Abstract
Haemophilus ducreyi is unable to synthesize heme and must acquire it from its only known host, humans. We cloned and sequenced a gene encoding an outer membrane receptor for heme. It was designated tdhA (for TonB-dependent heme receptor A) since it was related by sequence homology to the family of TonB-dependent receptors. TdhA was strikingly similar to open reading frame HI0113 from the genome of Haemophilus influenzae Rd and also shared homology with five other heme receptors, including HxuC, HemR, HmuR, ChuA, and ShuA, from gram-negative bacteria. An Escherichia coli hemA tonB mutant strongly expressing H. ducreyi tdhA grew on low levels of heme as a source of heme only when an intact H. ducreyi Ton system plasmid was present, formally demonstrating functional TonB dependence. tdhA was expressed poorly in vitro by H. ducreyi and only under conditions of heme limitation. A survey of H. ducreyi revealed that all tested strains but one synthesized small amounts of TdhA in vitro under heme-limiting conditions. Surprisingly, an isogenic mutant of tdhA as well as its parent, 35000, both required the same high levels of heme for growth (50 μg/ml [77 μM] on agar medium). This result, together with previous findings, suggests that in vitro, the uptake of heme by H. ducreyi is mediated by a TonB- and TdhA-independent mechanism, possibly diffusion.
Haemophilus ducreyi is the etiologic agent of chancroid, a genital ulcer disease transmitted by sexual contact (reviewed in references 1 and 49). Chancroid has gained importance recently because it has been implicated as an independent risk factor for the heterosexual transmission of human immunodeficiency virus in Africa (18, 24, 35, 49, 50).
H. ducreyi is characterized by its fastidious and slow-growing nature. H. ducreyi is unable to synthesize heme and must obtain heme compounds from its only known host, humans. In vitro, H. ducreyi is characterized by an inordinately high heme requirement (50 μg/ml; 77 μM) relative to other bacterial species which express heme receptors.
Host iron is sequestered by several mechanisms, and invading bacteria must gain access to these iron sources to survive and initiate disease. Some bacteria utilize iron-scavenging siderophore systems; others directly bind host iron or heme- or iron-containing compounds such as transferrin, lactoferrin, heme, and hemoglobin (9, 31). H. ducreyi does not make siderophores (26). Since H. ducreyi requires both heme and iron, differentiation between the requirements for both of these can be problematic.
It is well-established that hemoglobin, hemoglobin complexed to haptoglobin, catalase, and free hemin are able to fulfill the heme requirement of H. ducreyi (2, 26, 48). Extracellular heme is complexed by human serum hemopexin or human serum albumin. Whereas it is clear that H. ducreyi is unable to utilize heme-hemopexin (11, 20), it is unclear whether it can utilize heme-albumin (14, 26). With the possible exception of the H. influenzae system, the form(s) of heme recognized by heme receptors of gram-negative bacteria is not well understood. Heme sources for H. ducreyi could be released from host cells by the action of the H. ducreyi hemolysin (33, 34, 47) or cytotoxin (7, 25, 37). In acquiring heme from host heme sources, H. ducreyi also fulfills its iron requirement since the heme moiety contains iron.
Most receptors for host iron-binding compounds and siderophore receptors belong to a family of outer membrane receptors designated as TonB dependent because they require the cytoplasmic membrane protein TonB for internalization of bound ligand (3, 5, 43). TonB together with its accessory proteins ExbB and ExbD is believed to comprise a system for the transfer of energy from the cytoplasmic membrane to outer membrane receptors (4).
Initial studies from this laboratory on the heme and iron acquisition systems of H. ducreyi identified a 100-kDa hemoglobin binding protein termed HgbA (12). Molecular cloning of the hgbA locus revealed a deduced amino acid sequence similar to that of the family of TonB-dependent outer membrane receptors (13, 39). An hgbA isogenic mutant of H. ducreyi is unable to bind or utilize hemoglobin as a source of heme but can utilize free hemin, implying that HgbA is not required for the utilization of hemin. The hemoglobin acquisition system of H. ducreyi was reconstituted in an Escherichia coli heme biosynthetic mutant (IR754 hemA tonB aroB) by expressing hgbA from one plasmid and expressing the H. ducreyi Ton system from a second plasmid (14), which implied that the intact heme moiety is internalized. H. ducreyi isogenic mutants in either the Ton system or hgbA are unable to utilize hemoglobin. In contrast, growth by the H. ducreyi Ton system mutant in heme medium is not impaired relative to that of its parent, suggesting a TonB-independent system for heme uptake (14). The requirement for high levels of heme by H. ducreyi in vitro is also consistent with a TonB-independent mechanism of heme uptake.
The objective of this study was to determine if H. ducreyi contains a heme receptor. A heme receptor of H. ducreyi was cloned by transforming a plasmid library into an E. coli hemA strain expressing the H. ducreyi Ton system and plating on heme agar. We have named this H. ducreyi heme receptor TdhA for TonB-dependent heme receptor A.
MATERIALS AND METHODS
Strains and media.
Bacterial strains used in this study are shown in Table 1. For routine growth, H. ducreyi was maintained on chocolate agar plates prepared by following the instructions for gonococcal medium base (GCB) (Difco). Fetal bovine serum (FBS) was not utilized in chocolate agar plates used for routine passage. Plates containing the various heme sources were prepared with GCB and IsoVitaleX (BBL) enrichments (termed GCB-I). Human hemoglobin (catalog no. H7379; Sigma Chemical Co., St. Louis, Mo.) was dissolved in phosphate-buffered saline at a concentration of 10 mg/ml and rocked at room temperature for 2 h or overnight at 4°C prior to filter sterilization. Heme stock solutions (10 mg/ml) were made by dissolving bovine hemin chloride (Sigma) in 0.1 N NaOH and used without further sterilization.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli K-12 | ||
| EB53 | hemA aroB rpoB | 40 |
| IR754 | EB53 but tonB::Kanr | 42 |
| DH5αMCR | recA1 mcrA mcrBC | Bethesda Research Labs |
| H. ducreyi | ||
| 35000 | Wild type, parent strain; can utilize low levels of hemoglobin and high levels of heme | Stanley Spinola, Indiana University |
| FX504 | 35000 hgbA::CAT, unable to utilize hemoglobin | 13 |
| FX514 | 35000 Δ(exbB exbD tonB)::CAT, unable to utilize hemoglobin | 14 |
| FX515 | 35000 ΔtdhA::CAT, can utilize low levels of hemoglobin and high levels of heme | This work |
| Plasmids | ||
| pBluescript(pBS) | ColE1 replicon, Apr, high-copy cloning vector | Stratagene |
| pNC 40 | ColE1 replicon, Apr Cmr, source of CAT cassette for making insertional mutations | 45 |
| pMCL 200 | p15A replicon, Cmr, low-copy cloning vector | 30 |
| pHP 45 | Omega interposon, source of Strr/Spcr cassette for making insertional mutations | 36 |
| pUNCH 579 | hgbA in pBS, Apr, expresses functional hgbA from own promoter | 13 |
| pUNCH 563 | exbB exbD tonB in pBS, Apr, expresses functional H. ducreyi Ton system from own promoter | 14 |
| pUNCH 569 | exbB exbD tonB, 3.5-kb XbaI-XhoI of pUNCH 563 in pMCL200 | This work |
| pUNCH 1204 | Original tdhA clone, 2.9-kb Sau3A insert in pBluescript which confers ability of IR754 pUNCH 569 to grow on heme | This work |
| pUNCH 663 | ΔtdhA::CAT, CAT cassette of pNC40 (1-kb BglII, fill-in) between two MluI sites of pUNCH 1204 (tdhA) | This work |
| pUNCH 665 | ΔtdhA::Omega, 2-kb SmaI from Omega in between two MluI sites of pUNCH 1204 (tdhA) | This work |
| pUNCH 667 | tdhA, 2.9-kb XbaI-XmaI of pUNCH 1204 in pMCL200 | This work |
| pUNCH 676 | tdhA and exbB exbD tonB, 4-kb XhoI-PvuI of pUNCH 563 cloned between the SalI and PvuI sites of pUNCH 667 | This work |
E. coli hemA mutants EB53 and IR754 were maintained on Luria-Bertani (LB) agar plates containing 25 μM δ-aminolevulonic acid (δALA) with antibiotic selection where appropriate. The following antibiotics were used at the concentrations indicated for E. coli: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 30 μg/ml; spectinomycin, 50 μg/ml; and streptomycin, 50 μg/ml.
Growth conditions for testing phenotypes of recombinant E. coli.
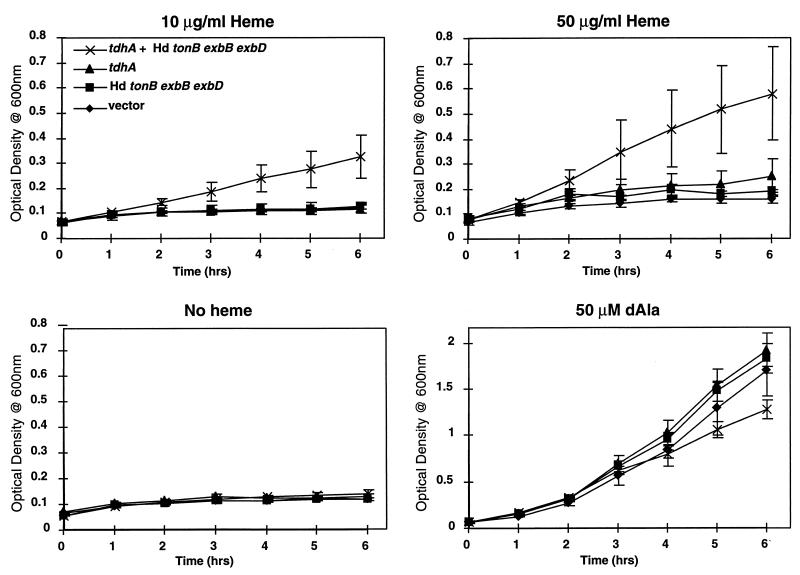
E. coli IR754 containing the H. ducreyi tdhA and/or Ton system plasmids was used to assess the ability of E. coli to utilize heme. After growth overnight at 37°C in LB broth with δALA and antibiotic selection, cultures were centrifuged and resuspended in LB medium with antibiotics but without δALA. Cultures were grown for 1 h to deplete intracellular heme stores and then diluted to an optical density at 600 nm of 0.075 in medium containing indicated amounts of heme or δALA (see Fig. 4).
FIG. 4.
Growth of recombinant E. coli IR754 expressing the H. ducreyi heme receptor. E. coli IR754 (hemA tonB aroB) containing pUNCH 676 (tdhA and exbB exbD tonB), pUNCH 667 (tdhA), pUNCH 569 (exbB exbD tonB), or pMCL200 (vector control) was prestarved for heme and inoculated into LB medium containing the indicated heme sources or heme precursors with selection. Note that the scale for the growth curve with the addition of δALA (dAla) differs from that for the other three panels.
To determine if internalized hemin could satisfy the iron requirement of IR754 containing the various relevant plasmids (see plasmid constructions below), a disc assay was utilized. IR754 containing either pUNCH 676 (tdhA and exbB exbD tonB), pUNCH 667 (tdhA), or pUNCH 569 (exbB exbD tonB) was depleted of heme as described above, 50 μl of this suspension was spread onto agar medium with or without chelators, sterile discs were placed on the agar surface, and heme (10 μl of a 10-mg/ml solution) was pipetted onto the discs. After 24 h at 37°C, growth was recorded. The agar medium used was LB containing 500 μM 2,2-dipyridal. Alternatively, nutrient agar containing either 50 to 200 μM 2,2-dipyridal or 100 μM apoferrichrome A was used.
Growth conditions for testing phenotypes of H. ducreyi.
H. ducreyi strains were heme starved by anaerobic growth on GCB-I plates without a heme source (14, 39). This condition reduced the possibility of heme carryover or differences in intracellular heme stores among the various strains. Heme-starved H. ducreyi inocula were standardized spectrophotometrically, and 10-fold dilutions were made for each strain in a microtiter tray. A Steers replicator was used to inoculate agar medium containing dilutions of each heme source. Thus, the inocula and heme sources were present in a checkerboard titration. The heme sources included free hemin, hemoglobin, human serum albumin at 50% heme saturation (assuming a single heme binding site per molecule), pooled normal human serum (pNHS) containing various dilutions of heme, heme containing various dilutions of pNHS, catalase, and FBS containing heme. Plates were incubated at 35°C in 5% CO2 for 48 h.
Outer membrane isolation, analysis, immunoblotting, and heme-affinity purification.
Large-scale cultures of H. ducreyi were performed in Fernbach flasks with 1 liter of GCB-I broth containing 5% FBS and 2 μg of heme per ml (heme limiting) or 50 μg of heme per ml (heme replete)(12). Large-scale cultures of E. coli were performed in Fernbach flasks containing LB broth and appropriate antibiotics. For induction of iron-stressed conditions in E. coli, 2,2-dipyridal (200 μM, final concentration) was added at early log phase, and growth was allowed to continue for 2 h. Outer membranes were harvested as previously described (12). Protein concentrations were determined by using the bicinchoninic acid kit from Pierce (Rockford, Ill.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as previously described (12).
The antiserum to TdhA was produced as follows. A predicted antigenic peptide was identified from the deduced amino acid sequence of tdhA and synthesized by standard fluoroenylmethoxycarbonyl (Fmoc) chemistry by the Microchemical Core Facility, Department of Microbiology and Immunology, University of North Carolina at Chapel Hill. The synthetic peptide sequence was CEFVRRQDRSPTDQD (C functioned for sulfhydral coupling and is not part of the TdhA protein sequence). After purification by reverse-phase high-performance liquid chromatography, the sequence was confirmed by fast atom bombardment-mass spectrophotometry. The peptide was conjugated to keyhole limpet hemocyanin, and rabbits were immunized a total of five times with 0.5 mg of peptide conjugate per immunization. Freunds complete adjuvant was used for the first immunization and incomplete for the remainder. Immune serum was affinity purified with the TdhA peptide immobilized on a thiopropyl agarose column as previously described (12).
Heme-affinity purification of recombinant TdhA was performed by the procedures of Lee (27, 28) and Yamomoto et al. (51). The difference between these two methods is that in the former method heme agarose from Sigma containing a C10 spacer was used and in the latter method heme agarose prepared with a C2 spacer was used. Outer membranes were used for affinity purification of recombinant TdhA (rTdhA) from recombinant E. coli strains (29).
Chemicals and reagents.
All chemicals and reagents, unless otherwise noted, were obtained from Sigma Chemical Co.
DNA manipulations.
Standard recombinant DNA methods were used as described in Sambrook et al. (38) or following the manufacturer’s instructions. The heme receptor was cloned by using a partially digested Sau3AI library from strain 35000 (14). To avoid restriction in subsequent hosts, E. coli DH5αMCR was transformed with the ligation mixture, the resulting ampicillin-resistant transformants were pooled, and plasmid DNA was isolated by the Magic Miniprep procedure (Promega). This plasmid pool was electroporated into IR754 (pUNCH 569, exbB exbD tonB), and selection was performed on LB agar containing heme (10 μg/ml), ampicillin (100 μg/ml) and chloramphenicol (30 μg/ml). Nine clones were obtained in this fashion, and they were related based on restriction analysis of sites within the inserts (data not shown).
Plasmid constructions.
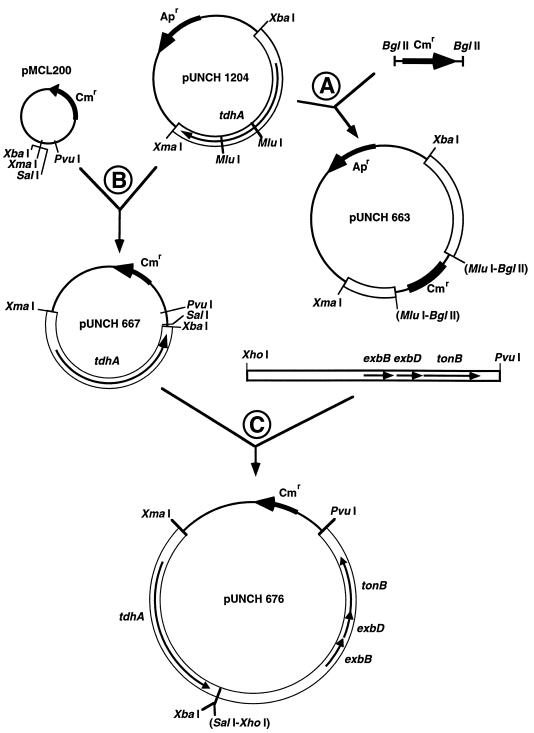
Diagrams of the cloning steps for construction of the new plasmids used in this study are shown in Fig. 1. The tdhA gene was disrupted in two separate deletion/insertion clones. The 542-bp MluI fragment in the coding sequence of tdhA was removed and replaced by blunt end ligation with either a chloramphenicol acetyltransferase (CAT) gene from pNC 40 to form pUNCH 663 (ΔtdhA::CAT) (step A of Fig. 1) or the omega interposon from pHP 45 to form pUNCH 665 (ΔtdhA::Omega). To reduce expression of tdhA and potential leakiness caused by overexpression, the XmaI-to-XbaI insert containing tdhA was moved to low-copy vector pMCL 200, resulting in pUNCH 667 (step B of Fig. 1). Since high-copy expression of the H. ducreyi Ton system resulted in reduced growth, the H. ducreyi Ton system was subcloned into low-copy plasmid pUNCH 667(tdhA) to form pUNCH 676 (tdhA and exbB exbD tonB) (step C of Fig. 1). This was accomplished by isolating the XhoI-PvuI fragment of pUNCH 563 (exbB exbD tonB) and ligating it to SalI-PvuI-digested pUNCH 667 (tdhA) (SalI and XhoI have compatible cohesive ends).
FIG. 1.
Construction of relevant H. ducreyi tdhA and Ton system (exbB exbD tonB) clones. Thick black arrows indicate antibiotic resistance markers; Apr and Cmr indicate the ampicillin resistance and chloramphenicol resistance markers, respectively; thin black arrows indicate relevant genes. Only the restriction sites used are shown; restriction sites in parentheses were predicted to be lost during cloning. The details of cloning steps A, B, and C are described in Materials and Methods.
DNA sequencing and analysis.
DNA sequence analysis was performed at the University of North Carolina at Chapel Hill Automated Sequencing Facility by utilizing Taq terminator chemistry. Both strands were completely sequenced and assembled with AssemblyLIGN software (IBI). Fur boxes were identified by using the Fur consensus oligonucleotide sequence GATAATGATAATCATTATC and the program DNA Strider. Amino acid alignments were done with the programs PILEUP (17) and SeqVu 1.01 (Garvin Institute, Sydney, Australia). The parameter settings for PILEUP were Blosum, 62; gap weight, 12; and gap length weight, 4. This alignment was then imported into SeqVu.
Construction of an H. ducreyi tdhA mutant.
An isogenic mutant (FX515 ΔtdhA::CAT [Table 1]) was constructed by allelic replacement of the wild-type locus of strain 35000 with the mutation in pUNCH 663 (ΔtdhA::CAT). Electroporation of strain 35000 was done with 1 μg of XbaI- and XhoI-digested pUNCH 663 (ΔtdhA::CAT), and selection was performed with 1 μg of chloramphenicol per ml in chocolate agar plates as previously described (13, 19). Chloramphenicol-resistant (Cmr) isolates were tested for the inability to make TdhA by Western blot analysis.
Southern blot analysis was used to confirm that a double crossover occurred in the generation of H. ducreyi mutant FX515 ΔtdhA::CAT. Chromosomal DNA was isolated, digested with NdeI and NruI, and subjected to electrophoresis and bidirectional transfer. The two blots were probed with either the insert from pUNCH 1204 (tdhA) or the BglII fragment from pNC 40 (CAT cassette). Digoxigenin-labeled, bound probe was detected with alkaline phosphatase-labeled antidigoxigenin antibody (Boehringer Mannheim) followed by detection with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate.
Nucleotide sequence accession number.
The DNA and deduced amino acid sequences of tdhA have been submitted to GenBank and assigned accession no. AF052977.
RESULTS
Cloning a TonB-dependent heme receptor.
Previously, we reconstructed the hemoglobin acquisition system of H. ducreyi in E. coli IR754 (hemA tonB aroB) (14). This strain of E. coli cannot synthesize heme due to the hemA mutation and is unable to internalize heme since it lacks hemoglobin and heme receptors. It was observed that the hemoglobin receptor required its homologous Ton system in multicopy in order to remove the heme from hemoglobin and to transport the heme across the outer membrane, demonstrating its TonB dependence. Several TonB-dependent heme receptors from enteric gram-negative bacteria have been cloned in E. coli strains containing mutations in heme and/or enterobactin synthesis by plating libraries on hemin agar. In those cases, the heterologous TonB-dependent heme receptor was able to function with the native E. coli Ton system. We reasoned that a putative TonB-dependent heme receptor from H. ducreyi, like the H. ducreyi hemoglobin receptor, would require its own Ton system. Thus, a partial Sau3AI library from H. ducreyi 35000 was transformed into E. coli IR754 (hemA tonB aroB) expressing the H. ducreyi Ton system from the compatible plasmid pUNCH 569 (exbB exbD tonB) (Table 1) and plated on low levels of heme (10 μg/ml) as the sole source of heme. Nine clones were obtained. To confirm that these clones required the H. ducreyi Ton system for growth on heme, plasmid DNA isolated from each clone was separately transformed into E. coli IR754 in the presence of pUNCH 569 (exbB exbD tonB) or pBluescript (vector control) and again plated onto heme agar plates. All nine grew in the presence of pUNCH 569 (exbB exbD tonB) but not pBluescript. This screen was designed to eliminate clones that allowed the heme to leak in nonspecifically or that directly complemented the hemA defect in IR754. One plasmid, pUNCH 1204 (tdhA), was chosen for further study (Table 1; Fig. 1).
DNA and deduced amino acid sequence of the H. ducreyi heme receptor tdhA.
The relevant region of pUNCH 1204 (tdhA) was sequenced and contained a single large open reading frame (ORF) of 2,217 bp which encoded a 739-amino-acid protein. A putative signal peptidase I cleavage site was identified 22 amino acids into TdhA, cleavage of which would result in a mature predicted protein of 81,758 Da. Consistent with the outer membrane localization, TdhA contained a carboxyl-terminal motif ending with a phenylalanine, which is found in the majority of integral outer membrane proteins (44). Beginning 69 and 45 nucleotides (nt) upstream of the tdhA start codon, sequences similar to −35 (TGATAA) and −10 (TATATT) consensus sequences were found; these sequences were separated by 18 nt. A putative ribosome-binding site (AAGGAA) beginning 13 nt upstream of the tdhA start codon was found. Three putative Fur boxes were found in the putative promoter region beginning 68, 74, and 77 nt upstream of the start codon. Each putative Fur box contained four or five mismatches and no gaps.
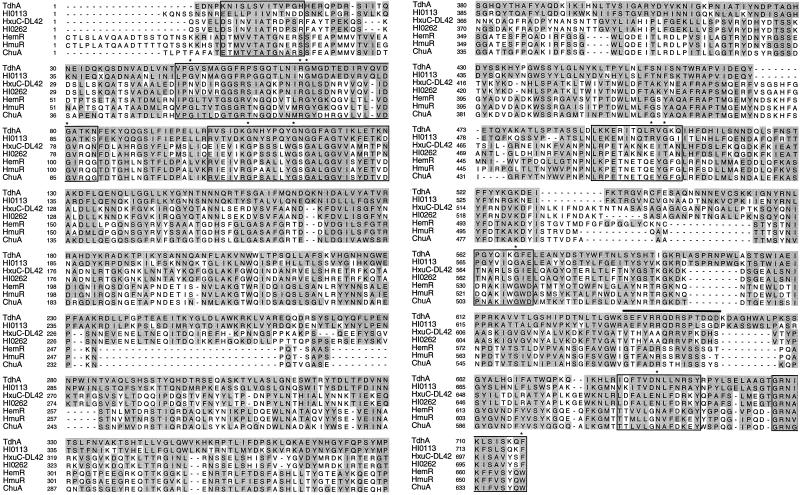
A search of the National Center for Biotechnology Information database with the TdhA sequence identified a number of related proteins. The most closely related proteins were a family of TonB-dependent heme receptors from other bacteria (Fig. 2). The amino acid sequence of H. ducreyi TdhA was most closely related to an unidentified ORF (HI0113) found in H. influenzae Rd. ORFs flanking tdhA were similar to HI0174 (5′) and HI0173 (3′) of the H. influenzae Rd genome sequence.
FIG. 2.
Comparison of H. ducreyi TdhA protein sequence with sequences from heme receptors from other gram-negative bacteria. The shaded boxes indicate residues that scored >75% by a GES matrix with respect to TdhA. The boxed regions indicate areas of conservation found in most TonB-dependent receptors (8). The asterisks indicate highly conserved residues present in TonB-dependent receptors (8). The overlined sequence indicates the residues used to make the peptide antiserum. HI0113, H. influenzae Rd ORF 0113 from the genome sequencing project (16); HxuC, heme-hemopexin binding protein from H. influenzae, HemR, Heme receptor from Yersinia enterocolitica; HmuR, heme receptor from Y. pestis; ChuA, heme receptor from enterohemorrhagic E. coli EDL933 O:157.
TdhA expression in H. ducreyi and E. coli.
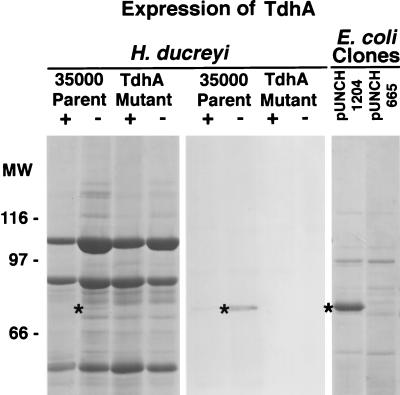
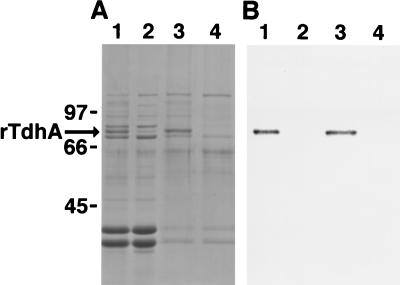
To identify the protein encoded by the tdhA locus, outer membrane proteins were isolated from H. ducreyi 35000, FX515 ΔtdhA::CAT, and E. coli IR754 containing either pUNCH 1204 (tdhA) or pUNCH 665 (ΔtdhA::Omega). After SDS-PAGE and Coomassie staining of the gel, parent strain 35000, but not FX515 ΔtdhA::CAT, expressed a 75-kDa protein that was barely detectable under low-heme growth conditions (Fig. 3). In contrast, in E. coli IR754, a relatively abundant comigrating band was expressed from pUNCH 1204 (tdhA) but not pUNCH 665 (ΔtdhA::Omega). A corresponding Western blot of the H. ducreyi samples probed with anti-TdhA peptide immunoglobulin G (IgG) indicated that these 75-kDa proteins were TdhA (Fig. 3).
FIG. 3.
SDS-PAGE and Western blotting with outer membranes from H. ducreyi or E. coli expressing recombinant TdhA. H. ducreyi strains were grown under heme-replete (+) or heme-limiting (−) conditions. E. coli cultures were grown in LB medium in the presence of excess iron. Outer membranes were isolated and subjected to SDS-PAGE (10% total polyacrylamide) and Coomassie staining (outer panels) or Western blotting (middle panel) with anti-TdhA peptide immunoglobulin G. The strains used were H. ducreyi parent strain 35000 (wild type), FX515 ΔtdhA::CAT, IR754 pUNCH 1204 (tdhA), and IR754 pUNCH 665 (ΔtdhA::Omega). Molecular weight markers (MW) are indicated in thousands. The asterisks indicate the position of the TdhA protein.
TdhA- and TonB-dependent heme utilization. (i) E. coli.
pUNCH 1204 (tdhA) and plasmid pUNCH 665 (ΔtdhA::Omega) were transformed into IR754 pUNCH 569 (exbB exbD tonB) and IR754 pMCL200 (vector control) and tested for the ability to grow on LB medium agar containing 10 μg of heme per ml. IR754 pUNCH 569 (exbB exbD tonB) was able to grow on heme agar when it contained pUNCH 1204 (tdhA) but not pUNCH 665 (ΔtdhA) (data not shown). E. coli IR754 required both pUNCH 1204 (tdhA) and pUNCH 569 (exbB exbD tonB) for growth on heme agar. If either plasmid pUNCH 1204 (tdhA) or pUNCH 569 (exbB exbD tonB) was replaced with a vector control, no growth occurred. E. coli EB53 (wild type at the tonB locus but hemA aroB) containing pUNCH 1204 (tdhA) did not grow on heme in the absence of pUNCH 569 (exbB exbD tonB) but did in its presence (data not shown). These results demonstrate the requirement for expression of both a functional TdhA heme receptor and a Ton system, both from H. ducreyi.
Compared to hemoglobin, heme is a relatively small molecule, which, in theory, could leak across the outer membrane if the outer membrane integrity of IR754 were disrupted due to expression of tdhA from pUNCH 1204. To reduce the level of tdhA expression and thereby minimize this possibility, low-copy plasmids, each containing tdhA and the H. ducreyi Ton system singly or in combination, were constructed (Table 1; Fig. 1). Growth curves of IR754 containing tdhA and Ton system plasmids singly or together were generated (Fig. 4). E. coli IR754 grew in heme broth only in the presence of the plasmid containing both the tdhA and Ton system; no growth was observed without heme. Thus, growth on heme was both TdhA and TonB dependent. All strains grew when δALA, which bypasses the hemA defect, was supplied, although a decreased rate of growth was observed in the strain expressing both tdhA and the Ton system gene products. In order to test for leakiness of the outer membrane to hydrophobic compounds as a potential explanation for growth on heme, discs containing crystal violet, erythromycin, or SDS were placed onto LB δALA agar inoculated with a lawn of IR754 containing tdhA and Ton system plasmids. The tdhA-expressing plasmids pUNCH 676 (tdhA and exbB exbD tonB) and pUNCH 667 (tdhA) showed zones of inhibition that were slightly larger than the vector control (pMCL200) for crystal violet and erythromycin (4 to 5 mm larger); no differences were observed for SDS. Zone sizes for all three of these compounds were identical for IR754 pUNCH 569 (exbB exbD tonB) and IR754 pMCL200 (vector). Thus, both strains which expressed tdhA were slightly leaky; however, only the strain which also expressed the H. ducreyi Ton system could grow on heme.
We determined whether internalized hemin could be utilized as a source of iron by IR754, pUNCH 667 (tdhA), and pUNCH 569 (exbB exbD tonB) by incorporating chelators (dipyridal or apoferrichrome A) in the medium and supplying heme on a disc. Growth was observed around the heme disc only in the absence of chelators, demonstrating that additional components may be necessary for efficient removal the iron from heme (data not shown).
(ii) H. ducreyi.
An H. ducreyi tdhA mutant was constructed to examine if heme acquisition was affected by mutagenesis of this locus. The parent strain H. ducreyi 35000 was electroporated with deletion mutant plasmid pUNCH 663 (ΔtdhA::CAT) (Table 1) and plated on chocolate agar containing chloramphenicol. One H. ducreyi transformant, FX515 ΔtdhA::CAT, was characterized. Southern blotting of chromosomal DNA confirmed that FX515 ΔtdhA::CAT contained the appropriately larger NruI-to-NheI fragment compared to parent 35000 (Fig. 1) (Southern blotting data not shown). This result indicated that FX515 ΔtdhA::CAT contained an allelic replacement of tdhA. The growth of H. ducreyi mutant FX515 ΔtdhA::CAT was compared to that of the parent strain, 35000, in several heme-containing media. These heme sources included free hemin, 50% heme-saturated heme-albumin, human hemoglobin, pNHS containing variable amounts of heme, catalase, and FBS. No differences in growth were noted (data not shown). Lack of a heme phenotype for mutant FX515 ΔtdhA::CAT was similar to that reported for Ton system mutant FX514 Δ(exbB exbD tonB)::CAT (14), with the exception that FX514 Δ(exbB exbD tonB)::CAT could not grow on hemoglobin.
Affinity purification of TdhA.
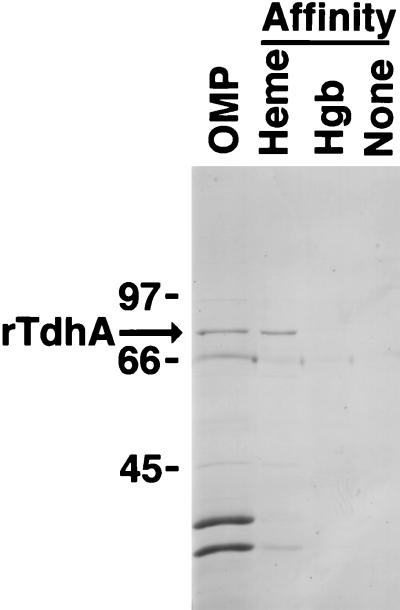
In several experiments, using a variety of published techniques, attempts to affinity purify a heme receptor from H. ducreyi were unsuccessful (data not shown). Since TdhA was expressed at low levels by H. ducreyi in vitro, but in relatively large amounts from pUNCH 1204 (tdhA) (Fig. 3), we attempted heme purification of rTdhA from E. coli. Outer membranes from IR754 containing pUNCH 1204 (tdhA) or pUNCH 665 (ΔtdhA::Omega) were subjected to heme agarose purification with use of a C2 spacer as described by Yamamoto et al. (51). Analysis of the heme-binding proteins by SDS-PAGE and Western blotting indicated that rTdhA was the predominant heme binding protein expressed from IR754 pUNCH 1204 (tdhA) (Fig. 5). Although other E. coli proteins also bound the heme matrix, TdhA was relatively enriched. Since heme is a hydrophobic molecule which could interact nonspecifically with hydrophobic residues found in membrane proteins, we investigated the specificity of TdhA binding to heme. We attempted to use free heme to compete for binding of TdhA to heme agarose. However, the insolubility of hemin at concentrations necessary to achieve heme excess precluded interpretation of these experiments. As an alternative to free heme competition to demonstrate specificity, we used additional alternative solid phases to affinity purify rTdhA (Fig. 6). Outer membranes were mixed with heme agarose, hemoglobin agarose, or agarose that did not contain any ligand and processed as described for heme-affinity purification (51). rTdhA bound well to heme agarose, bound poorly to hemoglobin agarose, and did not bind to agarose lacking a ligand (Fig. 6). Thus, binding of rTdhA to solid phase was specific for heme-containing agarose.
FIG. 5.
TdhA binds heme. Outer membranes from E. coli IR754 (pUNCH 1204, tdhA) and IR754 (pUNCH 665, ΔtdhA::Omega) grown under iron limitation were subjected to heme agarose-affinity purification (51) and analyzed by SDS-PAGE followed by Coomassie staining (A) or Western blotting (B) with anti-TdhA peptide immunoglobulin G. Lanes: 1 and 2, 10 μg of outer membrane protein from IR754 pUNCH 1204 (tdhA) and IR754 pUNCH 665 (ΔtdhA::Omega), respectively; 3 and 4, heme binding proteins from IR754 pUNCH 1204 (tdhA) and IR754 pUNCH 665 (ΔtdhA::Omega), respectively. The bands immediately above and below TdhA in lane 1 are iron-regulated E. coli proteins (compare with Fig. 6, lane OMP). Molecular sizes (in kilodaltons) are indicated on the left.
FIG. 6.
Binding specificity of rTdhA. Outer membranes from E. coli IR754 pUNCH 1204 (tdhA) grown under iron excess were subjected to heme agarose-, hemoglobin agarose (Hgb)-, or agarose (no ligand)-affinity purification (51) and analyzed by SDS-PAGE. In the original gel, there was a faint band of TdhA present in the hemoglobin agarose lane, but no band was present in the unsubstituted agarose (no ligand) lane. Molecular sizes (in kilodaltons) are indicated on the left.
Distribution of TdhA in other H. ducreyi strains.
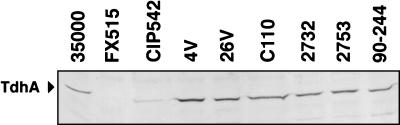
In order to determine if other strains of H. ducreyi expressed tdhA, Western blots of several geographically diverse laboratory and clinical isolates were probed with anti-TdhA peptide immunoglobulin G (Fig. 7). All strains, except the isogenic mutant FX515 ΔtdhA::CAT, expressed immunoreactive proteins with similar sizes when the strains were grown under low-heme conditions. In addition to the H. ducreyi strains presented in Fig. 7, an additional 26 of 27 strains expressed proteins with similar mobilities which were recognized by this antibody (data not shown).
FIG. 7.
Distribution of TdhA protein in H. ducreyi strains. Geographically diverse H. ducreyi strains grown in medium containing low heme were harvested, and total cellular protein (30 μg) was subjected to SDS-PAGE and Western blotting with anti-TdhA peptide immunoglobulin G. The names of strains are indicated above each lane.
DISCUSSION
In this paper we have presented three positive lines of evidence that H. ducreyi tdhA can function as a heme receptor: (i) reconstitution of a functional TonB-dependent heme uptake system in E. coli, (ii) sequence similarity of TdhA to five other TonB-dependent heme receptors, and (iii) binding of rTdhA to heme agarose. Additionally, TdhA is synthesized in H. ducreyi only under growth conditions of low heme, consistent with, but not proof of, a role in heme or iron uptake. Paradoxically, the tdhA mutant lacked a heme phenotype. Possible explanations for this paradox are that expression of tdhA in wild-type H. ducreyi in vitro is insufficient, that H. ducreyi lacks other components necessary for heme utilization present in E. coli, that TdhA functions in E. coli differently than in H. ducreyi, and that TdhA is not a receptor for heme.
Cloning and TonB dependence of tdhA.
Several TonB-dependent receptors for heme or hemoglobin have been cloned from gram-negative pathogens by utilizing a strategy based on the complementation of a heme synthesis defect (hemA) or iron acquisition (aroB ent) defect in E. coli (10, 21–23, 29, 40, 42, 43, 46). However, this powerful technique has the important limitation that if the outer membrane of the host is rendered permeable by expression of a heterologous protein, growth on heme may occur. Such leaky growth is characterized by TonB independence and the requirement for relatively high levels of heme. Using similar methods, we have cloned a gene, tdhA, encoding a novel heme receptor. Since TdhA required its homologous Ton system to support growth of E. coli IR754 on heme, we concluded that it was TonB dependent, at least in E. coli. Furthermore, in E. coli, TdhA-mediated heme uptake was seen at low (micromolar) heme concentrations, levels consistent with those of other TonB-dependent heme receptors. The leakiness observed for both of the tdhA-expressing strains (with and without the H. ducreyi Ton system) cannot account for the specific growth on heme by the strain expressing the Ton system, since the additional expression of the Ton system did not make the E. coli more leaky.
DNA and deduced amino acid sequence of the H. ducreyi heme receptor, tdhA.
Analysis of the region surrounding tdhA revealed that tdhA was the only gene implicated in heme transport. In several other systems involved in heme or iron transport, the TonB-dependent outer membrane receptor is flanked by genes encoding other members of the transport system such as a periplasmic binding protein, permease(s), and related ATPases (15, 32, 40, 41). The arrangement of such systems often constitute an operon. The DNA contiguous to tdhA did not contain sequences homologous to other genes involved in heme transport. The ORFs surrounding H. ducreyi tdhA were different from those surrounding the highly related H. influenzae Rd ORF HI0113. Heme utilization from heme or hemoglobin would also require transport across the cytoplasmic membrane. H. ducreyi and E. coli IR754 certainly must contain such a system, but it is yet to be described.
Examination of the deduced amino acid sequence of TdhA revealed homology to those of several TonB-dependent heme receptors from other genera. Homology was found in regions common to TonB-dependent receptors regardless of their ligand specificity, including a putative TonB box near the N terminus of tdhA and most of the so-called invariant residues. However, in addition to domains common to TonB-dependent receptors, there were certain residues or very short sequences scattered throughout the proteins that were common to all the heme receptors (Fig. 2). Longer stretches of sequence similarity were present among the TdhA and HxuC proteins from H. ducreyi and H. influenzae. We speculate that some of these common residues or sequences could be involved in heme binding or transport.
Expression of the TdhA protein is heme regulated in vitro.
In several experiments using H. ducreyi 35000, we found tdhA to be expressed only under heme-limiting conditions. In contrast to the major heme-regulated protein of H. ducreyi, HgbA, TdhA was a very minor protein in Coomassie-stained gels. We estimated that between 50 and 100 times more HgbA than TdhA is made (Fig. 3). Previously, we observed that a Ton system mutant of H. ducreyi produced increased amounts of HgbA and TdhA (labeled protein 3 in Fig. 5 of reference 14). These results are consistent with lower intracellular heme or iron levels in the Ton system mutant and subsequent derepression of Fur-regulated genes, as has previously been observed for E. coli Ton system mutants.
Three putative Fur boxes were observed in the promoter region of tdhA. We hypothesized that the poor expression of tdhA in H. ducreyi could be due to Fur repression. However, in E. coli IR754, tdhA expressed from high-copy plasmid pUNCH 1204 resulted in high levels of protein whether or not iron chelators were added to the medium used for growth (data not shown). Furthermore, experiments using E. coli fur mutants transformed with high-copy plasmid pUNCH 577 (H. ducreyi fur in pBluescript) (6) and low-copy plasmid pUNCH 667 (tdhA in pMCL200) failed to provide evidence for Fur regulation of tdhA (data not shown). Data suggesting possible H. ducreyi Fur regulation include the following: (i) that expression of both tdhA and hgbA is increased during growth in low levels of heme (iron), (ii) that both hgbA and tdhA genes contain putative Fur boxes in their promoters, and (iii) that the expression of both genes is increased in Ton system mutants. However, there is currently no direct evidence that Fur regulates these genes. Further studies are required to unravel this interesting regulatory network for tdhA, hgbA, and other heme regulated genes.
Distribution of TdhA among strains.
TdhA was found to be widely distributed in H. ducreyi by Western blotting with peptide-specific antisera. The conservation of TdhA among the various strains suggests that it serves an important role, yet its poor expression in vitro suggests that it may be expressed only under certain conditions in vivo. It has been reported that H. ducreyi has an extracellular and an intracellular life style and its heme requirement must be met in both environments. Upon lysis of host erythrocytes or nucleated cells by the H. ducreyi hemolysin or cytotoxin, it is likely that hemoglobin or heme could be acquired as extracellular sources of heme that could be taken up via their respective receptors. However, in an intracellular environment, it is unlikely that hemoglobin would serve as a source of heme. Thus we speculate that heme synthesis in the cytoplasm of eukaryotic cells could serve to fulfill the heme or iron requirement of intracellular H. ducreyi. Within this intracellular environment tdhA could be expressed and utilized to exploit this heme source. Studies are underway to address this possibility.
ACKNOWLEDGMENTS
We thank P. Frederick Sparling, members of the Sparling laboratory, Phillip Klebba, Eric Hansen, Leslie Cope, and Marcia Hobbs for helpful comments and for critiquing the manuscript and Annice Rountree for her expert technical assistance. We thank Igor Stojiljkovic for strains utilized in these experiments, S. Yamomoto for the generous gift of heme agarose, and Phillip Klebba for the gift of purified apoferrichrome A.
The work presented here was supported by a developmental grant from the North Carolina Sexually Transmitted Disease Infections Research Center (UO1-AI31496), University of North Carolina at Chapel Hill; WHO grants SDI/94/006 and R29-AI40263 to C.E.; and Public Health Service grant A126837 to P. Frederick Sparling.
REFERENCES
- 1.Albritton W L. Biology of Haemophilus ducreyi. Microbiol Rev. 1989;53:377–389. doi: 10.1128/mr.53.4.377-389.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albritton W L, Macklean I W, Bertram P D, Ronald A R. Haemin requirements in Haemophilus with special reference to H. ducreyi. In: Kilian M, Frederiksen W, Biberstein E L, editors. Haemophilus, Pasteurella and Actinobacillus. New York, N.Y: Academic Press, Inc.; 1981. pp. 75–82. [Google Scholar]
- 3.Biswas G D, Anderson J E, Sparling P F. Cloning, sequencing and genetic characterization of tonB-exbB-exbD genes of Neisseria gonorrhoeae. Mol Microbiol. 1997;24:169–179. doi: 10.1046/j.1365-2958.1997.3421692.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J Bacteriol. 1993;175:3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun V. Energy-coupled transport and signal transduction through the Gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 6.Carson S D B, Thomas C E, Elkins C. Cloning and sequencing of a Haemophilus ducreyi fur homolog. Gene. 1996;176:125–129. doi: 10.1016/0378-1119(96)00236-3. [DOI] [PubMed] [Google Scholar]
- 7.Cope L, Lumbley S, Latimer J, Klesney-Tait J, Stevens M, Johnson L, Purven M, Munson R J, Lagergard T, Radolf J, Hansen E. A diffusible cytotoxin of Haemophilus ducreyi. Proc Natl Acad Sci USA. 1997;94:4056–4061. doi: 10.1073/pnas.94.8.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelissen C N, Biswas G D, Tsai J, Paruchuri D K, Thompson S A, Sparling P F. Gonococcal transferrin-binding protein 1 is required for transferring utilization and is homologous to TonB-dependent outer membrane receptors. J Bacteriol. 1992;174:5788–5797. doi: 10.1128/jb.174.18.5788-5797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelissen C N, Sparling P F. Iron piracy: iron acquisition from transferrin by bacterial pathogens. Mol Microbiol. 1994;14:843–850. doi: 10.1111/j.1365-2958.1994.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 10.Daskaleros P A, Stoebner J A, Payne S M. Iron uptake in Plesiomonas shigelloides: cloning of the genes for the heme-iron uptake system. Infect Immun. 1991;59:2706–2711. doi: 10.1128/iai.59.8.2706-2711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins, C. Unpublished data.
- 12.Elkins C. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect Immun. 1995;63:1241–1245. doi: 10.1128/iai.63.4.1241-1245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkins C, Chen C J, Thomas C E. Characterization of the hgbA locus of Haemophilus ducreyi. Infect Immun. 1995;63:2194–2200. doi: 10.1128/iai.63.6.2194-2200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkins C, Totten P A, Olsen B, Thomas C E. Role of the Haemophilus ducreyi Ton system in internalization of heme from hemoglobin. Infect Immun. 1998;66:151–160. doi: 10.1128/iai.66.1.151-160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fecker L, Braun V. Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J Bacteriol. 1983;156:1301–1314. doi: 10.1128/jb.156.3.1301-1314.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischmann R D, Adams M D, White O, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 17.Genetics Computer Group. Wisconsin package version 9.1. University of Wisconsin, Madison; 1997. [Google Scholar]
- 18.Greenblatt R M, Lukehart S A, Plummer F A, Quinn T C, Critchlow C W, Ashley R L, DCosta L J, Ndinya A J O, Corey L, Ronald A R. Genital ulceration as a risk factor for human immunodeficiency virus infection. AIDS. 1988;2:47–50. doi: 10.1097/00002030-198802000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Hansen E J, Latimer J L, Thomas S E, Helminen M, Albritton W L, Radolf J D. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J Bacteriol. 1992;174:5442–5449. doi: 10.1128/jb.174.16.5442-5449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson M S, Pelzel S E, Latimer J, Muller-Eberhard U, Hansen E J. Identification of a genetic locus of Haemophilus influenzae type b necessary for the binding and utilization of heme bound to human hemopexin. Proc Natl Acad Sci USA. 1992;89:1973–1977. doi: 10.1073/pnas.89.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson D P, Payne S M. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol Microbiol. 1993;7:461–469. doi: 10.1111/j.1365-2958.1993.tb01137.x. [DOI] [PubMed] [Google Scholar]
- 22.Henderson D P, Payne S M. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression and homology to the family of TonB-dependent proteins. J Bacteriol. 1994;176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson D P, Payne S M. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect Immun. 1994;62:5120–5125. doi: 10.1128/iai.62.11.5120-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jessamine P G, Ronald A R. Chancroid and the role of genital ulcer disease in the spread of human retroviruses. Med Clin North Am. 1990;74:1417–1431. doi: 10.1016/s0025-7125(16)30488-6. [DOI] [PubMed] [Google Scholar]
- 25.Lagergard T, Purven M. Neutralizing antibodies to Haemophilus ducreyi cytotoxin. Infect Immun. 1993;61:1589–1592. doi: 10.1128/iai.61.4.1589-1592.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee B C. Iron sources for Haemophilus ducreyi. J Med Microbiol. 1991;34:317–322. doi: 10.1099/00222615-34-6-317. [DOI] [PubMed] [Google Scholar]
- 27.Lee B C. Isolation of an outer membrane hemin-binding protein of Haemophilus influenzae type b. Infect Immun. 1992;60:810–816. doi: 10.1128/iai.60.3.810-816.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee B C. Isolation of haemin-binding proteins of Neisseria gonorrhoeae. J Med Microbiol. 1992;36:121–127. doi: 10.1099/00222615-36-2-121. [DOI] [PubMed] [Google Scholar]
- 29.Mills M, Payne S M. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J Bacteriol. 1995;177:3004–3009. doi: 10.1128/jb.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano Y, Yoshida Y, Yamshita Y, Doga T. Construction of a series of pACYC-derived plasmid vectors. Gene. 1995;162:157–158. doi: 10.1016/0378-1119(95)00320-6. [DOI] [PubMed] [Google Scholar]
- 31.Otto B R, van Vught A M J J V, MacLaren D M. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 32.Ozenberger B, Nahlik M, McIntosh M. Genetic organization of multiple fep genes encoding ferric enterobactin transport functions in Escherichia coli. J Bacteriol. 1989;169:3638–3646. doi: 10.1128/jb.169.8.3638-3646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer K L, Grass S, Munson R S. Identification of a hemolytic activity elaborated by Haemophilus ducreyi. Infect Immun. 1994;62:3041–3043. doi: 10.1128/iai.62.7.3041-3043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer K L, Munson R S. Cloning and characterization of the genes encoding haemolysin of Haemophilus ducreyi. Mol Microbiol. 1995;18:821–830. doi: 10.1111/j.1365-2958.1995.18050821.x. [DOI] [PubMed] [Google Scholar]
- 35.Plummer F A, Wainberg M A, Plourde P, Jessamine P, DCosta L J, Wamola I A, Ronald A R. Detection of human immunodeficiency virus type 1 (HIV-1) in genital ulcer exudate of HIV-1-infected men by culture and gene amplification [letter] J Infect Dis. 1990;161:810–811. doi: 10.1093/infdis/161.4.810. [DOI] [PubMed] [Google Scholar]
- 36.Prentki P, Kirsch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 37.Purven M, Lagergard T. Haemophilus ducreyi, a cytotoxin-producing bacterium. Infect Immun. 1992;60:1156–1162. doi: 10.1128/iai.60.3.1156-1162.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Stevens M K, Porcella S, Klesney-Tait J, Lumbley S, Thomas S E, Norgard M V, Radolf J D, Hansen E J. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect Immun. 1996;64:1724–1735. doi: 10.1128/iai.64.5.1724-1735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stojiljkovic I, Hantke K. Haemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in Gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stojiljkovic I, Hantke K. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 42.Stojiljkovic I, Hwa V, Martin L D S, O’Gaora P, Nassif X, Heffron F, So M. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 43.Stojiljkovic I, Srinivasan N. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in neisseriae. J Bacteriol. 1997;179:805–812. doi: 10.1128/jb.179.3.805-812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Struyve M, Moons M, Tommassen J. Carboxyl-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 45.Thomas C E, Carbonetti N H, Sparling P F. Pseudo-transposition of a Tn5 derivative in Neisseria gonorrhoeae. FEMS Microbiol Lett. 1996;145:371–376. doi: 10.1111/j.1574-6968.1996.tb08603.x. [DOI] [PubMed] [Google Scholar]
- 46.Torres A G, Payne S M. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1996;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 47.Totten P A, Norn D V, Stamm W E. Characterization of the hemolytic activity of Haemophilus ducreyi. Infect Immun. 1995;63:4409–4416. doi: 10.1128/iai.63.11.4409-4416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Totten P A, Stamm W E. Clear broth and plate media for culture of Haemophilus ducreyi. J Clin Microbiol. 1994;32:2019–2023. doi: 10.1128/jcm.32.8.2019-2023.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trees D L, Morse S A. Chancroid and Haemophilus ducreyi: an update. Clin Microbiol Rev. 1995;8:357–375. doi: 10.1128/cmr.8.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wasserheit J N. Interrelationships between human immunodeficiency virus infection and other sexually transmitted diseases. Sex Transm Dis. 1991;19:61–77. [PubMed] [Google Scholar]
- 51.Yamamoto S, Hara Y, Tomochicka K-I, Shinoda S. Utilization of hemin and hemoglobin as iron sources by Vibrio parahaemolyticus and identification of an iron-repressible hemin-binding protein. Microbiol Lett. 1995;128:195–200. doi: 10.1111/j.1574-6968.1995.tb07522.x. [DOI] [PubMed] [Google Scholar]