wanted to examine the effect of a random distribution of LPS. Therefore, a mutation in the rol gene, responsible for the bimodal distribution of LPS, was made (3).
Construction of mutations in GR-12.
A brief description of the construction of the mutants follows; details are given in Materials and Methods and Tables 1, 2, and 3. The kfiC, rfbD, and rfbKM genes, to be mutated by insertion of an antibiotic resistance gene, were initially subcloned onto a plasmid. For the kfiC and rfbKM genes, a portion of the gene was deleted. A cat gene was inserted in the same orientation as the wild-type gene to disrupt the gene(s) and allow for the selection of mutants. The aphA gene was used for creation of the kfiC mutation in the double mutant (kfiC-rfbD mutant). The rol gene was completely deleted by insertion of a cat gene between the upstream and downstream sequences of the rol gene. The mutated gene in each case was further subcloned into the suicide vector (pGP704) or a derivative of the suicide vector. The suicide vector was placed into a donor strain, SM10, and conjugated with GR-12 or SMB20 (for the double mutant) (Fig. 2). Transconjugants were selected on minimal medium with appropriate antibiotics and confirmed by PCR (except for the rfbKM mutant) and Southern analysis. The mutants were designated as follows: SMB20 (rfbD mutant), SMB43 (rfbKM mutant), SMB122 (rol deletion mutant), SMB213 (kfiC mutant), and SMB316 (kfiC-rfbD mutant).
FIG. 2.
Diagram of construction of chromosomal mutants. Genes deleted or mutated are indicated. Only relevant restriction sites are included: A, SalI; C, ClaI; D, NdeI; E, EcoRV; H, HpaI; K, KpnI; M, MunI; N, NruI; P, PstI; and S, SnaBI. Plasmids are described in Table 2. Crossed lines denote recombination.
Phenotypic analyses of GR-12 and mutants.
The LPS phenotype of a wild-type strain, such as GR-12, is characterized by a bimodal distribution of O-antigen subunits visualized on a polyacrylamide gel (23, 43). Strains possessing a smooth LPS phenotype have long O-antigen chains; a rough strain has only core oligosaccharide. A semirough strain has also been described; the LPS is characterized by the substitution of only one O-antigen subunit on the core oligosaccharide. An example is an rfc mutant (35) in which the loss of O polymerase function does not allow polymerization of the O-antigen subunits.
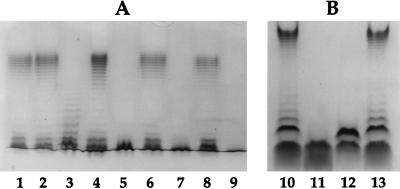
LPSs of GR-12 and the five mutants were prepared and analyzed. GR-12, SMB20, SMB43, SMB122, SMB213, and SMB316 were analyzed by glycine-SDS-PAGE (Fig. 3A). GR-12 and SMB213 (kfiC mutant) produced a wild-type bimodal distribution of LPS. SMB122 (rol mutant) had a random distribution of LPS. SMB20 (rfbD mutant) and SMB316 (kfiC-rfbD mutant) had a rough phenotype with only the core and no O-antigen chains attached. The LPS of SMB43 (rfbKM mutant) appeared rough, similar to that of SMB20 on glycine-SDS-PAGE. When SMB43 was analyzed on a Tricine-SDS buffer system, which allows better resolution of the core and the core plus one O antigen (33), it was apparent that SMB43 had not only the core but also the core plus a partial O antigen (Fig. 3B). The O antigen attached to the core of SMB43 migrated faster on the gel than the core plus one O antigen of GR-12.
FIG. 3.
Visualization of LPSs of GR-12, mutants, and complemented mutants. (A) SDS-PAGE. Lanes: 1, GR-12 (wild type); 2, SMB213 (kfiC mutant); 3, SMB122 (rol mutant); 4, SMB122(pRAB10); 5, SMB43 (rfbKM mutant); 6, SMB43(pSMB109); 7, SMB20 (rfbD mutant); 8, SMB20(pGH1610); 9, SMB316 (kfiC-rfbD double mutant). (B) Tricine-SDS buffer system. Lanes: 10 and 13, GR-12; 11, SMB20 (rfbD mutant); 12, SMB43 (rfbKM mutant).
GR-12 and the five mutants were tested for sensitivity to K5-specific phage. GR-12, SMB20, and SMB43 were sensitive to K5-specific phage. SMB213 and SMB316 were resistant to K5-specific phage.
GR-12 and the five mutants were also tested for agglutination of erythrocytes. GR-12 has P pili, which bind the P blood group (19). Upon addition to whole blood, GR-12 agglutinates erythrocytes. All mutants were tested for hemagglutination. One of the two double mutants initially isolated did not agglutinate in whole blood and was not used for further analysis.
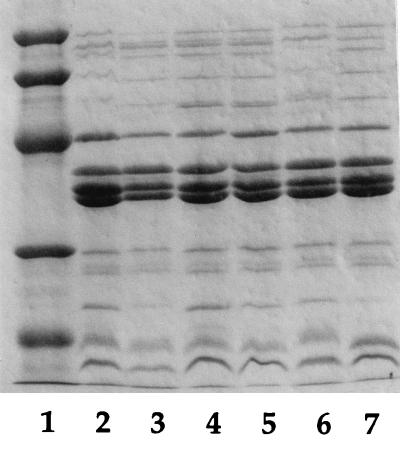
In order to confirm that the loss of the wild-type expression of O or K antigens had no effect on the expression of proteins in the outer membrane, outer membrane proteins were prepared from GR-12 and the mutants and analyzed by SDS-PAGE as shown in Fig. 4. No differences from the wild-type strain, GR-12, were found. Distortions of the upper bands of GR-12 and SMB213 were apparently due to the presence of long-chain O antigens, as visualized by silver staining of the samples on SDS-PAGE (data not shown).
FIG. 4.
Analysis of outer membrane protein profiles of GR-12 and mutants. A 5-μl sample of each strain was loaded in each lane. Lanes: 1, protein standard (from top: molecular weight of 94,000 [94K], 67K, 43K, 30K, and 20.1K); 2, GR-12 (wild type); 3, SMB122 (rol mutant); 4, SMB43 (rfbKM mutant); 5, SMB20 (rfbD mutant); 6, SMB213 (kfiC mutant); 7, SMB316 (kfiC-rfbD mutant).
Therefore, mutants were made with five different phenotypes for analysis: SMB122, with a random distribution of LPS; SMB43, with the core plus one partial O-antigen subunit; SMB20, with a rough LPS phenotype (O75− K5+); SMB213, lacking K5 antigen (O75+ K5−); and SMB316, lacking both O75 and K5 antigens (O75− K5−).
Serum assays of GR-12 and mutants.
GR-12 is serum resistant. The sensitivity of the mutants in serum can be compared to that of GR-12. Since isogenic mutants were constructed, any loss of resistance could be attributed to mutation of a particular gene.
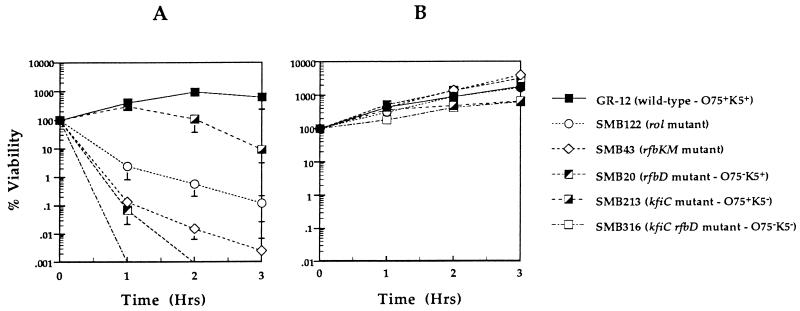
Serum assays were performed on all five mutants and parental strain GR-12 with 80% serum and 80% heat-inactivated serum. Bacteria at the exponential phase of growth were diluted and added to serum to obtain approximately 3 × 105 CFU/ml. The mixture was incubated for 3 h. Viable counts were determined in duplicate at the beginning of the experiment and at each hour for 3 h.
The results are shown in Fig. 5A as percent viability. As expected, GR-12 was resistant to 80% serum, increasing in numbers by 500% in the first hour, with growth reaching a plateau by the second hour of incubation. The K5− mutant (SMB213) was not affected by serum in the first hour, increasing in numbers similar to GR-12; however, an 80% decrease in viability occurred by the third hour of incubation. The rol deletion mutant (SMB122) was more sensitive to serum, exhibiting a decrease in viability of almost 99%, or 2 log units of killing. The rfbKM mutant (SMB43) with only a partial O-antigen subunit attached to the core, was even more sensitive, exhibiting a 99.9% decrease in numbers in the first hour of incubation (3 log units of killing). The O75− mutant (SMB20) was similar to SMB43 in viability in the first hour but was undetectable by the end of the second hour of incubation. Both SMB122 and SMB20 steadily decreased in numbers by the third hour of incubation. The O75− K5− mutant (SMB316) was the most sensitive, exhibiting more than 4 log units of killing within the first hour of incubation. All five mutants and the parental strain grew equally well in 80% heat-inactivated serum (Fig. 5B).
FIG. 5.
Serum assays of wild-type strain GR-12 and mutants in 80% serum (A) or 80% heat-inactivated serum (B) with an inoculum of 3 × 105 CFU/ml. The assays were performed for 3 h; samples were taken in duplicate at the start and at each hour. The graphs are a result of at least two different assays for each strain. Results are shown as percent viability. Error bars indicate standard deviations.
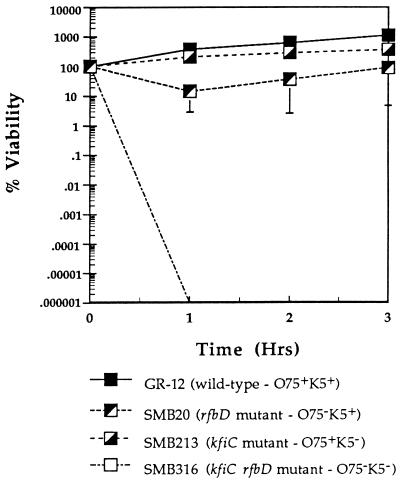
To measure the difference in the sensitivities of the O75− mutant (SMB20) and the double mutant (SMB316) to serum, the inoculum was increased to obtain approximately 3 × 107 CFU/ml at the start of the assay (Fig. 6). GR-12 numbers increased by 1 log unit in the third hour of incubation. At this inoculum, SMB213 was completely resistant to serum, also exhibiting growth. SMB20 viability decreased by approximately 80% in the first hour, but survivors grew during the second and third hours of incubation, possibly indicating that lytic factors had been depleted. SMB316 was extremely sensitive to serum, being undetectable by the end of the first hour of incubation.
FIG. 6.
Serum assays (80% serum) of GR-12 and O75−, K5−, and O75− K5− mutants at an inoculum of 3 × 107 CFU/ml. The assays were performed for 3 h. Samples were taken in duplicate at the start and at each hour. The graph is a result of at least two different assays for each strain. Results are shown as percent viability. Error bars indicate standard deviations.
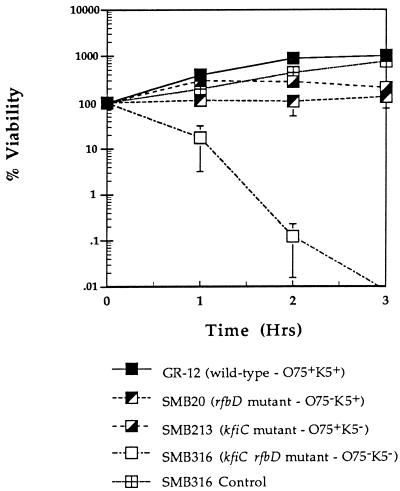
The membrane attack complex can be activated via two different complement pathways, the classical and alternative pathways. These pathways can be distinguished by the addition of EGTA to serum to inactivate the classical pathway of complement. Thus, only the alternative pathway is active to kill bacteria. To determine which pathway was more important in killing of the O75−, K5−, and O75− K5− mutants, serum assays were done with EGTA plus MgCl2 added (Fig. 7). As expected, GR-12 was resistant to 80% serum with EGTA, exhibiting an increase in numbers even greater than that observed in 80% serum without EGTA. SMB213 (O75+ K5−) viability increased by 300% and was maintained in the second hour, with a slight decline in the third hour of incubation. SMB20 (O75− K5+) was inhibited but not killed during the 3-h incubation, while the numbers of the double mutant (SMB316) decreased by 90% in the first hour and by 99.9% in the second hour of incubation. A control reaction was performed with the double mutant in 80% heat-inactivated serum with EGTA, showing that the sensitivity of the double mutant was not due to the presence of EGTA.
FIG. 7.
Serum assays (80% serum) with 10 mM EGTA plus MgCl2 of GR-12 and O75−, K5−, and O75− K5− mutants at an inoculum of 3 × 105 CFU/ml. The assays were performed for 3 h. Samples were taken in duplicate at time zero and at each hour. A control assay was performed with 80% heat-inactivated serum plus EGTA and the double mutant (SMB316). The graph is a result of at least two different assays for each strain. Results are shown as percent viability. Error bars indicate standard deviations.
Complementation of mutants.
In order to confirm that the loss of function of the mutated gene caused the failure to produce a wild-type phenotype and the loss of serum resistance, the wild-type gene was transferred to the mutants in trans on a plasmid. The wild-type gene on a plasmid was introduced into the mutants via electroporation, and complemented mutants were selected on the basis of resistance to tetracycline. Complementation of mutants is described in Materials and Methods. All single mutants were also electroporated with the cloning vector alone; i.e., SMB122 was electroporated with pACYC184 and SMB20, SMB43, and SMB213 were electroporated with pBR322 to confirm that the vector plasmid itself was not complementing the capacity of the mutants to resist the bactericidal effect of serum.
All complemented mutants were analyzed by visualization of LPS by PAGE and by serum assays. In each complemented mutant, the characteristic bimodal distribution of LPS was restored (Fig. 3A). The complemented K5− mutant regained sensitivity to K5-specific phage. All complemented mutants regained serum resistance in 80% serum similar to that of GR-12, increasing in viable counts in the first hour of incubation (Fig. 8). Complemented mutants also grew in 80% heat-inactivated serum (data not shown). The mutants with the vector plasmid alone were not resistant to serum (data not shown).
FIG. 8.
Serum assays (80% serum) of GR-12 and each complemented mutant at an inoculum of 3 × 105 CFU/ml. The assays were performed for 3 h. Samples were taken in duplicate at time zero and at each hour. The graph is a result of at least two different assays for each strain. Results are shown as percent viability. Error bars indicate standard deviations.
DISCUSSION
Many previous studies, in which a role in serum resistance was assigned to LPS or capsules, used mutants in which the exact genetic lesion was unknown. Furthermore, the potential for secondary mutations in these strains was not always investigated. Therefore, although correlations were made as to the role of LPS and capsules in serum resistance, the evidence was not conclusive. Through creation of a defined mutant strain from an otherwise wild-type pathogenic strain, it is possible to definitively point to the mutation as the cause for the loss or gain of a specific attribute.
GR-12 and the five mutants were analyzed phenotypically for the K5 capsule and LPS. K5-specific phage sensitivity testing confirmed that GR-12 and LPS single mutants were sensitive to K5-specific phage, while SMB213 (kfiC mutant) and SMB316 (double mutant) were resistant. GR-12 had the typical bimodal distribution of LPS; SMB122 (rol mutant) had a random distribution of LPS, as expected; and SMB20 (rfbD mutant) was rough, with only the core being present. We predicted that the rfbKM mutant would have one of several phenotypes: rough, core plus one partial O-antigen subunit, or O-antigen chains with no side-chain sugars present. Previous studies with E. coli reported the identification of an O4 strain which had O-antigen chains without the side-chain sugars normally present (24). The rfbKM mutant (SMB43) appeared rough on SDS-PAGE but, after analysis with a Tricine-SDS buffer system, SMB43 was discovered to have the core plus one partial O-antigen subunit. The partial O-antigen subunit, consisting of N-acetylglucosamine, rhamnose, and galactose, was ligated to the core without the side-chain sugar mannose, but polymerization of the partial O-antigen subunit was not apparent. Thus, the side-chain sugar mannose is not necessary for ligation of the O-antigen subunit to the core but is required for polymerization of the O-antigen subunits.
Analyzing the mutants with serum assays proved that the O75 antigen is more important than the K5 antigen in serum resistance. Rough mutant SMB20 was completely killed in serum by the second hour of incubation. In comparison, K5− mutant SMB213 increased in numbers in the first hour and then slowly decreased in viability by the third hour of incubation in serum. A complete loss of the O75 antigen made the bacterium sensitive to serum. This work is in contrast to work done by Russo et al (48). They made an isogenic mutant lacking O antigen in an O4 pathogenic strain. Contrary to the expected results, the O4 antigen played only a minor role in serum resistance in comparison to the K54 antigen. However, our work did agree with that of Russo et al. in that the loss of both O and K antigens was correlated with a synergistic effect on serum sensitivity (48). The O75− K5− mutant was undetectable by the end of the first hour of incubation at both inoculum sizes tested, while the O75− mutant was not as sensitive to serum when the inoculum size was increased. The K5− mutant, in contrast, was resistant to serum at the larger inoculum size.
Complement may be activated by either the classical or the alternative pathway. The classical pathway can be blocked by the addition of a calcium chelator, such as EGTA. GR-12 and all mutants, except for the double mutant, were resistant to serum when the classical pathway was blocked. The alternative pathway was not sufficient to kill the rough mutant (O75− K5+ mutant) but was able to kill the double mutant over a 3-h incubation. Thus, the O75 antigen is important in resistance to the classical pathway, and the K5 antigen may provide resistance to the alternative pathway, as indicated by the difference in sensitivity between the O75− K5+ mutant and the O75− K5− mutant in serum plus EGTA. These findings agree with studies done by Devine and Roberts (14) in which O75 strains were affected by the classical complement pathway and K5 strains showed a delayed sensitivity to serum and were resistant to the alternative complement pathway.
The rfbKM mutant was also very sensitive to serum. Although the serum sensitivity was similar to that of the rough mutant in the first hour, the rfbKM mutant was not completely killed by the third hour of incubation. Substitution of the partial O-antigen subunit on the core oligosaccharide must have given the rfbKM mutant some protection against serum complement components in the second and third hours of incubation in serum.
Several studies have indicated the importance of long-chain LPS (18, 27). Smooth strains of Salmonella and E. coli have been shown to interact with terminal components of the complement cascade, yet insertion of the membrane attack complex never occurs (30). The membrane attack complex, formed by the activation of long O-antigen side chains at points distal to the vulnerable lipid layer, is sterically hindered from insertion into the outer membranes of smooth strains (27, 28). Although smooth strains are less susceptible to serum, the degree of susceptibility also depends on the coverage of LPS (59). Grossman et al. (17) showed that an increase in LPS coverage correlated with an increase in survival in serum. The present study demonstrated that, in the case of the O75 antigen, a bimodal distribution of O75 O antigens is crucial in protecting the bacteria from complement-mediated lysis. The rol deletion mutant could still produce long-chain LPS but in fewer full-length chains than the wild-type strain. Loss of the majority of long O-antigen chains made the rol deletion mutant sensitive to serum but to a lesser degree than the rough mutant. The rol deletion mutant exhibited almost 2 log units of death in the first hour and continued to decrease in numbers in the second and third hours of incubation, while GR-12 increased in numbers. These studies showed the importance of regulation of the length of the O75 O antigens for survival in serum.
Through restoration of the wild-type phenotype with the wild-type gene in trans, the mutations causing the defect in the synthesis of LPS were demonstrated to be responsible for the loss of serum resistance. All complemented mutants regained a wild-type bimodal distribution of LPS or K5-specific phage sensitivity and serum resistance.
When compared to the studies of Russo et al. (48), our results established the fact that the roles of the O and K antigens may be different in different strains. Our finding was that the O75 antigen plays a more important role in serum resistance than does the K5 antigen. However, Russo et al. found these roles to be reversed in an O4:K54 strain (48). Although one antigen may not be critical in serum resistance, it may play a more important role in another aspect of virulence, such as resistance to phagocytosis or to phagocytic killing. Thus, the antigens may have a complementary relationship in virulence—one antigen providing resistance to one aspect of the host defense system and the other antigen providing resistance to another part of the host defense system. Testing of other isogenic mutants of different O:K pathogenic strains is necessary to prove this point.
This is the first study in which defined LPS and capsular mutants were created for a pathogenic strain, tested in serum resistance assays, and complemented, showing restoration of the wild-type phenotype along with serum resistance. The mutants from this study will allow us to further understand the capacity of the O75 and K5 antigens in protection against the host defense system and to address important aspects of LPS synthesis.
ACKNOWLEDGMENT
This research was supported by U.S. Public Health Service grant NIAID21009 to S.I.H.
REFERENCES
- 1.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver R P. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun. 1983;39:315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen P M, Roberts I, Boulnois G J, Saunders J R, Hart C A. Contribution of capsular polysaccharide and surface properties to virulence of Escherichia coli K1. Infect Immun. 1987;40:359–368. doi: 10.1128/iai.55.11.2662-2668.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batchelor R, Haraguchi G E, Hull R A, Hull S I. Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol. 1991;173:5699–5704. doi: 10.1128/jb.173.18.5699-5704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batchelor R A. Ph.D. thesis. Houston, Tex: Baylor College of Medicine; 1992. [Google Scholar]
- 5.Batchelor R A, Alfina P, Biffali E, Hull S I, Hull R. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J Bacteriol. 1992;174:5228–5236. doi: 10.1128/jb.174.16.5228-5236.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulnois G J, Roberts I S. Genetics of capsular polysaccharide production in bacteria. Curr Top Microbiol Immunol. 1990;150:1–18. doi: 10.1007/978-3-642-74694-9_1. [DOI] [PubMed] [Google Scholar]
- 7.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross A S. The biological significance of bacterial encapsulation. Curr Top Microbiol Immunol. 1990;150:87–98. doi: 10.1007/978-3-642-74694-9_5. [DOI] [PubMed] [Google Scholar]
- 9.Cross A S, Gemski P, Sadoff J C, Ørskov F, Ørskov I. The importance of the K1 capsule in invasive infections caused by Escherichia coli. J Infect Dis. 1984;149:184–193. doi: 10.1093/infdis/149.2.184. [DOI] [PubMed] [Google Scholar]
- 10.Cross A S, Kim K S, Wright D C, Sadoff J C, Gemski P. Role of lipopolysaccharide and capsule in the serum resistance of bacteremic strains of Escherichia coli. J Infect Dis. 1986;154:497–503. doi: 10.1093/infdis/154.3.497. [DOI] [PubMed] [Google Scholar]
- 11.Curtiss R. Bacterial conjugation. Annu Rev Microbiol. 1969;23:69–136. doi: 10.1146/annurev.mi.23.100169.000441. [DOI] [PubMed] [Google Scholar]
- 12.Curtiss R, III, Charamella L J, Berg C M, Harris P E. Kinetic and genetic analysis of d-cycloserine inhibition and resistance in Escherichia coli. J Bacteriol. 1965;90:1238–1250. doi: 10.1128/jb.90.5.1238-1250.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis B D, Mingioli E S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950;60:17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devine D A, Roberts A P. K1, K5 and O antigens of Escherichia coli in relation to serum killing via the classical and alternative complement pathways. J Med Microbiol. 1994;41:139–144. doi: 10.1099/00222615-41-2-139. [DOI] [PubMed] [Google Scholar]
- 15.Erbing C, Svensson S, Hammarstrom S. Structural studies on the O-specific side-chains of the cell wall lipopolysaccharide from E. coli O75. Carbohydr Res. 1975;44:259–265. doi: 10.1016/s0008-6215(00)84169-5. [DOI] [PubMed] [Google Scholar]
- 16.Gemski P, Cross A S, Sadoff J C. K1 antigen-associated resistance to the bactericidal activity of serum. FEMS Microbiol Lett. 1980;9:193–197. [Google Scholar]
- 17.Grossman N, Joiner K A, Frank M M, Leive L. C3b binding but not its breakdown is affected by the structure of the O-antigen polysaccharide in lipopolysaccharide from Salmonella. J Immunol. 1986;136:2208–2212. [PubMed] [Google Scholar]
- 18.Grossman N, Schmetz M A, Foulds J, Klima E, Jimenez-Lucho V, Leive L, Joiner K A. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J Bacteriol. 1987;169:856–863. doi: 10.1128/jb.169.2.856-863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagberg L, Hull R, Hull S, Falkow S, Freter R, Svanborg Edén C. Contribution of adhesion to bacterial persistence in the mouse urinary tract. Infect Immun. 1983;40:265–272. doi: 10.1128/iai.40.1.265-272.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haraguchi G E, Ulrich Z, Jann B, Jann K, Hull R A, Hull S. Genetic characterization of the O4 polysaccharide gene cluster from Escherichia coli. Microb Pathog. 1991;10:351–361. doi: 10.1016/0882-4010(91)90080-t. [DOI] [PubMed] [Google Scholar]
- 21.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hull, S. I., K. L. Yoas, and R. A. Hull. Unpublished data.
- 23.Jann B, Reske K, Jann K. Heterogeneity of lipopolysaccharides. Analysis of chain length by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Eur J Biochem. 1975;60:239–246. doi: 10.1111/j.1432-1033.1975.tb20996.x. [DOI] [PubMed] [Google Scholar]
- 24.Jann B, Shashkov A S, Kochanowski H, Jann K. Structural comparison of the O4 specific polysaccharides from E. coli O4:K6 and E. coli O4:K52. Carbohydr Res. 1993;248:241–250. doi: 10.1016/0008-6215(93)84131-o. [DOI] [PubMed] [Google Scholar]
- 25.Jann K, Jann B. Polysaccharide antigens of Escherichia coli. Rev Infect Dis. 1987;9:516–526. doi: 10.1093/clinids/9.supplement_5.s517. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X-M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:2285–2292. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 27.Joiner K A, Grossman N, Schmetz M, Leive L. C3 binds preferentially to long-chain lipopolysaccharide during alternative pathway activation by Salmonella montevideo. J Immunol. 1986;136:710–715. [PubMed] [Google Scholar]
- 28.Joiner K A. Complement evasion by bacteria and parasites. Annu Rev Microbiol. 1988;42:201–230. doi: 10.1146/annurev.mi.42.100188.001221. [DOI] [PubMed] [Google Scholar]
- 29.Joiner K A. Role of complement in infectious diseases. In: Ross G D, editor. Immunobiology of the complement system: an introduction for research and clinical medicine. Orlando, Fla: Academic Press, Inc.; 1986. pp. 183–195. [Google Scholar]
- 30.Joiner K A, Hammer C H, Brown E J, Cole R J, Frank M M. Studies on the mechanism of bacterial resistance to complement-mediated killing. I. Terminal complement components are deposited and released from Salmonella minnesota S218 without causing bacterial death. J Exp Med. 1982;155:797–804. doi: 10.1084/jem.155.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kushner S. Useful host strains and techniques for recombinant DNA experiments. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1190–1219. [Google Scholar]
- 32.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Lesse A J, Campagnari A A, Bittner W E, Apicella M A. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Immunol Methods. 1990;126:109–117. doi: 10.1016/0022-1759(90)90018-q. [DOI] [PubMed] [Google Scholar]
- 34.Leying H, Suerbaum S, Kroll H-P, Stahl D, Opferkuch W. The capsular polysaccharide is a major determinant of serum resistance in K-1-positive blood culture isolates of Escherichia coli. Infect Immun. 1990;58:222–227. doi: 10.1128/iai.58.1.222-227.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukomski S, Hull R A, Hull S I. Identification of the O antigen polymerase (rfc) gene in Escherichia coli O4 by insertional mutagenesis using a nonpolar chloramphenicol resistance cassette. J Bacteriol. 1996;178:240–247. doi: 10.1128/jb.178.1.240-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 37.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navia J L, Riesenfeld J, Vann W R, Lindahl U, Roden L. Assay of N-acetylheparosan deacetylase with a capsular polysaccharide from Escherichia coli K5 as substrate. Anal Biochem. 1983;135:134–140. doi: 10.1016/0003-2697(83)90741-8. [DOI] [PubMed] [Google Scholar]
- 40.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ørskov I, Ørskov F. Escherichia coli and extra-intestinal infections. J Hyg Camb. 1985;95:551–575. doi: 10.1017/s0022172400060678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ørskov I, Ørskov F, Jann B, Jann K. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol Rev. 1977;41:667–710. doi: 10.1128/br.41.3.667-710.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palva E T, Mäkelä P H. Lipopolysaccharide heterogeneity in Salmonella typhimurium analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Eur J Biochem. 1980;107:137–143. doi: 10.1111/j.1432-1033.1980.tb04634.x. [DOI] [PubMed] [Google Scholar]
- 44.Petit C, Rigg G, Pazzani C, Smith A, Sieberth V, Stevens M, Boulnois G J, Jann K, Roberts I S. Analysis of region 2 of the Escherichia coli K5 capsule gene cluster: a region encoding proteins for the biosynthesis of the K5 polysaccharide. Mol Microbiol. 1985;17:611–620. doi: 10.1111/j.1365-2958.1995.mmi_17040611.x. [DOI] [PubMed] [Google Scholar]
- 45.Petit C, Rigg G P, Pazzani C, Smith A, Sieberth V, Stevens M, Boulnois G, Jann K, Roberts I S. Region 2 of the Escherichia coli K5 capsule gene cluster encoding proteins for the biosynthesis of the K5 polysaccharide. Mol Microbiol. 1995;17:611–620. doi: 10.1111/j.1365-2958.1995.mmi_17040611.x. [DOI] [PubMed] [Google Scholar]
- 46.Pluschke G, Mayden J, Achtman M, Levine R P. Role of the capsule and the O antigen in resistance of O18:K1 Escherichia coli to complement killing. Infect Immun. 1983;42:907–913. doi: 10.1128/iai.42.3.907-913.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russo T A, Singh G. An extraintestinal, pathogenic isolate of Escherichia coli (O4/K54/H5) can produce a group 1 capsule which is divergently regulated from its constitutively produced group 2, K54 capsular polysaccharide. J Bacteriol. 1993;175:7617–7623. doi: 10.1128/jb.175.23.7617-7623.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russo T A, Sharma G, Brown C R, Campagnari A A. Loss of the O4 antigen moiety from the lipopolysaccharide of an extraintestinal isolate of Escherichia coli has only minor effects on serum sensitivity and virulence in vivo. Infect Immun. 1995;63:1263–1269. doi: 10.1128/iai.63.4.1263-1269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 50.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schweizer H P. The pUC18CM plasmids: a chloramphenicol resistance gene cassette for site-directed insertion and deletion mutagenesis in Escherichia coli. BioFeedback. 1990;8:614–616. [PubMed] [Google Scholar]
- 52.Sieberth V, Rigg G P, Roberts I S, Jann K. Expression and characterization of UDPGlc dehydrogenase (KfiD), which is encoded in type-specific region 2 of the Escherichia coli K5 capsule genes. J Bacteriol. 1995;177:4562–4565. doi: 10.1128/jb.177.15.4562-4565.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 54.Stevens P, Huang S N-Y, Welch W D, Welch W L. Restricted complement activation by Escherichia coli with the K-1 capsular serotype: a possible role in pathogenicity. J Immunol. 1978;121:1216–1217. [PubMed] [Google Scholar]
- 55.Svanborg-Edén C, Hagberg L, Hull R, Hull S, Magnusson K-E, Ohman L. Bacterial virulence versus host resistance in the urinary tracts of mice. Infect Immun. 1987;55:1224–1232. doi: 10.1128/iai.55.5.1224-1232.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svanborg-Edén C, Hagberg L, Briles D, McGhee J, Michalek S. Susceptibility to Escherichia coli urinary tract infection and LPS responsiveness. In: Skamene E, editor. Genetic control of host resistance to infection and malignancy. New York, N.Y: Alan R. Liss, Inc.; 1985. pp. 385–391. [Google Scholar]
- 57.Svanborg-Edén C, Hagberg L, Hanson L A, Korhonen T, Leffler H, Olling S. Target cell specificity of wild type E. coli and mutants and clones with genetically defined adhesins. Prog Food Nutr Sci. 1983;7:75–89. [PubMed] [Google Scholar]
- 58.Taylor P W. Bactericidal and bacteriolytic activity of serum against gram-negative bacteria. Microbiol Rev. 1983;47:46–83. doi: 10.1128/mr.47.1.46-83.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor P W, Messner P, Parton R. Effect of the growth environment on cell envelope components of Escherichia coli in relation to sensitivity to human serum. J Med Microbiol. 1981;14:9–19. doi: 10.1099/00222615-14-1-9. [DOI] [PubMed] [Google Scholar]
- 60.Taylor P W. Measurement of the bactericidal activity of serum. In: Sussman M, editor. The virulence of Escherichia coli. London, England: Academic Press Ltd.; 1985. pp. 445–457. [Google Scholar]
- 61.Timmis K N, Boulnois G J, Bitter-Suermann D, Cabello F C. Surface components of Escherichia coli that mediate resistance to the bactericidal activities of serum and phagocytes. Curr Top Microbiol Immunol. 1985;118:197–218. doi: 10.1007/978-3-642-70586-1_11. [DOI] [PubMed] [Google Scholar]
- 62.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharide in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 63.Valvano M A. Pathogenicity and molecular genetics of O-specific side-chain lipopolysaccharides of Escherichia coli. Can J Microbiol. 1992;38:711–719. doi: 10.1139/m92-117. [DOI] [PubMed] [Google Scholar]
- 64.Vermeulen C, Cross A, Byrne W R, Zollinger W. Quantitative relationship between capsular content and killing of K1-encapsulated Escherichia coli. Infect Immun. 1988;56:2723–2730. doi: 10.1128/iai.56.10.2723-2730.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]