Abstract
Hepatic fibrosis is a wound healing response that results in excessive extracellular matrix (ECM) accumulation in response to chronic hepatic injury. Signal transducer and activator of transcription 3 (STAT3) is an important transcription factor associated with the pathogenesis of liver fibrosis. Though a promising potential therapeutic target, there are no specific drug candidates for STAT3. Exosomes are extracellular vesicles generated by all cell types with a capacity to efficiently enter cells across different biological barriers. Here, we utilize exosomes as delivery conduit to specifically target STAT3 in liver fibrosis. Exosomes derived from clinical grade fibroblast-like mesenchymal stem cells (MSCs) were engineered to carry siRNA or antisense oligonucleotide (ASO) targeting STAT3 (iExosiRNA-STAT3 or iExomASO-STAT3). Compared to scrambled siRNA control, siRNA-STAT3, or ASO-STAT3, iExosiRNA-STAT3 or iExomASO-STAT3 showed enhanced STAT3 targeting efficiency. iExosiRNA-STAT3 or iExomASO-STAT3 treatments suppressed STAT3 levels and ECM deposition in established liver fibrosis in mice, and significantly improved liver function. iExomASO-Stat3 restored liver function more efficiently when compared to iExosiRNA-STAT3. Our results identify a novel anti-fibrotic approach for direct targeting of STAT3 with exosomes with immediate translational potential.
Keywords: engineered exosomes, liver fibrosis, mesenchymal stromal cells, STAT3
Introduction
Liver fibrosis is defined as excessive deposition of extracellular matrix (ECM) in the liver, replacing the functional parenchyma and severely impacting health worldwide1,2. Currently, there are no effective anti-fibrosis therapies3. Effective treatments for liver fibrosis urgently need innovative new approaches. Among the critical regulators of liver fibrosis, signal transducer and activator of transcription 3 (STAT3) signaling is centrally implicated, driving the activation of fibroblasts as well as hepatic stellate cells (HSCs), and their conversion into myofibroblast-like phenotype4–6. STAT3 is phosphorylated in response to cytokines and growth factors by Janus tyrosine kinases (JAK). Upon activation, phosphorylated STAT3 dimerizes and translocates to the nucleus to induce the transcription of cytokine-responsive downstream genes4. Cytokines that activate STAT3 include TGFβ17, the interleukin (IL)-6 family cytokines, and growth hormone (GH). STAT3 activation has been reported in liver fibrosis observed in patients and mouse models5,8, and indirect STAT3 inhibition using sorafenib or other inhibitors partially ameliorates CCl4-induced liver fibrosis in mice8,9. Although STAT3 has emerged as an important vulnerability for liver fibrosis, therapeutic targeting of STAT3 remains a challenge due to a lack of STAT3specific inhibitors10.
The current experimental anti-fibrotic therapies are directed against hepatic inflammation or the initiating injury. Direct targeting of activated stellate cells (myofibroblasts) and fibrogenic mediators remains an attractive therapeutic strategy against liver fibrosis11. Nanoparticle-based drug delivery is purported to circumvent limitations related to circulation half-life, and drug localization and biodistribution12. In this regard, exosomes are a class of extracellular vesicles with a size range of 40–150 nm and a lipid bilayer membrane reflecting the cells they originate from13,14. Exosomes can be readily collected from the conditioned media of cultured cells, including mesenchymal stromal cells (MSCs)15,16. Some investigators have also referred to exosomes as small EVs17.
Our previous study showed that exosomes engineered to deliver an siRNA payload (iExosomes) can specifically target oncogenic Kras in pancreatic cancer, inhibit tumor progression and significantly prolong overall survival of mice as a single agent18. GMP (Good Manufacturing Practice) certified production of exosomes from MSCs16 are FDA (Food & Drug Administration) approved as suitable for clinical testing, and this strategy is currently undergoing clinical testing (NCT03608631). Exosomes are important mediators of intercellular communications, enabling exchange of proteins and nucleic acids between cells and regulating intracellular signaling pathways. Exosomes interact with recipient cells, which subsequently can undergo gene expression and phenotypic changes16,18. The properties of exosomes in efficiently entering cells have underlined their potential role as novel drug delivery vehicles, especially in modulating gene expression in vivo18–22.
Here, we evaluated whether an exosomes-based therapeutic strategy could be employed to specifically target STAT3 in liver fibrosis. We engineered exosomes containing siRNA or antisense oligonucleotide (ASO) targeting STAT3 (siSTAT3) and tested their therapeutic role in an established model of liver fibrosis2,23,24. We show that therapeutic administration of STAT3-targeting iExosomes effectively suppress liver fibrosis in mice. Our studies identify that targeting STAT3 mRNA significantly impacts fibrotic liver transcriptome and provide a proof of concept for the use of exosomes-based therapies in the treatment of chronic fibrotic diseases.
Material and Methods
HSCs isolation and α-SMA staining
Mouse primary HSCs were isolated from female mice (Balb/c, 8-week-old; Jackson Laboratories) as previously described25. HSCs were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM, Corning) containing 20% FBS (Gemini) and 1% antibiotics (penicillin-streptomycin, PS; Corning). The culture-activated primary HSCs (on the day 7) were immunostained with Cy3-α-SMA antibody (Sigma, C6198, 1:200) overnight. Slides were coverslipped with Fluoroshield mounting medium containing DAPI (Sigma-Aldrich, F6057). HSC phase contrast images were taken using a BZ-X710 microscope (Keyence). Images of immunolabeled cells were taken with Zeiss LSM800 using ZEN software (Zeiss) at 200x magnification.
Real-time PCR analyses
Total RNA was isolated from liver with TRIzol™ (Invitrogen, 15596018) and High-Capacity cDNA Reverse Transcriptase Kit (Life Technology, 4368814) was used for cDNA synthesis according to the manufacturers’ instructions. Quantitative RT-PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems, 4385612). Total amount of mRNA of the target genes was normalized to Gapdh expression. The primer sequences were: Gapdh Forward (5’-CTGGAGAAACCTGCCAAGTA-3’), Reverse (5’- AAGAGTGGGAGTTGCTGTTG-3’). Stat3 Forward (5’-AGAACCTCCAGGACGACTTTG-3’), Reverse (5’-TCACAATGCTTCTCCGCATCT-3’); Col1a1 Forward (5’-CATGTTCAGCTTTGTGGACCT-3’), Reverse (5’GCAGCTGACTTCAGGGATGT-3’); Acta2 Forward (5’-GTCCCAGACATCAGGGAGTAA-3’), Reverse (5’- TCGGATACTTCAGCGTCAGGA-3’). Primer BLAST results detailing product size and target templates are listed in Table S1. Statistical analyses for variance were performed based on the ΔCt values. The fold change is presented and normalized to the control group, setting the control comparative group to 1.
Purification and electroporation of exosomes
MSCs derived from bone marrow were obtained from the Cell Therapy Laboratory at MD Anderson Cancer Center (MDACC) and cultured in αMEM (Corning) with 20% FBS, 1% PS, 1% non-essential amino acids (NEAA, Gibco) and 1% L-glutamine (Corning). Passage 4 to 6 MSCs were used for exosomes collections. Exosomes were purified by differential centrifugation processes according to our established protocols16,18. Briefly, cells were washed with 1X PBS (Corning) when grown to 70–80% confluency and cultured in serum-free media (αMEM with 1% L-glutamine, 1% PS, and 1% NEAA) for 48 hours. Supernatant was collected, followed by centrifugation at 800 x g for 5 minutes and 2,000 x g for 10 minutes, and filtered using a 0.2 μm filter (Thermo Fisher). Filtered supernatant was centrifuged at 4°C with the speed of 100,000 x g for 3 hours in a SW 32 Ti rotor (Beckman). The pellet was resuspended in 100 μL of 1X PBS. Exosomes concentration and size were verified by nanoparticle tracking analysis (NTA, NanoSight LM10, Malvern). One-dose mixture contained 1 billion of total exosomes according to NTA and 1 μg of siRNA or antisense oligos (ASO) in 100 μl of PlasmaLyte (Medline, BHL2B2544XH), while the other dose mixture contained 2 billion of total exosomes and 5 μg of siRNA or ASO in 100 μl of PlasmaLyte. 400 μl of the siRNA/ASO-exosomes mixture was loaded in the cuvette, electroporated at 400V, 125μF and ∞ ohms, and immediately transferred to ice.
The siSTAT3 sequence was: sense strand 5′- GUUGAAUUAUCAGCUUAAA-3′, anti-sense 5′- UUUAAGCUGAUAAUUCAAC-3′ (Sigma-Aldrich). The mASO Scrbl sequence was 5′- mG*mG*mC*mU*mA*C*U*A*C*G*C*mC*mG*mU*mC*mA-3’. The umASO STAT3 sequence was 5’-CTATTTGGATGTCAGC-3’. The mASO STAT3 sequence was 5′-mC*mU*mA*mU*mU*U*G*G*A*U*G*mU*mC*mA*mG*mC-3’. ‘m’ denotes 2’ O-methoxy-ethyl bases, * denotes phosphorothioate bonds. The siCntrl was obtained from Sigma-Aldrich (SIC001, Sigma-Aldrich). The ASOs were synthesized by Integrated DNA Technologies, Inc. The siRNA was designed with equal potential efficiency to target mouse and human STAT3. The ASO was also designed to target mouse and human STAT3, but with potentially lower efficacy against mouse STAT3 due to a 3 nucleotides mismatch with the mouse sequence.
Visualization of exosomes biodistribution in vivo
Mice were treated with CCl4 to induce liver fibrosis (as detailed below). For the biodistribution of MSC-derived exosomes, 8 billion exosomes labeled with XenoLight DiR (1,1′-dioctadecyltetramethyl indotricarbocyanine iodide, Perkin Elmer, catalog 125964) were injected i.p (100 μl) in healthy (sham) and fibrotic Balb/c mice according to our previous study16. Briefly, 1 μL DiR was added per 5 billion MSC-derived exosomes, followed by incubation at 37°C for 1 hour, and then at 4°C for 15 minutes, and then ultracentrifuged at 4°C for 3 hours at 40,000 x g in 10 ml of PBS16. The labeled exosomes (8 billion total) were then resuspended in 100 μl of PBS. For control samples (DiR only), the aforementioned labeling and wash procedure was performed in the absence of exosomes. Six hours after injection, the mice were euthanized, and tissues were harvested and imaged immediately. The fluorescence of organs was imaged with the In Vivo Imaging Systems (IVIS) 200 small animal imaging system (PerkinElmer) with the emission filter at 780 nm and the excitation filter at 710 nm. Fluorescence intensity was quantified using Living Image Software (PerkinElmer).
Mice
Liver fibrosis was induced in female Balb/c mice (8-weeks old unless noted otherwise; Jackson Laboratories) with i.p. injections of CCl4 (Sigma–Aldrich, 56–23-5) at a dosage of 10% (v/v) in 100 μl olive oil twice a week for 37 days. The mice were randomly assigned into groups 9 days after initiation of CCl4 treatment. Analysis was performed without blinding to group allocation. Control mice were administered with olive oil devoid of CCl4 (Fig. S2A). Mice were also administered with 1 μg siRNA/ASO of 1 billion engineered exosomes or 5 μg siRNA/ASO of 2 billion engineered exosomes i.p. in 100μl volume of PlasmaLyte (Medline), or 5 μg siRNA-STAT3/mASO-STAT3 alone i.p. in 100μl PBS every other day. The mice were euthanized within 24 hours of the last treatment. All protocols and procedures were approved by the MDACC Institute for Animal Care and Use Committee.
Detection of exosomes in liver sections
Six months old wild type mice (Balb/c) were intraperitoneally injected with CCl4 in olive oil at a dosage of 10% twice a week for 1.5 month to induce liver fibrosis. Prior to euthanasia, 100 μl of PKH67-labeled exosomes were administred intraperitoneally. Exosomes were labeled with PKH67 (Sigma-Aldrich) according to the manufacturer’s protocol without electroporation of siRNA/ASO. Organs were obtained from these mice 24 hours after injection and embeded in O.C.T mounting media. 5 μm cryostat sections of liver were immunostained with α-SMA antibody (Sigma Aldrich, A5228, 1:200), CD31 antibody (Dianova, DIA 310, 1:50), F4/80 antibody (Abcam, ab6640, 1:100), CD45 (Cell Signaling Technology 70257, 1:100), or collagen I (Southern Biotech 1310–01, 1:100) at 4°C overnight. The liver sections were incubated with secondary antibodies: Alexa Fluor 647 goat anti-mouse IgG (H+L) (Invitrogen, A21235, 1:400), Alexa Fluor 568 goat anti-rat lgG (H+L) (Invitrogen, A11077, 1:400), Alexa Fluor 594 goat anti-rabbit lgG (H+L) (Invitrogen A11037, 1:250), and Alexa Fluor 594 donkey anti-goat IgG (H+L) (Invitrogen A11058, 1:250) for 1 hour at room temperature, and they were washed three times with 1X PBS. Slides were coverslipped with Fluoroshield mounting medium containing DAPI. Representative images at 200x magnification were taken with confocal laser scanning microscope (Zeiss LSM800). The number of PKH+ exosomes per cell were quantified from 3 visual fields per liver section.
Visualization of labeled siSTAT3 and mASO STAT3 localization in liver tissue
Mice were treated with CCl4 to induce liver fibrosis (as detailed above in Mice section). Exosomes were electroporated with AF647-tagged siRNA and mASO (Integrated DNA Technologies) prior to injection. AF647-labeled iExosomes and AF647 tagged siRNA or mASO alone were injected i.p. into wild-type (WT) Balb/c mice and Balb/c mice with liver fibrosis. Sectioned liver tissues at a thickness of 10 μm were mounted with Fluoroshield mounting medium with DAPI (Sigma–Aldrich, F6057). Liver sections were imaged at 200x by confocal laser scanning microscope (Zeiss LSM800) and then quantified by counting the cells positive for AF647 (3 visual fields per liver section).
Sirius red staining and quantification
Formalin fixed and paraffin embedded liver sections (5 μm) were used for Sirius red staining. After being rinsed for three times and stained with Weigert’s haematoxylin for 8 min, the slides were counterstained by picrosirius red (Sigma–Aldrich, 365548) for 1 hour. A counting grid was used to analyze three independent Sirius red-stained sections from each mouse. The percent area of fibrosis was measured as previously described26.
Immunohistochemistry and immunofluorescence
Formalin fixed and paraffin embedded liver sections (5 μm) were processed with a heat-mediated antigen retrieval for 1 hour in Tris-EDTA (10mM Tris, 1 mM EDTA; pH 9.0). Liver sections were blocked for 1 hour with 4% Aurion cold water fish skin gelatin prior to overnight incubation with primary collagen I antibody (Southern Biotech, 1310–01, 1:200), vimentin antibody (Cell Signaling Technology, 5471, 1:200), phosphorylated STAT3 antibody (Tyr705, Cell Signaling Technology 9145, 1:100), and α-SMA antibody (Dako M0851, 1:100). The liver sections were incubated with secondary antibodies for 1 hour: biotinylated anti–goat (Vector Laboratories, BA9500, 1:400) and biotinlated anti-rabbit (Vector Laboratories, BA-1000–1.5, 1:400), reacted with ABC for 30 minutes, and then incubated with DAB. For immunofluorescence staining of pSTAT3 and α-SMA, liver sections were incubated with Alexa Fluor 488 goat anti-rabbit IgG (H+L) (Invitrogen A11034, 1:250) and Alexa Fluor 495 goat anti-mouse IgG (H+L) (Invitrogen A11032, 1:250). Slides were coverslipped with Fluoroshield mounting medium containing DAPI (Sigma-Aldrich, F6057).
Mouse livers were fixed at 4°C in 4% paraformaldehyde for 24 hours and equilibrated 4°C in 30% sucrose for 24 hours and changed fresh 30% sucrose for another 24 hours. Livers were then embedded in OCT compound, and frozen sections (5 μm) were blocked for one hour with 4% Aurion cold water fish skin gelatin prior to overnight incubation with anti–α-SMA–Cy3 (Sigma–Aldrich, C6198, 1:200) and fibronectin (Abcam, ab206928, 1:200). Slides were coverslipped with Fluoroshield mounting medium containing DAPI (Sigma-Aldrich, F6057). Quantification of immunohistochemistry and immunofluorescence was based on established methods in the laboratory27. Immunohistochemistry was quantified by ImageJ as percent of positive area and immunofluorescence was quantified as percent of positive cells or number of positive cells/exosomes, as indicated in the figure legends.
Liver function evaluation
Mouse blood was collected from the retro-orbital plexus. Serum was then immediately isolated by centrifugation at 4°C for 10 min at 6,000 rpm. Measurements of ALT and AST were performed by the MD Anderson Cancer Center Veterinary Pathology Core.
Haematoxylin and eosin staining and histopathology assessment
Formalin fixed and paraffin embedded liver sections (5 μm thickness) were processed with haematoxylin and eosin (H&E). For each slide, three to five distinct 200x visual fields were selected randomly and the numbers of necrotic and degenerated hepatocytes were counted manually by the count tool of Adobe Photoshop 7.0. Hepatocytes were determined as necrotic according to condensation and dark staining of the cytoplasm and absence of nucleus28. Hepatocytes degeneration was determined by cell swelling and enlargement as previously reported29. The data was expressed the percentage of necrotic or degenerated hepatocytes out of all hepatocytes in the visual field.
RNA sequencing
Livers were homogenized in TRIzol™ (Invitrogen, 15596018). Total RNA was purified according to manufacturer’s instructions (Zymo Research, 11–331). RNA integrity was determined using RNA 6000 Nano Assay and sequencing was performed using Illumina TrueSeq stranded mRNAseq by the MDACC Sequencing and ncRNA Program core. TopHat software (v2.0.9; https://ccb.jhu.edu/software/tophat/index.shtml) was used for genome mapping. The Cufflinks algorithm was used for identification of transcripts and DESeq2 was used for differentially expressed genes (https://bioconductor.org/packages/release/bioc/html/DESeq2.html). Significant expressed genes were determined by an adjusted p-value less than 0.05. Over-representation analysis and gene annotation were conducted by WebGestalt 2019 (http://www.webgestalt.org/) 30. The STAT3–ECM genes interaction regulatory network was constructed using NetworkAnalyst 3.0 (https://www.networkanalyst.ca/)31.
Data availability
RNA-Seq data has been deposited at Gene Expression Omnibus, accession number: GSE151851, reviewer token: uvalcucednqdhqf. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151851).
Statistical analyses
Statistical analyses used are detailed in the figure legends. Data are expressed as mean ± SD. p < 0.05 was considered to be statistically significant. Sample size was chosen based on an effect size of 4.5 to detect statistically meaningful differences between the 9 groups with a power of 0.95 and α error probability of 0.005 (0.05/9, with Bonferroni correction). Statistical significance was established using GraphPad Prism (GraphPad Software). Normal distribution of data was confirmed using Shapiro-Wilk normality test. For comparison between two groups, F-test was performed to compare variances. For comparisons between three or more groups, Brown-Forsythe and Bartlett’s tests were performed to compare SDs. For comparison of 3 or more groups, one-way ANOVA with Sidak’s post–hoc analysis was used for data with similar SDs or Brown-Forsythe and Welch ANOVA with Dunnett’s T3 post–hoc analysis was used for data with significantly different SDs. Kruskall-Wallis ANOVA with Dunn’s multiple comparison test was used to compare 3 or more groups that were not normally distributed. For comparison of two groups, unpaired two–tailed Student’s t–test or Welch’s unpaired two-tailed t-test for data with significantly different SDs were used. Exact p-values are reported in the graphs.
Results
Exosomes enable efficient STAT3 targeting in HSCs and and show liver tropism
Primary hepatic stellate cells (HSCs) isolated from wild-type (WT) mouse were cultured for 7 days, which led to their spontaneous activation (Fig. S1A)32–34. The activation of HSCs is characterized by gain of expression of alpha-smooth muscle actin (α-SMA, Fig. S1B). Vesicles isolated from MSCs that were used to generate iExo containing siRNA and ASOs displayed the size distribution and expression of tetraspanin markers indicative of exosomes (Fig. S1C-D). iExosiRNA-STAT3 or iExomASO-STAT3 treatment significantly reduced Stat3 mRNA levels in HSCs (Fig. 1A-B) with similar efficiency compared to commonly used lipid-based transfection (Fig. S1E).
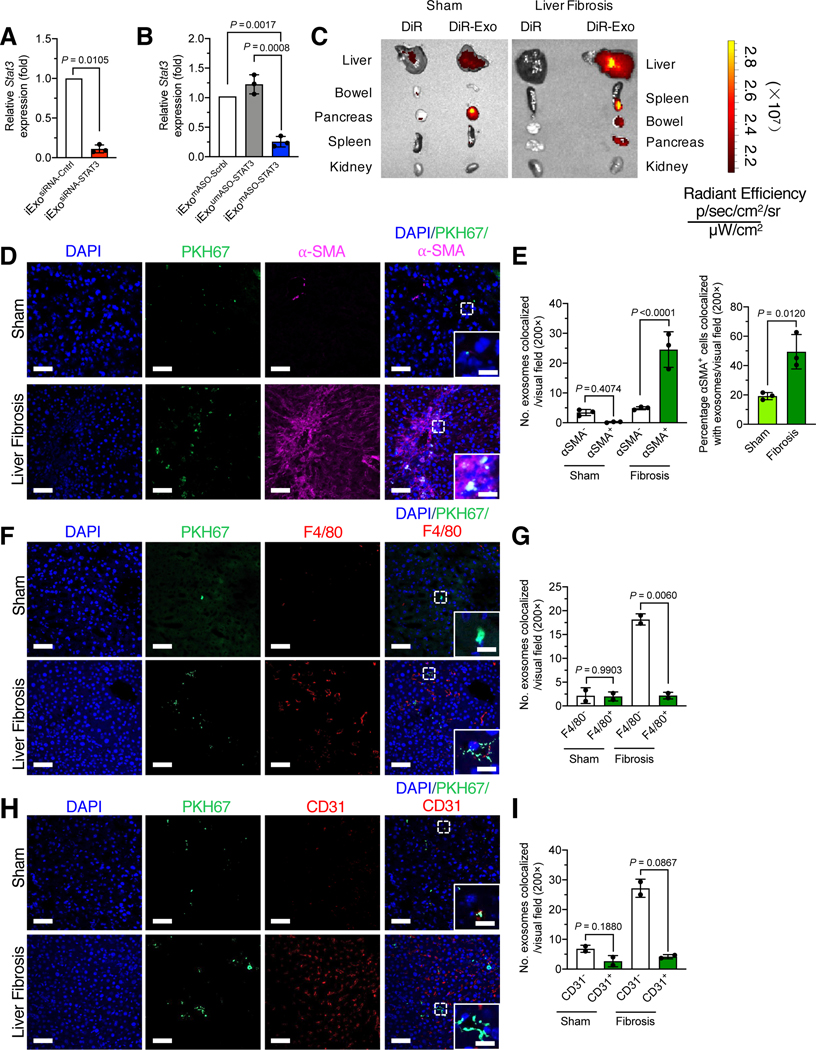
Figure 1. Exosomes enable efficient STAT3 targeting in HSCs and show liver tropism.
(A-B) Relative Stat3 expression in HSC treated with 5 μg/2 billion iExosiRNA-STAT3 (A) or iExomASO-STAT3 (B). n=3 independent experiments.
(C) Representative images of organs analyzed for presence of DiR-labeled exosomes in nonfibrotic (sham) (left panel) and fibrotic mice (right panel). n = 3 mouse per group.
(D-E) Immunofluorescence staining for PKH67+ exosomes (green), α-SMA (pink) and DAPI (blue) of frozen liver tissues from CCl4-treated mice (D). Scale bar: 100 μm (inset scale bar: 25 μm). Quantification of number of PKH+ exosomes colocalized with α–SMA- and α–SMA+ cells (left panel) and the percentage of PKH+α–SMA+ cells (right panel) in nonfibrotic (sham) and fibrotic livers. 1–3 visual fields for each tissue analyzed. n=3 mice per group.
(F-G) Immunofluorescence staining for PKH67+ exosomes (green), F4/80 (red) and DAPI (blue) of frozen liver tissues from CCl4-treated mice (F). Scale bar: 100 μm (inset scale bar: 25 μm). Quantification of number of PKH+ exosomes colocalized with F4/80- and F4/80+ cells in nonfibrotic (sham) and fibrotic livers (G). At least 3 visual fields for each tissue analyzed. n=2 mice per group.
(H-I) Immunofluorescence staining for PKH67+ exosomes (green), CD31 (red) and DAPI (blue) of frozen liver tissues from CCl4-treated mice (H). Scale bar: 100 μm (inset scale bar: 25 μm). Quantification of number of PKH+ exosomes colocalized with CD31- and CD31+ cells in nonfibrotic (sham) and fibrotic livers (I). At least 3 visual fields for each tissue analyzed. n=2 mice per group.
Data are represented as mean ± SD. A, unpaired Welch’s two–tailed t–test. B, E left panel, One–way ANOVA with Sidak’s post–hoc analysis. E right panel, unpaired two-tailed t-test. G, I, Brown-Forsythe and Welch ANOVA with Dunnett’s T3 post-hoc analysis. Exact p-values are indicated in all of the graphs.
Organotropism of exogenously administered exosomes in mice was examined and the liver tropism of exosomes derived from human mesenchymal stromal cells (MSCs) confirmed, along with other peritoneal organs in healthy mice (WT, Fig. 1C)16,18. To further investigate biodistribution of exosomes in fibrotic tissue, DiR labeled exosomes were administered intraperitoneally (i.p.) into mice with healthy liver as well as mice with liver fibrosis induced by exposure to carbon tetrachloride (CCl4). The results again revealed a specific accumulation of exosome-associated DiR signal in the liver and pancreas of WT mice, and lower amounts of signal detected in the kidney, bowel and spleen (Fig. 1C). Notabley, the fibrotic liver exhibited higher enrichment of DiR-labeled exosomes compared to the healthy liver (Fig. 1C). To further investigate the cellular localization of MSC exosomes, PKH labeled exosomes were injected into mice with and without liver fibrosis. Compared to control livers, fibrotic livers accumulated more exosomes and displayed increased colocalization of exosomes with α-SMA+ activated HSCs (Fig. 1D-E). In contrast, exosomes minimally colocalized with macrophages (Fig. 1F-G) and endothelial cells (Fig. 1H-I), as evidenced by costaining of PKH67 with macrophages marker F4/80 or with endothelial cells marker CD31. Staining for CD45 and collagen I revealed minimal overlap between PKH67 and immune cells and extracellular matrix, respectively (Fig. S1F-G). Furthermore, we injected mice intraperitoneallly with exosomes containing Alexa Fluor 647 (AF647)-tagged siSTAT3 or modified ASO (mASO) STAT3 (iExosiRNA647-STAT3 or iExomASO647-STAT3), and microscopic evalution of the liver showed enhanced accumulation of fluorescently labeled siRNA and when compared to mice administered naked siRNA or ASO (Fig. S1H).
iExosomes targeting STAT3 ameliorate liver fibrosis
In order to investigate the in vivo therapeutic efficacy of iExosomes as anti-fibrotic agent, WT mice were administrated with CCl4 twice weekly (i.p., injection) to induce chronic liver fibrosis (Fig. S2A). The mice were also treated with STAT3 targeting siRNA or ASO alone, or iExosiRNA-STAT3 or iExomASO-STAT3 on day 9, after fibrosis was established in the liver (Fig. S2A). iExosomes with siRNA or ASO targeting STAT3 were administered at two different dosages, 1 billion exosomes electroporated with 1 μg siRNA or ASO (1 μg/1 billion iExo) and 2 billion exosomes electroporated with 5 μg siRNA or ASO (5 μg/2 billion iExo). The siRNA and ASOs were designed to target human STAT3 with ability to target mouse STAT3 (Fig. 1A-B). ASO design included an unmodified (umASO) and modified 2’ O-methoxy-ethyl bases at the 5’ and 3’ end of the sequence and phosphorothioate modified bases (mASO). Untreated mice and mice treated with exosomes containing non-targeting control siRNA (siCntrl), modified scrambled ASO (mASO Scrbl), or unmodified ASO (umASO STAT3) were included as controls for comparison with the experimental arms of the study.
iExosomes targeting STAT3 using siRNA or mASO reduced Stat3 expression in fibrotic livers upon treatment with both 1 μg/1 billion iExosiRNA-STAT3 and 5 μg/2 billion iExosiRNA-STAT3 or iExomASO-STAT3 (Fig. 2A-B). Superior efficacy was observed at 5 μg/2 billion iExosiRNA-STAT3 and iExomASO-STAT3 compared to siRNA (siRNA-STAT3) or ASO (mASO-STAT3) alone (Fig. 2A-B). umASO did not significantly suppress Stat3 in vivo, possibly as a result of diminished stability of the ASO in this setting, whereas the enhanced stability of the mASO correlated with robust targeting of Stat3 (Fig. 2B). iExosomes containing both siRNA and mASO showed similar efficacy in suppressing Stat3 expression in fibrotic liver (Fig. 2A-B).
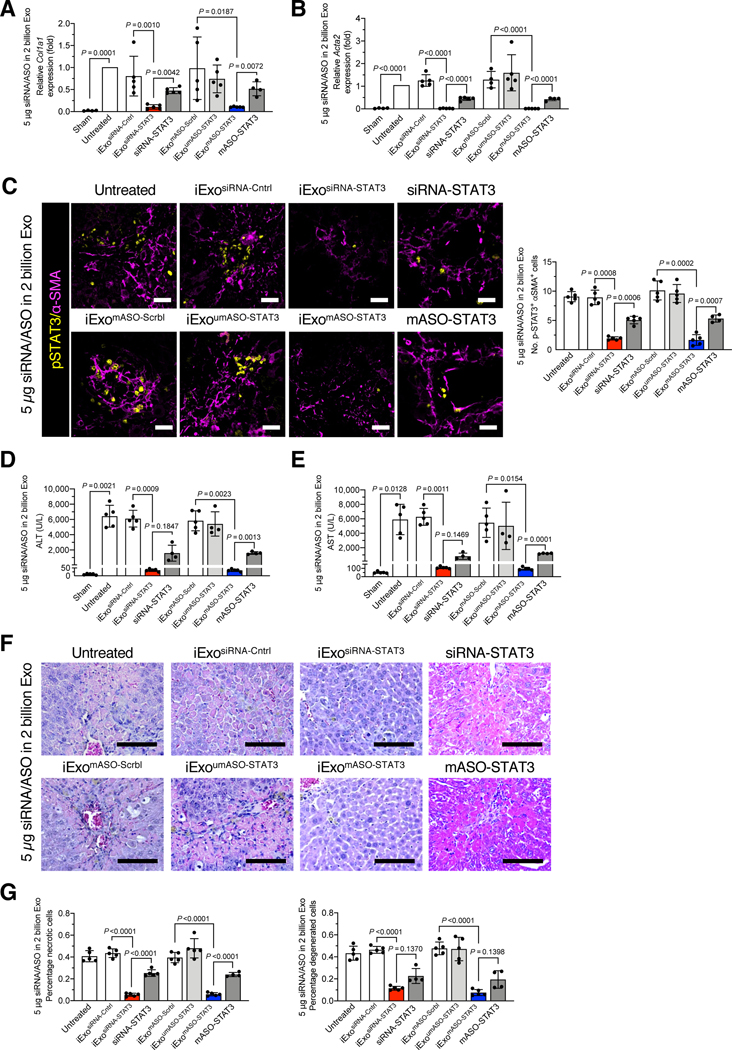
Figure 2. iExosomes targeting STAT3 reduce liver fibrosis.
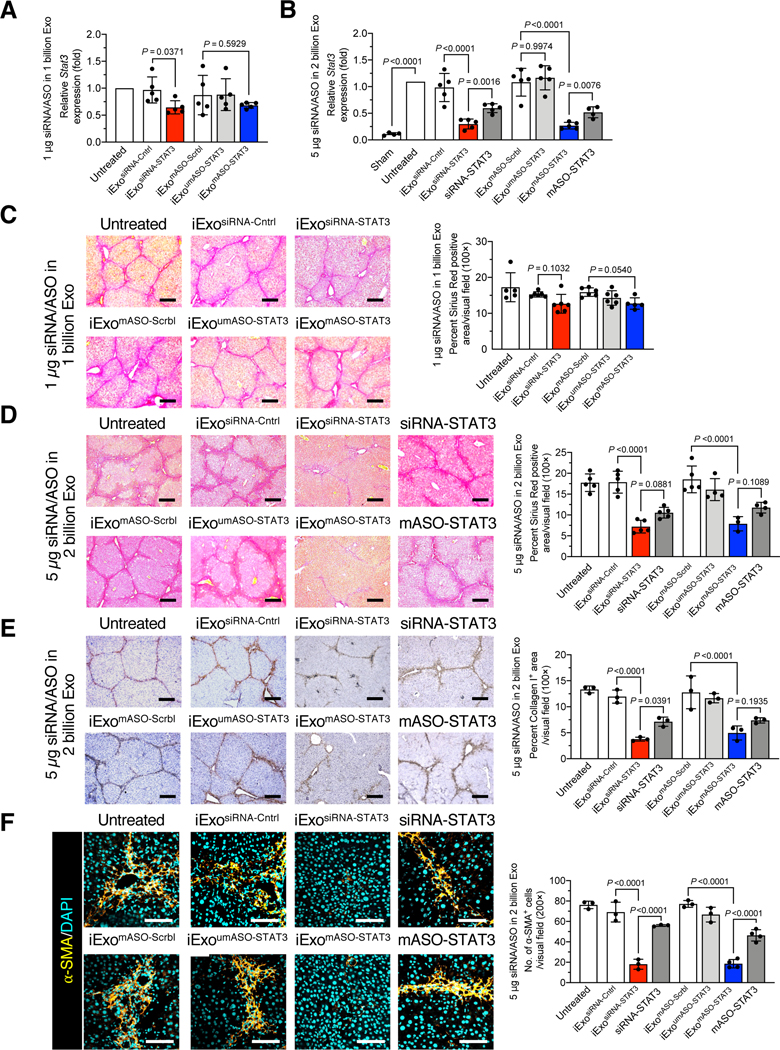
(A-B) Relative Stat3 mRNA expression in liver of mice treated with 1 μg/1 billion (A) or 5 μg/2 billion (B) iExosiRNA-STAT3 or iExomASO-STAT3 of the indicated treatments. A, n = 5 mice per group. B, sham, untreated, and mASO-STAT3: n=4 mice per group; n=5 mice per group for all other groups. (C-D) Representative Sirius red staining of liver sections from the 1 μg/1 billion (C) or 5 μg/2 billion (D) iExosiRNA-STAT3 or iExomASO-STAT3 treatment group (3 visual fields for each tissue analyzed). C, untreated and iExomASO-STAT3: n=5 mice per group; n=6 mice per group for all other groups. D, iExoumASO-STAT3 and mASO-STAT3: n=4 mice per group; iExomASO-STAT3: n=3 mice; n=5 mice per group for all other groups. Scale bar: 200 μm.
(E) Representative images (3 visual fields were analyzed per tissue) of immunohistochemical staining for collagen I in mice treated with 5 μg/2 billion iExosiRNA-STAT3 or iExomASO-STAT3. n=3 mice per group. Scale bar: 200 μm.
(F) α-SMA immunofluorescence staining in mice treated with 5 μg/2 billion iExosiRNA-STAT3 or iExomASO-STAT3 (3 visual fields for each tissue analyzed). iExomASO-STAT3 and mASO-STAT3: n=4 mice per group; n = 3 mice per group for all other groups; Scale bar: 200 μm.
The data are represented as mean ± SD. Individual dots in graphs depict distinct mice. A, Brown-Forsythe and Welch ANOVA with Dunnett’s T3 post–hoc analysis. B-F, One–way ANOVA with Sidak’s post–hoc analysis. Exact p-values are indicated in all of the graphs.
Repetitive exposure to the hepatotoxin CCl4 induces prominent inflammation and liver damage, which drive progressive fibrosis and accumulation of activated HSCs or myofibroblasts35. Sirius red staining and collagen I staining of liver was used to assess extracellular matrix (ECM) deposition. Our results revealed a significant reduction in ECM in mice treated with 5 μg/2 billion iExosiRNA-STAT3 or iExomASO-STAT3, whereas a modest reduction in fibrosis was observed in mice treated with 1 μg/1 billion iExosiRNA-STAT3 or iExomASO-STAT3 (Fig. 2C-E, Fig S2B), which was futher evidenced by vimentin (Fig. S2C), fibronectin (Fig. S2D), and α-SMA expression (Fig. S2E). Type I collagen deposition (Fig. 2E), vimentin expression (Fig. S3A), and fibronectin expression (Fig. S3B) were significantly reduced with 5 μg/2 billion iExosiRNA-STAT3 and iExomASO-STAT3 treatment compared to control groups. The expression of α-SMA, a well-established marker of activated HSCs (aHSCs) in fibrotic livers (Fig. S1B), was significantly reduced in mice treated with 5 μg/2 billion iExosiRNA-STAT3 and iExomASO-STAT3 compared to mice treated with siRNA-STAT3 or mASO-STAT3 alone (Fig. 2F).
iExosomes targeting STAT3 preserve liver function
In addition to the downregulation of STAT3 transcripts (Fig. 2A-B), treatment with 5 μg/2 billion iExosiRNA-STAT3 or iExomASO-STAT3 resulted in a significant transcriptional downregulation of alpha 1 chain of type I collagen (Col1a1) and smooth muscle actin (Acta2) compared to treatment with siRNA-STAT3 or mASO-STAT3 (Fig. 3A-B). Moreover, costaining revealed significant downregulation of pSTAT3 in α-SMA cells after treatment with iExosiRNA-STAT3 or iExomASO-STAT3 (Fig. 3C), indicating successful downregulation of STAT3 signaling. Liver function, ascertained with alanine aminotransferase (ALT) and aspartate aminotransferase (AST) serum levels, was significantly improved in mice treated with 5 μg/2 billion iExosiRNA-STAT3 and iExomASO-STAT3, as well as with siRNA-STAT3 or mASO-STAT3, albeit to a lesser extent (Fig. 3D–E). Treatment with 5 μg/2 billion iExosiRNA-STAT3 or iExomASO-STAT3 restored ALT and AST to levels nearing those of healthy control mice (Fig. 3D-E). Histopathological evaluation of CCl4-induced hepatic fibrosis showed hepatocytes degeneration, focal bridging necrosis, and significant structural disruption of the lobule architecture (Fig. 3F–G, untreated group). The percentage of hepatocyte necrosis and degeneration was significantly reduced when mice were administered 5 μg/2 billion iExosiRNA-STAT3 and iExomASO-STAT3 compared to control groups, including treatment with siRNA-STAT3 or mASO-STAT3 (Fig. 3F-G). No significant changes in Col1a1 and Acta2 expression were observed in mice administered 1 μg/1 billion iExosiRNA-STAT3 or iExomASO-STAT3 (Fig. S4A-B). At this low dose (1 μg/1 billion iExosiRNA-STAT3 or iExomASO-STAT3), no significant improvement in liver function, measured with ALT and AST levels (Fig. S4C-D) and liver histopathology (Fig. S4E-F) were observed. Importantly, neither low or high dose (5 μg/2 billion iExosiRNA-STAT3 or iExomASO-STAT3) of iExosomes produced measureable toxicity in kidney, pancreas, heart, lung or spleen (Fig. S5).
Figure 3. iExosomes targeting STAT3 restore liver functional parenchyma.
(A-B) Relative Col1a1 (A) and Acta2 (B) expression in livers with the indicated treatments. Untreated, sham, and mASO-STAT3: n=4 mice per group; n = 5 mice for all other groups.
(C) Immunofluorescence staining for pSTAT3 (yellow) and α–SMA (pink) in mice treated with 5 μg/2 billion iExosiRNA-STAT3 or iExomASO-STAT3 (3 visual fields for each tissue analyzed). mASOSTAT3: n=4 mice per group; n = 5 mice per group for all other groups; Scale bar: 200 μm.
(D-E) Serum ALT (D) and AST (E) level in mice with the indicated treatments. siRNA-STAT3, iExoumASO-STAT3, mASO-STAT3: n=4 mice per group; n=5 mice per group for all other groups.
(F) H&E staining of paraffin-embedded liver sections. Scale bar: 100 μm.
(G) Percentage of necrotic and degenerated hepatocytes. 3–5 visual fields for each tissue analyzed. mASO-STAT3, n=4 mice; n=5 mice per group for all other groups.
The data are expressed as mean ± SD. Individual dots in graphs depict distinct mice. A, C, D, E, G, Brown-Forsythe and Welch ANOVA with Dunnett’s T3 post-hoc analysis. B, one-way ANOVA with Sidak’s post–hoc analysis. Exact p-values are indicated in all of the graphs.
iExosomes targeting STAT3 reprogram the fibrotic liver transcriptome
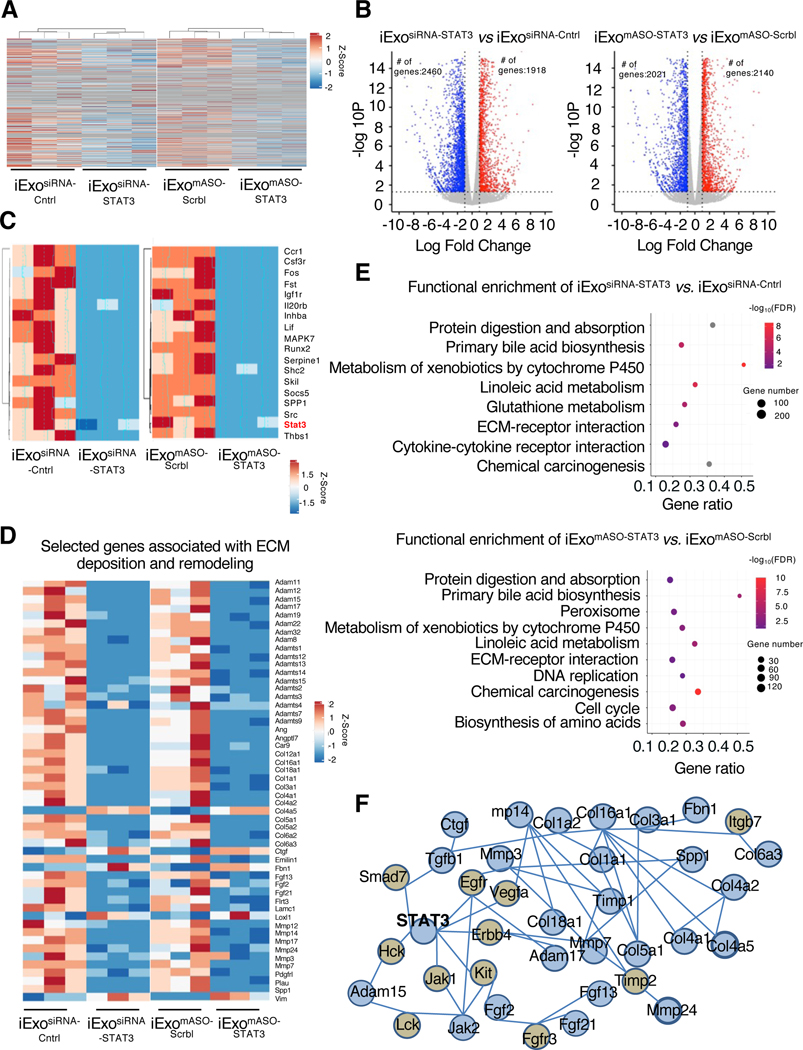
To investigate the impact of iExosomes treatment on liver transcriptome, we carried out RNA sequencing of the livers from 5 μg/2 billion iExosiRNA-Cntrl, iExo siRNA-STAT3, iExomASO-Scrbl, and iExomASO-STAT3 treated mice. Differentially expressed genes (DEGs) in each experimental group are shown as a heat map (Fig. 4A), wherein significant change in expression of a given gene was defined by a ratio greater than two-fold increase or decrease and an adjusted p-value < 0.05. The heat maps and volcano plots indicated that iExosiRNA-STAT3 and iExomASO-STAT3 treatment resulted widespread gene expression changes when compared to their respective controls (Fig. 4A–B). Liver transcript analyses from the iExosiRNA-STAT3 group showed increased expression of 1,918 genes and decreased expression of 2,460 genes compared to relevant controls, whereas liver transcript analyses in iExomASO-STAT3 group showed an increase in expression of 2,140 genes and a decrease in expression of 2,021 genes (Fig. 4B). STAT3 was found to be signficantly downregulated with exosomes-based STAT3 targeting (Fig. 4C). A cluster of genes involved in STAT3 signaling were downregulated following iExosomes treatment, and it included Spp1 and Thbs1 genes, which are known to play a critical role in liver fibrosis (Fig. 4C)36,37. Deregulated genes related to STAT3 signaling in liver fibrosis also included genes associated with ECM deposition and remodeling (Fig. 4D).
Figure 4. iExosomes reprogram the fibrotic liver transcriptome.
(A) Heat map depicting relative intensity of genes amongst the experimental groups. n=3 mice per group.
(B) Volcano plots depicting the number of differentially regulated genes in the livers of the listed experimental groups.
(C) Heat map of STAT3 signaling.
(D) Expression levels of selected genes associated with ECM deposition and remodeling. (E) Representation of differences in target genes by using over-representation analysis (WebGestalt) enrichment.
(F) Interaction network generated by NetworkAnalyst for STAT3 signaling and ECM-associated genes.
iExosomes treatment was associated with a reduction in the expression of canonical fibrosis-associated genes, including Col1a1, Col1a2, and Vim (Fig. 4D), supporting the role of STAT3 as a key mediator of liver fibrosis. Over-representation analysis showed that the DEGs were mainly enriched in ECM-receptor interaction pathway and also indicated that the downregulated genes were enriched for pathways involved in metabolism of xenobiotics by cytochrome P450, protein digestion and absorption, primary bile acid biosynthesis, linoleic acid metabolism, and chemical carcinogenesis (Fig. 4E). A similar set of altered downstream pathways was observed for both iExosiRNA-STAT3 and iExomASO-STAT3 treatment (Fig. 4E), and supports our dataset as a useful tool for further inquiry into STAT3 regulated pathways in liver fibrosis. To further investigate the association between STAT3 signaling and targeted ECM genes associated with liver fibrosis, an ECM regulatory network associated with STAT3 mRNA and liver fibrosis was constructed based on DEGs (Fig. 4A). As shown in Figure 4F, the generated network displays a connection between 24 ECM-associated genes. Our network analysis identifies STAT3 as an important node of regulation for ECM deposition in liver fibrosis, and successful suppression of these networks is noted with iExosiRNA-STAT3 and iExomASO-STAT3 treatment.
Discussion
Our studies collectively support previous reports underlining the critical role of STAT3 in promoting liver fibrosis38. STAT3 deregulation in liver fibrosis is complex, with a protective function in hepatocytes, and a pro-fibrotic function in aHSCs/myofibroblasts4,38,39. The anti-fibrotic outcome of the iExosomes approach to target STAT3 may reflect a preferential uptake by aHSCs/myofibroblasts. This observation is also in accordance with previous reports using exosomes from adipose-derived mesenchymal stem cells, which were shown to prevent liver fibrosis via exosomal miR-181–5p, with one its many targets being STAT340. Although various inhibitors have shown efficacy in mice, their specificity in targeting STAT3 remain unexplored or not validated41. Critically, while mASO-STAT3 alone displayed some efficacy in restoring liver function, iExomASO-STAT3 treatment further improved liver function, suggesting that enhanced drug delivery capacity of exosomes for the targeting of STAT3. Compared to siSTAT3 and ASO alone, iExosomes may be more stable in the blood as they avoid coagulation factors, antibody responses, complement and opsonins, possibly enhancing the delivery of embedded siRNAs and ASOs42–44. Additionally, exosomes are associated with a significant increase in drug accumulation in target organs such as the liver18, potentially further improving cargo delivery.
We observe targeting of α-SMA+ HSCs in fibrotic livers with iExosomes, with minimal interactions of iExosomes into immune and endothelial cells and collagen ECM. The isolation methods utilized in our studies yielded EVs with size and surface marker expression indicative of exosomes; however, other subsets of extracellular vesicles (EVs) may be present. A number of endocytic pathways have been identified as meditors of EV entry into cells45, but the precise mechanisms regulating EV organotropism and entry into specific cell types remain to be fully elucidated. Our previous studies showed that loading of siRNA into exosomes via electroporation is highly efficient and loaded siRNA is protected from RNase degradation18, which may further aid in cargo delivery in vivo. Future studies unraveling the mechanisms of EV/exosome targeting of cell subsets and regulators of nucleic acid loading into exosomes may provide additional routes to further improve iExosome efficacy and specificity.
Our iExosomes approach offers targeting specificity and may be used in combination with additional siRNA/ASOs. Furthermore, the use of exosomes in therapeutic approaches for liver cancer is also being considered46. Transformed hepatocytes in liver cancer rely on STAT3 expression39,47, and iExosomes targeting STAT3 could also provide benefits in limiting liver cancer progression. Our previous studies reporting the development of GMP certified clinical grade exosomes with siRNA for cancer therapy offers rapid translational potential for the therapy candidates identified in this study (NCT03608631)16,18. Collectively, our study supports the potential use of iExosomes as a therapeutic option for liver fibrosis.
Supplementary Material
Acknowledgements:
This work was primarily supported by funds from MD Anderson Cancer Center to R. Kalluri. M. Tang and C. Yang are supported by National Natural Science Foundation of China grants 81820108006 and 81670571. We thank P.E. Phillips and M.L. Kirtley for assistance with biodistribution experiments.
Nonstandard abbreviations:
- ALT
alanine aminotransferase
- α-SMA
alpha-smooth muscle actin
- ASO
antisense oligonucleotide
- AST
aspartate aminotransferase
- CCl4
carbon tetrachloride
- DEGs
differentially expressed genes
- ECM
extracellular matrix
- EV
extracellular vesicle
- GMP
Good manufacturing practice
- H&E
haematoxylin and eosin
- HSCs
hepatic stellate cells
- JAK
Janus tyrosine kinases
- MSCs
mesenchymal stromal cells
- STAT3
Signal transducer and activator of transcription 3
Footnotes
Conflict of interest: MD Anderson Cancer Center and R. Kalluri hold patents in the area of exosome biology and are licensed to Codiak Biosciences, Inc. MD Anderson Cancer Center and R. Kalluri are stock equity holders in Codiak Biosciences, Inc. R. Kalluri is a consultant and scientific adviser for Codiak Biosciences, Inc. V.S. LeBleu was a paid consultant for Codiak Biosciences, Inc. The other authors declare no competing interests.
References
- 1.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. [DOI] [PubMed] [Google Scholar]
- 2.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115(2):209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeybel M, Luli S, Sabater L, et al. A Proof-of-Concept for Epigenetic Therapy of Tissue Fibrosis: Inhibition of Liver Fibrosis Progression by 3-Deazaneplanocin A. Mol Ther. 2017;25(1):218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty D, Sumova B, Mallano T, et al. Activation of STAT3 integrates common profibrotic pathways to promote fibroblast activation and tissue fibrosis. Nat Commun. 2017;8(1):1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang DM, Sun W, Ning BF, et al. The HLF/IL-6/STAT3 feedforward circuit drives hepatic stellate cell activation to promote liver fibrosis. Gut. 2018;67(9):1704–1715. [DOI] [PubMed] [Google Scholar]
- 6.Pechkovsky DV, Prele CM, Wong J, et al. STAT3-mediated signaling dysregulates lung fibroblast-myofibroblast activation and differentiation in UIP/IPF. Am J Pathol. 2012;180(4):1398–1412. [DOI] [PubMed] [Google Scholar]
- 7.Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol. 2016;12(6):325–338. [DOI] [PubMed] [Google Scholar]
- 8.Choi S, Jung HJ, Kim MW, et al. A novel STAT3 inhibitor, STX-0119, attenuates liver fibrosis by inactivating hepatic stellate cells in mice. Biochem Biophys Res Commun. 2019;513(1):49–55. [DOI] [PubMed] [Google Scholar]
- 9.Su TH, Shiau CW, Jao P, et al. Sorafenib and its derivative SC-1 exhibit antifibrotic effects through signal transducer and activator of transcription 3 inhibition. Proc Natl Acad Sci U S A. 2015;112(23):7243–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartneck M, Warzecha KT, Tacke F. Therapeutic targeting of liver inflammation and fibrosis by nanomedicine. Hepatobiliary Surg Nutr. 2014;3(6):364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuppan D, Ashfaq-Khan M, Yang AT, Kim YO. Liver fibrosis: Direct antifibrotic agents and targeted therapies. Matrix Biol. 2018;68–69:435–451. [DOI] [PubMed] [Google Scholar]
- 12.Busatto S, Pham A, Suh A, Shapiro S, Wolfram J. Organotropic drug delivery: Synthetic nanoparticles and extracellular vesicles. Biomed Microdevices. 2019;21(2):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. [DOI] [PubMed] [Google Scholar]
- 14.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164(6):1226–1232. [DOI] [PubMed] [Google Scholar]
- 16.Mendt M, Kamerkar S, Sugimoto H, et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lotvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. [DOI] [PubMed] [Google Scholar]
- 20.Borges FT, Melo SA, Ozdemir BC, et al. TGF-beta1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J Am Soc Nephrol. 2013;24(3):385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeBleu VS, Kalluri R. Exosomes as a Multicomponent Biomarker Platform in Cancer. Trends Cancer. 2020. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. 2020;21(10):585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogiso H, Ito H, Ando T, et al. The Deficiency of Indoleamine 2,3-Dioxygenase Aggravates the CCl4-Induced Liver Fibrosis in Mice. PLoS One. 2016;11(9):e0162183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kisseleva T, Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2020. [DOI] [PubMed] [Google Scholar]
- 25.Vinas O, Bataller R, Sancho-Bru P, et al. Human hepatic stellate cells show features of antigen-presenting cells and stimulate lymphocyte proliferation. Hepatology. 2003;38(4):919–929. [DOI] [PubMed] [Google Scholar]
- 26.Whittaker P, Kloner RA, Boughner DR, Pickering JG. Quantitative assessment of myocardial collagen with picrosirius red staining and circularly polarized light. Basic Res Cardiol. 1994;89(5):397–410. [DOI] [PubMed] [Google Scholar]
- 27.Lovisa S, LeBleu VS, Tampe B, et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med. 2015;21(9):998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishna M. Patterns of necrosis in liver disease. Clin Liver Dis (Hoboken). 2017;10(2):53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lackner C, Gogg-Kamerer M, Zatloukal K, Stumptner C, Brunt EM, Denk H. Ballooned hepatocytes in steatohepatitis: the value of keratin immunohistochemistry for diagnosis. J Hepatol. 2008;48(5):821–828. [DOI] [PubMed] [Google Scholar]
- 30.He W, Fu L, Yan Q, et al. Gene set enrichment analysis and meta-analysis identified 12 key genes regulating and controlling the prognosis of lung adenocarcinoma. Oncol Lett. 2019;17(6):5608–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–W241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhai X, Wang W, Dou D, et al. A novel technique to prepare a single cell suspension of isolated quiescent human hepatic stellate cells. Sci Rep. 2019;9(1):12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Lv F, Kong M, et al. A cAbl-MRTF-A Feedback Loop Contributes to Hepatic Stellate Cell Activation. Front Cell Dev Biol. 2019;7:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong XH, Chen C, Hou P, et al. Overexpression of miR-126 inhibits the activation and migration of HSCs through targeting CRK. Cell Physiol Biochem. 2014;33(1):97–106. [DOI] [PubMed] [Google Scholar]
- 35.Mederacke I, Hsu CC, Troeger JS, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arriazu E, Ge X, Leung TM, et al. Signalling via the osteopontin and high mobility group box-1 axis drives the fibrogenic response to liver injury. Gut. 2017;66(6):1123–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breitkopf K, Sawitza I, Westhoff JH, Wickert L, Dooley S, Gressner AM. Thrombospondin 1 acts as a strong promoter of transforming growth factor beta effects via two distinct mechanisms in hepatic stellate cells. Gut. 2005;54(5):673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Lafdil F, Kong X, Gao B. Signal transducer and activator of transcription 3 in liver diseases: a novel therapeutic target. Int J Biol Sci. 2011;7(5):536–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Lafdil F, Wang L, et al. Hepatoprotective versus oncogenic functions of STAT3 in liver tumorigenesis. Am J Pathol. 2011;179(2):714–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu Y, Zhang Q, Cai X, et al. Exosomes derived from miR-181–5p-modified adipose-derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. J Cell Mol Med. 2017;21(10):2491–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beebe JD, Liu JY, Zhang JT. Two decades of research in discovery of anticancer drugs targeting STAT3, how close are we? Pharmacol Ther. 2018;191:74–91. [DOI] [PubMed] [Google Scholar]
- 42.Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles - endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta. 2014;1846(1):75–87. [DOI] [PubMed] [Google Scholar]
- 43.Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular vesicles for drug delivery. Adv Drug Deliv Rev. 2016;106(Pt A):148–156. [DOI] [PubMed] [Google Scholar]
- 44.van den Boorn JG, Schlee M, Coch C, Hartmann G. SiRNA delivery with exosome nanoparticles. Nat Biotechnol. 2011;29(4):325–326. [DOI] [PubMed] [Google Scholar]
- 45.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lou G, Chen L, Xia C, et al. MiR-199a-modified exosomes from adipose tissue-derived mesenchymal stem cells improve hepatocellular carcinoma chemosensitivity through mTOR pathway. J Exp Clin Cancer Res. 2020;39(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung KH, Yoo W, Stevenson HL, et al. Multifunctional Effects of a Small-Molecule STAT3 Inhibitor on NASH and Hepatocellular Carcinoma in Mice. Clin Cancer Res. 2017;23(18):5537–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data has been deposited at Gene Expression Omnibus, accession number: GSE151851, reviewer token: uvalcucednqdhqf. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151851).