Abstract
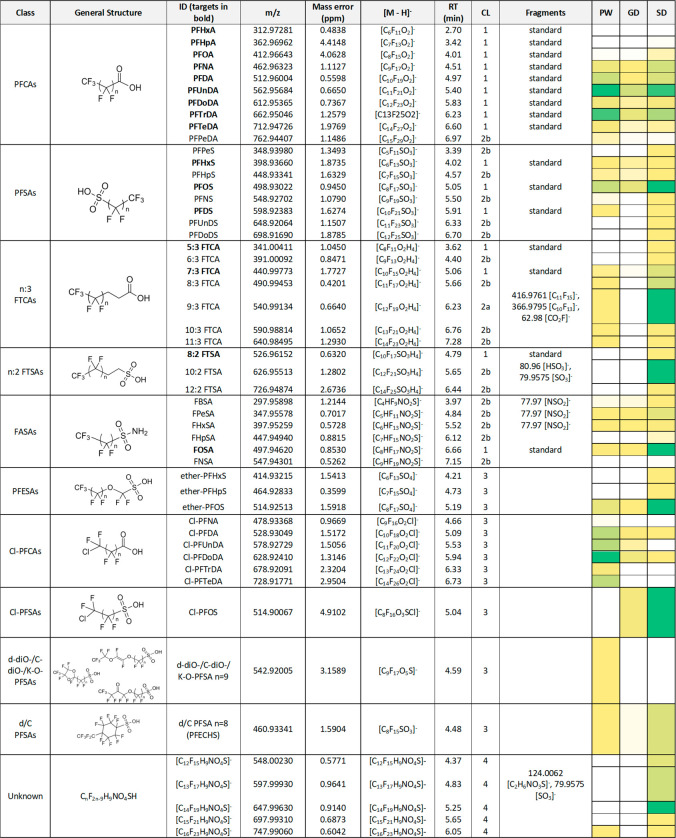
High-resolution mass spectrometry (HRMS)-based suspect and nontarget screening has identified a growing number of novel per- and polyfluoroalkyl substances (PFASs) in the environment. However, without analytical standards, the fraction of overall PFAS exposure accounted for by these suspects remains ambiguous. Fortunately, recent developments in ionization efficiency (IE) prediction using machine learning offer the possibility to quantify suspects lacking analytical standards. In the present work, a gradient boosted tree-based model for predicting log IE in negative mode was trained and then validated using 33 PFAS standards. The root-mean-square errors were 0.79 (for the entire test set) and 0.29 (for the 7 PFASs in the test set) log IE units. Thereafter, the model was applied to samples of liver from pilot whales (n = 5; East Greenland) and white beaked dolphins (n = 5, West Greenland; n = 3, Sweden) which contained a significant fraction (up to 70%) of unidentified organofluorine and 35 unquantified suspect PFASs (confidence level 2–4). IE-based quantification reduced the fraction of unidentified extractable organofluorine to 0–27%, demonstrating the utility of the method for closing the fluorine mass balance in the absence of analytical standards.
Keywords: Combustion ion chromatography, high resolution mass spectrometry, suspect screening, ionization efficiency-based quantification, dolphins, cetaceans
Short abstract
A machine-learning log IE prediction model was trained and validated using a suite of analytical PFAS standards; thereafter, the model was applied to quantify PFAS suspects observed in marine mammal livers.
Introduction
Per- and polyfluoroalkyl substances (PFASs) are defined as chemicals containing at least one fully fluorinated methyl (−CF3) or methylene (−CF2−) group without hydrogen, chlorine, bromine, or iodine atoms.1 According to PubChem, > 7 million substances fall under this definition, although the commercial relevance of all of these chemicals remains unclear.2 PFASs are widely used in industrial or consumer applications, with over 200 different uses for more than 1400 individual substances.3 While many PFASs are hazardous4 and/or prone to large-range transport,5,6 persistence is considered a common property of the entire class and the principal cause for concern.7
Due to their large number and diversity of structures, PFAS contamination in the environment is highly complex. Recent studies involving organofluorine mass balance and suspect screening have determined that exposure to PFASs in marine mammals using targeted analytical methods can be considerably underestimated.8−11 Given their position at the top of the marine food web, marine mammals are used globally for monitoring persistent and bioaccumulative pollutants in the marine environment.12,13 Moreover, since they partly share their diet with humans (e.g., fish, crustaceans, cephalopods) and are regularly consumed by Arctic communities , marine mammals can be used as early indicators of emerging contaminants relevant for human exposure through diet and other environmental exposure media.14
The large number of suspect PFASs detected by liquid chromatography high-resolution mass spectrometry (LC-HRMS) and relatively few analytical standards for quantification has provided considerable impetus for development of alternative quantitative approaches. Traditionally, analytical standards are required for quantification in LC-HRMS in order to compensate for the wide range of ionization efficiency (IE) among chemicals, i.e., the number of ions produced in the ionization source from a given concentration of analyte. IE is highly variable even within one group of structurally similar chemicals.15 For example, when measured under the same conditions and concentrations, 2,4-dinitrophenol can produce a 380-fold higher signal compared to 2-nitrophenol.16 For PFAS analysis, it is common to quantify substances that lack analytical standards using a homologue that is close in carbon chain length. For example, the concentration of perfluoroheptanesulfonic acid (PFHpS) has been estimated based on the calibration curve of perfluorohexanesulfonic acid (PFHxS).17 In recent years, machine learning-based models that incorporate molecular and eluent descriptors have shown considerable potential for predicting IE. Compared to the homologue quantification approach, machine learning models have a clear advantage: they can be used for any carbon number as well as for chemicals for which no analytical standards are available in the homologue series. The application of such tools depends on the chemical space covered by the data used for training these models; therefore, they need to be retrained and revalidated to expand the application domain for new chemical classes.
The overarching goal of this study was to develop a quantification tool to determine the organofluorine mass balance in environmental samples in the absence of analytical standards. To achieve this goal, we increased the scope of a previously published quantification model by including 33 additional PFASs in model training. Thereafter, the model was applied to quantify PFASs lacking an analytical standard detected via suspect screening in marine mammal livers, thereby reducing the fraction of unidentified extractable organofluorine (UEOF) in these samples. To the best of our knowledge, this is the first time a machine learning-based quantitative approach has been used to help close the organofluorine mass balance in environmental samples.
Materials and Methods
Sample Collection
Liver samples were collected from 5 long-finned pilot whales (Globicephala melas) from East Greenland and 5 white-beaked dolphins (Lagenorhynchus albirostris) from West Greenland in 2018 and 3 white-beaked dolphins from the West coast of Sweden between 2007 and 2009. All animals from Greenland were obtained as part of subsistence hunting, while individuals from Sweden were found stranded. Samples will be referred to in relation to their sampling location, but it is important to note that these animals can travel long distances during their lives. All samples were stored at −20 °C until analysis. Portions of liver from one pilot whale and one dolphin were analyzed in triplicate, and their relative standard deviation (RSD) was used for the other individuals of the same species. Further details on sample collection, including shipping permits from the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), can be found in Table S1.
Chemicals and Reagents
Both native- and isotopically labeled internal standards (ISs) included in the targeted analysis were purchased from Wellington Laboratories (Guelph, Canada) and are listed in Table S2. Among the 33 target PFASs were 11 perfluoroalkyl carboxylic acids (PFCAs, C5–14,16), 4 perfluoroalkyl sulfonic acids (PFSAs, C4,6,8,10), 3 fluorotelomer carboxylic acids (3:3, 5:3, and 7:3 FTCAs), 3 fluorotelomer sulfonic acids (4:2, 6:2, and 8:2 FTSAs), 3 (N-alkyl) perfluoroalkane sulfonamides (FASAs; FOSA, MeFOSA, and EtFOSA), 3 (N-alkyl) perfluoroalkane sulfonamidoacetic acids (FASAAs; FOSAA, MeFOSAA, and EtFOSAA), 3 polyfluoroalkyl phosphoric acid diesters (6:2, 6:2/8:2, and 8:2 diPAP), 2 chlorinated perfluorinated ether sulfonates (Cl-PFESAs; 9Cl-PF3ONS [also known as 6:2 Cl-PFESA] and 11Cl-PF3OUdS [also known as 8:2 Cl-PFESA]), and 4,8-dioxa-3H-perfluorononanoic acid (ADONA). Other chemicals and reagents are provided in the SI.
Sample Preparation for Organofluorine Mass Balance
A schematic overview of the organofluorine mass balance approach used in this study can be found in Figure S1. Liver samples were thawed at room temperature, and subsampling was carried out with a stainless-steel knife precleaned with methanol. Extraction was carried out using a previously published method,9 which is described in detail in the SI. Briefly, ∼0.5 g of liver was extracted twice with acetonitrile and bead blending followed by a dispersive carbon cleanup. The final extract was split into two aliquots of 250 μL each. The first aliquot (destined for HRMS-based target and suspect screening) was transferred to another Eppendorf tube and fortified with ISs and buffer. The second aliquot, destined for extractable organofluorine (EOF) analysis by Combustion Ion Chromatography (CIC), was transferred to a new Eppendorf tube. All extracts were stored at −20 °C. Upon analysis, the extracts were adjusted to room temperature, vortexed, and transferred to LC vials.
Instrumental Analysis
EOF Analysis
CIC measurements were carried out with a Thermo-Mitsubishi CIC and based on previously described methods (see Table S3 and the Supporting Information section on EOF analysis for details).9,18 Briefly, samples were combusted for 5 min at 1100 °C under a flow of gases, which were absorbed in Milli-Q water. A portion of this water was injected onto an IC for determination of fluoride. The mean fluoride concentration from procedural blanks was subtracted from the samples before quantification and the limit of quantification (LOQ; 29.3 ng F/g) was calculated using 3 times the standard deviation of fluoride in the procedural blanks (n = 3 for each batch).
Target Analysis and Suspect Screening
Target and suspect screening analyses were carried out simultaneously using a previously validated method,9,19 described in detail in the SI. Briefly, sample extracts were injected onto a Dionex ultra high-performance liquid chromatograph (UHPLC) equipped with a C18 column and coupled to a Q-Exactive OrbitrapTM mass spectrometer via an electrospray ionization (ESI) source. The instrument was operated in negative ionization, full-scan (200–1800 m/z, resolution 120 000) data-dependent MS2 acquisition (DDA, resolution 15 000) mode, based on an inclusion list of molecular ions for 324 known PFASs. The list was established from prior publications reporting novel PFASs and monofluorinated substances in marine mammals, birds, and fish and is provided as Table S4.9,10,20−23
Quantification of targets was carried out using Thermo Scientific TraceFinderTM Software version 4.1. Relative response factors were used for quantification in the linear range with 1/x weighting. A second data processing method was created in TraceFinderTM for suspects, and those with peak heights > 10 000 cps were considered significant. Using the Thermo Excalibur Qual browser, each detected suspect’s MS2 spectra were checked for fragments which could confirm their identity by comparing with literature information. Confidence Levels (CLs) were assigned to suspects according to the scale proposed by Schymanski et al. (see overview in the SI).24
Quality Control
A suite of QC samples was used to ensure the accuracy and precision of EOF and target PFAS data. This included: spiked liver (with native nonisotopically labeled PFASs and with inorganic fluorine, both n = 3), procedural blanks (n = 3), and analysis of certified reference materials (fluorine in clay, n = 3). Spiked liver and procedural blanks were analyzed in the same manner as other samples, and background subtraction (either using unspiked liver for spiked QC samples and procedural blanks for liver samples) was performed prior to calculations. Results of these experiments, which are described in detail in the SI, demonstrated acceptable accuracy and precision across all analyses.
Developing Ionization Efficiency Prediction Model for Quantification
Data Preparation
Calibration curves based on molar concentration vs peak area (weighting: 1/concentration; including intercept) were constructed for 33 target PFASs using experimental data of the calibration mix from the targeted analysis of the samples. To account for all ions formed through the same ionization mechanism, peak areas from [M-H]− ions and their in-source fragments were summed to provide the signal for each compound in the calibration mix. The in-source fragments were identified by a careful manual evaluation of the full-scan spectra. Additionally, the detected monoisotopic mass peak area was corrected for uncounted isotope peaks by calculating the isotope distribution from the molecular formula. Absolute response factors (RFs) were obtained as the slopes of the calibration graph for each compound. The linearity was checked based on relative residuals and considered acceptable if the relative residual with highest absolute value was ≤20%.
The RFs were used to expand a previously combined data set by Liigand et al.16 For this, the RFs of 33 target PFASs were converted to relative IE values by anchoring the data using the response factor of PFOS that was present in both data sets. To calculate the IE value for any of the 33 remaining PFASs (IEM), the following equation was used
| 1 |
where RFM is the experimental response factor of the respective PFAS, RFPFOS is the experimental response factor of PFOS measured in the same sequence as the respective PFAS, and IEPFOS is the IE value of PFOS from the data set of Liigand et al.16 Experimental conditions such as organic modifier percentage during elution, pH, and additives used in the measurements affect the IE of a chemical. In the Liigand et al. data set, experimental conditions were as follows: pH of 7.8 with an ammonium acetate buffer and an organic modifier content of 80% acetonitrile at time of elution, resulting in the relative ionization efficiency value of log IEPFOS = 2.59 log-units. For the experimental response factor of PFOS obtained in the target analysis of this study, the conditions were as follows: organic modifier content 52% acetonitrile at time of elution and pH of 7.0 with 1 mM ammonium acetate buffer resulting in RFPFOS = 11.56 log-units. These conditions were considered sufficiently similar to assume differences in ionization efficiency as insignificant. Additionally, all IE values in this study are relative.
Modeling
Original Data Set for Modeling
The first model was trained using 100 unique chemicals measured in negative mode under different experimental conditions such as organic modifier percentage, mobile phase pH, and additive type, resulting in 1286 data points.16 Out of the 100 chemicals, 19 were fluorinated: PFOS, perfluoro-tert-butanol, 11 aromatic PFASs, and 6 aromatic organofluorine compounds that do not fall under the PFAS definition.
Data Set with Additional PFASs
The ESI negative mode prediction model developed in this study was trained on a set of 132 unique chemicals (including 33 PFASs measured for this study and 13 that were present in the original data set compiled by Liigand et al.)16 and altogether 1319 data points measured under different eluent conditions.
Descriptors for Modeling
For training the model, the chemical structures were translated into numerical molecular descriptors. Pharmaceutical Data Exploration Laboratory (PaDEL)25 molecular descriptors have shown good predicting power for IEs16,26,27 and were therefore used in the present work. These molecular descriptors include both structural information such as atom and bond count descriptors as well as more complex descriptors such as Topological Distance Matrix and Electrotopological State Atom Type descriptors, which were calculated off-line from the Simplified Molecular-Input Line-Entry System (SMILES) notation of a chemical with code provided by the Chemical Development Kit. In addition to PaDEL descriptors, five eluent descriptors were used (aqueous pH, polarity index, viscosity, surface tension, and presence of NH4+) to account for the effect of the mobile phase composition.
Prior to modeling, the data set was cleaned to remove correlated or noninformative columns to avoid overtraining (details in the SI). For data processing, feature calculations and modeling, R version 4.1.1 was used. The code, data, and models are provided on GitHub page https://github.com/kruvelab/PFAS_quantification_model.
Model Performance Evaluation Parameters
The model’s prediction power was assessed based on the root-mean-square error (RMSE) of the training and test sets, as well as calculating the fold prediction error for IE and concentration predictions using the following equation:
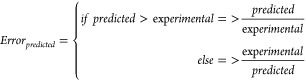 |
2 |
The mean, median, and geometric mean of fold prediction errors were used to compare model performances.
Ionization Efficiency Prediction Model Training
To evaluate the model’s performance after adding PFASs to the training of the model, the data was divided into training (80%) and test (20%) sets with stratified sampling so that 80% of PFASs would be in the training set. The regression model for log IE predictions was trained using the extreme gradient boosting tree (xgbTree) algorithm. The hyperparameters were optimized with the bootstrap resampling method (boot) using cross-validation with five sets. As chemicals in the original data set had multiple log IE values that were measured under different experimental conditions, all data points were used in training and testing the model; however, cross-validation sets were generated using chemical names with the groupKFold() function to avoid the same chemical ending up in both training and test subsets. For the final model, the optimized hyperparameters were used to train a model based on all data in the combined data set. More detailed model analysis and the 10 most impactful descriptors in the final model can be found in Table S5.
Leave-one-out Modeling Approach for Quantification
To compare the performance of prediction for individual PFASs between the model trained on the original data set and the model trained with the additional PFAS data, the leave-one-out approach was used. For this, 33 models were trained - one for each of the added PFASs. In these models, the individual PFAS being evaluated was left out from the training data set, while all other data points were used for modeling. Then the model was used to predict the log IE for the PFAS that was left out, and the results were compared to the experimental log IE of the respective PFAS.
Homologue Series Approach for Quantification
One approach for quantifying a PFAS without an analytical standard is to use another PFAS within the same homologue series (i.e., with a different carbon chain length). Therefore, we compared the performance of quantification using predicted log IE values versus using the homologue series approach. For this, we determined all close homologues present in our data set and used the response factors of these chemicals for quantification. In total, there were 10 homologues that had a carbon chain difference of −CF2– and 26 with a difference of −C2F4–. In case smaller and larger homologues were present, both were used for quantification, and the results were averaged. The overview of all homologues and comparison between model and −C2F4– homologue quantification can be found in Tables S6–S7 and Figure S2.
Calculating Concentration Using Predicted Ionization Efficiencies
For quantification approaches using a homologue (i.e., homologue series approach), the RF of a structurally similar PFAS is assumed to be insignificantly different from the detected chemical, and therefore, the RF of the homologue is used instead to calculate the concentration:
| 3 |
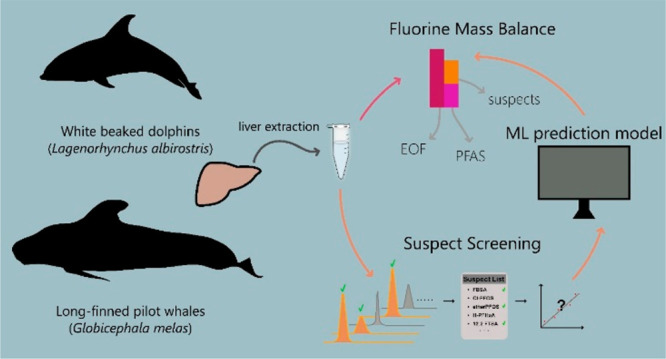
In the case of the IE-prediction model, the predicted log IE indicates how the RF of a respective chemical relates to other chemicals and is instrument independent. However, RFs are instrument- and lab-specific, meaning that the magnitude of RF depends on the source design, instrument vendor, and to some extent even on the software used for data integration (units, peak picking, integration, etc.). Therefore, predicted IE needs to be converted to measurement-specific RF before quantification. For this, the 33 target PFASs could be used as calibrants because they were measured together with the suspects we aimed to quantify. For the target PFAS, a calibration graph between measurement-specific response factors and predicted IEs was constructed. Using this calibration graph, the log IE was predicted for suspect chemicals detected in liver samples, converted into predicted RF, and used to calculate the concentration (Figure 1).
Figure 1.
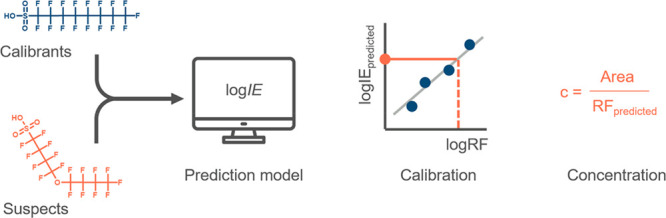
Workflow for obtaining predicted concentrations for suspects detected in liver samples using predicted log IE values. The 33 target PFASs were used as calibrants for converting the predicted log IE of the suspect chemicals to the predicted response factor, which was used in concentration estimations.
Statistical Significance
Due to non-normal and asymmetrical distribution of the fold prediction errors, the statistical significance between errors arising from different quantification approaches was tested with paired two-sided Wilcoxon signed rank tests using a level of significance of α = 0.05.
Fluorine Mass Balance Calculations
To facilitate comparisons with EOF measurements, PFASs quantified using analytical standards or the IE-prediction model were converted to fluorine equivalent concentrations (i.e., CF_PFAS in ng F/g):
| 4 |
CPFAS is a PFAS concentration in ng/g, nF is the number of fluorine atoms on the compound, AF is the average atomic weight of one fluorine atom in g/mol, and MWPFAS is its molecular weight. The sum of concentrations of target PFASs (i.e., ∑PFAS) or suspects (i.e., ∑Suspects) was obtained by summing CF_PFAS values of a sample for individual targets or suspects, respectively.
Results and Discussion
Targeted Analysis and Organofluorine Mass Balance
A total of 16 out of 33 target PFASs were quantified in one or more samples (Table S8). Concentrations are displayed here with up to 2 significant figures, and raw concentrations can be found in the SI (Tables S8 and S9). The highest average ∑PFAS concentrations were observed in dolphins from Sweden (620 ± 220 ng/g wet weight [ww]; 410 ± 150 ng F/g ww), followed by East Greenland pilot whale (220 ± 64 ng/g ww; 150 ± 43 ng F/g ww) and West Greenland dolphin (78 ± 14 ng/g ww; 53 ± 10 ng F/g ww). Across all Greenlandic samples, PFAS profiles were dominated by PFOS (up to 94 ng/g), PFUnDA (up to 66 ng/g), and PFTriDA (up to 58 ng/g), which collectively accounted for ∼72% of ∑PFAS (and ∼90% when including all PFCAs), consistent with prior observations in cetaceans from the Nordic environment.9,28 Swedish dolphins also displayed high PFOS concentrations (up to 300 ng/g) but with significant contributions from 7:3 FTCA (up to 220 ng/g) and FOSA (up to 250 ng/g), collectively accounting for ∼80% of ∑PFAS (Figure S3). 7:3 FTCA is a stable transformation product of fluorotelomer alcohols29−31 and has been previously measured in marine mammals and seabirds globally,9−11,20,32−34 while FOSA has been previously reported at elevated concentrations in cetaceans9,10,20,35,36 due to the limited capacity of these species to biotransform FOSA into PFOS.37,38
EOF concentrations were also highest in dolphins from Sweden (1200 ± 600 ng F/g ww) followed by pilot whales from East Greenland (210 ± 78 ng F/g ww) and dolphins from West Greenland (33 ± 4 ng F/g ww; Figure 2, Table S9). The significantly higher EOF concentrations in Swedish dolphins might be due to proximity to more industrialized coastal regions, but considering differences in sampling time periods (the Swedish samples are ∼10 years older than the Greenlandic samples), geographical comparisons should be interpreted cautiously.
Figure 2.
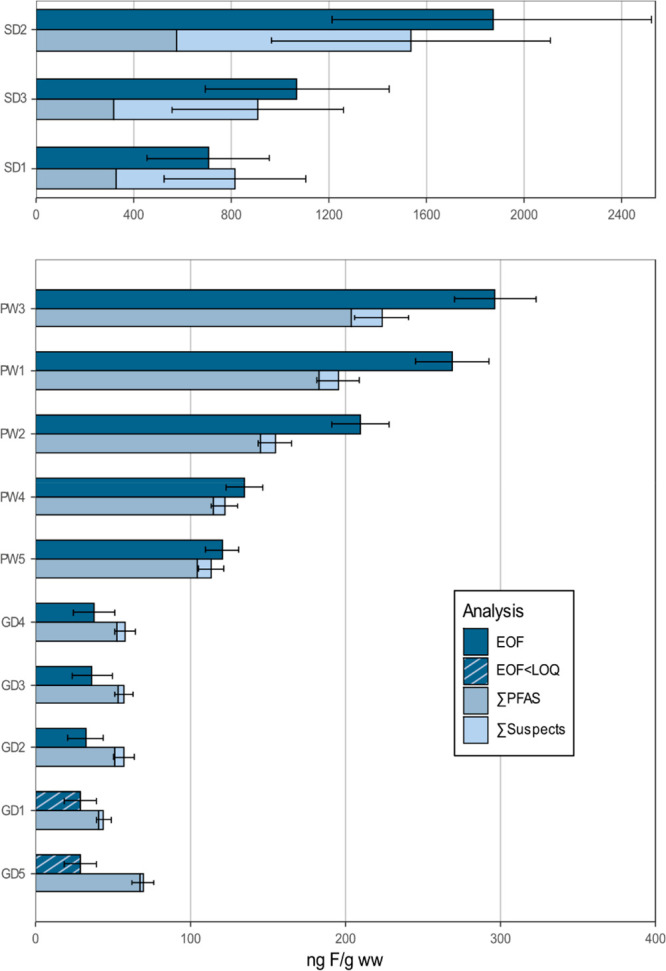
Fluorine mass balance for dolphins from Sweden (SD), pilot whales from East Greenland (PW), and dolphins from West Greenland (GD) in decreasing order of EOF concentrations from top to bottom. Model error is not plotted here, error bars are based on relative standard deviation observed for triplicate measurements of GD3 (for dolphins) and PW-3 (for pilot whales), and the standard deviations of ∑PFAS and ∑Suspects are combined. GD1 and -5 have EOF < LOQ; therefore, the LOQ value (29.3) was plotted here.
A comparison of target PFASs to EOF concentrations revealed a significant gap in the fluorine mass balance in Swedish dolphins (53–70% UEOF), while long-finned pilot whales from East Greenland displayed a modest fraction of UEOF (13–32%) and dolphins from West Greenland had a closed fluorine mass balance. Similar observations have been made previously in marine mammals from the Nordic and Baltic environments. For example, Spaan et al. observed largely closed fluorine mass balances in marine mammals from Sweden and Greenland, with the exception of some apex predators (polar bears and killer whales).9 Similarly, Kärrman et al. reported a white beaked dolphin from East Greenland (2016) with EOF below LOQ (158 ng F/g) and only one out of five studied pilot whales from the Faroe Islands (sampled 2017) with EOF above the LOQ (42 ng F/g) and a significant quantity of UEOF (80%).28 Additionally, a pooled sample of 6 white beaked dolphins from East Greenland (2016) had a concentration of ∼250 ng F/g,28 similar to pilot whales in the present study from the same region. Overall, EOF and fluorine mass balances were similar to those reported previously.
Suspect Screening
A total of 35 suspects from 11 classes were detected between CLs 2 to 4. A summary of targets and suspects observed in this study can be found in Table 1, along with details on their m/z, formula, retention times, assigned CL, and observed fragments ions. Of the 35 suspects, 18 were part of a homologue series for which at least one homologue was included as a target (i.e., PFCAs, PFSAs, n:3 FTCAs, n:2 FTSAs, and FASAs). Three additional homologue series (encompassing 14 suspects), for which no homologue was included as part of the targeted analysis, were also discovered: chlorine substituted perfluorocarboxylic acids (C9 to C14), ether-PFSAs (C6 to C8), and a homologue series with formula CnF2–9H9NO4SH (C12 to C16) but unknown structure. The 3 remaining suspects were not part of a homologue series. Suspects are discussed by class in more detail below.
Table 1. Heatmap of PFASs Identified by Target (in Bold) and Suspect Screening in Pilot Whales (PW, n = 5), West Greenland’s Dolphins (GD, n = 5), and Swedish Dolphins (SD, n = 3)a.
White represents nondetect and dark green represents the highest detect among sample groups. A different heatmap was created for each homologue series, except for the three compounds not part of a series, for which a joint heatmap was created. RT = retention time and CL= confidence level.
Classes 1 and 2: Perfluoroalkyl Acids
An additional PFCA (perfluoropentadecanoic acid, PFPeA) and five additional PFSAs were identified and assigned CL 2b, given MS2 data were unavailable, but RTs fitted the homologue series (Figures S4 and S5).
Classes 3 and 4: Fluorotelomer n:3 Carboxylic and n:2 Sulfonic Acids (n:3 FTCAs and n:2 FTSAs)
Five FTCAs in addition to the targets 5:3 and 7:3 FTCA were identified, of which 9:3 was assigned CL 2a from its MS2 matching the literature, and the others were assigned level 2b (Figures S6 and S7). Two FTSAs in addition to 8:2 FTSA were identified at level 2b: 10:2 and 12:2 FTSA, no MS2 was collected for 10:2 FTSA, while 12:2 FTSA showed [HSO3]− and [SO3]− fragments (Figures S8 and S9).
Class 5: Perfluoroalkane Sulfonamides (FASAs)
Five FASAs in addition to the targeted FOSA were identified at level 2b given the increasing RTs with an increasing chain length (Figure S10). Perfluorobutane, -pentane, and -hexane sulfonamide (FBSA, FPeSA, and FHxSA) MS2 spectra had the characteristic [NSO2]− fragment (Figure S11), while MS2 was not triggered for perfluoroheptane and -nonane sulfonamide (FHpSA and FNSA).
Class 6: Chlorine Substituted PFCAs (Cl-PFCAs)
Six Cl-PFCAs (C9 to C14) were identified at level 3 since RTs increased with increasing chain length, but no MS2 data were acquired (Figure S12); therefore, the position of the chlorine substitution is not known, and only a probable structure is assigned. The presence of chlorine substitution is confirmed by the observation of the [37ClCnF2n–2O2]− isotopologues, with approximately 30% relative abundance of [35ClCnF2n–2O2]−, for all homologues in the series (an example is shown in Figure S13).
Class 7: Ether PFSAs (PFESAs)
Ether-PFOS was found in samples of Swedish dolphins and Greenlandic pilot whales as well as in one Greenlandic dolphin; ether-PFHxS and ether-PFHpS were found in one sample of Swedish dolphins with increasing RT with increasing chain-length, but no MS2 data was obtained for this homologue series (Figure S14).
Classes 8 to 10
Three additional suspects, not part of a homologue series, were also discovered with CL 3: chlorine substituted perfluorooctanesulfonic acid (Cl-PFOS, from the Cl-PFSAs class), double bond-diether/cyclic-diether/ketone-ether perfluorononanoic sulfonic acid (d-diO/C-diO/K–O PFSA n = 9), and double bond/cyclic PFOS (d/C PFSA n = 8 or PFECHS).
Class 11: CnF2n–9H9NO4SH
An unknown homologue series, previously reported in marine mammals9 and in white-tailed sea eagle eggs,34 was also observed here, with molecular formula CnF2–9H9NO4SH (n = 12 to 16). It was not possible to assign a probable structure to this class (Figures S15 and S16).9
With the exception of the chlorine substituted substances and d-diO/C-diO/K–O PFSA n = 9, most of the aforementioned suspects were previously identified in various marine mammals from Greenland and Sweden by Spaan et al.9 Cl-PFCAs have been previously detected in wastewater from fluorochemical manufacturing parks,39,40 fish from the Yangtze river,41 and eggs of white-tailed sea eagle.34 Cl-PFSAs have also been previously identified in wastewater from a fluorochemical manufacturing park40 and in firefighters,42 and Cl-PFOS specifically has been identified in samples of marine mammals from the south China sea.10 D-diO/C-diO/K–O PFSA n = 9 has been previously identified in fish samples from Tangxun Lake, China.41
Ionization Efficiency Model Development and Validation
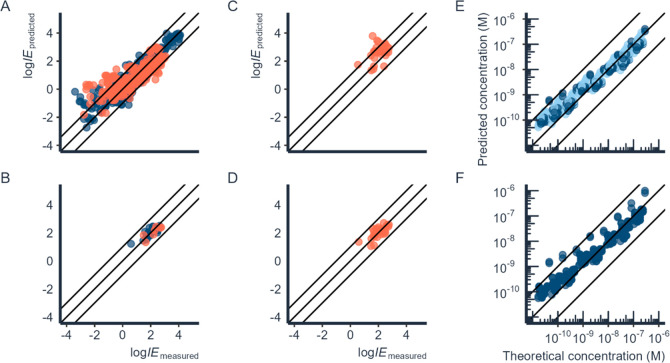
The new IE model was developed on a combined data set including 33 PFASs and 100 compounds measured previously by Liigand et al.16 with one chemical - PFOS - occurring in both data sets. The RMSEs over all data points in training and test sets were 0.43 and 0.79 log IE units, respectively. This corresponds to fold errors of 2.7 and 6.2 (Figure 3A). The mean, geometric mean, and median prediction errors for the test set were 19.6×, 3.6×, and 2.3×, respectively. For the added PFAS data, the RMSEs of the training and test sets were 0.26 (1.8×) and 0.29 (2.0×) log IE units, respectively (Figure 3B). The mean error was 1.9×, the geometric mean error was 1.8×, and the median error was 2.1× on the test set. Out of seven PFASs in the test set, the highest fold prediction error of 2.3× was obtained for 11Cl-PF3OUdS. In comparison, the RMSE obtained by Liigand et al.16 for negative mode was 2.0× and 2.3× on training and test sets, respectively. The highest prediction errors for the whole test set were observed for 2-methoxyphenol (831.8×), 4-phenylphenol (563.0×), and 2-nitrophenol (465.8×). These chemicals have low IEs (<0 log IE units) and are potentially more difficult to model. The training and test set chemicals were joined into one data set, and a new model was trained with previously optimized hyperparameters to further use the model for quantification of suspect PFASs.
Figure 3.
Training (dark blue) and test (orange) sets of the ESI negative mode prediction model for A) non-PFAS chemicals and B) PFASs. The correlation plots for experimental log IE values for PFASs compared to C) predicted log IE values using a model without added PFASs and D) predicted log IE values obtained from the leave-one-out approach. The correlation between spiked concentrations and E) concentration estimations using the response factor of a homologue chemical that is one −CF2– unit smaller (light blue) or larger (dark blue) compared to the respective suspect and F) concentration estimations using predicted ionization efficiency for the same set of chemicals with a leave-one-out approach.
To investigate the improvement in model predictions for PFASs, we trained a model based on the original data set used by Liigand et al.16 and predicted log IEs for each PFAS. The Liigand et al.16 data set contained 19 fluorinated compounds but only PFOS from the perfluoroalkyl acids subclass. For comparison, we trained 33 models using the leave-one-out approach to predict log IEs for PFASs. The model built on data without adding PFASs resulted in an RMSE of 0.82 log-units (6.6×) on the data set of 33 target PFASs, while using the leave-one-out approach resulted in an RMSE of 0.29 log-units (2.0×). The mean, geometric mean, and median obtained were 11.1×, 4.7×, and 4.8× for the model without PFASs and 2.1×, 1.9×, and 1.6× with the leave-one-out approach, respectively (Figures 3C and 3D). Therefore, a significant improvement was observed in model performance by adding more PFASs to the negative mode IE-prediction model (Wilcoxon signed rank test; p = 7.54 × 10–5). A similar improvement in prediction accuracy has also been reported previously for hydroxylated polychlorinated biphenyls.43
Comparing predicted log IE to using a response factor of a structurally similar homologue series compound for quantification, the performance of the two approaches was statistically indistinguishable (Wilcoxon signed rank test; p = 0.25; Figures 3E and 3F). For additional comparison of quantification results with the machine learning model and homologues with a difference of −C2F4–, we direct the reader to Figure S2 in the Supporting Information. Nevertheless, predicting IE allows us to evaluate the concentration in cases where no homologues are present or if the detected feature cannot be identified at a high level of confidence but multiple candidate structures are present.
Model Validation
Quantification in Spiked Biota Samples
To evaluate the performance of the ionization efficiency predictions in real samples, the models from the leave-one-out approach were used to quantify target PFASs in three liver samples of PW-4, which were spiked with 5 ng of the native PFAS mixture (see Materials and Methods). The predicted concentrations for the targets were compared with the concentrations obtained by quantification using the calibration curve of the respective analytical standards. The mean fold difference between model and target quantification was 2.1×, and in 73% of the cases, quantification with an analytical standard resulted in higher concentrations compared to model quantification (see Figure S17)
Prediction for Different Isomers
To assess the effect of isomerization on predicted concentrations, we applied the model to quantify different linear isomers of two CL 3-suspects: Cl-PFNA, using all possible positions of the chlorine substitution along the chain, and ether-PFOS, using all possible positions of the ether linkage. For both substances, we observed that predicted concentrations remain within the same order of magnitude for different linear isomers (Figures S18 and S19); however, branched isomers were not investigated and might show a higher variability in predicted log IE (and by extension concentrations) due to inductive effects on the polar headgroup which could lead to changes in pKa.44
Suspect Quantification with the Final Model
The final model trained on all available IE data was used for quantification of CL 2–3 suspects, and the predicted concentrations can be found in Table S10. In the case of CL3 suspects, the structure of the linear isomer has been used for quantification (except for PFECHS). Additionally, in the case of a chlorine substitution, the structure with Cl on the last carbon was used, and for PFESAs, the structure with ether between the first and second carbon was used. For the d-diO/C-diO/K–O PFSA n = 9 suspect, a double bond/diether structure has been selected, and its SMILE is available in Table S10. The applicability of the IE-prediction model was confirmed visually with principal component analysis and t-distributed stochastic neighbor embedding analysis for all suspects (Figure S20).
The highest average ∑Suspect concentrations were observed in Swedish dolphins (1000 ± 370 ng/g ww/680 ± 250 ng F/g ww), followed by East Greenland pilot whales (17 ± 8 ng/g ww/12 ± 5 ng F/g ww) and West Greenland dolphins (6 ± 3 ng/g ww/4 ± 2 ng F/g ww). One suspect class, n:3 FTCAs, displayed predicted concentrations considerably higher than all the others, with the highest concentrations in Swedish dolphins (average ∑n:3 FTCAs 970 ng/g) and lowest concentrations in pilot whales (average ∑n:3 FTCAs 7.6 ng/g) and absent in Greenlandic dolphins. In this class, the predominant PFAS was 9:3 FTCA (up to 1000 ng/g), followed by 8:3 FTCA (up to 300 ng/g and which had similar concentrations to the target 7:3 FTCA) and 10:3 FTCA (up to 67 ng/g). FTCAs in Swedish dolphins made up ∼95% of the ∑Suspect concentrations. In pilot whale samples, the highest predicted concentrations were observed for 9:3 and 11:3 FTCA (up to 9.2 and 4.8 ng/g, respectively), followed by FHxSA (up to 3.5 ng/g) and perfluoropentadecanoic acid (PFPeDA, up to 2.4 ng/g). In West Greenlandic dolphins, the highest predicted concentrations were found for FPeSA (up to 6.7 ng/g), FHxSA (up to 2 ng/g), and PFPeDA (up to 0.5 ng/g).
Closing the Fluorine Mass Balance with Machine Learning-Based Quantification
IE-based quantification of suspects helped explain significant additional fractions of the EOF (Figure 2, Table S11). The highest predicted concentrations were determined for samples of Swedish dolphins based on the summed fluorine equivalent concentrations of suspects, where an average additional 680 ng F/g could be attributed to suspects. Here, the UEOF after machine learning-based quantification decreased from 53 to 70% to 0–18%. Suspects in pilot whale samples explained an additional 12 ng F/g on average and decreased the UEOF from 13 to 32% to 6–27%. Samples of Greenlandic dolphins already had a closed organofluorine mass balance; nevertheless, some suspects were detected at low concentrations. This is not surprising given the variability of EOF measurements (83% recovery of organofluorine [RSD: 8%]).
The present work offers the possibility of closing the fluorine mass balance in environmental samples in the absence of analytical standards by combining suspect screening with IE-based quantification. Nevertheless, a fraction of EOF in some samples remains unexplained. This may arise from poor sensitivity of some PFASs to LC-ESI-MS-based detection, or alternatively, the absence of a tentative structural.
Acknowledgments
We would like to thank the local subsistence hunters in Tasiilaq and Maniitsoq for help sampling their catches. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under Marie Skłodowska-Curie Action Grant Agreement 860665 (PERFORCE3), Ministry of Environment of Denmark (#MST-113-00054), and the Swedish Research Council for Sustainable Development grant 2020-01511.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.3c07220.
Additional information on samples, chemicals and reagents, instrumental analysis, PFAS profiles, EICs of homologue series, MS2 of suspects, the evaluation of the homologue series approach for quantification, information on molecular descriptors, model evaluation in spiked biota samples and model application domain (PDF)
Table S4 (inclusion list for suspect screening) and Tables S8–S11 (EOF, target and suspect concentrations, fluorine mass balance calculations) (XLSX)
Author Contributions
MZL and HS contributed equally and shared the first authorship. AK and JPB contributed equally and share the last authorship.
The authors declare no competing financial interest.
Supplementary Material
References
- OECD . Reconciling Terminology of the Universe of Per- and Polyfluoroalkyl Substances: Recommendations and Practical Guidance; OECD Series on Risk Management, Ed.; OECD Publishing: Paris, 2021; No. 61. https://www.oecd.org/chemicalsafety/portal-perfluorinated-chemicals/terminology-per-and-polyfluoroalkyl-substances.pdf (accessed 2021–09–03).
- Schymanski E. L.; Zhang J.; Thiessen P. A.; Chirsir P.; Kondic T.; Bolton E. E. Per- and Polyfluoroalkyl Substances (PFAS) in PubChem: 7 Million and Growing. Environ. Sci. Technol. 2023, 57 (44), 16918–16928. 10.1021/acs.est.3c04855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glüge J.; Scheringer M.; Cousins I. T.; DeWitt J. C.; Goldenman G.; Herzke D.; Lohmann R.; Ng C. A.; Trier X.; Wang Z. An Overview of the Uses of Per- and Polyfluoroalkyl Substances (PFAS). Environ. Sci. Process Impacts 2020, 22 (12), 2345–2373. 10.1039/D0EM00291G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton S. E.; Ducatman A.; Boobis A.; DeWitt J. C.; Lau C.; Ng C.; Smith J. S.; Roberts S. M. Per- and Polyfluoroalkyl Substance Toxicity and Human Health Review: Current State of Knowledge and Strategies for Informing Future Research. Environ. Toxicol. Chem. 2021, 40 (3), 606–630. 10.1002/etc.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evich M. G.; Davis M. J. B.; McCord J. P.; Acrey B.; Awkerman J. A.; Knappe D. R. U.; Lindstrom A. B.; Speth T. F.; Tebes-Stevens C.; Strynar M. J.; Wang Z.; Weber E. J.; Henderson W. M.; Washington J. W. Per- and Polyfluoroalkyl Substances in the Environment. Science (1979) 2022, 375 (6580), eabg9065. 10.1126/science.abg9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevedouros K.; Cousins I. T.; Buck R. C.; Korzeniowski S. H. Sources, Fate and Transport of Perfluorocarboxylates. Environ. Sci. Technol. 2006, 40 (1), 32–44. 10.1021/es0512475. [DOI] [PubMed] [Google Scholar]
- Cousins I. T.; DeWitt J. C.; Glüge J.; Goldenman G.; Herzke D.; Lohmann R.; Ng C. A.; Scheringer M.; Wang Z. The High Persistence of PFAS Is Sufficient for Their Management as a Chemical Class. Environ. Sci. Process Impacts 2020, 22 (12), 2307–2312. 10.1039/D0EM00355G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett H.; Du X.; Houde M.; Lair S.; Verreault J.; Peng H. Suspect and Nontarget Screening Revealed Class-Specific Temporal Trends (2000–2017) of Poly-and Perfluoroalkyl Substances in St. Lawrence Beluga Whales. Environ. Sci. Technol. 2021, 55 (3), 1659–1671. 10.1021/acs.est.0c05957. [DOI] [PubMed] [Google Scholar]
- Spaan K. M.; van Noordenburg C.; Plassmann M. M.; Schultes L.; Shaw S.; Berger M.; Heide-Jørgensen M. P.; Rosing-Asvid A.; Granquist S. M.; Dietz R.; Sonne C.; Rigét F.; Roos A.; Benskin J. P. Fluorine Mass Balance and Suspect Screening in Marine Mammals from the Northern Hemisphere. Environ. Sci. Technol. 2020, 54 (7), 4046–4058. 10.1021/acs.est.9b06773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Ruan Y.; Jin L.; Zhang X.; Li J.; He Y.; Wei S.; Lam J. C. W.; Lam P. K. S. Target, Nontarget, and Suspect Screening and Temporal Trends of Per- and Polyfluoroalkyl Substances in Marine Mammals from the South China Sea. Environ. Sci. Technol. 2021, 55 (2), 1045–1056. 10.1021/acs.est.0c06685. [DOI] [PubMed] [Google Scholar]
- Schultes L.; van Noordenburg C.; Spaan K. M.; Plassmann M. M.; Simon M.; Roos A.; Benskin J. P. High Concentrations of Unidentified Extractable Organofluorine Observed in Blubber from a Greenland Killer Whale (Orcinus Orca. Environ. Sci. Technol. Lett. 2020, 7 (12), 909–915. 10.1021/acs.estlett.0c00661. [DOI] [Google Scholar]
- Mössner S.; Ballschmiter K. Marine Mammals as Global Pollution Indicators for Organochlorines. Chemosphere 1997, 34 (5–7), 1285–1296. 10.1016/S0045-6535(97)00426-8. [DOI] [PubMed] [Google Scholar]
- Bossart G. D. Marine Mammals as Sentinel Species for Oceans and Human Health. Vet Pathol 2011, 48 (3), 676–690. 10.1177/0300985810388525. [DOI] [PubMed] [Google Scholar]
- Pauly D.; Trites A. W.; Capuli E.; Christensen V. Diet Composition and Trophic Levels of Marine Mammals. ICES Journal of Marine Science 1998, 55 (3), 467–481. 10.1006/jmsc.1997.0280. [DOI] [Google Scholar]
- Khabazbashi S.; Engelhardt J.; Möckel C.; Weiss J.; Kruve A. Estimation of the Concentrations of Hydroxylated Polychlorinated Biphenyls in Human Serum Using Ionization Efficiency Prediction for Electrospray. Anal Bioanal Chem. 2022, 414 (25), 7451–7460. 10.1007/s00216-022-04096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liigand J.; Wang T.; Kellogg J.; Smedsgaard J.; Cech N.; Kruve A. Quantification for Non-Targeted LC/MS Screening without Standard Substances. Sci. Rep 2020, 10 (1), 5808. 10.1038/s41598-020-62573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Yu N.; Du L.; Shi W.; Yu H.; Song M.; Wei S. Transplacental Transfer of Per- and Polyfluoroalkyl Substances Identified in Paired Maternal and Cord Sera Using Suspect and Nontarget Screening. Environ. Sci. Technol. 2020, 54 (6), 3407–3416. 10.1021/acs.est.9b06505. [DOI] [PubMed] [Google Scholar]
- Schultes L.; Vestergren R.; Volkova K.; Westberg E.; Jacobson T.; Benskin J. P. Per- and Polyfluoroalkyl Substances and Fluorine Mass Balance in Cosmetic Products from the Swedish Market: Implications for Environmental Emissions and Human Exposure. Environ. Sci. Process Impacts 2018, 20 (12), 1680–1690. 10.1039/C8EM00368H. [DOI] [PubMed] [Google Scholar]
- Miaz L. T.; Plassmann M. M.; Gyllenhammar I.; Bignert A.; Sandblom O.; Lignell S.; Glynn A.; Benskin J. P. Temporal Trends of Suspect- and Target-per/Polyfluoroalkyl Substances (PFAS), Extractable Organic Fluorine (EOF) and Total Fluorine (TF) in Pooled Serum from First-Time Mothers in Uppsala, Sweden, 1996–2017. Environ. Sci. Process Impacts 2020, 22 (4), 1071–1083. 10.1039/C9EM00502A. [DOI] [PubMed] [Google Scholar]
- Barrett H.; Du X.; Houde M.; Lair S.; Verreault J.; Peng H. Suspect and Nontarget Screening Revealed Class-Specific Temporal Trends (2000–2017) of Poly- and Perfluoroalkyl Substances in St. Lawrence Beluga Whales. Environ. Sci. Technol. 2021, 55 (3), 1659–1671. 10.1021/acs.est.0c05957. [DOI] [PubMed] [Google Scholar]
- Baygi S. F.; Fernando S.; Hopke P. K.; Holsen T. M.; Crimmins B. S. Nontargeted Discovery of Novel Contaminants in the Great Lakes Region: A Comparison of Fish Fillets and Fish Consumers. Cite This: Environ. Sci. Technol. 2021, 55, 3765. 10.1021/acs.est.0c08507. [DOI] [PubMed] [Google Scholar]
- Liu Y.; D’Agostino L. A.; Qu G.; Jiang G.; Martin J. W. High-Resolution Mass Spectrometry (HRMS) Methods for Nontarget Discovery and Characterization of Poly- and per-Fluoroalkyl Substances (PFASs) in Environmental and Human Samples. TrAC Trends in Analytical Chemistry 2019, 121, 115420 10.1016/j.trac.2019.02.021. [DOI] [Google Scholar]
- Robuck A. R.; McCord J. P.; Strynar M. J.; Cantwell M. G.; Wiley D. N.; Lohmann R. Tissue-Specific Distribution of Legacy and Novel Per- and Polyfluoroalkyl Substances in Juvenile Seabirds. Environ. Sci. Technol. Lett. 2021, 8 (6), 457–462. 10.1021/acs.estlett.1c00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski E. L.; Jeon J.; Gulde R.; Fenner K.; Ruff M.; Singer H. P.; Hollender J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- Yap C. W. PaDEL-Descriptor: An Open Source Software to Calculate Molecular Descriptors and Fingerprints. J. Comput. Chem. 2011, 32 (7), 1466–1474. 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- Lowe C. N.; Isaacs K. K.; McEachran A.; Grulke C. M.; Sobus J. R.; Ulrich E. M.; Richard A.; Chao A.; Wambaugh J.; Williams A. J. Predicting Compound Amenability with Liquid Chromatography-Mass Spectrometry to Improve Non-Targeted Analysis. Anal Bioanal Chem. 2021, 413 (30), 7495–7508. 10.1007/s00216-021-03713-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aalizadeh R.; Nikolopoulou V.; Alygizakis N.; Slobodnik J.; Thomaidis N. S. A Novel Workflow for Semi-Quantification of Emerging Contaminants in Environmental Samples Analyzed by LC-HRMS. Anal Bioanal Chem. 2022, 414 (25), 7435–7450. 10.1007/s00216-022-04084-6. [DOI] [PubMed] [Google Scholar]
- Kärrman A.; Wang T.; Kallenborn R.; Langseter A. M.; Ræder E. M.; Lyche J. L.; Yeung L.; Chen F.; Eriksson U.; Aro R.; Fredriksson F.. PFASs in the Nordic Environment; TemaNord; Nordic Council of Ministers: Copenhagen, 2019; 10.6027/TN2019-515. [DOI] [Google Scholar]
- Hamid H.; Li L. Y.; Grace J. R. Aerobic Biotransformation of Fluorotelomer Compounds in Landfill Leachate-Sediment. Science of The Total Environment 2020, 713, 136547 10.1016/j.scitotenv.2020.136547. [DOI] [PubMed] [Google Scholar]
- Butt C. M.; Muir D. C. G.; Mabury S. A. Biotransformation Pathways of Fluorotelomer-Based Polyfluoroalkyl Substances: A Review. Environ. Toxicol. Chem. 2014, 33 (2), 243–267. 10.1002/etc.2407. [DOI] [PubMed] [Google Scholar]
- Nilsson H.; Kärrman A.; Rotander A.; van Bavel B.; Lindström G.; Westberg H. Biotransformation of Fluorotelomer Compound to Perfluorocarboxylates in Humans. Environ. Int. 2013, 51, 8–12. 10.1016/j.envint.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Jouanneau W.; Léandri-Breton D.-J.; Corbeau A.; Herzke D.; Moe B.; Nikiforov V. A.; Gabrielsen G. W.; Chastel O. A Bad Start in Life? Maternal Transfer of Legacy and Emerging Poly- and Perfluoroalkyl Substances to Eggs in an Arctic Seabird. Environ. Sci. Technol. 2022, 56 (10), 6091–6102. 10.1021/acs.est.1c03773. [DOI] [PubMed] [Google Scholar]
- Lee K.; Alava J. J.; Cottrell P.; Cottrell L.; Grace R.; Zysk I.; Raverty S. Emerging Contaminants and New POPs (PFAS and HBCDD) in Endangered Southern Resident and Bigg’s (Transient) Killer Whales (Orcinus Orca): In Utero Maternal Transfer and Pollution Management Implications. Environ. Sci. Technol. 2023, 57 (1), 360–374. 10.1021/acs.est.2c04126. [DOI] [PubMed] [Google Scholar]
- Haque F.; Soerensen A. L.; Sköld M.; Awad R.; Spaan K. M.; Lauria M. Z.; Plassmann M. M.; Benskin J. P. Per- and Polyfluoroalkyl Substances (PFAS) in White-Tailed Sea Eagle Eggs from Sweden: Temporal Trends (1969–2021), Spatial Variations, Fluorine Mass Balance, and Suspect Screening. Environ. Sci. Process Impacts 2023, 25 (9), 1549–1563. 10.1039/D3EM00141E. [DOI] [PubMed] [Google Scholar]
- López-Berenguer G.; Bossi R.; Eulaers I.; Dietz R.; Peñalver J.; Schulz R.; Zubrod J.; Sonne C.; Martínez-López E. Stranded Cetaceans Warn of High Perfluoroalkyl Substance Pollution in the Western Mediterranean Sea. Environ. Pollut. 2020, 267, 115367 10.1016/j.envpol.2020.115367. [DOI] [PubMed] [Google Scholar]
- Stockin K. A.; Yi S.; Northcott G. L.; Betty E. L.; Machovsky-Capuska G. E.; Jones B.; Perrott M. R.; Law R. J.; Rumsby A.; Thelen M. A.; Graham L.; Palmer E. I.; Tremblay L. A. Per- and Polyfluoroalkyl Substances (PFAS), Trace Elements and Life History Parameters of Mass-Stranded Common Dolphins (Delphinus Delphis) in New Zealand. Mar. Pollut. Bull. 2021, 173, 112896 10.1016/j.marpolbul.2021.112896. [DOI] [PubMed] [Google Scholar]
- Letcher R. J.; Chu S.; McKinney M. A.; Tomy G. T.; Sonne C.; Dietz R. Comparative Hepatic in Vitro Depletion and Metabolite Formation of Major Perfluorooctane Sulfonate Precursors in Arctic Polar Bear, Beluga Whale, and Ringed Seal. Chemosphere 2014, 112, 225–231. 10.1016/j.chemosphere.2014.04.022. [DOI] [PubMed] [Google Scholar]
- Galatius A.; Bossi R.; Sonne C.; Rigét F. F.; Kinze C. C.; Lockyer C.; Teilmann J.; Dietz R. PFAS Profiles in Three North Sea Top Predators: Metabolic Differences among Species?. Environmental Science and Pollution Research 2013, 20 (11), 8013–8020. 10.1007/s11356-013-1633-x. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Pereira A. D. S.; Martin J. W. Discovery of C5–C17 Poly- and Perfluoroalkyl Substances in Water by In-Line SPE-HPLC-Orbitrap with In-Source Fragmentation Flagging. Anal. Chem. 2015, 87 (8), 4260–4268. 10.1021/acs.analchem.5b00039. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Yu N.; Zhu X.; Guo H.; Jiang J.; Wang X.; Shi W.; Wu J.; Yu H.; Wei S. Suspect and Nontarget Screening of Per- and Polyfluoroalkyl Substances in Wastewater from a Fluorochemical Manufacturing Park. Environ. Sci. Technol. 2018, 52 (19), 11007–11016. 10.1021/acs.est.8b03030. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Qian M.; Ma X.; Zhu L.; Martin J. W. Nontarget Mass Spectrometry Reveals New Perfluoroalkyl Substances in Fish from the Yangtze River and Tangxun Lake, China. Environ. Sci. Technol. 2018, 52 (10), 5830–5840. 10.1021/acs.est.8b00779. [DOI] [PubMed] [Google Scholar]
- Rotander A.; Kärrman A.; Toms L.-M. L.; Kay M.; Mueller J. F.; Gómez Ramos M. J. Novel Fluorinated Surfactants Tentatively Identified in Firefighters Using Liquid Chromatography Quadrupole Time-of-Flight Tandem Mass Spectrometry and a Case-Control Approach. Environ. Sci. Technol. 2015, 49 (4), 2434–2442. 10.1021/es503653n. [DOI] [PubMed] [Google Scholar]
- Khabazbashi S.; Engelhardt J.; Möckel C.; Weiss J.; Kruve A. Estimation of the Concentrations of Hydroxylated Polychlorinated Biphenyls in Human Serum Using Ionization Efficiency Prediction for Electrospray. Anal Bioanal Chem. 2022, 414 (25), 7451–7460. 10.1007/s00216-022-04096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benskin J. P.; De Silva A. O.; Martin J. W. Isomer Profiling of Perfluorinated Substances as a Tool for Source Tracking: A Review of Early Findings and Future Applications. Reviews of Environmental Contamination and Toxicology 2010, 208, 111–160. 10.1007/978-1-4419-6880-7_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.