Abstract

Vital to the treatment of influenza is the use of antivirals such as Oseltamivir (Tamiflu) and Zanamivir (Relenza); however, antiviral resistance is becoming an increasing problem for these therapeutics. The RNA-dependent RNA polymerase acidic N-terminal (PAN) endonuclease, a critical component of influenza viral replication machinery, is an antiviral target that was recently validated with the approval of Baloxavir Marboxil (BXM). Despite its clinical success, BXM has demonstrated susceptibility to resistance mutations, specifically the I38T, E23K, and A36 V mutants of PAN. To better understand the effects of these mutations on BXM resistance and improve the design of more robust therapeutics, this study examines key differences in protein–inhibitor interactions with two inhibitors and the I38T, E23K, and A36 V mutants. Differences in inhibitor binding were evaluated by measuring changes in binding to PAN using two biophysical methods. The binding mode of two distinct inhibitors was determined crystallographically with both wild-type and mutant forms of PAN. Collectively, these studies give some insight into the mechanism of antiviral resistance of these mutants.
Introduction
The influenza virus is responsible for a significant burden of illness, causing approximately 35.5 million cases, 500,000 hospitalizations, and 35,000 deaths in the United States during the 2018/19 season alone.1 Children and elderly populations are particularly vulnerable to complicated cases of influenza and make up the largest percentage of hospitalizations and deaths.2 During the COVID-19 pandemic, nonpharmaceutical interventions (NPIs) such as stay-at-home orders, masking, social distancing, and increased disinfection measures were put into practice in public spaces to prevent the spread of SARS-CoV-2. This also led to a substantial decrease of global influenza infections through the 2020/21 and 2021/22 seasons,3 which has consequences for the annual reformulation of the influenza vaccine. Reformulation is heavily dependent on data from prior infectious seasons of circulating strains to predict the most effective vaccine composition for the coming influenza season.3 Therefore, there were less data to predict the optimal 2022/23 vaccines, which explains the substantial resurgence of influenza observed for the 2022/23 season.3,4 The use of antivirals for influenza is a crucial second line of defense, especially during times of resurgence, epidemics, and pandemics.
Vital to the treatment of influenza is the use of neuraminidase inhibitors (NAIs) such as Oseltamivir (Tamiflu) and Zanamivir (Relenza). Although antiviral resistance remains low for these medications, resistance has been on a steady rise and there is an urgent need for new medications with novel mechanisms of action.5 Baloxavir marboxil (BXM) and its active metabolite baloxavir acid (BXA) is a first-in-class inhibitor targeting the influenza virus polymerase PA N-terminal endonuclease domain (PAN) that was approved in Japan and the United States in 2018 (Figure 1).6 The RNA-dependent RNA polymerase of the influenza virus cannot synthesize its own 5′-mRNA cap, which is necessary for eukaryotic translation. To overcome this, the polymerase engages in a unique “cap snatching” mechanism, in which the polymerase subunit PB2 captures host pre-mRNA and PAN cleaves 10–13 nucleotides to serve as a primer for viral mRNA synthesis by the PB1 subunit.7,8 PAN is a metalloenzyme that is essential for viral replication, is highly conserved, and is not naturally occurring in humans, making it an attractive antiviral target.6 BXM has been shown to reduce fever in an average of 24 h and is an oral monotherapy that can be taken at the time of diagnosis, which ensures higher patient compliance than medications taken over several days. Despite its clinical success, BXM has also demonstrated a high susceptibility to treatment-emergent resistance and viral rebound.6,9
Figure 1.
(a) Wild type (WT) PAN endonuclease with all α helices labeled (PDB: 8T5Z). (b) Baloxavir marboxil (BXM), its active metabolite baloxavir acid (BXA), and compound 23. (c) PAN active site with Mn2+ ions shown and coordinating residues labeled. The protein backbone (gray) is shown as a gray ribbon. Mn2+ ions and water molecules are shown as purple and red spheres, respectively. Residues coordinating the Mn2+ ions are shown in sticks.
In a resistance monitoring study following the 2014 approval of BXM in Japan, many treatment-emergent resistance mutations were found. The highest change in susceptibility and most prevalent resistance mutations found were in isoleucine 38 with I38T/M/F/L/N/S mutations. I38T has been reported to decrease the efficacy of the BXM by 30- to 50-fold.6,10 Other emergent mutations of interest are E23K and A36 V mutations, like I38T, which were identified as resistance mutations resulting from BXM treatment.6 The probability of widespread transmission of these variants depends on the replication capacity and onward transmission. Recent transmissibility studies in animal models of these mutations suggest that the I38T mutation on its own has a reduced fitness compared to WT viruses; however, I38T in combination with other stabilizing mutations can recover.11 These fast emergent resistance mutations highlight the need for additional members of this class of antivirals and a better understanding of the mechanisms of resistance for rational drug design.
Unlike NAI resistance mechanisms that have been studied extensively for over 20 years, the resistance landscape of PAN endonuclease inhibitors is largely unknown, and the monitoring of resistance markers (I38X and E23X) is still in its infancy. Several structural studies of the I38T variant with BXA have been published, and it is largely agreed that the reduced activity of BXA is due to less favorable van der Waals packing and induced fit changes within the PAN active site.6,12−14 To date, the structures of E23K and A36V have not been published; however, in silico studies for the E23K/G mutations hinted at the possibility of the destabilization of the α2 helix (Figure 1) and disruption of other important interactions for RNA recognition by BXA.15−17 Several other potent inhibitors for PAN endonuclease have been developed. A group of hydroxypyridinone heterocycle compounds have been reported to bind to the PAN endonuclease active site metals with a similar metal-binding pharmacophore (MBP) to BXA.18,19 A majority of these compounds are known to make van der Waals contacts with Ile38 and could be at a similar disadvantage as BXA.19 Of these hydroxypyridinone heterocycles, compound 23 is among the most potent and is included here to examine the effects of BXA resistance mutants on binding to an alternative inhibitor type (Figure 1).
In the present study, seven co-crystal structures of BXA and a second PAN inhibitor (23) with WT, I38T, E23K, and A36V PAN, as well as the apo (ligand-free) structures of WT, I38T, and E23K constructs are reported. The E23K and A36 V structures are the first reported structural data for these mutants and may aid in studies to develop a structure–activity relationship (SAR) for these variants. The structural data obtained are complimented by inhibitor affinity determined by two independent biophysical methods to quantify the impact of the changes in binding observed in the co-crystal structures.
Results and Discussion
Biophysical Evaluation
Due to the high affinity of the inhibitors used in this study, determining the inhibitory activity by measuring IC50 values using conventional FRET-based enzymatic assays is not particularly informative, as many inhibitors will simply display activity at concentrations below the sensitivity threshold of the assay. Therefore, inhibitor binding in this study was evaluated using biophysical methods to determine binding constants (Kd) and changes in the thermal stability (TM) of the enzyme. Although Kd is fundamentally different, and not directly comparable to an IC50 value, overall trends between Kd and IC50 are typically comparable.20
Spectral shift methods (SpS) are based on the well-known phenomenon that organic fluorophores can report changes in their chemical environment via perturbations in their emission spectrum, e.g., changes in their overall fluorescence intensity or lmax (red or blue spectral shifts). In the studies performed here, fluorescence was recorded at two wavelengths (650 and 670 nm), and the change in emission intensity between the 670/650 nm fluorescence signal was monitored as a function of ligand concentration (see the SI for experimental details). This method allows for the determination of binding constants (Kd) over a large dynamic range from low nanomolar (nM) to high millimolar (mM) values.21,22 In this study, purified PAN endonuclease was used (see the SI for experimental details).19 Purified PAN is known to bind substrates and inhibitors with poorer affinity than the complete PA subunit or heterotrimer polymerase complex, which may explain why the Kd values reported here (see below) are weaker than reported elsewhere.23 Using this method, binding of BXA to WT PAN proved to be the tightest binding interaction measured in this study (343 nM, Table 1). Binding of BXA to the I38T mutant showed a 27-fold reduction in affinity that is consistent with previously reported studies of BXA affinity for this mutant (30- to 50-fold reduction).6 There are fewer data available on the binding of BXA to the E23K mutant, but the SpS experiments showed an even larger 79-fold reduction in binding affinity.
Table 1. Dissociation Constants (Kd, μM) and Relative Fold Change in Binding Affinity (FC) of PAN Endonuclease Variants with BXA and 23.
| BXA |
23 |
|||
|---|---|---|---|---|
| Kd (μM) | relative affinity | Kd (μM) | relative affinity | |
| WT | 0.343 ± 0.106 | 277 ± 30.6 | ||
| I38T | 9.3 ± 4.2 | 27-fold | 384 ± 19.4 | 1.4-fold |
| E23K | 26.1 ± 11.2 | 79-fold | 328 ± 26.6 | 1.2-fold |
Compound 23 is known to have a poorer affinity than BXA for WT PAN. This was verified by SpS, with 23 giving a Kd value of 277 μM for WT PAN (∼660-fold weaker than BXA).6,12,19 Although the affinity of 23 is not as high as BXA for WT PAN, the relative change in affinity of 23 for the I38T and E23K variants is much smaller compared to BXA (<2-fold for 23, compared to 27- and 79-fold for BXA). This suggests that inhibitors that access different parts of the active site may be less susceptible to mutations generated against BXA. However, it is important to note that BXA still has a substantially greater absolute affinity for I38T and E23K when compared to compound 23 (Table 1). Of note, the A36V construct was found to denature and precipitate out of solution when left at room temperature for prolonged periods of time, including the time required for incubation in the SpS assay. Thus, the A36V construct was not stable enough for the determination of an accurate Kd value by the SpS method and is not reported here.
The thermal stability of a protein generally increases upon formation of favorable protein–ligand contacts, with tighter-binding interactions typically resulting in larger changes in thermal shift (ΔTM) compared to those of an apo (ligand-free) control. Differential scanning fluorometry (DSF) experiments were performed with both BXA and compound 23 with WT PAN and all three mutants. WT, I38T, and E23K PAN displayed an average melting temperature of between 58 and 61 °C, while A36 V proved to be much less thermally stable with a melting temperature of 50–52 °C (Table 2). DSF experiments were performed at 200 μM for both BXA and 23. A concentration of 200 μM is sufficient for the low Kd values of BXA, but it is lower than the Kd values for compound 23 (≥200 μM). To compare these two compounds with a sufficient excess of compound, an additional measurement at 1 mM was performed for compound 23. As described below, the melting temperatures for experiments run with 1 mM compound 23 showed an expected shift to higher values. BXA bound to WT PAN produced the largest ΔTM at 22.9 °C (Table 2). The difference in ΔTM for BXA bound to I38T compared to WT was ∼8 °C lower with a ΔTM of 14.9 °C. This indicates the interaction between BXA and I38T is not as stable as WT. This finding is consistent with previous studies that reported ΔTM melting value between the WT and I38T mutants with BXA to be between 8 and 10 °C.6,12 Binding of BXA to the E23K mutant produced a ΔTM of 19.8 °C, which is <3 °C difference when compared to binding to WT PAN. This suggests some loss in stability when BXA is bound to E23K when compared to WT PAN, but still results in significant stabilization of the E23K mutant enzyme. Interestingly, a larger change in ΔTM was observed when BXA is bound to the E23K mutant versus the I38T mutant, contradicting the SpS experiments, which show the opposite trend in affinity. This may indicate that the E23K mutation promotes dissociation of BXA without affecting protein stability. This E23K mutant may increase flexibility of α2 encouraging BXA to dissociate and preventing new binding events. The melting temperature of the A36V mutant is significantly lower than its counterparts, adding to the evidence that the construct itself is not as stable as the other mutations in this study.24 However, the ΔTM of the A36 V mutant with BXA is 21.7 °C, nearly identical to that of BXA with WT, suggesting the binding of BXA to the A36 V mutation is strong despite the instability of this mutant.
Table 2. Thermal Melting Temperature Change of PAN Endonuclease Variants with BXA and 23 (ΔTM, °C)a.
| DMSO | BXA (200 μM) | 23 (200 μM) | 23 (1 mM) | |
|---|---|---|---|---|
| TM (°C) | ΔTM (°C) | ΔTM (°C) | ΔTM (°C) | |
| WT | 58.7 ± 0.11 | 22.9 ± 0.08 | 17.8 ± 0.10 | 19.6 ± 0.16 |
| I38T | 59.6 ± 0.08 | 14.9 ± 0.04 | 12.9 ± 0.09 | 15.6 ± 0.28 |
| E23K | 61.5 ± 0.08 | 19.8 ± 0.06 | 10.2 ± 0.08 | 14.3 ± 0.09 |
| A36V | 51.3 ± 0.20 | 21.7 ± 0.11 | 16.4 ± 0.14 | 17.1 ± 0.12 |
Experiments with BXA and 23 were averaged from eight independent measurements.
Compound 23 bound to WT PAN showed a ΔTM of 17.8 °C at 200 μM and 19.6 °C at 1 mM (Table 2), which is <5 °C less than BXA bound to WT and consistent with the large difference in Kd values between these compounds (Table 1). The ΔTM for 23 bound to I38T was found to be 12.9 °C at 200 μM and 15.6 °C at 1 mM, which is around 4 °C less than the WT. Like BXA, this indicates the interaction between 23 and I38T is not as stable as with WT PAN. E23K incubated with 23 showed a ΔTM of 10.2 °C at 200 μM and 14.3 °C at 1 mM, ∼ 8 °C lower than with WT and 2 °C lower than the value obtained with I38T. This indicates the interaction between 23 and E23K is not as stable as WT or I38T. The difference between the ΔTM values for 23 with E23K and I38T (>2 °C) is relatively small, suggesting that binding of 23 to these two mutants might be rather comparable, which is consistent with the SpS experiments (Table 1). The A36V mutant with compound 23 shows a trend similar to that of BXA with the ΔTM being comparable to that of the WT, suggesting the binding of compound 23 to A36V is also largely unaffected by this mutation.
X-ray Crystallography
To determine the mode of binding of BXA and compound 23 with WT, I38T, E23K, and A36V constructs, the complexes were crystallized, and the co-crystal structures determined. In addition to the co-crystal structures, the apo structures of WT, I38T, and E23K PAN endonuclease were determined. In all apo structures, the active site metal cations Mn2+ are coordinated by His41, Glu80, Asp108, Glu 119, and Ile120 and a total of five water molecules, producing an octahedral coordination geometry at each metal center (Figure 1). No major structural changes were observed in the I38T apo structure compared with apo WT (Figure S2). When the apo structure of E23K is compared with that of WT, the terminus of α2, where the E23K mutation is located, is distorted (Figure S2). In WT PAN, Glu23 forms hydrogen bonds with Arg84, stabilizing the terminus of α2 which promotes the correct position of Tyr24 for base stacking. In the E23K structure, the change in charge that accompanies the change from a glutamate to a lysine results in a loss of hydrogen bonding with Arg84 and no stabilization. Lys23, Tyr24, and Arg84 become more flexible, causing disorder of α2 (Figure 3).
Figure 3.
(a) Co-crystal structure of WT PAN (gray PDB: 6FS6) overlaid with E23K mutant (pink, Lys23 in bright pink PDB: 8T6Z) with BXA coordinated to the Mn2+ ions. (b) Alternate view of WT PAN (gray) overlaid with E23K mutant (pink) with BXA (RMSD: 0.9 Å for pocket residues). The E23K mutant shows a distortion in the terminus of α2 due to loss of stabilizing hydrogen-bonding interactions, which allows Tyr24 further into the active site causing a shift in BXA binding by 1.1 Å and 23 binding by 0.3 Å. (c) Co-crystal structure of WT PAN (gray PDB: 8T94) overlaid with E23K mutant (pink PDB: 8T81) with 23. (d) Alternate view of the co-crystal structure of WT PAN (gray) overlaid with E23K mutant (pink) with 23 (RMSD: 0.3 Å for pocket residues). In this structure, differences in pi–cation interactions involving the 2′-trifluoro-substituted benzene ring and subsequently the position of the 4′-tetrazole are impacted. The ring is rotated in this structure to maintain the pi–cation interaction in the new position of Tyr24. The protein backbone (WT gray and E23K pink) is shown as a cartoon, and Mn2+ ions are shown as purple spheres.
Upon binding either inhibitor, three water molecules are displaced and the octahedral geometry is maintained at each metal center for all constructs for which structures were obtained. BXA binds to the active site metal ions through a triad of oxygen donor atoms, including oxygen donor atoms from the ketone and carboxylic acid groups, while the “butterfly” shape of the remainder of the molecule provides extensive interactions with hydrophobic pockets of the active site in WT PAN endonuclease (Figure 2). Significant interactions contributing to the high affinity of BXA include hydrophobic interactions between the aromatic rings and Ile38, hydrogen bonding between the hydroxyl group of Tyr24 and the BXA morpholine oxygen, as well as pi-stacking between the fluorinated BXA ring and Tyr24. Extended hydroxypyridinone heterocycle inhibitors including compound 23 coordinate to the active site metal ions with good affinity through hydroxypyridinone MBPs, and make two types of additional contacts with WT PAN.19 The addition of a 2′- or 3′-substituted benzene ring and a 4′-tetrazole drives the affinity for PAN even lower. Compound 23 was previously reported to have an IC50 of 47 pM against WT PAN endonuclease in an enzyme-based assay, making contacts with the active site wall via a 2′-trifluoro-substituted benzene ring, a weak van der Waals interaction with Ile38, and a 4′-tetrazole engaged in favorable hydrogen-bonding interactions shown to be critical in RNA binding with Lys34 and Arg124 (Figure 2).
Figure 2.
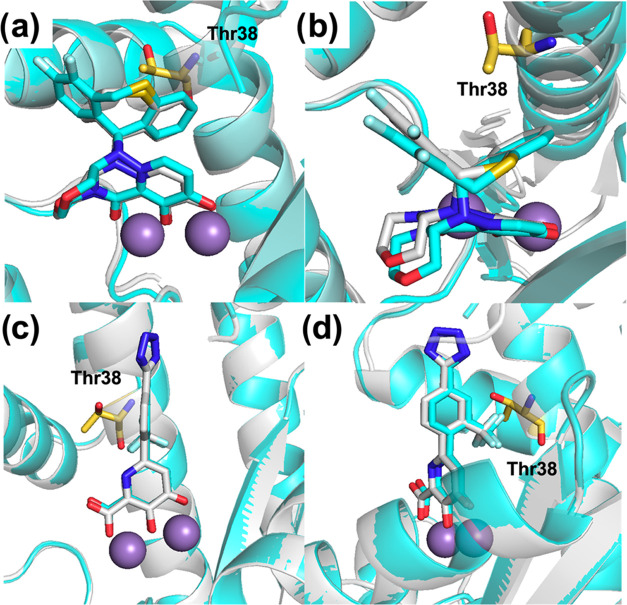
(a) Co-crystal structure of WT PAN (gray PDB: 6FS6) overlaid with I38T mutant (cyan, Thr38 in yellow PDB: 8T5W) with BXA. (b) Alternate view of the co-crystal structure of WT PAN (gray) overlaid with I38T mutant (cyan) with BXA (RMSD: 0.8 for pocket residues). The binding pose of BXA to I38T is virtually unaffected when compared to WT. Although BXA binding is largely unchanged, Thr38 must undergo a large rotamer change to accommodate BXA. (c) Co-crystal structure of WT PAN (gray) overlaid with I38T mutant (cyan) with 23. (d) Alternate view of the co-crystal structure of WT PAN (gray) overlaid with I38T mutant (cyan) with 23 (RMSD: 0.2 for pocket residues). The change in binding of compound 23 with the I38T mutant shows a slight change in binding angle of the compound toward Thr38 so that the trifluoro group makes similar van der Waals contacts to that observed in the WT enzyme. The protein backbone (WT gray and I38T cyan) is shown as a cartoon, and Mn2+ ions are shown as purple spheres.
The I38T mutation has been the most extensively studied mutation with several reported co-crystal structures with bound inhibitors (e.g., bound to RO-5, a BXA analogue).12−14 The reduced susceptibility of BXA for the I38T mutant has been attributed to less favorable van der Waals packing and induced fit changes.6 The butterfly shape of BXA provides near-perfect packing with hydrophobic pockets of the active site of WT PAN. Ile38 packs directly behind the V-shaped difluoro-dihydro-dibenzothiepine tail group, and this packing is slightly less effective with the change to the more polar, smaller Thr38. When comparing the BXA bound structure and the apo structure of I38T, a large rotamer change in Thr38 is observed (Figure S3). This rotamer change can also be observed when comparing the position of Thr38 in structures of BXA (Figure 2a) and 23 (Figure 2c) where the hydroxyl of Thr38 in the BXA structure is pointing toward the protein backbone and the hydroxyl of Thr38 in the structure of compound 23 is pointing toward the compound. No major local distortions are observed in α3 surrounding I38T in either structure (Figure S3). The change in the binding of compound 23 with the I38T mutant shows a slight change in the binding angle of the compound toward Thr38. The change to threonine removes a methyl group, making the slight tilt in 23 necessary so that the trifluoro group makes similar van der Waals contacts to that observed in the WT enzyme (Figure 2). However, small changes from the optimal binding angle are known to distort the octahedral coordination geometry around the metal centers and cause a decrease in affinity.25,26 The observed change in Kd and ΔTM is most likely due to this sacrifice in favorable, highly enthalpic metal-binding interactions when the interaction with I38T is maintained. No rotamer changes were observed in Thr38 upon binding of compound 23, unlike upon binding of BXA.
From the biophysical assay data, it was expected that changes in BXA binding would be less significant with the E23K mutant when compared to the I38T mutant and there would be very slight, if any change at all in the binding of BXA with A36V. This was indeed the case, but significant changes to the position of Tyr24 and pi–cation interactions were observed. In WT PAN, Tyr24 makes a crucial base stacking interaction for RNA recognition in the active site. The RNA bases stack with Tyr24 and forms hydrogen-bonding interactions with Glu26 that, in turn, forms a salt bridge with Lys34.15,24 Glu23 contributes to this complex by hydrogen bonding with Arg84 and stabilizing the terminus of α2 and promoting the correct position of Tyr24 for base stacking.15 Comparing the co-crystal structures of WT and E23K with BXA bound, E23K disrupts the end terminus of α2 and allows Tyr24 to extend further into the active site, forcing BXA closer to α3 (Figure 3). The shift in BXA toward α3 disrupts hydrogen bonding between the hydroxyl group of Tyr24 and the BXA morpholine oxygen increasing the distance from 4.3 to 4.8 Å, as well as pi-stacking between the fluorinated BXA ring and Tyr24, increasing the distance from 5.2 to 6.3 Å. The change in position of BXA away from Tyr24 increases the bond distances, decreasing the strength of both the hydrogen bonding and pi-stacking (Figure 3). Unlike the BXA binding to I38T, no rotamer changes were found when comparing the BXA bound and unbound in the E23K structure. The disordered terminus of α2 and the change in position of Tyr24 could explain the relative change in binding affinity being greater for this mutant than for the I38T mutant. In solution, Lys23, Tyr24, and Arg84 are free to move and make a wide variety of transient protein–protein contacts and protein–inhibitor contacts that could lead to the destabilization of the binding of BXA without greatly affecting the overall protein stability. The E23K mutant in complex with compound 23 shows interesting changes in Lys23 and Tyr24 that have clearly defined electron density. In this structure, differences in pi–cation interactions involving the 2′-trifluoro-substituted benzene ring and subsequently the position of the 4′-tetrazole are impacted. The ring is rotated by 0.3 in this structure to maintain the pi–cation interaction in the new position of Tyr24 (Figure 3). This rotation causes changes in hydrogen bonding in the 4′-tetrazole with Lys34 and Arg124. There is also a slight change in MBP coordination, causing distortions from optimal octahedral geometry (Figure 3).
Although it was hypothesized that the A36V mutant would disrupt BXA binding by disrupting α3, the DSF experiments revealed that the A36V treated with BXA did not show a significant change in ΔTM. This suggests that the A36V mutation causes very minimal or no change at all to the binding of BXA. The structure of A36V in complex with BXA indeed shows no notable changes in the binding mode of BXA to the protein when compared to WT (Figure 4). No large changes to α3 such as rotamer changes, or changes to the overall structure of the helix were observed (Figure 4). The change from alanine to valine does change the side chain from a methyl to an isopropyl which changes both the conformational entropy and hydrophobicity of α3.27 A36V is partially surface exposed so it is possible the mutation promotes local or global unfolding.28 If this is the case, the crystal structure of A36V would be very similar to the WT structure, and assays such as DSF which measure unfolding through reporter binding to hydrophobic residues would give lower values than the WT due to A36V promoting unfolding of the protein. Additional studies to determine the ΔG between the unfolded and folded states would be needed to unambiguously determine if the resistance mechanism for this mutation is linked to the instability of the protein.29 Attempts to grow crystals of A36V bound to compound 23 were not successful. This is likely because compound 23 does not sufficiently stabilize the A36V mutant (Table 1) to enable crystallization.
Figure 4.
(a) Co-crystal structure and overlay of BXA bound to WT (gray PDB: 6FS6) and A36V mutant (green, Val36 in bright pink PDB: 8T5 V) PAN. (b) Alternate view of co-crystal structure and overlay of BXA bound to WT (gray) and A36V mutant (green) PAN. The structure of A36V in complex with BXA shows essentially no changes in the binding conformation of the inhibitor.
Materials and Methods
Mutation Generation
Point mutations were generated in the PAN endonuclease pET 28a plasmid by QuikChange mutagenesis (Agilent). Each PCR reaction of 50 μL contained 50 ng of template, 125 ng of primer pair, 200 μM dNTPs, and 3 units of Pfu DNA polymerase. The PCR cycles were initiated at 95 °C for 1 min to denature the template DNA, followed by 12 amplification cycles. Each amplification cycle consisted of 95 °C for 50 s, 60 °C for 1 min, and 68 °C for 6 min. The PCR cycles were finished with an extension step at 68 °C for 7 min. The PCR products were treated with 5 units of DpnI at 37 °C for 1 h and transformed into ultra-competent cells and plasmid extracted. All mutations were first verified by Sanger Sequencing (Eton Biosciences) and then full, Non-Sanger plasmid sequencing (Primordium Laboratories) to ensure no other mutations occurred during the QuikChange prior to expression and purification.
Protein Expression and Purification
Expression and purification of PAN endonuclease were performed with slight modification (J. Med. Chem. 2019, 62, 9438–9449) and described in detail in the Supporting Information. Expression was induced by the addition of IPTG to a final concentration of 0.1 mM. The cultures were grown with vigorous shaking (250 rpm) overnight at room temperature. After ∼18 h, the cells were harvested by centrifuging at 2000g for 30 min at 4 °C. The resulting paste was stored at −80 °C prior to lysis.
Protein Crystallography
Purified protein for crystallization was stored at 2.2–4.3 mg/mL at −80 °C after flash freezing in buffer consisting of 150 mM sodium chloride, 20 mM HEPES (pH 7.5), 2 mM MgCl2, and 2 mM MnCl2. Co-crystallization and crystal soaking methods were used to obtain co-crystal structures of inhibitors bound to PAN endonuclease. BXA was purchased from Fisher and used without further purification. Compound 23 was previously synthesized according to a prior procedure (J. Med. Chem. 2019, 62, 9438–9449). For co-crystallization, protein was incubated with 0.5 mM inhibitor for 1 h on ice prior to setting the crystallization drops. For crystal soaking, fully formed holo crystals were transferred to a new drop containing 5 μL of reservoir solution and 1 μL of a 50 mM DMSO inhibitor stock solution (final concentration 8.3 mM). Crystals were left undisturbed overnight and either stored in liquid nitrogen or collected on an in-house X-ray diffractometer the following day. In both crystallization methods, crystals were grown using hanging drop and set in 24-well pregreased plates (Hampton HR3-171) with siliconized glass slides (Hampton HR3-231). A 5:1 ratio of purified protein to reservoir solution at room temperature was found to be the optimal ratio for the largest crystal formation. Reservoir solution consisted of 22–34% PEG (MW = 4000 g/mol), 100 mM Tris (pH 8.35), and 220 mM sodium acetate. Colorless crystals with hexagonal bipyramidal morphology appeared within 2 days and reached full size after 1–2 weeks. Crystals were typically 50–200 μm in diameter. Crystals were cryoprotected with perfluoroether (Hampton HR2-814) prior to flash freezing in liquid nitrogen. Crystals were stored in liquid nitrogen until data collection.
Biophysical Assays
DSF experiments were performed identically to (J. Med. Chem. 2019, 62, 9438–9449) and described in detail in the Supporting Information. SpS experiments were performed following manufacturer’s recommendation with slight modification. Protein was labeled using the Monolith X Protein Labeling Kit RED-NHS second Generation (NanoTemper Technologies, MO-L011) following the recommended procedure by the manufacturer. Labeled protein was frozen in 50 μL aliquots and stored at −80 °C to be used in future experiments. Prior to experiments, aliquots were thawed on ice and centrifuged for 10 min at 20 °C and 13 000 rpm to remove protein aggregates. Serial dilutions were prepared in assay buffer (1× MST buffer 50 mM Tris pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.05% Tween- 20, and 10% DMSO for BXA and 1X PBS pH 7.4 (Thermo Fisher, J62036-K7) with 0.1% pluronic F-127 for compound 23) in 384-well plates (Greiner) Reactions were initialized by gently mixing 20 μL of 40 nM protein to yield a final 1:1 solution and a volume of 40 μL per reaction. The reaction mixture was incubated on a plate shaker at room temperature, protected from light, at 300 rpm for 30 min. The reaction mixtures were loaded into premium capillaries (NanoTemper Technologies, MO-K025) and analyzed by Monolith X between 60 and 80% power. Compound 23 showed a ligand-induced fluorescent change and required an SD test to confirm the change in fluorescence. The SD test was performed according to the manufacturer’s instructions, and the remaining reaction after the run was used for the test.
Conclusions
This work describes the first crystal structures of BXA in complex with E23K and A36V PAN mutants and additional structures of BXA with WT and I38T. This study also examines another potent PAN endonuclease inhibitor (23). Compound 23 has been previously reported to make hydrophobic interactions with Ile38 and could be susceptible to active site mutations and loss of inhibition similar to BXA. New information regarding the consequences of these mutations on inhibitor binding to PAN endonuclease has revealed disruptions in crucial hydrophobic contacts, base stacking, and pi–cation interactions. The changes to active site binding of BXA with the I38T mutant have been replicated in this truncation model, and the weaker binding of BXA is attributed to less favorable van der Waals packing and induced fit changes. The change in binding of compound 23 with I38T was minimally disturbed, and only a small change in the position of the inhibitor in the active site was observed. In the E23K construct, Lys23, Tyr24, and Arg84 are more flexible and lead to destabilization of the binding of BXA without affecting the overall stability of the construct. Changes to the binding affinity of compound 23 with E23K were smaller, resulting in changes to the binding angle and changes in hydrogen bonding. E23K disrupts the end terminus of α2 and allows Y24 to extend further into the active site, disrupting important hydrogen-bonding, pi–cation, and base stacking interactions made by inhibitors or natural substrates such as RNA. E23K was found to interrupt hydrogen-bonding and cation interactions upon binding of both BXA and 23. The A36V mutant melts at around 10 °C lower than its WT, I38T, and E23K counterparts. Despite this, A36 V treated with BXA did not have a significant change in ΔTM. This suggested that the A36V mutation makes the endonuclease unstable and causes very minimal, or no change at all, to the binding of BXA. The structure of A36V with BXA was obtained and indeed showed no notable changes in the binding mode of BXA to the protein when compared to BXA binding to WT. Overall, the findings here may prove useful for the design of more efficient inhibitors against these resistant mutants by providing new insight into what drives resistance.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01 AI149444). This work was in part supported by beamlines 5.0.3 and 8.2.1 of the Advanced Light Source, a U.S. DOE Office of Science User Facility under Contract No. DE-AC02-05CH11231, is supported in part by the ALS-ENABLE program funded by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169-01. The authors acknowledge Camille Lawrence, Tufan Aydogdu, and Zackary Prag (NanoTemper Technologies) for their help with demo opportunities, resources, and help with data analysis using their Monolith X (a.k.a., spectral shift, SpS) technology. The authors acknowledge Lydia Madura and Giuseppe Destito (Kyowa Kirin) for their help with Monolith X training, data collection, and the use of their facilities. The authors acknowledge Jake Bailey and Milan Gembicky (U.C. San Diego, X-ray Crystallography Facility) for their help with data collection and analysis of X-ray diffraction data.
Glossary
Abbreviations
- FDA
Food and Drug Administration
- IC50
half-maximal inhibitory concentration
- PAN
N-terminal domain of the RNA-dependent RNA polymerase
- SAR
structure–activity relationship
- BXM
Baloxavir Marboxil
- BXA
Baloxavir Acid
- NPI
nonpharmaceutical intervention
- NAI
neuraminidase inhibitor
- MBP
metal-binding pharmacophore
- WT
wild type
- Kd
dissociation binding constant
- TM
thermal melting
- SpS
spectral shift
- FC
fold change
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.3c00536.
Generation of mutants, protein expression, purification, and crystallographic details (including crystallography tables and electron density maps); detailed experimental procedure for differential scanning fluorimetry and spectral shift/microscale thermophoresis (Figures S1–S8) (Tables S1–S3) (PDF)
Accession Codes
The UNIPROT entry for the N-terminal domain of the RNA-dependent RNA polymerase, PAN is C3W5S0.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported by a grant from the National Institutes of Health (R01 AI149444).
The authors declare the following competing financial interest(s): S.M.C. is a co-founder, has an equity interest, and receives income as member of the Scientific Advisory Board for Forge Therapeutics; is a co-founder, has an equity interest, and is a member of the Scientific Advisory Board for Blacksmith Medicines; and is a co-founder and has an equity interest Cleave Therapeutics (formerly Cleave Biosciences). These companies may potentially benefit from the research results of certain projects in the laboratory of S.M.C. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies.
Supplementary Material
References
- Xu X.; Blanton L.; Abd Elal A. I.; Alabi N.; Barnes J.; Biggerstaff M.; Brammer L.; Budd A. P.; Burns E.; Cummings C. N.; et al. Update: influenza activity in the United States during the 2018–19 season and composition of the 2019–20 influenza vaccine. Morb. Mortal. Wkly. Rep. 2019, 68, 544. 10.15585/mmwr.mm6824a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzillé G.; Poirier C.; Campillo-Gimenez B.; Aubert M.-L.; Chabot M.; Chazard E.; Lavenu A.; Cuggia M. Leveraging hospital big data to monitor flu epidemics. Comput. Methods Programs Biomed. 2018, 154, 153–160. 10.1016/j.cmpb.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Merced-Morales A.; Daly P.; Abd Elal A. I.; Ajayi N.; Annan E.; Budd A.; Barnes J.; Colon A.; Cummings C. N.; Iuliano A. D.; DaSilva J. Influenza activity and composition of the 2022–23 influenza vaccine–United States, 2021–22 season. Morb. Mortal. Wkly. Rep. 2022, 71 (29), 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D.-Y.; Milde J.; Ham Y.; Ramos Calderón J. P.; Wedde M.; Dürrwald R.; Duwe S. C. Preparing for the Next Influenza Season: Monitoring the Emergence and Spread of Antiviral Resistance. Infect. Drug Resist. 2023, 16, 949–959. 10.2147/IDR.S389263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampejo T. Influenza and antiviral resistance: an overview. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1201–1208. 10.1007/s10096-020-03840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S.; Speranzini V.; Hashimoto T.; Noshi T.; Yamaguchi H.; Kawai M.; Kawaguchi K.; Uehara T.; Shishido T.; Naito A.; Cusack S. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 2018, 8, 9633 10.1038/s41598-018-27890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin S.; Cusack S.; Ruigrok R. W.; Hart D. J. Influenza A virus polymerase: structural insights into replication and host adaptation mechanisms. J. Biol. Chem. 2010, 285, 28411–28417. 10.1074/jbc.R110.117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias A.; Bouvier D.; Crépin T.; McCarthy A. A.; Hart D. J.; Baudin F.; Cusack S.; Ruigrok R. W. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 2009, 458, 914–918. 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- Ince W. L.; Smith F. B.; O’Rear J. J.; Thomson M. Treatment-emergent influenza virus polymerase acidic substitutions independent of those at I38 associated with reduced baloxavir susceptibility and virus rebound in trials of baloxavir marboxil. J. Infect. Dis. 2020, 222, 957–961. 10.1093/infdis/jiaa164. [DOI] [PubMed] [Google Scholar]
- Govorkova E. A.; Takashita E.; Daniels R. S.; Fujisaki S.; Presser L. D.; Patel M. C.; Huang W.; Lackenby A.; Nguyen H. T.; Pereyaslov D.; et al. Global update on the susceptibilities of human influenza viruses to neuraminidase inhibitors and the cap-dependent endonuclease inhibitor baloxavir, 2018–2020. Antiviral Res. 2022, 200, 105281 10.1016/j.antiviral.2022.105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. Y.; Zhou J.; Koszalka P.; Frise R.; Farrukee R.; Baba K.; Miah S.; Shishido T.; Galiano M.; Hashimoto T.; et al. Evaluating the fitness of PA/I38T-substituted influenza A viruses with reduced baloxavir susceptibility in a competitive mixtures ferret model. PLoS Pathog. 2021, 17, e1009527 10.1371/journal.ppat.1009527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. C.; Kumar G.; Barman S.; Najera I.; White S. W.; Webby R. J.; Govorkova E. A. Identification of the I38T PA substitution as a resistance marker for next-generation influenza virus endonuclease inhibitors. MBio 2018, 9, 10–128. 10.1128/mBio.00430-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavish P. J.; Cuypers M. G.; Rimmer M. A.; Abdolvahabi A.; Jeevan T.; Kumar G.; Jarusiewicz J. A.; Vaithiyalingam S.; Jones J. C.; Bowling J. J.; et al. Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential. Eur. J. Med. Chem. 2023, 247, 115035 10.1016/j.ejmech.2022.115035. [DOI] [PubMed] [Google Scholar]
- Meng X.; Wang Y. Drug Repurposing for Influenza Virus Polymerase Acidic (PA) Endonuclease Inhibitor. Molecules 2021, 26, 7326. 10.3390/molecules26237326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G.; Cuypers M.; Webby R. R.; Webb T. R.; White S. W. Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism. Nucleic Acids Res. 2021, 49, 1609–1618. 10.1093/nar/gkaa1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflug A.; Guilligay D.; Reich S.; Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 2014, 516, 355–360. 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- Jones J. C.; Zagribelnyy B.; Pascua P. N. Q.; Bezrukov D. S.; Barman S.; Okda F.; Webby R. J.; Ivanenkov Y. A.; Govorkova E. A. Influenza A virus polymerase acidic protein E23G/K substitutions weaken key baloxavir drug-binding contacts with minimal impact on replication and transmission. PLoS Pathog. 2022, 18, e1010698 10.1371/journal.ppat.1010698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beylkin D.; Kumar G.; Zhou W.; Park J.; Jeevan T.; Lagisetti C.; Harfoot R.; Webby R. J.; White S. W.; Webb T. R. Protein-structure assisted optimization of 4, 5-dihydroxypyrimidine-6-carboxamide inhibitors of influenza virus endonuclease. Sci. Rep. 2017, 7, 17139 10.1038/s41598-017-17419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Credille C. V.; Morrison C. N.; Stokes R. W.; Dick B. L.; Feng Y.; Sun J.; Chen Y.; Cohen S. M. SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease. J. Med. Chem. 2019, 62, 9438–9449. 10.1021/acs.jmedchem.9b00747. [DOI] [PubMed] [Google Scholar]
- Naqui A. What does I50 mean?. Biochem. J. 1983, 215, 429. 10.1042/bj2150429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiveri C. M.; López-Méndez B.; Huecas S.; Alfonso C.; Luque-Ortega J. R.; Campos-Olivas R. A Biophysical Toolkit for Molecular Interactions. eLS 2017, 1–16. 10.1002/9780470015902.a0027015. [DOI] [Google Scholar]
- López-Méndez B.; Baron B.; Brautigam C. A.; Jowitt T. A.; Knauer S. H.; Uebel S.; Williams M. A.; Sedivy A.; Abian O.; Abreu C.; et al. Reproducibility and accuracy of microscale thermophoresis in the NanoTemper Monolith: a multi laboratory benchmark study. Eur. Biophys. J. 2021, 50, 411–427. 10.1007/s00249-021-01532-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavagnoli L.; Cucuzza S.; Campanini G.; Rovida F.; Paolucci S.; Baldanti F.; Maga G. The novel influenza A virus protein PA-X and its naturally deleted variant show different enzymatic properties in comparison to the viral endonuclease PA. Nucleic Acids Res. 2015, 43, 9405–9417. 10.1093/nar/gkv926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C.; Lou Z.; Guo Y.; Ma M.; Chen Y.; Liang S.; Zhang L.; Chen S.; Li X.; Liu Y.; et al. Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center. J. Virol. 2009, 83, 9024–9030. 10.1128/JVI.00911-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Credille C. V.; Chen Y.; Cohen S. M. Fragment-based identification of influenza endonuclease inhibitors. J. Med. Chem. 2016, 59, 6444–6454. 10.1021/acs.jmedchem.6b00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karges J.; Stokes R. W.; Cohen S. M. Computational Prediction of the Binding Pose of Metal-Binding Pharmacophores. ACS Med. Chem. Lett. 2022, 13, 428–435. 10.1021/acsmedchemlett.1c00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig A. J.; Sternberg M. J. Side-chain conformational entropy in protein folding. Protein Sci. 1995, 4, 2247–2251. 10.1002/pro.5560041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn P. M.; Janovick J. A. A New Understanding of Protein Mutation Unfolds: Long believed to be nonfunctional, mutant proteins that cause disease can often behave normally when refolded with the help of a pharmacological template. Am. Sci. 2005, 93, 314–321. 10.1511/2005.54.967. [DOI] [Google Scholar]
- Lattman E. E.; Rose G. D. Protein folding--what’s the question?. Proc. Natl. Acad. Sci. U.S.A. 1993, 90, 439–441. 10.1073/pnas.90.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.