Abstract
Members of the family of surface adhesins of oral streptococci, including P1 of Streptococcus mutans, contain two highly conserved repeat domains, one rich in alanine (A region) and the other rich in proline (P region). To assess the contribution of the P region to the biological properties of P1, an internal deletion in spaP was engineered. In addition, the P region was subcloned and expressed as a fusion partner with the maltose binding protein of Escherichia coli and liberated by digestion with factor Xa. Results of Western blot experiments in which recombinant polypeptides were probed with a panel of 11 monoclonal antibodies indicated that the P region is a necessary component of conformational epitopes within the central portion of P1. Antibodies reactive with the P region were detected in a polyclonal rabbit antiserum generated against whole S. mutans cells but not in two rabbit antisera generated against purified P1 (Mr ∼ 185,000), suggesting that this domain is immunogenic on the surface of intact bacteria but not as part of a soluble full-length molecule. Finally, transformation of a spaP-negative mutant with a shuttle vector containing an internally deleted spaP lacking P-region DNA resulted in a complete absence of surface-localized P1 and substantially less P1 in sonicated cells compared to the case for the mutant complemented with the full-length gene. These results suggest that the P region is an integral component contributing to the conformation of the central region of P1 and indicate that its presence is necessary for surface expression of the molecule on S. mutans.
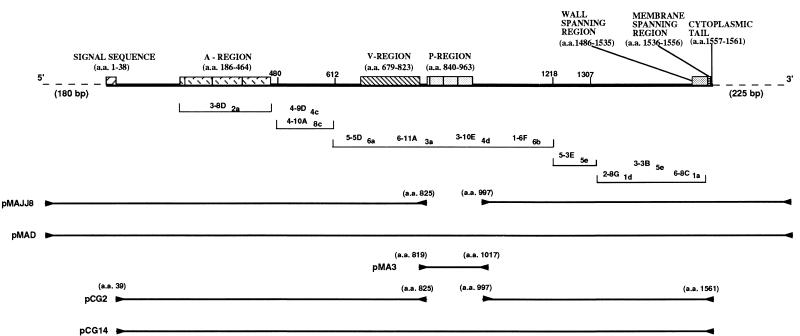
Streptococcus mutans is a major etiologic agent of dental caries (25, 39). The organism possesses a number of virulence factors that enable it to colonize and eventually dominate its niche in the oral cavity. The ∼185,000-Mr surface molecule variously referred to as P1 (18), SAI/II (58), PAc (49), antigen B (59), MSL-1 (15), and SR (1) is believed to participate in the adherence of the organism to teeth via its interaction with the salivary pellicle (6, 16, 24, 38, 45, 46, 48, 61). The gene encoding P1 of S. mutans serotype c, called spaP or pac, has been cloned and sequenced by two groups (31, 37, 49, 50). Notable features include a 38-residue amino-terminal signal sequence, a series of three 82-residue alanine-rich repeats within the amino-terminal third of the molecule, a 150-residue variable region where 20 of the 36 amino acid sequence substitutions identified between the spaP- and pac-encoded proteins are clustered, a series of three 39-residue proline-rich repeats in the central portion of the molecule, and carboxy-terminal sequences characteristic of wall- and membrane-spanning domains of streptococcal surface proteins. The carboxy-terminal sequences include a second proline-rich segment believed to span the cell wall, a hydrophobic transmembrane domain, and a charged tail. Also observed is the carboxy-terminal LPXTG consensus motif, which represents the substrate for a thiol-dependent membrane anchor-cleaving enzyme which modifies proteins posttranslationally, allowing subsequent attachment to a membrane-wall anchor site (17, 28, 51). A panel of 11 anti-P1 monoclonal antibodies (MAbs) has been evaluated for reactivity with a number of truncated P1 polypeptides, and approximate binding sites within the primary amino acid sequence have been deduced (5). A linear map of P1 showing the features described above as well as the putative binding sites of the anti-P1 MAbs is illustrated in Fig. 1.
FIG. 1.
Schematic representation of the linear structure of P1. A linear map showing salient features of P1 is at the top. Approximate binding sites of the anti-P1 MAbs 3-8D2a, 4-10A8c, 4-9D4c, 5-5D6a, 6-11A3a, 3-10E4d, 1-6F6b, 5-3E5e, 2-8G1d, 3-3B5e, and 6-8C1a are shown below the linear map. The order in which these antibodies are listed within each indicated segment is arbitrary and does not reflect their exact binding locations. The locations of the forward and reverse PCR primers used to amplify spaP DNA are indicated by forward and reverse arrowheads, while the amplified DNA sequences contained in the indicated plasmids are represented by solid lines in the bottom half. Additional information regarding these plasmids is given in Table 1. a.a., amino acids.
Secondary-structure predictions based on the deduced amino acid sequence for spaP suggest that the alanine-rich repeat domain would form an α-helical structure typical of coiled-coil proteins, including the M protein of Streptococcus pyogenes, and that the central proline-rich repeat domain would form an extended, pleated β-turn structure (31). There is, however, scant experimental evidence regarding the actual physical structure of P1 or immunologically related molecules expressed by other oral streptococci, such as SpaA of Streptococcus sobrinus (35, 69) or SSP-5 of Streptococcus gordonii (14).
DNA sequences homologous to spaP have been demonstrated in S. mutans serotypes c, e, and f, S. sobrinus serotype d, S. cricetus serotype a (37), and S. gordonii (15). In addition, Ma et al. (41) demonstrated by Southern analysis that chromosomal DNAs from viridans and nonviridans streptococci which did not hybridize with full-length spaP were reactive when the probe was limited to a much shorter gene fragment consisting primarily of DNA encoding the central proline-rich repeat domain. This suggested that similar sequences are present in other streptococci. Subsequently, open reading frames (ORFs) of unknown function encoding deduced amino acid sequences homologous to the P region of P1 have been identified within plasmid pDB101 of S. pyogenes (ORF iota [9]) and within plasmid pCF10, containing regulatory and structural genes involved in pheromone-inducible conjugation in Enterococcus faecalis (prgC, ORF15 [30]). The cell-bound fructosyltransferase of S. salivarius also includes a homologous carboxy-terminal proline-rich domain believed to be involved in cell surface localization in the absence of a conserved LPXTG motif (56). In addition, the fibronectin binding proteins of Staphylococcus aureus and S. pyogenes contain proline-rich tandem repeat segments just carboxy terminal to their fibronectin binding domains which include the amino-acid sequence motif PTPPT common to the P region of P1 (29, 62, 64). Repetitive proline-rich segments are also present in the published sequences encoded by genes encoding intermedilysin (a cytolytic factor of Streptococcus intermedius) (47), pneumococcal surface protein A (PspA) of S. pneumoniae (72), immunogenic secreted protein of S. pyogenes (44), the immunoglobulin A (IgA) Fc binding protein of group B streptococci (27), and numerous proteins expressed by a wide variety of bacterial species (10, 21, 26, 36, 52, 53, 63, 66). While the function of most microbial proline-rich repeat domains is not understood, it has been suggested that such sequences may be involved in protein-protein interactions (21, 55, 71).
The function of the P region of P1 is not yet known. Nakai et al. (48) reported that the P region can bind to the PAc (P1) molecule itself and proposed that this segment may contribute to spontaneous self-aggregation of the molecule. Munro et al. (46) demonstrated that a fragment of P1 (amino acid residues 816 to 1213) which includes the P region was inhibitory to adherence of S. mutans to saliva-coated hydroxyapatite. Kelly et al. (32) subsequently identified a specific segment (amino acid residues 1005 to 1044) carboxy terminal to the P region as being involved in adhesion to salivary components. Other investigators have implicated amino-terminal sequences including the alanine-rich repeats (A region) in the adhesion function of P1 (11, 24, 48). A second manifestation of the interaction of P1 and related molecules with salivary components is cell-cell aggregation (13, 15). In a previous study, anti-P1 MAbs were used to inhibit adherence and aggregation mediated by the high-molecular-weight salivary agglutinin glycoprotein. MAbs specific for different epitopes demonstrated marked differences in their relative abilities to inhibit each of these processes (6), suggesting that P1 possesses adherence-specific and aggregation-specific functional domains which are not confined within discrete contiguous segments of P1.
To understand the relative contribution of the P region to the biological properties of P1, a dual strategy was undertaken. A spaP gene devoid of the DNA encoding the P region was engineered, and the DNA encoding the P region itself was subcloned. Recombinant polypeptides expressed in Escherichia coli were evaluated for reactivity with a panel of 11 anti-P1 MAbs and three polyclonal antisera. The results suggest that the P region is an integral component of conformational epitopes within the central portion of P1. The internally deleted spaP gene was also subcloned into a shuttle vector and introduced into a spaP-negative mutant of S. mutans. Absolutely no cell surface P1 and substantially decreased levels of cytoplasmic protein were detectable in the transformed mutant, with no decrease in the production of spaP-specific mRNA. The P region is postulated to play a critical role in the three-dimensional conformation (tertiary structure) of P1 and is apparently essential in S. mutans for expression, intracellular stability, and/or translocation of the molecule to the cell surface.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Serotype c S. mutans NG8 and the spaP-negative mutant PC3370 and its derivatives (Table 1) were grown for 16 h at 37°C in Todd-Hewitt broth (BBL, Cockeysville, Md.) supplemented with 0.3% yeast extract (THBYE). E. coli strains used in these experiments included MC1061, SURE (Stratagene, La Jolla, Calif.), INVαF′ (Invitrogen), Top Ten (Invitrogen), and M15(pREP4) (QIAGEN, Santa Clarita, Calif.). E. coli was grown aerobically at 37°C with vigorous shaking in Luria-Bertani broth (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 1% [wt/vol] NaCl, pH 7.0) supplemented with ampicillin (50 to 100 mg/ml) or kanamycin (25 to 50 mg/ml) as appropriate. Plasmids pUC18, pCRII (Invitrogen), pMal-p (New England BioLabs, Inc. [NEB], Beverly, Mass.), pQE30 (QIAGEN), and pDL289 (kindly provided by D. LeBlanc) (7) were used as cloning and expression vectors.
TABLE 1.
Plasmids and bacterial strains
| Plasmid or strain | Description |
|---|---|
| Plasmids | |
| pDL289 | E. coli-streptococcal shuttle vector (7) |
| pMAJJ8 | pDL289-derived plasmid containing internally deleted spaP encoding amino acids 1 to 825 and 997 to 1561 (31) of P1 |
| pMAD | pDL289-derived plasmid containing PCR-amplified spaP encoding full-length P1 |
| pMal-p | Vector for expression of MBP fusions (NEB) |
| pMA3 | pMal-p-derived plasmid containing PCR-amplified DNA encoding amino acids 819 to 1017 (31) of P1 |
| pQE30 | Vector for expression of histidine-tagged polypeptides (QIAGEN) |
| pCG2 | pQE30-derived plasmid containing internally deleted spaP encoding amino acids 39 to 825 and 997 to 1561 (31) of P1 |
| pCG14 | pQE30-derived plasmid containing PCR-amplified spaP encoding amino acids 39 to 1561 (31) of P1 |
| Strains | |
| M15(pREP4) | Host strain of E. coli for pQE30-derived plasmids (QIAGEN) |
| CG2 | M15(pREP4) harboring pCG2 |
| CG14 | M15(pREP4) harboring pCG14 |
| PC3370 | spaP-negative mutant derived from S. mutans NG8 (12) |
| PC3370A | PC3370 transformed with pDL289 |
| PC3370B | PC3370 transformed with pMAJJ8 |
| PC3370C | PC3370 transformed with pMAD |
Preparation of chromosomal and plasmid DNAs.
S. mutans NG8 chromosomal DNA was prepared as described previously (4). Plasmid DNA was purified by using a modified alkaline lysis-polyethylene glycol precipitation procedure (1a) (Applied Biosystems, Inc., Foster City, Calif.).
PCRs and construction of recombinant molecules.
PCR amplification of spaP was undertaken so that desired fragments containing convenient restriction sites for cloning into appropriate vectors could be engineered. Fidelity of the reactions was confirmed by restriction and sequence analysis. The locations of spaP sequences amplified by PCR and cloned to create plasmids pMAJJ-8, pMAD, pMA3, pCG2, and pCG14 are illustrated in Fig. 1 (also see Table 1).
Forward and reverse primers 5′-GCGTCGACGTTGGATAAAGTGTGGAG and 5′-GCATCGATAGGAAGATTAACGCGACG were designed based on the published spaP (31) and pac (69) sequences and used to amplify DNA upstream of the P region, including the spaP promoter. Underlining indicates engineered SalI and ClaI restriction sites, respectively. Primers 5′-GCATCGA T-AAACTAGCTGTTCAGCCG and 5′-GCGTCGACGCAGTGCGAAGTACCTTA were used to amplify spaP DNA downstream of the P region. Underlining indicates engineered ClaI and SalI restriction sites, respectively. Reactions were carried out with a UNO thermoblock thermocycler (Biometra, Tampa, Fla.) with S. mutans NG8 chromosomal DNA as the template and Taq polymerase (Promega, Madison, Wis.), under the following conditions: (i) denaturation at 94°C for 2 min; (ii) denaturation at 94°C for 45 s, primer annealing at 53°C for 1 min 30 s, and primer extension at 72°C for 3 min for 30 cycles; and (iii) primer extension at 72°C for an additional 7 min. The resulting 2,663- and 1,982-bp gene fragments were cloned into the TA cloning vector pCRII (Invitrogen) with E. coli INVαF′ as the host strain. Two clones with inserts in opposite orientations with respect to the Plac promoter of pCRII were used. Each purified plasmid was digested with NcoI (Promega) (one site in pCRII, no site in spaP) and ClaI (GIBCO BRL, Gaithersburg, Md.) (no site in pCRII, engineered sites in PCR primers internal to spaP). Appropriate-sized fragments were recovered by gel purification, religated, and used to transform E. coli MC1061. This strategy resulted in reconstitution of the vector and in-frame ligation of an internally deleted gene lacking 519 bp (bp 2571 to 3090 of spaP [31]) encoding amino acids 825 to 997. This plasmid was called pJJ-1. As a positive control, the same 5′ and 3′ flanking primers were used to amplify full-length spaP with NG8 chromosomal DNA as the template by using the Gene Amp XL PCR kit (Perkin-Elmer/Roche, Branchburg, N.J.) according to the manufacturer’s instructions and under the following conditions: (i) denaturation at 93°C for 1 min and (ii) denaturation at 93°C for 1 min, primer annealing at 60°C for 10 min, and primer extension at 72°C for 10 min for 30 cycles. The amplified product (bp 20 to 5167 of pac [69]) was cloned into pCRII to create pDC-4.
The spaP inserts in pJJ-1 and pDC-4 were excised with SalI (Promega) and ligated into SalI-digested pUC18 to create pDC-9 and pDC-20, respectively. This intermediate cloning step was undertaken so that internally deleted and full-length spaP could be cloned directionally into the streptococcal shuttle vector pDL289 (7). Inserts were excised by digestion with SphI (Promega) and SmaI (Promega) and ligated into SphI-SmaI-digested pDL289 with transformation into the E. coli strain SURE. The resultant plasmids were designated pMAJJ8 and pMAD and contained internally deleted and full-length spaP DNA, respectively (Fig. 1 and Table 1).
DNA (bp 2554 to 3150 of spaP [31]) encoding the P region itself (amino acids 819 to 1017) was amplified by PCR for expression as a fusion product with maltose binding protein (MBP). Forward and reverse primers were GGGAGTAC-TCGTGCGGTTAATCTTCCT and GGGAATTCTCAGTCAGTCATGCCACCAAAGT-TCTGTC, respectively. Underlining indicates engineered ScaI and EcoRI restriction sites, respectively. Boldface indicates the reverse and complement of stop codons engineered in all three reading frames. The reaction was carried out with pSM2949 containing the original cloned spaP insert (37) as the template and Taq polymerase (Promega) under the following conditions: (i) denaturation at 94°C for 2 min; (ii) denaturation at 94°C for 45 s, primer annealing at 53°C for 1 min 30 s, and primer extension at 72°C for 3 min for 30 cycles; and (iii) primer extension at 72°C for an additional 7 min. The amplified product was cloned into pCRII (Invitrogen), and the insert was excised by digestion with ScaI (Promega) and EcoRI (Promega) and ligated into StuI (Promega)- and EcoRI-linearized pMal-p (NEB). This resulted in an in-frame fusion of the P-region insert with the vector containing the malE gene. The recombinant plasmid was designated pMA-3 (Fig. 1 and Table 1).
Finally, internally deleted and full-length spaP were reamplified by PCR for directional cloning into the His6-tagged expression system (QIAGEN). Forward and reverse primers were 5′-GGCATGCGATGAA-ACGACCACTAC and 5′-GGGTACCGTAATGTCTATGCTGTC, respectively. Underlining indicates engineered SphI and KpnI restriction sites, respectively. This forward primer was designed to eliminate the 38-residue N-terminal signal sequence of P1 so its cleavage would not result in removal of the histidine tag. Reactions were carried out with VENT polymerase (NEB) and pMAJJ8 and pMAD (Fig. 1 and Table 1) as the templates under the following conditions: (i) denaturation at 94°C for 5 min; (ii) denaturation at 94°C for 1 min, primer annealing at 49°C for 2 min, and primer extension at 72°C for 5 min for 30 cycles; and (iii) primer extension at 72°C for an additional 10 min. The VENT-amplified blunt-end products were cloned by using the Zero Blunt (Invitrogen) PCR cloning kit with E. coli Top Ten (Invitrogen) as the host. Inserts were excised by digestion with SphI (Promega) and KpnI (NEB) and ligated into SphI-KpnI-restricted pQE30 (QIAGEN), and the recombinant plasmids were transformed into E. coli M15(pREP4) (QIAGEN) according to the manufacturer’s instructions. Plasmids containing internally deleted and full-length spaP were designated pCG2 and pCG14, and the recombinant E. coli strains were designated CG2 and CG14, respectively (Fig. 1 and Table 1).
Construction of spaP-negative mutant PC3370.
The spaP-negative mutant strain PC3370 was constructed as described in detail elsewhere (12, 12a). Briefly, a 480-bp DNA fragment located upstream of the spaP promoter and a 275-bp DNA fragment located immediately downstream of and including the translational stop codon for spaP were amplified by PCR with S. mutans NG5 chromosomal DNA as the template. The amplified products, into which appropriate restriction enzyme recognition sequences had been engineered for cloning, were ligated as a single fragment with the two amplified sequences in opposite orientations into the PstI site of pVA981 (67). The resulting recombinant plasmid was digested with BamHI to linearize it at a unique site engineered to lie between the amplified sequences and was used to transform S. mutans NG8. The spaP-negative mutants of NG8 generated via allelic exchange with the linearized construct were selected on THBYE agar containing 10% sucrose and 15 μg of tetracycline per ml. Mutant PC3370 was selected for further evaluation. Southern analysis demonstrated that the spaP gene had been eliminated, and radioimmunoassay demonstrated a complete lack of reactivity of PC3370 with anti-P1 MAbs and polyclonal antibodies.
Introduction of recombinant spaP plasmids into spaP-negative mutant PC3370.
The pDL289 shuttle vector (7), pMAJJ8, and pMAD (Fig. 1 and Table 1) were introduced into the NG8-derived spaP-negative mutant PC3370 by electrotransformation to create PC3370A, PC3370B, and PC3370C, respectively (Table 1). Briefly, 1,250 ml of an overnight culture of PC3370 was used to inoculate 50 ml of THBYE supplemented with 10% heat-inactivated horse serum (Sigma Chemical Co., St. Louis, Mo.), and the culture was grown at 37°C to an optical density at 600 nm (OD600) of 0.13 to 0.25. Cells were harvested by centrifugation at 800 × g for 10 min, washed three times with cold 10% glycerol, and resuspended in 0.1 volume of 10% glycerol. One milligram of plasmid DNA was added to 25 ml of cells in a chilled cuvette (0.1 cm; Bio-Rad, Hercules, Calif.) and allowed to stand on ice for 1 min. Electroporation was performed with a Gene Pulsar (Bio-Rad) and settings of 200 Ω, 25 mF, and 1.25 kV. Pulse times were between 4.5 and 5.0 ms. One milliliter of THBYE was immediately added to the cells, which were incubated at 37°C for 90 min. Transformants were selected on THBYE agar containing 500 mg of kanamycin per ml.
Purification of MBP–P-region fusion polypeptide and cleavage with factor Xa.
An overnight culture of E. coli harboring pMA3 (Fig. 1 and Table 1) was diluted 1:100 into fresh Luria-Bertani broth and grown to an OD600 of ∼0.4. The medium was supplemented with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the culture was incubated for an additional 2 h at 37°C. Periplasmic contents were extracted by osmotic shock (57). Affinity purification of the MBP–P-region fusion protein was performed by passage of the periplasmic fraction through a column of amylose resin (NEB). The fusion protein was eluted with 10 mM maltose by a standard protocol (57). Dissociation of the P region from the purified fusion protein was accomplished by cleavage with the proteolytic enzyme factor Xa (NEB), also by standard methods (57).
MAbs and polyclonal antibodies.
Previously prepared immunological reagents available for use in this study included 11 murine MAbs (2) and three rabbit polyclonal antisera. Murine hybridoma ascites fluids served as the source of the MAbs. Antiserum 209 was generated by immunizing a female New Zealand White rabbit with P1 isolated by ion-exchange and gel filtration chromatography from S. mutans serotype c strain Ingbritt 175 as previously described (6). Antiserum 230 was generated against recombinant P1 encoded on plasmid pSMI/II (46) and isolated in a similar manner. Antiserum 218 was generated by immunizing a female New Zealand White rabbit intravenously with ∼109 CFU of S. mutans NG8 harvested from a stationary-phase culture grown in chemically defined medium (65) and resuspended in 0.1 ml of phosphate-buffered saline, pH 7.2 (PBS). The rabbit was boosted with 2 × 109 CFU on day 14 and 3 × 109 CFU on day 21, rested for 5 months, and then immunized subcutaneously with 1 mg of protein extract harvested from NG8 cells that had been broken with glass beads in a tissue homogenizer (3). The rabbit was boosted with 2 mg of protein on day 7 and 3 mg of protein on day 14 and exsanguinated 5 months later. Antiserum 218 was rendered monospecific for P1 by adsorption with spaP-negative mutant PC3370 until all reactivity against PC3370 cells was eliminated. Rabbit polyclonal anti-MBP was purchased from NEB and used at a dilution of 1:104. Mouse anti-β-galactosidase was purchased from Sigma Chemical Co. Peroxidase-conjugated goat anti-mouse and goat anti-rabbit IgGs were purchased from Cappel, Organon Teknika Corp. (West Chester, Pa.). All primary and secondary antibodies were used at a dilution of 1:103 unless otherwise noted.
Dot blot analysis for detection of P1 surface expression by mutant PC3370 and derivatives.
S. mutans NG8, PC3370, PC3370A, PC3370B, and PC3370C (Table 1) were grown for 16 h at 37°C, and the cells were harvested by centrifugation and washed twice with PBS. Cells were resuspended in PBS to a density of 109 CFU/ml. Twofold serial dilutions of cell suspensions were made in PBS, and 50 ml of each dilution was applied in duplicate to a nitrocellulose filter (Bio-Rad) by using a 96-well dot blot manifold (Schleicher and Schuell, Keene, N.H.). Wells were washed twice with 200 ml of PBS, and the filter was removed from the apparatus and blocked with PBS containing 0.25% gelatin and 0.25% Tween 20. Cell surface P1 was detected with antiserum 209 as the primary antibody, peroxidase-conjugated goat anti-rabbit IgG as the secondary antibody, and development with 4-chloro-1-naphthol solution (7 ml of PBS, 1 ml of 4-chloro-1-naphthol [Sigma; 3 mg/ml in ice-cold methanol], and 8 ml of 30% hydrogen peroxide).
SDS-polyacrylamide gel electrophoresis and Western blotting.
E. coli M15(pREP4) harboring the pQE30 His-tagged vector (QIAGEN), CG2, and CG14 (Fig. 1 and Table 1) were induced with 1 mM IPTG, harvested, and lysed according to the manufacturer’s suggested protocol. The MBP–P-region fusion polypeptide and isolated P region were prepared as described above. S. mutans sonic extracts were prepared by harvesting the cells from 250 ml of an overnight culture by centrifugation and resuspension in 2 ml of PBS. Approximately 1 g of glass spheres (P-080; Potters Industries, Inc., Hasbrouk Heights, N.J.) was added to the suspension, and the mixture was sonicated on ice with a SONIC300 Dismembrator (ARTEK Systems, Farmingdale, N.Y.) for three 30-s bursts at 60% output. Cell debris was removed by centrifugation at 10,000 × g for 15 min. Approximate protein concentrations were determined by using the bicinchoninic acid protein assay (Sigma) or by measurement of A280. Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on 7.5% acrylamide gels by the method of Laemmli (34), and slab gels were stained with Coomassie brilliant blue G (Sigma). Approximately 1 mg of protein was loaded per lane. Prestained high-molecular-weight standards (Sigma) were included on each gel. Proteins were electroblotted onto nitrocellulose filters for 3 h at 70 V by the method of Towbin et al. (70). Immunoblots were blocked and developed as described above for the dot blot assay. In the case of preparatory gels, blots were cut into ∼0.25-cm strips and reactions were performed in individual troughs of an Incutray (Schleichter and Schuell).
RNA isolation and RNA dot blotting.
To isolate S. mutans RNA, 18-h THBYE cultures were subcultured 1:10 into 50 ml of prewarmed THBYE medium supplemented with 20 mM dl-threonine. Cells were grown to an OD600 of between 0.4 and 0.5. The method of Lunsford (40) was used for isolation of total streptococcal RNA, except that glycine was omitted and 500 U of mutanolysin (Sigma) per ml was used instead of lysozyme. Lysates were subjected to selective RNA isolation with the TRIZOL (GIBCO BRL) reagent according to the manufacturer’s instructions. spaP mRNA levels were detected by RNA dot blotting with the Genius nonradioactive nucleic acid labeling and detection system (Boehringer Mannheim, Indianapolis, Ind.) according to the manufacturer’s instructions. Cloned DNA encoding the alanine-rich A region (11) was gel purified, labeled with digoxigenin-dUTP, and used as the probe. Twofold serial dilutions of total cellular RNA, starting with 5 mg, were applied to the filter.
RESULTS
Expression of P1 with the P-region deleted in E. coli.
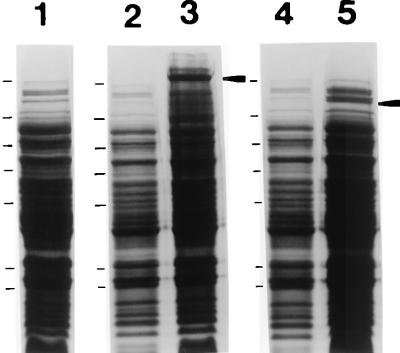
A spaP gene devoid of DNA encoding the P region and full-length positive control spaP were engineered by PCR amplification and a series of cloning steps described in Materials and Methods. The expression of P1 with the P-region deleted was detectable in cell extracts of recombinant E. coli (INVaF′, MC1061, and SURE) by Western blotting with three different anti-P1 polyclonal antisera; however, substantially reduced immunoreactivity compared to the full-length gene product was observed (data not shown). Because there were too many background bands expressed by negative control E. coli to clearly identify a P-region deletion P1 band on stained SDS-polyacrylamide gels, it was unclear whether the difference in band intensity on Western blots resulted from decreased P1 expression or stability, altered antigenicity of the internally deleted recombinant polypeptide, or a combination of multiple factors. To resolve this difficulty, both internally deleted and full-length control spaP were cloned into the His6-tagged vector pQE30 (QIAGEN) so that recombinant molecules could be isolated on nickel resin, and Western blot analysis was repeated with equivalent amounts of purified protein. An SDS-polyacrylamide gel of lysates of E. coli expressing both full-length (CG14) and internally deleted (CG2) P1 is shown in Fig. 2. Full-length P1 is clearly visible following IPTG induction (lane 3), as is the product of the internally deleted gene (lane 5). The identity of the P1 bands indicated in these lanes was confirmed by Western blotting with the three anti-P1 antisera. Lanes 2 and 4 show lysates of the recombinant E. coli prior to IPTG induction. Lane 1 is a negative control showing the E. coli host strain harboring the His-tagged vector only. The predicted molecular weights of full-length and internally deleted P1 are 166,124 and 146,994, respectively. The full-length gene product migrates as a ∼185,000-Mr band (lane 3), the same as has been reported for native and recombinant P1 in previous studies. The larger-than-predicted apparent molecular weight of P1 on SDS-polyacrylamide gels has been attributed to aberrant migration caused by the proline-rich repeat region (31). It is not surprising, therefore, that elimination of the P region would result in a larger-than-predicted decrease in apparent molecular weight. The apparent Mr of ∼140,000 of the internally deleted polypeptide (Fig. 2, lane 5) is closer to the predicted value than is that of the full-length molecule.
FIG. 2.
SDS-polyacrylamide gel electrophoresis of E. coli containing plasmids with internally deleted and full-length spaP. Lane 1, cell lysate following IPTG induction of E. coli M15(pREP4) containing the His-tagged pQE30 vector only. Lanes 2 and 3, lysates of CG14 (full-length spaP) before and after IPTG induction, respectively. Lanes 4 and 5, lysates of CG2 (spaP with the P region deleted) before and after IPTG induction, respectively. The IPTG-induced P1 products are indicated by arrowheads to the right of lanes 3 and 5. The migrations of molecular weight standards are indicated to the left of lanes 1, 2, and 3 and correspond to Mrs of 180,000, 116,000, 84,000, 58,000, 48,500, 36,000, and 26,500.
Histidine-tagged full-length P1 was easily isolated on nickel resin, but purification of the P-region deletion product was problematic. The internally deleted product bound to the nickel resin but could not be eluted with imidizole under any of the manufacturer’s suggested conditions (data not shown). This result indicated to us that removal of the P region had substantially altered the properties of the P1 molecule. Since the P-region deletion polypeptide was clearly identifiable as a separate band with this particular vector and strain of E. coli, which had not been the case with other combinations of vectors and strains used previously, Western blot experiments were performed with E. coli lysates instead of purified protein.
Reactivity of anti-P1 MAbs with full-length and P-region deletion P1.
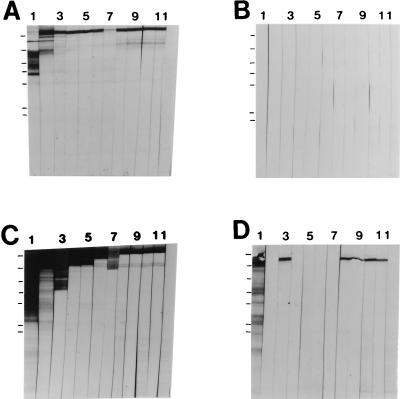
To evaluate the effect of the P-region deletion on the antigenicity of P1, the recombinant proteins described above were analyzed by Western blotting with a panel of 11 anti-P1 MAbs. These results are shown in Fig. 3. The approximate binding sites of the MAbs are illustrated in Fig. 1. Figure 3A shows a positive control and demonstrates the reactivities of the MAbs against purified recombinant P1 encoded on plasmid pSMI/II (46). MAb 3-8D2a (lane 1) reacts specifically with the alanine-rich repeat domain of P1 (11) and, as seen previously, is more reactive with P1 breakdown products. This suggests that at least a portion of the A region is not well exposed in the context of full-length P1. Figure 3B shows a negative control and demonstrates the lack of reactivity of the MAbs with proteins expressed by E. coli harboring the vector alone. Figure 3C shows a positive control and demonstrates that each of the 11 MAbs is reactive with the product of PCR-amplified full-length spaP. Figure 3D shows the reactivities of the anti-P1 MAbs with P1 lacking the P region. The reactivities of 5 of 11 MAbs, i.e., 4-10A8c (Fig. 3D, lane 2), 5-5D6a (lane 4), 6-11A3a (lane 5), 3-10E4d (lane 6), and 1-6F6b (lane 7), were clearly destroyed by this manipulation of the P1 molecule. Binding of the A-region-specific MAb 3-8D2a (Fig. 3D, lane 1) and the four MAbs which map to the carboxy terminus of P1, 5-3E5e (lane 8), 2-8G1d (lane 9), 3-3B5e (lane 10), and 6-8C1a (lane 11), was unaffected by the deletion. Lane 12 of each panel of Fig. 3 shows an isotype-matched negative control MAb directed against Actinobacillus actinomycetemcomitans.
FIG. 3.
Western blot analysis of recombinant full-length P1 and P1 lacking the P region. Lanes 1 through 11, reaction with anti-P1 MAbs 3-8D2a, 4-10A8c, 4-9D4c, 5-5D6a, 6-11A3a, 3-10E4d, 1-6F6b, 5-3E5e, 2-8G1d, 3-3B5e, and 6-8C1a, respectively (Fig. 1). Lane 12, reaction with the isotype-matched negative control MAb directed against A. actinomycetemcomitans. (A) Recombinant P1 purified from clone SMI/II (46). (B) Cell lysate of E. coli M15(pREP4) containing the His-tagged vector pQE30 only. (C) Cell lysate of CG14 (full-length spaP). (D) Cell lysate of CG2 (spaP with the P region deleted). The migrations of molecular weight standards are indicated to the left of lane 1 in each panel and correspond to Mrs of 180,000, 116,000, 84,000, 58,000, 48,500, 36,000, and 26,500.
Western blot reactivities of anti-P1 MAbs and polyclonal antibodies with the isolated P region.
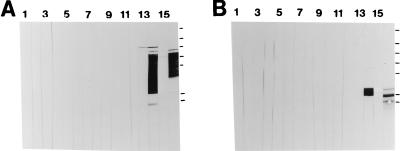
To test whether the anti-P1 MAbs whose reactivities were destroyed by removal of the P region recognized epitopes contained directly within that sequence, the P region was expressed as a fusion protein with MBP, purified by amylose resin chromatography, and analyzed by Western blotting. These results are shown in Fig. 4A. None of the MAbs (lanes 1 to 11) or the isotype-matched negative control MAb (lane 12) was reactive with the fusion protein. Lane 13 was reacted with polyclonal mouse anti-β-galactosidase. In the absence of insert DNA in pMal-p (Table 1), the MBP is expressed as a fusion with β-galactosidase. Therefore, negligible reactivity with this antiserum indicates that translation into β-galactosidase was interrupted by the stop codons engineered into the 3′ terminus of the PCR-amplified P-region DNA. Lane 14 of Fig. 4A was probed with polyclonal rabbit anti-MBP, which was reactive with a number of polypeptide bands, presumably the fusion protein and breakdown products. The molecular weight of MBP is approximately 42,000, and the predicted molecular weight of the subcloned P region is 22,000. The fusion moiety was not reactive with antiserum 209 or 230, made against purified streptococcal P1 (Fig. 4A, lane 15) or purified recombinant P1 (data not shown), respectively. The fusion moiety was reactive, however, with antiserum 218, made against NG8 whole and mechanically broken cells (lane 16).
FIG. 4.
Western blot analysis of the P region of P1. Lanes 1 through 11, reaction with MAbs 3-8D2a, 4-10A8c, 4-9D4c, 5-5D6a, 6-11A3a, 3-10E4d, 1-6F6b, 5-3E5e, 2-8G1d, 3-3B5e, and 6-8C1a, respectively (Fig. 1). Lane 12, reaction with the negative control anti-A. actinomycetemcomitans MAb 1-5F2a. Lane 13, reaction with mouse anti-β-galactosidase. Lane 14, reaction with rabbit anti-MBP. Lane 15, reaction with rabbit antiserum 209 made against purified P1 from S. mutans. Lane 16, reaction with rabbit antiserum 218 made against NG8 cells. (A) MBP–P-region fusion polypeptide recovered from recombinant E. coli periplasmic fraction by amylose resin chromatography. (B) MBP–P-region fusion polypeptide digested with factor Xa. The migrations of molecular weight standards are indicated to the right of lane 16 in each panel and correspond to Mrs of 180,000, 116,000, 84,000, 58,000, 48,500, 36,000, and 26,500.
To evaluate whether P1 epitopes were masked within the MBP–P-region moiety, the P region was liberated from the MBP by digestion with factor Xa and the cleavage products were analyzed by Western blotting. These results are shown in Fig. 4B. None of the anti-P1 MAbs (lanes 1 to 11), the negative control anti-A. actinomycetemcomitans (lane 12), or anti-β galactosidase (lane 13) was reactive with MBP or the isolated P region. Anti-MBP (lane 14) reacted with the predicted ∼42,000-Mr MBP product. The anti-P1 antiserum prepared against NG8 whole cells (antiserum 218) was reactive with the isolated P region (lane 16), but antisera 209 (lane 15) and 230 (data not shown), made against solubilized P1 protein, were not. There was a slight cross-reactivity of antiserum 209 with MBP (Fig. 4B, lane 15). In addition, anti-MBP was found to be slightly cross-reactive with full-length P1 (data not shown). The predominant P-region band migrated with an apparent Mr of ∼36,000 (Fig. 4B, lane 16), larger than the predicted size. Because this band was not reactive with antisera 209 and 230, its identity as the P region was verified by N-terminal sequence analysis. To exclude the possibility that lack of reactivity of the isolated P-region with those two polyclonal antisera and the anti-P1 MAbs was a consequence of destruction of epitopes during the SDS-polyacrylamide gel electrophoresis and Western blotting procedures, an enzyme-linked immunosorbent assay was performed as well (data not shown). Again, the only immunological reagent which demonstrated reactivity with the P-region polypeptide was rabbit antiserum 218.
Transformation of mutant PC3370 with full-length and internally deleted plasmid-encoded spaP.
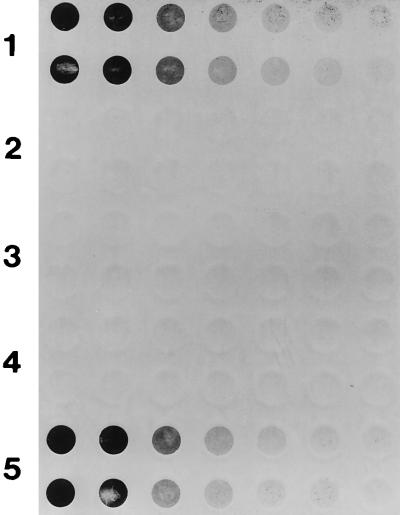
The spaP-negative mutant PC3370 was used as the host into which plasmid-encoded variants of spaP were introduced (Table 1). Dot blot analysis with polyclonal anti-P1 antisera was used to evaluate the surface expression of P1 by transformants. The results with antiserum 209 are shown in Fig. 5. Row 1 is a positive control and shows the reactivity of twofold serial dilutions (in duplicate) of the parent strain NG8 used to create mutant PC3370. Rows 2 and 3 are negative controls and show no reactivity of the antiserum with the mutant or the mutant harboring the vector only (PC3370A). Rows 4 and 5 correspond to PC3370B and PC3370C, respectively. These results clearly demonstrate that the degree of surface expression of P1 by PC3370C, which contains the full-length gene (Fig. 5, row 5), is similar to that by wild-type NG8 (row 1). No P1 is present on the surface of PC3370B, which contains the internally deleted gene (Fig. 5, row 4). Identical results were obtained with the other two anti-P1 polyclonal antisera (data not shown). To determine whether there was a problem with cell surface attachment of P1 on PC3370B versus PC3370C, dot blots of culture supernatants were also performed. No P1 was detected in the spent culture medium of PC3370B, although P1 was readily detected in spent culture medium of NG8 as well as PC3370C (data not shown).
FIG. 5.
Dot blot analysis of P1 expression by S. mutans NG8, spaP-negative mutant PC3370, and derivatives. Twofold serial dilutions of bacterial cells, beginning at 5 × 106 CFU/well, were applied in duplicate to the membrane. Rabbit antiserum 209 made against purified P1 from S. mutans was used as the primary antibody. Rows 1 through 5, NG8, spaP-negative mutant PC3370, PC3370A (shuttle vector only), PC3370B (spaP with the P region deleted), and PC3370C (full-length spaP), respectively.
To determine whether any P1 at all was expressed by PC3370B, sonicated cell extracts were analyzed by Western blotting. These results are shown in Fig. 6. Cell sonicates of S. mutans NG8 (lane 1), PC3370C (lane 2), PC3370B (lane 3), PC3370A (lane 4), and PC3370 containing no plasmid (lane 5) were reacted with a cocktail of the six anti-P1 MAbs shown to be reactive with P-region deletion P1 expressed in E. coli (Fig. 4). Full-length P1 (Mr of ∼185,000) and numerous breakdown products were present in sonicated cell extracts of NG8 (Fig. 6, lane 1) and PC3370C (lane 2). A low level of P-region deletion P1 was observed in the sonicated cell extract of PC3370B (lane 3), but the amount of immunoreactive material was substantially decreased compared to that of full-length P1 expressed by PC3370C (lane 2). No P1 was observed in the sonicated extracts of PC3370A (lane 4) or PC3370 (lane 5). The visible bands in lanes 4 and 5 of Fig. 6 represent background binding by the peroxidase-conjugated goat anti-mouse IgG secondary antibody. Subtle differences in the banding patterns between lanes 1 and 2 do not reflect true differences, since these samples were electrophoresed on separate slab gels. The indicated positions of the molecular weight standards are therefore approximate.
FIG. 6.
Western blot analysis of P1 expression by S. mutans NG8, spaP-negative mutant PC3370, and derivatives. Lanes 1 through 5, sonicated cell extracts prepared from NG8, PC3370C (full-length spaP), PC3370B (spaP with the P region deleted), PC3370A (shuttle vector only), and PC3370 with no plasmid, respectively. The filter was reacted with a cocktail of MAbs including 3-8D2a, 4-9D4c, 5-3E5e, 2-8G1d, 3-3B5e, and 6-8C1a (Fig. 1 and 3). The migrations of molecular weight standards are indicated to the left of lane 1 and correspond to Mrs of 180,000, 116,000, 84,000, 58,000, and 48,500. Arrowheads (from top to bottom) indicate full-length and internally deleted P1.
Detection of spaP-specific mRNA in PC3370 derivatives.
To test whether spaP-specific mRNA was transcribed in PC3370 transformed with the shuttle vector constructs, an RNA dot blot was performed. These results are shown in Fig. 7. Dilutions of total cellular RNA were reacted with a dUTP-digoxigenin-labeled spaP probe corresponding to the N-terminal alanine-rich repeat gene segment (11). This region was unaffected during the construction of pMAJJ8 (Fig. 1 and Table 1). Row 1 of Fig. 7 shows the lack of reactivity of the probe with mRNA prepared from PC3370A. Rows 2 and 3 show nearly identical levels of spaP mRNA in PC3370B and PC3370C, respectively. Row 4 was a positive control for the probe and contains purified pMAD DNA (Table 1). To ensure that the transcript detected in PC3370B was not degraded from the 3′ end, the dot blot was repeated with a probe corresponding to the 3′ terminus of spaP (bp 3703 to 4817). Again, comparable levels of spaP mRNA were detected in PC3370B and PC3370C (data not shown). Taken together, these results indicate that there was no problem with transcription of the internally deleted spaP gene or stability of the mRNA from PC3370B compared with PC3370C.
FIG. 7.
RNA dot blot analysis of spaP-specific mRNA levels in spaP-negative mutant PC3370 derivatives. Twofold serial dilutions of total cellular RNA, beginning with 5 mg, were probed with DNA encoding the A region of spaP. Rows 1 through 3, PC3370A (shuttle vector only), PC3370C (full-length spaP), and PC3370B (spaP with the P region deleted), respectively. Row 4, twofold serial dilutions of purified pMAD (Table 1), beginning with 100 ng.
DISCUSSION
The fact that different proteins with dissimilar functions from a variety of microorganisms have similar internal proline-rich sequences is curious and is not likely to be coincidental. Proline is unique among the amino acids in that the end of the side chain is covalently bound to the preceding peptide bond nitrogen; therefore, proline residues are recognized as being of particular importance in their effect on chain conformation and protein folding (43). Proline-rich sequences are frequently repetitive and generally form extended structures and flexible regions (71). cis-trans isomerizations of prolyl peptide bonds are important in the folding of many proteins (60), and a number of peptidyl-prolyl isomerases (PPIases) have been identified as being important components of the intracellular chaperonin machinery of both prokaryotes and eukaryotes (20). The importance of the P region of P1 is highlighted by several studies. As stated above, Munro et al. (46) used a fragment of P1 (amino acid residues 816 to 1213) including the central proline-rich repeats to inhibit adherence of S. mutans to saliva-coated hydroxyapatite. The same fragment was recognized by two MAbs which were protective against recolonization with S. mutans in human passive immunization experiments (42, 46). Burnie et al. (8) identified an epitope directly within the P region of the Streptococcus oralis homolog of P1 as being protective in a mouse model of lethal infection. The P region of S. mutans P1 has been reported to contain predominantly B-cell, rather than T-cell, epitopes in studies of naturally sensitized humans (32) and immunized inbred mice (68). Whether the P region itself represents a functional domain or whether it plays a supportive structural role has not been clearly established. The fact that the fibronectin binding proteins of S. aureus and S. pyogenes have very similar proline-rich sequences adjacent to their fibronectin binding domains (29, 62, 64) suggests the latter. If the P region is a necessary component of a functional conformation of P1, protective antibodies may alter that conformation rather than directly block an adhesin domain.
To evaluate the contribution of the P region to the antigenicity and structure of P1, both a P-region subclone and a P-region deletion construct were generated. None of 11 anti-P1 MAbs prepared against mutanolysin-digested S. mutans cell walls and boosted with purified P1 (2) or two polyclonal rabbit antisera prepared against purified streptococcal and recombinant P1 were reactive by Western immunoblotting with an MBP–P-region fusion polypeptide or with the isolated P region. Only a polyclonal rabbit antiserum made against whole S. mutans cells was reactive (Fig. 4). This result was somewhat surprising in light of the results of Todryk et al. (68), who demonstrated the presence of B-cell epitopes within the P region. However, those investigators utilized whole S. mutans cells or an internal P1 fragment containing the proline-rich repeats as their immunogens. The results presented in this study suggest that while the P region is exposed and immunogenic on the surface of whole S. mutans cells, it does not appear to be accessible when solubilized full-length P1 is used as the immunogen. Also of interest was the finding that deletion of the P region obliterated binding of five of the six anti-P1 MAbs which map to the central portion of P1 (Fig. 1 and 3). None of the five affected MAbs bound to the isolated P region (compare Fig. 3 and 4), indicating that they recognize conformational epitopes dependent on the presence of the P-region but not linear epitopes directly within the P-region sequence. Several of these antibodies were inhibitory to agglutinin-mediated adherence or aggregation or both in a previous study (6). The profound effect of the P-region deletion on the reactivities of most of the MAbs which map to the central portion of P1 substantiates the conclusion of that report, namely, that the functional properties of P1 are mediated in a large part by overall conformation rather than by linear sequence. Binding of one MAb which mapped to the alanine-rich A region and of four MAbs which mapped to the carboxy terminus of the molecule was unaffected by the deletion of the P region. These results confirmed that the two spaP gene fragments used to engineer the internal deletion were joined in frame and indicated that these antibodies recognize epitopes which are not conformationally dependent on the presence of an internal proline-rich segment.
To evaluate the contribution of the P region to the function of P1 when it is expressed on the surface of S. mutans, the spaP construct devoid of P-region DNA was introduced via a shuttle vector into a spaP-negative mutant. It was reasoned that the internally deleted polypeptide would be anchored to the cell wall via normal mechanisms involving carboxy-terminal sequences, because this segment of the gene was not affected by the amplification and cloning strategy. As stated above, all four anti-P1 MAbs which map to the carboxy terminus of P1 were unaffected by the deletion. Transformation of mutant PC3370 with a plasmid containing the full-length spaP gene resulted in complementation of wild-type levels of P1 expressed on the cell surface. However, as illustrated in Fig. 5, transformation with the internally deleted gene gave a surprising result: absolutely no cell surface P1 was detectable. Analysis of culture supernatants demonstrated no internally deleted polypeptide in spent culture media, indicating that the lack of the molecule on the cell surface was not reflective of a problem with attachment.
Comparable message levels were detected in PC3370B and PC3370C by using both 5′ and 3′ spaP probes, indicating that both the internally deleted and full-length genes were transcribed and that the transcripts were equally stable (Fig. 7). To determine whether any P1 polypeptide at all was produced by PC3370B, Western blotting was performed with sonicated cell extracts. As shown in Fig. 6, a low level of internally deleted P1 was observed; however, the amount was substantially reduced compared to that of the ∼185,000-Mr P1 expressed by S. mutans NG8 or PC3370C. Taken together, these results suggest that P1 lacking the proline-rich repeat domain is produced but that this form of the molecule is unstable and most likely is degraded intracellularly before it can be translocated to the cell surface. While lower levels of P1 with the P region deleted than of full-length P1 were observed in recombinant E. coli, the relative difference in detectable protein was not nearly as pronounced as in S. mutans. Reactivity of the same anti-P1 MAbs was affected by deletion of the P region irrespective of whether the polypeptide was expressed in S. mutans or E. coli.
The substantial effect of the P-region deletion on the overall conformation of the central region of the molecule as evidenced by the abrogation of binding of five distinct MAbs has led to the hypothesis that the deletion interfered with proper folding of P1, which presumably is a necessary prerequisite for stability and translocation of the protein from the cytosol to the cell surface. It is now recognized that correct tertiary and quaternary structures are important determinants of efficient intracellular protein transport (reviewed in reference 20). Enzymes and chaperonins involved in folding, assembly, rearrangement, and degradation of proteins include protein disulfide isomerases; PPIases, which by virtue of cis-trans isomerization of proline act as “conformases” catalyzing steps in the initial folding or rearrangement of protein structures; and the chaperonins or heat shock proteins, including members of the Hsp60 (including GroEL and GroES), Hsp70 (including DnaK and DnaJ), and Hsp90 families. The P region may be a substrate of a putative PPIase, and cis-trans isomerization of prolines within the P region may confer a conformation enabling interaction with intracellular chaperonins and appropriate cell surface localization. In eukaryotes two non-sequence-related families of proteins, cyclophilins and FK506 binding proteins (collectively called immunophilins), possess PPIase activity (reviewed in references 19 and 23). Cyclophilins appear to be present in all microbial species investigated, and FK506 binding proteins have been found in yeast and a number of pathogenic bacteria as well. Recently the existence of a third type of PPIase in E. coli was reported (54). This protein was designated parvulin for its small size (92 amino acids), and homology to the PrtM protein of Lactococcus lactis (22) and the PrsA lipoprotein of Bacillus subtilis (33), both of which are known to be involved in protein transport and maturation, was found. Experiments to identify cytoplasmic molecules which may interact with the P region (or with P1 containing it) and potentially mediate the correct folding or localization of the adhesin molecule to the cell surface are planned. This information is expected to be more broadly applicable to molecules expressed by other gram-positive organisms which, although functionally distinct, share the property of containing internal repetitive proline-rich sequences similar to the P region of P1.
ACKNOWLEDGMENTS
We thank C. Alford for technical assistance.
This research was supported by NIH grant DE08007-12.
REFERENCES
- 1.Ackermans S F, Klein J P, Ogier J A, Bazin H, Cromont F, Franck R M. Purification and characterization of a saliva interacting cell wall protein from Streptococcus mutans serotype f by using monoclonal antibody immunoaffinity chromatography. Biochem J. 1985;228:211–217. doi: 10.1042/bj2280211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Applied Biosystems, Inc. User bulletin 18. Piscataway, N.J: Applied Biosystems, Inc., Pharmacia Biotech; 1991. [Google Scholar]
- 2.Ayakawa G Y, Boushell L W, Crowley P J, Erdos G W, McArthur W P, Bleiweis A S. Isolation and characterization of monoclonal antibodies specific for antigen P1, a major surface protein of mutans streptococci. Infect Immun. 1987;55:2759–2767. doi: 10.1128/iai.55.11.2759-2767.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bleiweis A S, Karakawa W W, Krause R M. Improved technique for preparation of streptococcal cell walls. J Bacteriol. 1964;88:1198–1200. doi: 10.1128/jb.88.4.1198-1200.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady L J, Crowley P J, Ma J K-C, Kelly C, Lee S F, Lehner T, Bleiweis A S. Restriction fragment length polymorphisms and sequence variation within the spaP gene of Streptococcus mutans serotype c. Infect Immun. 1991;59:1803–1810. doi: 10.1128/iai.59.5.1803-1810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady L J, Piacentini D A, Crowley P J, Bleiweis A S. Identification of monoclonal antibody-binding domains within antigen P1 of Streptococcus mutans and cross-reactivity with related surface antigens of oral streptococci. Infect Immun. 1991;59:4425–4435. doi: 10.1128/iai.59.12.4425-4435.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brady L J, Piacentini D A, Crowley P J, Oyston P C F, Bleiweis A S. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect Immun. 1992;60:1008–1017. doi: 10.1128/iai.60.3.1008-1017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckley N, Lee L N, LeBlanc D J. Use of a novel mobilizable vector to inactivate the srcA gene of Streptococcus sobrinus by allelic replacement. J Bacteriol. 1995;177:5028–5034. doi: 10.1128/jb.177.17.5028-5034.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnie J P, Brooks W, Donohoe M, Hodgetts S, Al-Ghamdi A, Matthews R C. Defining antibody targets in Streptococcus oralis infection. Infect Immun. 1996;64:1600–1608. doi: 10.1128/iai.64.5.1600-1608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceglowski P, Alonso J C. Gene organization of the Streptococcus pyogenes plasmid pDB101: sequence analysis of the orf eta-copS region. Gene. 1994;145:33–39. doi: 10.1016/0378-1119(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 10.Chen R, Schmidmayr C, Krämer C, Chen-Schmeisser U, Henning U. Primary structure of major outer membrane protein II (omp A protein) of Escherichia coli K-12. Proc Natl Acad Sci USA. 1980;77:4592–4596. doi: 10.1073/pnas.77.8.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley P J, Brady L J, Piacentini D A, Bleiweis A S. Identification of a salivary agglutinin-binding domain within cell surface adhesin P1 of Streptococcus mutans. Infect Immun. 1993;61:1547–1552. doi: 10.1128/iai.61.4.1547-1552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowley P J, Michalek S M, Bleiweis A S. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. 1997. Virulence of an isogenic spaP mutant of Streptococcus mutans in the germ-free rat, abstr. D-168. [Google Scholar]
- 12a.Crowley, P. J., et al. Unpublished data.
- 13.Demuth D R, Berthold P, Leboy P S, Golub E E, Davis C A, Malamud D. Saliva-mediated aggregation of Enterococcus faecalis transformed with a Streptococcus sanguis gene encoding the SSP-5 surface antigen. Infect Immun. 1989;57:1470–1475. doi: 10.1128/iai.57.5.1470-1475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demuth D R, Golub E E, Malamud D. Streptococcal-host interactions. Structural and functional analysis of a Streptococcus sanguis receptor for a human salivary glycoprotein. J Biol Chem. 1990;265:7120–7126. [PubMed] [Google Scholar]
- 15.Demuth D R, Lammey M S, Huck M, Lally E T, Malamud D. Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin. Microb Pathog. 1990;9:199–211. doi: 10.1016/0882-4010(90)90022-i. [DOI] [PubMed] [Google Scholar]
- 16.Douglas C W I, Russell R R B. Effect of specific antisera upon Streptococcus mutans adherence to saliva-coated hydroxylapatite. FEMS Microbiol Lett. 1984;25:211–214. [Google Scholar]
- 17.Fischetti V A, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol Microbiol. 1990;4:1603–1615. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 18.Forester H, Hunter N, Knox K W. Characteristics of a high molecular weight extracellular protein of Streptococcus mutans. J Gen Microbiol. 1983;129:2779–2788. doi: 10.1099/00221287-129-9-2779. [DOI] [PubMed] [Google Scholar]
- 19.Galat A. Peptidylproline cis-trans-isomerases: immunophilins. Eur J Biochem. 1993;216:689–707. doi: 10.1111/j.1432-1033.1993.tb18189.x. [DOI] [PubMed] [Google Scholar]
- 20.Gething M-J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 21.Gilkes N R, Henrissat G, Kilburn D G, Miller J R C, Warren R A J. Domains in microbial β1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haandrikman A J, Kok J, Laan H, Soemitro S, Ledeboer A M, Konings W N, Venema G. Identification of a gene required for maturation of an extracellular lactococcal serine proteinase. J Bacteriol. 1989;171:2789–2794. doi: 10.1128/jb.171.5.2789-2794.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hacker J, Fisher G. Immunophilins: structure-function relationship and possible role in microbial pathogenicity. Mol Microbiol. 1993;10:445–456. doi: 10.1111/j.1365-2958.1993.tb00917.x. [DOI] [PubMed] [Google Scholar]
- 24.Hajishengallis G, Koga T, Russell M W. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J Dent Res. 1994;73:1493–1502. doi: 10.1177/00220345940730090301. [DOI] [PubMed] [Google Scholar]
- 25.Hamada S, Slade H D. Biology, immunology, and cariogenicity of Streptococcus mutans in human dental decay. Microbiol Rev. 1980;50:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han N, Whitlock J, Progulske-Fox A. The hemagglutinin gene A (hagA) of Porphyromonas gingivalis 381 contains four large, contiguous, direct repeats. Infect Immun. 1996;64:4000–4007. doi: 10.1128/iai.64.10.4000-4007.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heden L-O, Frithz E, Lindahl G. Molecular characterization of an IgA receptor from group B streptococci: sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA binding capacity. Eur J Immunol. 1991;21:1481–1490. doi: 10.1002/eji.1830210623. [DOI] [PubMed] [Google Scholar]
- 28.Homonylo-McGavin M K, Lee S F. Role of the C terminus in antigen P1 surface localization in Streptococcus mutans and two related cocci. J Bacteriol. 1996;178:801–807. doi: 10.1128/jb.178.3.801-807.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jönsson K, Signäs C, Müller H-P, Lindberg M. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J Biochem. 1991;202:1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x. [DOI] [PubMed] [Google Scholar]
- 30.Kao S-M, Olmsted S B, Viksnins A S, Gallo J C, Dunny G M. Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis. J Bacteriol. 1991;173:7650–7664. doi: 10.1128/jb.173.23.7650-7664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly C, Evans P, Bergmeier L, Lee S F, Progulske-Fox A, Harris A C, Aitken A, Bleiweis A S, Lehner T. Sequence analysis of the cloned streptococcal cell surface antigen I/II. FEBS Lett. 1989;258:127–132. doi: 10.1016/0014-5793(89)81632-1. [DOI] [PubMed] [Google Scholar]
- 32.Kelly C, Todryk S, Kendal H, Munro G, Lehner T. T-cell, adhesion, and B-cell epitopes of the cell surface Streptococcus mutans protein antigen I/II. Infect Immun. 1995;63:3649–3658. doi: 10.1128/iai.63.9.3649-3658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kontinen V P, Sarvas M. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol Microbiol. 1993;8:727–737. doi: 10.1111/j.1365-2958.1993.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.LaPolla R J, Haron J A, Kelly C G, Taylor W R, Bohart C, Hendricks M, Pyati J, Graff R T, Ma J K-C, Lehner T. Sequence and structural analysis of surface proteins antigen I/II (SpaA) of Streptococcus sobrinus. Infect Immun. 1991;59:2677–2685. doi: 10.1128/iai.59.8.2677-2685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsen R A, Wood G E, Postle K. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol Microbiol. 1993;10:943–953. doi: 10.1111/j.1365-2958.1993.tb00966.x. [DOI] [PubMed] [Google Scholar]
- 37.Lee S F, Progulske-Fox A, Bleiweis A S. Molecular cloning and expression of a Streptococcus mutans major surface protein antigen, P1 (I/II), in Escherichia coli. Infect Immun. 1988;56:2114–2119. doi: 10.1128/iai.56.8.2114-2119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S F, Progulske-Fox A, Erdos G W, Piacentini D A, Ayakawa G Y, Crowley P J, Bleiweis A S. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II) Infect Immun. 1989;57:3306–3313. doi: 10.1128/iai.57.11.3306-3313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loesche W J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lunsford R D. Recovery of RNA from oral streptococci. BioTechniques. 1995;18:412–413. [PubMed] [Google Scholar]
- 41.Ma JK-C, Kelly C G, Munro G, Whiley R A, Lehner T. Conservation of the gene encoding streptococcal antigen I/II in oral streptococci. Infect Immun. 1991;59:2686–2694. doi: 10.1128/iai.59.8.2686-2694.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma JK-C, Hunjan M, Smith R, Lehner T. Specificity of monoclonal antibodies in local passive immunization against Streptococcus mutans. Clin Exp Immunol. 1989;77:331–337. [PMC free article] [PubMed] [Google Scholar]
- 43.MacArthur M W, Thornton J M. Influence of proline residues on protein conformation. J Mol Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 44.McIver K S, Subbarao S, Kellner E M, Heath A S, Scott J R. Identification of isp, a locus encoding an immunogenic secreted protein conserved among group A streptococci. Infect Immun. 1996;64:2548–2555. doi: 10.1128/iai.64.7.2548-2555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moisset A, Schatz M, Lepoivre Y, Amadio S, Wachsmann D, Schöller M, Klein J P. Conservation of salivary glycoprotein-interacting and human immunoglobulin G cross-reactive domains of antigen I/II in oral streptococci. Infect Immun. 1994;62:184–193. doi: 10.1128/iai.62.1.184-193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munro G H, Evans P, Todryk S, Buckett P, Kelly G C, Lehner T. A protein fragment of streptococcal cell surface antigen I/II which prevents adhesion of Streptococcus mutans. Infect Immun. 1993;61:4590. doi: 10.1128/iai.61.11.4590-4598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagamune H, Ohnishi C, Katsuura A, Fushitani K, Whiley R A, Tsuji A, Matsuda Y. Intermedilysin, a novel cytotoxin specific for human cells, secreted by Streptococcus intermedius UNS46 isolated from a human liver abscess. Infect Immun. 1996;64:3093–3100. doi: 10.1128/iai.64.8.3093-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakai M, Okahashi N, Ohta H, Koga T. Saliva-binding region of Streptococcus mutans surface protein antigen. Infect Immun. 1993;61:4344–4349. doi: 10.1128/iai.61.10.4344-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okahashi N, Sasakawa C, Yoshikawa M, Hamada S, Koga T. Cloning of a surface protein antigen gene from serotype c Streptococcus mutans. Mol Microbiol. 1989;3:221–228. doi: 10.1111/j.1365-2958.1989.tb01811.x. [DOI] [PubMed] [Google Scholar]
- 50.Okahashi N, Sasakawa C, Yoshikawa M, Hamada S, Koga T. Molecular characterization of a surface protein antigen gene from serotype c Streptococcus mutans, implicated in dental caries. Mol Microbiol. 1989;3:673–678. doi: 10.1111/j.1365-2958.1989.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 51.Pancholi V, Fischetti V A. Identification of an endogenous membrane anchor-cleaving enzyme for group A streptococcal M protein: its implication for the attachment of surface proteins in gram-positive bacteria. J Exp Med. 1989;170:2119–2133. doi: 10.1084/jem.170.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pistor S, Chakaborty T, Niebuhr K, Domann E, Wehland J. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 1994;13:758–763. doi: 10.1002/j.1460-2075.1994.tb06318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Proft T, Hilbert H, Layh-Schmit G, Herrmann R. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J Bacteriol. 1995;177:3370–3378. doi: 10.1128/jb.177.12.3370-3378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahfeld J-U, Rücknagel K P, Schelbert B, Ludwig B, Hacker J, Mann K, Fischer G. Confirmation of the existence of a third family among peptidyl-prolyl cis/trans isomerases. Amino acid sequence and recombinant production of parvulin. FEBS Lett. 1994;352:180–184. doi: 10.1016/0014-5793(94)00932-5. [DOI] [PubMed] [Google Scholar]
- 55.Rajendrakumar C S V, Reddy B D B, Reddy A R. Proline-protein interactions: protection of structural and functional integrity of M4 lactate dehydrogenase. Biochem Biophys Res Commun. 1994;201:957–963. doi: 10.1006/bbrc.1994.1795. [DOI] [PubMed] [Google Scholar]
- 56.Rathsam C, Giffard P M, Jacques N A. The cell-bound fructosyltransferase of Streptococcus salivarius: the carboxyl terminus specifies attachment in a Streptococcus gordonii model system. J Bacteriol. 1993;175:4520–4527. doi: 10.1128/jb.175.14.4520-4527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riggs P. Expression and purification of maltose-binding protein fusions. In: Ausubel F M, Brent R, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1992. pp. 16.6.1–16.6.12. [DOI] [PubMed] [Google Scholar]
- 58.Russell M W, Mansson-Rahemtulla B. Interaction between surface protein antigens of Streptococcus mutans and human salivary components. Oral Microbiol Immunol. 1989;4:106–111. doi: 10.1111/j.1399-302x.1989.tb00107.x. [DOI] [PubMed] [Google Scholar]
- 59.Russell R R B. Wall-associated antigens of Streptococcus mutans. J Gen Microbiol. 1979;114:109–115. doi: 10.1099/00221287-114-1-109. [DOI] [PubMed] [Google Scholar]
- 60.Schmid F X. Prolyl isomerase: enzymatic catalysis of slow protein-folding reactions. Annu Rev Biophys Biomol Struct. 1993;22:123–143. doi: 10.1146/annurev.bb.22.060193.001011. [DOI] [PubMed] [Google Scholar]
- 61.Senpuku H, Takako M, Hanada N, Nisizawa T. An antigenic peptide inducing cross-reacting antibodies inhibiting the interaction of Streptococcus mutans PAc with human salivary components. Infect Immun. 1995;63:4695–4703. doi: 10.1128/iai.63.12.4695-4703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Signäs C, Raucci G, Jönsson K, Lindgren P-E, Anantharamaiah G M, Höök M, Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:669–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Southwick F S, Purich D L. Intracellular pathogenesis of listeriosis. N Engl J Med. 1996;334:770–776. doi: 10.1056/NEJM199603213341206. [DOI] [PubMed] [Google Scholar]
- 64.Talay S R, Valentin-Weigand P, Timmis K N, Chatwal G S. Domain structure and conserved epitopes of Sfb protein, the fibronectin-binding adhesin of Streptococcus pyogenes. Gene. 1994;126:123–128. doi: 10.1111/j.1365-2958.1994.tb00448.x. [DOI] [PubMed] [Google Scholar]
- 65.Terleckyj B, Willet N P, Shockman G D. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun. 1975;11:649–655. doi: 10.1128/iai.11.4.649-655.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Timoney J F, Walker J, Zhou M, Ding J. Cloning and sequence analysis of a protective M-like protein gene from Streptococcus equi subsp. zooepidemicus. Infect Immun. 1995;63:1440–1455. doi: 10.1128/iai.63.4.1440-1445.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tobian J A, Cline M L, Macrina F L. Characterization and expression of a cloned tetracycline resistance determinant from the chromosome of Streptococcus mutans. J Bacteriol. 1984;160:556–563. doi: 10.1128/jb.160.2.556-563.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Todryk S M, Kelly G C, Munro G H, Lehner T. Induction of immune responses to functional determinants of a cell surface streptococcal antigen. Immunology. 1996;87:55–63. [PMC free article] [PubMed] [Google Scholar]
- 69.Tokuda M, Okahashi N, Takahashi I, Nakai M, Nagaoka S, Kawagoe M, Koga T. Complete nucleotide sequence of the gene for a surface protein antigen of Streptococcus sobrinus. Infect Immun. 1991;59:3309–3312. doi: 10.1128/iai.59.9.3309-3312.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yother J, Briles D E. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J Bacteriol. 1992;174:601–609. doi: 10.1128/jb.174.2.601-609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]