Abstract
Although polymer-based lipid-nanodiscs are increasingly used in the structural studies of membrane proteins, the charge of the belt-forming polymer is a major limitation for functional reconstitution of membrane proteins possessing an opposite net charge to that of the polymer. This limitation also rules out the reconstitution of a protein-protein complex composed of oppositely-charged membrane proteins. In this study, we report the first successful functional reconstitution of a membrane-bound redox complex constituting a cationic cytochrome P450 (CYP450) and an anionic cytochrome P450 reductase (CPR) in non-ionic inulin-based lipid-nanodiscs. The gel-to-liquid-crystalline phase-transition temperature (Tm) of DMPC:DMPG (7:3 w/w) lipids in polymer-nanodiscs was determined by differential scanning calorimetry (DSC) and 31P NMR experiments. The CYP450-CPR redox complex reconstitution in polymer-nanodiscs was characterized by size-exclusion chromatography (SEC), and the electron transfer kinetics was carried out using the stopped-flow technique under anaerobic conditions. The Tm of DMPC:DMPG (7:3 w/w) in polymer-nanodiscs measured from 31P NMR agrees with that obtained from DSC, and was found to be higher than that for liposomes due to decreased cooperativity of lipids present in the nanodiscs. The stopped-flow measurements revealed the CYP450-CPR redox complex reconstituted in nanodiscs to be functional, and the electron transfer kinetics was found to be temperature-dependent. Based on the successful demonstration of the use of non-ionic inulin-based polymer-nanodiscs reported in this study, we expect them to be useful in studying the function and structures of a variety of membrane proteins/complexes irrespective of the charge of the molecular components. Since the polymer-nanodiscs were shown to align in an externally applied magnetic field, they can also be used to measure residual dipolar couplings (RDCs) and residual quadrupolar couplings (RQCs) for various molecules ranging from small molecules to soluble proteins and nucleic acids.
Graphical Abstract
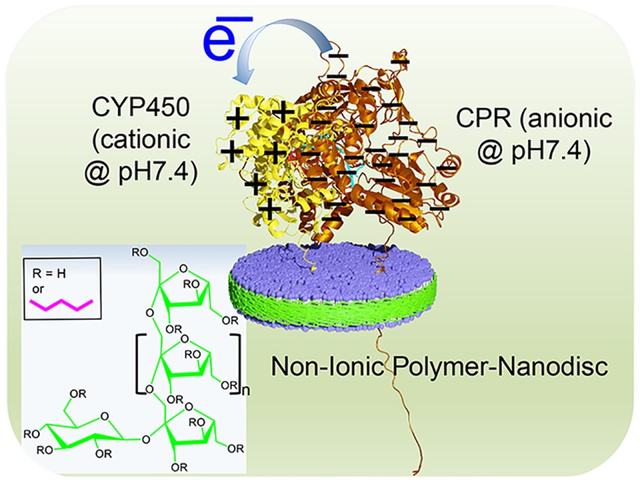
The compatibility of non-ionic polymer-based lipid-nanodiscs to study functional reconstitution of membrane protein complexes composed of cationic cytochrome P450 (CYP450) and anionic cytochrome P450 reductase (CPR) is demonstrated.
INTRODUCTION
Although recent studies have shown the importance of solving atomic-resolution structures of membrane proteins in a lipid membrane environment, membrane proteins still pose major challenges to most biophysical approaches due to their intrinsic instability in non-native membrane environments. Some of the major difficulties have been overcome by the use of membrane mimetics such as bicelles and nanodiscs.1–6 While both bicelles and nanodiscs are frequently used in membrane protein structural biology; recent studies have shown the major impact of the use of different types of nanodiscs.4, 7 These nanodiscs are made up of a planar lipid bilayer surrounded by a belt forming membrane scaffold proteins (MSPs) or short amphipathic peptides.4, 8–10 Similarly, synthetic amphipathic polymers such as poly(diisobutylene-alt-maleic acid (DIBMA) or poly(styrene-co-maleic acid) (SMA) forms soluble lipid-bilayer discs with lipids, which are called as polymer-nanodiscs.11, 12 As studies continue to develop and characterize these nanodiscs, each type of these nanodiscs has been shown to exhibit unique advantages and some limitations. The availability of a library of different types of nanodiscs-forming synthetic amphipathic polymers, and the stability of some of the polymer-based nanodiscs against pH and divalent metal ions, have expanded the scope of the nanodisc technology.2–4, 6, 11, 13–19 Additionally, several of these polymers have been successfully used to extract membrane proteins along with local native lipids from the cell membrane without the use of detergents.6, 12, 19–23 The nanodisc-forming polymers are charged molecules; therefore, they can electrostatically interact with oppositely charged molecules. As a result, the high charge-density of the polymers present in the belt of the nanodiscs is a limitation in studying membrane proteins and their complexes that possess opposite net charges.24, 25 For example, the interference of a charged polymer belt has been shown to destabilize the reconstituted anionic cytochrome-b5 (cytb5) or cationic cytochrome P450 (CYP450).24 The impediment to the structure and function of these proteins in SMA-based nanodiscs is due to the formation of non-specific charge-charge interactions between the cytochrome proteins and the oppositely charged SMA from nanodiscs.24 Therefore, polymers with high charge-densities are not suitable for studying (or extracting) differently charged membrane proteins and membrane protein complexes.24, 25 Although high-salt concentrations (~500 mM) can be used to suppress the non-specific charge-charge interactions between the polymer and the target membrane protein,24 such conditions are physiologically irrelevant (~150 mM) in general, may not be applicable to all membrane proteins and are also not desirable for NMR-based structural studies as shown for cytb5.24 Thus, probing the structure and dynamics of oppositely-charged membrane proteins and protein-protein complexes in a charged-polymer nanodiscs is not feasible. Therefore, to overcome these difficulties, non-ionic polymers that exhibit excellent lipid solubilizing and stable nanodiscs-forming properties are needed.
The recently developed non-ionic pentyl-functionalized inulin polymer is shown to form nanodiscs when mixed with liposomes;26 hence, the nanodiscs of this polymer can be suitable to study differently charged membrane proteins/complexes. Additionally, unlike the SMA-polymers, the pentyl-inulin does not contain an aromatic moiety, and, therefore, it is less likely to interfere with protein characterization by UV-based spectroscopic methods.27, 28 Here, we report the first successful demonstration of the functional reconstitution of a redox complex constituting a cationic CYP450 (55.7-kDa) and an anionic CYP450-reductase (CPR) (76.8-kDa) in non-ionic inulin-based nanodiscs. CYP450 and CPR used in this study are full-length (wild-type) proteins; each contains a single transmembrane domain. CYP450 and CPR are large soluble-domain containing membrane-anchored proteins with opposite net charges under physiological conditions (Figure 1; Table S1). Microsomal CYP450 is a key enzyme in the oxidative metabolism of drugs and xenobiotics.29, 30 CPR, via FAD and FMN prosthetic groups, donates two electrons one at a time to prosthetic heme iron of CYP450 during the drug metabolism. The lipid membrane has been shown to be important for the function of the CYP450-CPR redox complex.31–37 In this study, an investigation of the electron transfer kinetics in the redox CYP450-CPR complex38 reconstituted in pentyl-inulin nanodiscs containing 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-dimyristoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (sodium salt) (DMPG) lipids is reported (Figure 1).
Figure 1.
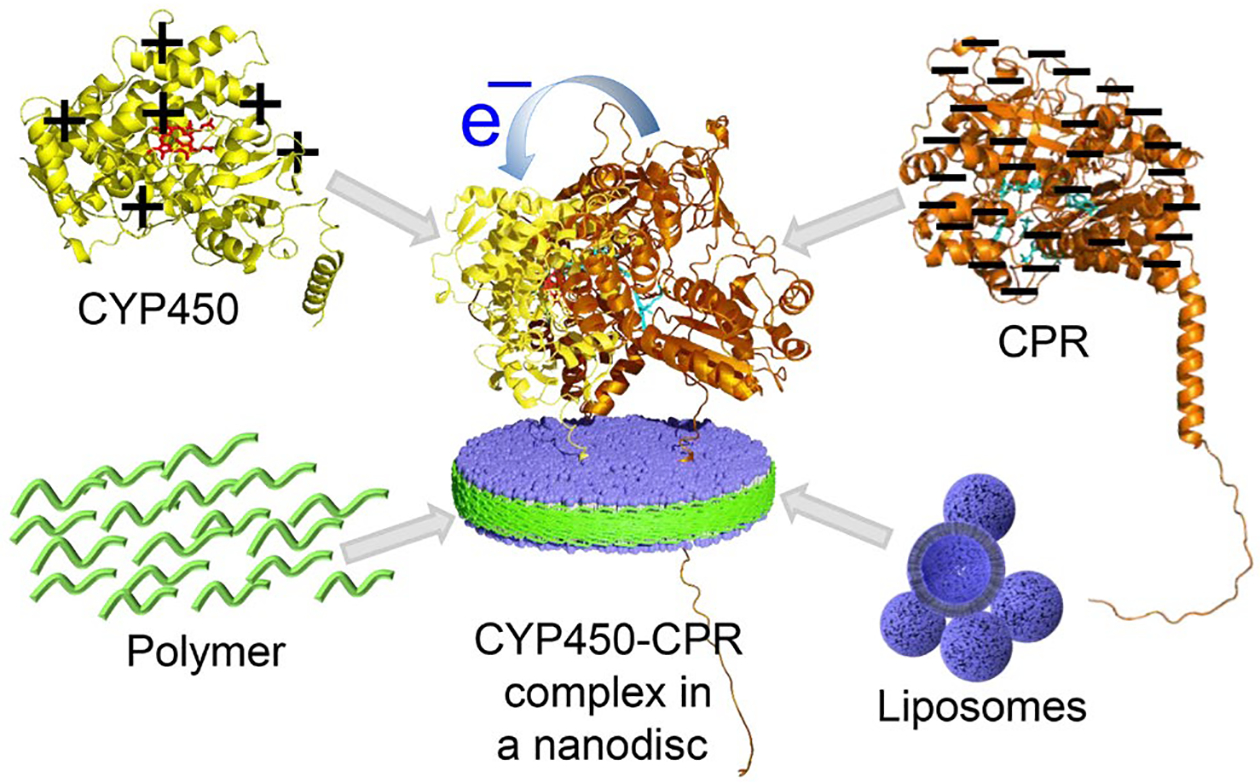
Schematic illustration of the functional reconstitution of the oppositely charged CYP450 (yellow-red) and CPR (Orange-cyan) redox proteins in non-ionic pentyl-inulin-based polymer-nanodiscs (blue-green). The net charge of the proteins is indicated with +/− signs. Protein structure models were taken from the AlphaFold protein structure database.39
EXPERIMENTAL SECTION
Pentyl-inulin synthesis.
Inulin extracted from chicory roots was purchased from Sigma-Aldrich (St. Louis, Missouri, USA). Pentyl-inulin (average molecular weight (MW) of ~3 kDa) polymer used in this study was synthesized by functionalizing inulin with hydrophobic pentyl groups using pentyl bromide (Sigma-Aldrich), purified, and characterized as described elsewhere.26
Expression and purification of CYP450 2B4 and CPR.
CYP450 2B4 and CPR were expressed from E.coli, purified, and characterized as reported in the literature.40–45
Preparation of DMPC:DMPG liposomes.
The lipids were purchased from Avanti Polar Lipids (Alabaster, USA). 7 mg of DMPC and 3 mg of DMPG (7:3 w/w) taken in separate 1.5 mL centrifuge tubes were dissolved in a solvent mixture containing 1:1 v/v CH3O:CHCl3 (50 μL each) (Sigma-Aldrich). The dissolved lipids were then mixed by transferring them into a single 1.5 mL centrifuge tube. Next, the organic solvents were evaporated by applying a low-pressure N2-gas (20–30 min) onto the lipid-solvents mixture. The lipid-mixture was further dried under vacuum for 1 hour to remove all the residual solvents. Finally, the liposome solution was prepared by resuspending the solvent-free dry lipid-mixture in 10 mM potassium phosphate buffer (pH 7.4) containing 50 mM NaCl and by subjecting it to freeze-thaw cycles (using liquid N2 and hot water [~70 °C]) three times.
Preparation of DMPC:DMPG (7:3 w/w) polymer-nanodiscs.
The liposomes were mixed with pentyl-inulin at a 1:1 w/w ratio, and the solution was incubated overnight at 4 °C. Then, the polymer-nanodiscs solution was purified in 10 mM potassium phosphate buffer (pH 7.4) containing 50 mM NaCl by size-exclusion chromatography (SEC) column (10×300 Superdex 200 [GE Healthcare, Chicago, USA]) using fast protein liquid chromatography (FPLC [GE Healthcare, Chicago, USA]). The fractions (detected at 214 nm) containing polymer-nanodiscs were combined, and 900 μL of it was used for protein reconstitution.
Transmission electron microscopy (TEM).
TEM micrographs were recorded using a Technai® T-20® machine (FEI®, Netherlands) with a 60 kV operating voltage. The polymer-nanodiscs were prepared in 10 mM Tris buffer (pH 7.4) containing 50 mM NaCl and used for TEM studies without any purification. A dilute sample was dropped on a carbon-coated copper grid and dried overnight at room temperature in a desiccator before using in TEM experiments.
Phase-transition of DMPC:DMPG (7:3 w/w) polymer-nanodiscs by Differential Scanning Calorimetry (DSC).
DSC measurements were performed on liposomes (5 mg/mL), and on the polymer-nanodiscs that were purified using SEC. The experiments were performed on a Differential Scanning Calorimeter (DSC Nano, TA instruments, New Castle, DE, USA) using a constant pressure of 3 atm (44 psi) and a scan rate of 1 °C/min between 10 and 50 °C. NanoAnalyze™ software was used for background subtraction and baseline correction.
31P NMR of DMPC:DMPG (7:3 w/w) polymer-nanodiscs.
Temperature-dependent 31P NMR spectra were recorded under static conditions on a 400 MHz Bruker solid-state NMR spectrometer (Billerica, MA, USA) equipped with a 5 mm triple-resonance HXY MAS NMR probe, operating at a resonance frequency of 400.11 MHz for protons and 161.97 MHz for 31P. A 5 mm glass tube was used for the sample. Each 31P NMR spectrum was acquired using a 5 μs 90° pulse followed by a 25 kHz TPPM (two-pulse phase-modulated) proton decoupling,46 using 1024 scans, and relaxation/recycle delay of 3 s. The data were processed and analyzed in Bruker TopSpin (3.6.2). The samples were prepared at 100 mg/mL concentration in 10 mM Tris buffer (pH 7.4) containing 50 mM NaCl, and the free polymer/impurities were removed using Amicon centricon filter (50-kDa cut-off).
Reconstitution, purification, and UV measurements of CYP450 2B4 and CPR proteins in DMPC:DMPG (7:3 w/w) polymer-nanodiscs.
100 μL of 60 μM CYP450 2B4 was added to 900 μL of polymer-nanodiscs (final protein concentration was 6 μM) and carefully mixed with a micropipette. The solution was incubated at 4 °C or at room temperature overnight (without mixing). The sample was centrifuged at 8000 rpm for 1 min and purified by SEC (detected at 280 nm) in a 10 mM potassium phosphate buffer (pH 7.4) containing 50 mM NaCl. The fractions containing nanodisc-incorporated CYP450 2B4 were combined, concentrated to 900 μL, and quantitated using the carbon monoxide (CO)-binding assay (4.5 μM; ε=91 mM−1 cm−1). 1 molar equivalent of CPR was added to the solution and carefully mixed with a micropipette. The solution was incubated at 4 °C overnight without any mixing. The sample was centrifuged to remove any insoluble material and purified by SEC in a 10 mM Tris (pH 7.4) buffer containing 50 mM NaCl. The protein fractions were concentrated to 1 mL final volume, and the protein concentration was measured using CO-assay (3 μM). The protein solution with and without a substrate (benzphetamine) was saturated with CO-gas just before starting stopped-flow measurements.
UV-visible spectroscopy.
The absorption spectra were recorded on a UV/vis spectrophotometer (DeNovix DS-11+[M/C], Wilmington, DE, USA) using a 1 cm light path quartz cuvette.
Dynamic light scattering (DLS).
DLS experiments were performed using Wyatt Technology® DynaPro® NanoStar® using a 1 μL quartz MicroCuvette. The DLS data were collected on the SEC purified polymer-nanodiscs with and without redox complex in 10 mM Tris buffer pH 7.4 containing 50 mM NaCl.
CO-assay on CYP450 2B4.
CO-assay was performed to estimate the functional protein concentration.47, 48 CYP450 2B4 was reduced (Fe3+ [low-spin state] to Fe2+ [high-spin state]) by adding 1–2 μL of sodium dithionate (10 mg/mL), and a reference UV spectrum was recorded. Then the solution was saturated with CO gas. The protein concentration was measured from the absorption peak at 451 nm using an extinction coefficient of 91 cm−1 mM−1.
Electron transfer measurements using stopped-flow experiments.
The electron transfer measurement was performed under anaerobic conditions49–53 using a Hi-Tech SF61DX2 stopped-flow spectrophotometer (Bradford-on-Avon, UK) housed in an anaerobic Belle Technology glove box (Weymouth, UK). The 10 mM Tris buffer (pH 7.4) containing 50 mM NaCl was purged with N2 gas for 1 hour to remove the dissolved oxygen before being transferred to the glove box. A 1 mM stock solution of NADPH was prepared under anaerobic conditions by dissolving NADPH powder in 10 mM Tris buffer (pH 7.4) containing 50 mM NaCl. The working solution was prepared (in a separate 1.5 mL centrifuge tube) such that the final NADPH concentration in the reaction mixture was 20 molar equivalents to protein concentration. The CO-saturated protein and NADPH solutions were injected into two different valves of the stopped-flow using two new syringes, mixed, and UV-vis spectra were recorded for 60 s at different temperatures ranging from 11 to 38 °C to measure the electron transfer rates. The absorbance was detected using a Photodiode Array (PDA) detector. The redox reaction rate constants and the amplitudes were calculated by fitting the reaction kinetics data to a double-exponential equation:
| [eq. 1] |
Where a1 and a2 are amplitudes, and k1 and k2 are rate constants.
RESULTS AND DISCUSSION
First, the pentyl-inulin polymer-nanodiscs containing DMPC and DMPG lipids were prepared, purified using SEC, and characterized using DLS and TEM. The purified nanodiscs were then used in DSC and 31P NMR experiments to measure the phase-transition behavior of lipids in nanodiscs. 31P NMR spectra also revealed the magnetic-alignment of nanodiscs. After these characterizations of nanodiscs, as discussed in detail below, they were used to reconstitute a redox complex composed of 55.7-kDa rabbit CYP450 2B4 and 76.8-kDa rat-CPR (Table S1) and characterized using SEC and CO-binding assay. Electron transfer kinetics in the redox complex was evaluated by conducting stopped-flow measurements under anaerobic conditions.
Inulin-based polymers form monodispersed nanodiscs.
Inulin-based polymer-nanodiscs containing pentyl-inulin, zwitterionic DMPC, and anionic DMPG lipids (Figure 2(a, b)) were prepared as described in the Materials and Methods section above and purified using SEC using a 0.75 mL/min flow rate. Specifically, pentyl-inulin polymer and 7:3 w/w DMPC:DMPG mixture was used at a 1:1 (w/w) polymer:lipid ratio to prepare the nanodiscs. In SEC, DMPC:DMPG (7:3 w/w) polymer-nanodiscs were eluted as a single major peak between 7.7 and 11 mL, indicating good homogeneity of the polymer-nanodiscs sample (Figure 2c). The DLS measurements of DMPC:DMPG (7:3 w/w) polymer-nanodiscs showed a hydrodynamic radius (RHYD) of ~14 nm, and TEM revealed the presence of polymer-nanodiscs with a diameter of 30 ±5 nm (Figure 2(d, e)). These observations are in good agreement with previously reported results.26
Figure 2.
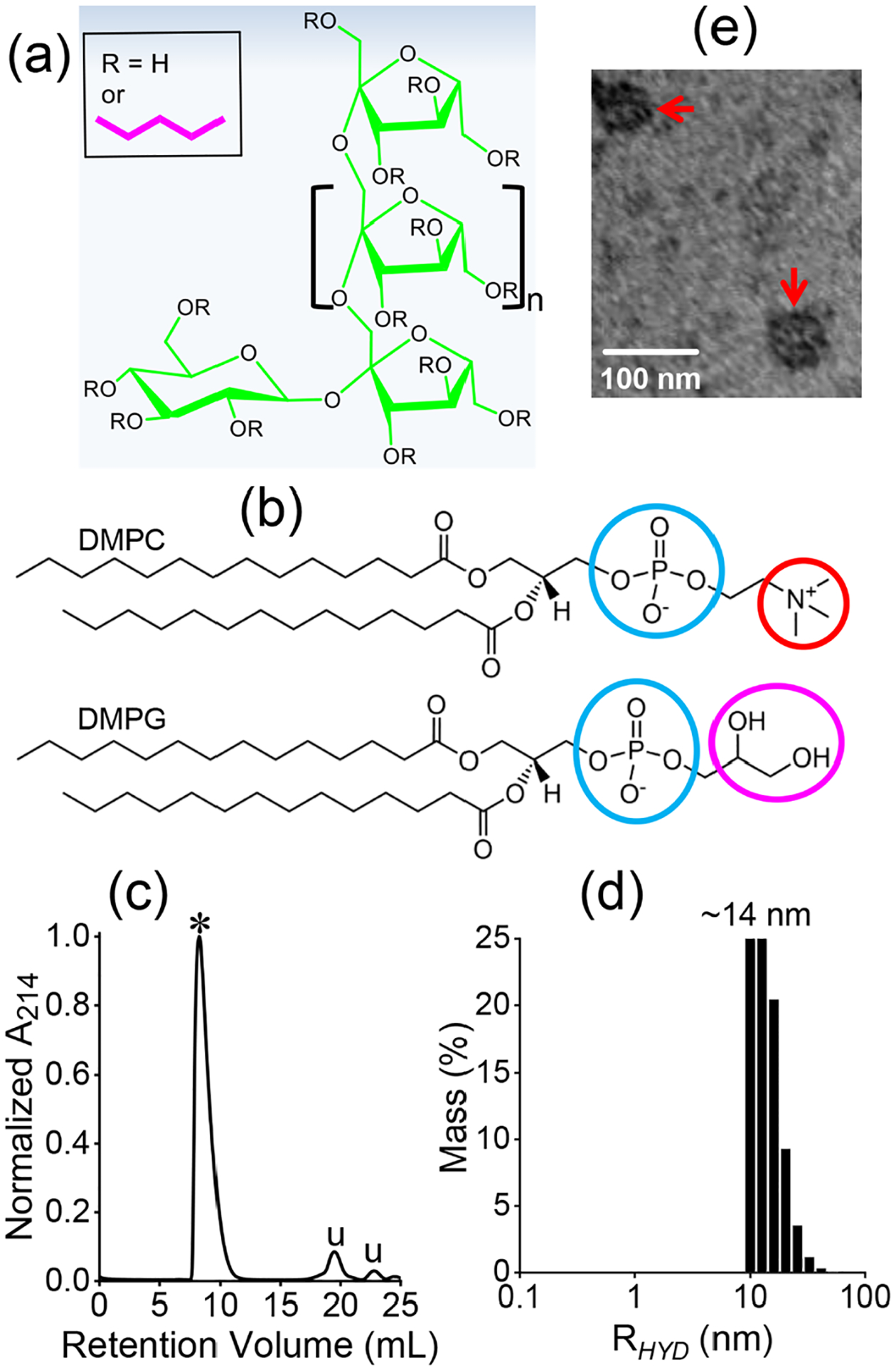
Molecular structures of non-ionic pentyl-inulin polymer (a), zwitterionic DMPC, and the anionic DMPG lipids (b) used in this study. Circles highlight the charge on the lipids: cyan (negative), red (positive), and magenta (neutral, polar). (c) SEC chromatogram of pentyl-inulin 7:3 w/w DMPC:DMPG nanodiscs. ‘⁎’ indicates the polymer-nanodiscs, whereas ‘u’ indicates uncharacterized peaks likely arising from free polymers and small molecule impurities. (d) DLS profile of SEC-purified pentyl-inulin DMPC:DMPG (7:3 w/w) nanodiscs with a hydrodynamic radius of ~14 nm. (e) TEM image of pentyl-inulin DMPC:DMPG (7:3 w/w) nanodiscs. Nanodiscs are indicated with red arrows.
DSC experiments revealed a decreased cooperativity in the gel-to-liquid-crystalline phase-transition of lipids.
Previous studies have shown the effect of belt-forming synthetic polymers on the phase-transition of lipids present in polymer nanodiscs. In this study, to evaluate the effect of pentyl-inulin polymer on the encased lipid bilayer, the gel-to-liquid-crystalline phase-transition temperature (Tm) of 7:3 w/w DMPC:DMPG nanodiscs was measured using DSC and 31P NMR experiments (Figure 3). In DSC experiments, the gel to liquid-crystalline lamellar phase-transition peak for the DMPC:DMPG (7:3 w/w) bilayer in polymer-nanodiscs is broadened substantially compared to that observed for liposomes (Figure 3a). The Tm of 7:3 (w/w) DMPC:DMPG polymer-nanodiscs was measured to be ~26.5 °C, whereas it is 24 °C for the same lipids present in liposomes (Figure 3a). These observations indicate a decreased cooperativity in the phase-transition and possibly a more ordered packing of DMPC:DMPG lipids in polymer-nanodiscs.
Figure 3.
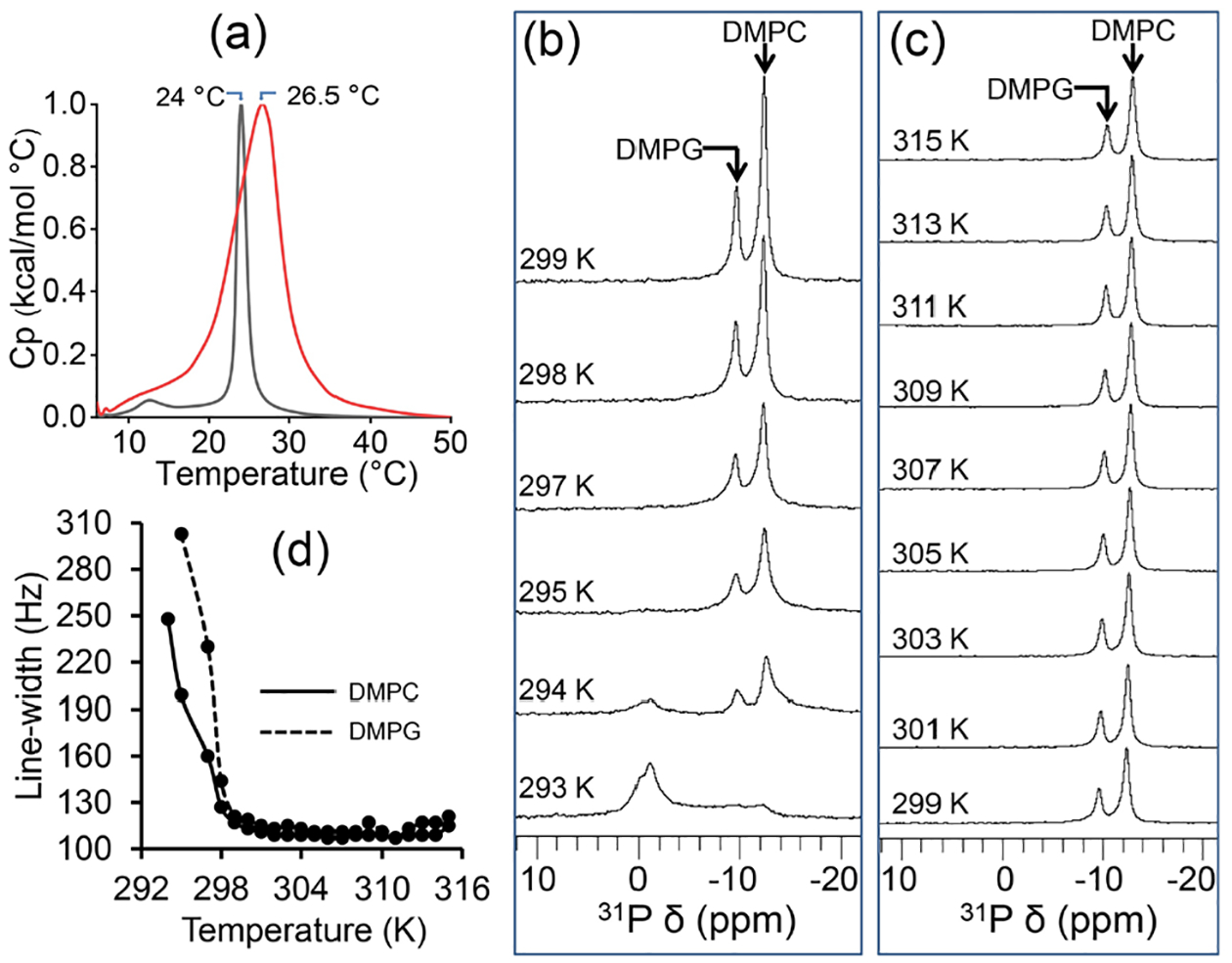
(a) DSC profiles of lipid melting in pentyl-inulin 7:3 w/w DMPC:DMPG nanodiscs (red) and 7:3 w/w DMPC:DMPG liposomes (black); heat capacity (Cp) values are normalized. (b, c) Variable temperature 31P NMR spectra of pentyl-inulin 7:3 w/w DMPC:DMPG nanodiscs. (d) Temperature dependence of 31P-DMPC and DMPG line-widths measured from the spectra (b, c) of pentyl-inulin 7:3 w/w DMPC:DMPG nanodiscs.
Magnetic-alignment of nanodiscs observed from 31P NMR experiments.
31P NMR spectra were recorded at different temperatures ranging from 293 to 315 K. Since the DMPC:DMPG lipids are mostly in the gel phase at 293 K, a combination of a major peak near 0 ppm arising from isotropic nanodiscs and a broad low-intensity peak (between −8 and −14 ppm) due to perpendicular component of the axially symmetric powder pattern arising from randomly oriented lipids (either lipid aggregates like liposomes or randomly oriented nanodiscs) was observed (Figures 3b and S1). With the increase in sample temperature, lipids transitioning to the lamellar phase contributed to the magnetic-alignment of nanodiscs as indicated by the presence of two peaks in the high-field region: ~−10 ppm from DMPG and ~−13 ppm from DMPC; peak assignment was based on the ratio of peak intensities resembling the concentration ratio between the lipids present in the sample. The line-width of these peaks decreased (or the intensity increased) with temperature, and the isotropic peak near 0 ppm and the broad low-intensity peak (293 K) disappeared completely when the temperature was increased above 298 K (Figure 3(b, c)); whereas the frequency of these peaks varied within 0.6 ppm (for DMPC) and 0.8 ppm (for DMPG) with the sample temperature increasing from 299 to 315 K. The line-width for the DMPC peak decreased from 248 Hz (at 294 K) to 113 Hz (at 300 K) (Figure 3d). Similarly, the line-width for the DMPG peak decreased from 303 Hz (295 K) to 115 Hz (301 K). No substantial change was observed either in line-width (109 ± 4 Hz) or in peak intensity for both peaks between 300 and 315 K (Figure 3d). The observed changes in line-width/intensity for 293 to 300/301 K in NMR spectra can be attributed to the change in the physical state of lipids, i.e., gel to the liquid-crystalline phase, which occurred at a broad temperature range and is in good agreement with the observations from DSC experiments (Figure 3(a, d)). The enhancement in the extent of magnetic-alignment of nanodiscs at a higher temperature also contributes to the increased 31P signal intensity. Thus, the broad temperature-dependent phase-transition of lipids in nanodiscs suggests that the packing of lipids in the center of the nanodisc may be different from those regions located closer to the polymer belt.54 The spectra also reveal the magnetic-alignment of the polymer-nanodiscs between 295 to 315 K, a desired property for data acquisition using multidimensional solid-state NMR experiments.
Electron transfer kinetics measured from the CYP450-CPR complex reconstituted in polymer-nanodiscs.
The non-ionic DMPC:DMPG (7:3 w/w) polymer-nanodiscs are used to demonstrate their compatibility in studying the CYP450-CPR redox membrane-bound protein-protein complex. At physiological pH 7.4, CPR is negatively charged (z=−27), and CYP450 2B4 is positively charged (z=+6) (Figure 1, and Table S1). The nanodiscs with the CYP450-CPR complex were purified using SEC, where the redox complex in nanodiscs was eluted between 7.6–11 mL (Figures 4a and S2). The purified redox complex in polymer-nanodiscs was analyzed by SDS-PAGE, TEM, and DLS (Figures 4(b, c), and S2, S3). Within the experimental limits, the measured size of nanodiscs with redox complex was similar to that of protein-free nanodiscs (Figures 2(d, e), 4c, and S3), indicating that the reconstitution of proteins does not affect the size of pentyl-inulin-based lipid-nanodiscs. No change in the nanodisc size may be because both CYP450 and CPR are single transmembrane domain containing proteins and the size of the nanodisc is sufficiently large to accommodate the complex. The concentration of CYP450 in nanodiscs was determined using a CO-binding assay in the presence of a reducing agent, sodium dithionate.47 The estimated reconstitution in nanodiscs was 75% for CYP450 alone (4.5 μM) and 50% for the CYP450-CPR complex (3 μM). The electron transfer experiments were performed under anaerobic conditions using a stopped-flow instrument at temperatures ranging from 11 to 38 °C in the presence of a substrate benzphetamine for CYP450 2B4 (Figure 5). The absorption peak (Soret band) at 451.25 nm was observed for the CYP450-CPR complex in CO-saturated buffer upon mixing with NADPH solution, indicating the formation of CO-CYP450 complex due to the reduction of Fe3+ (high-spin state) to Fe2+ (low-spin state) by the first electron transfer from CPR (Figure 5a).
Figure 4.
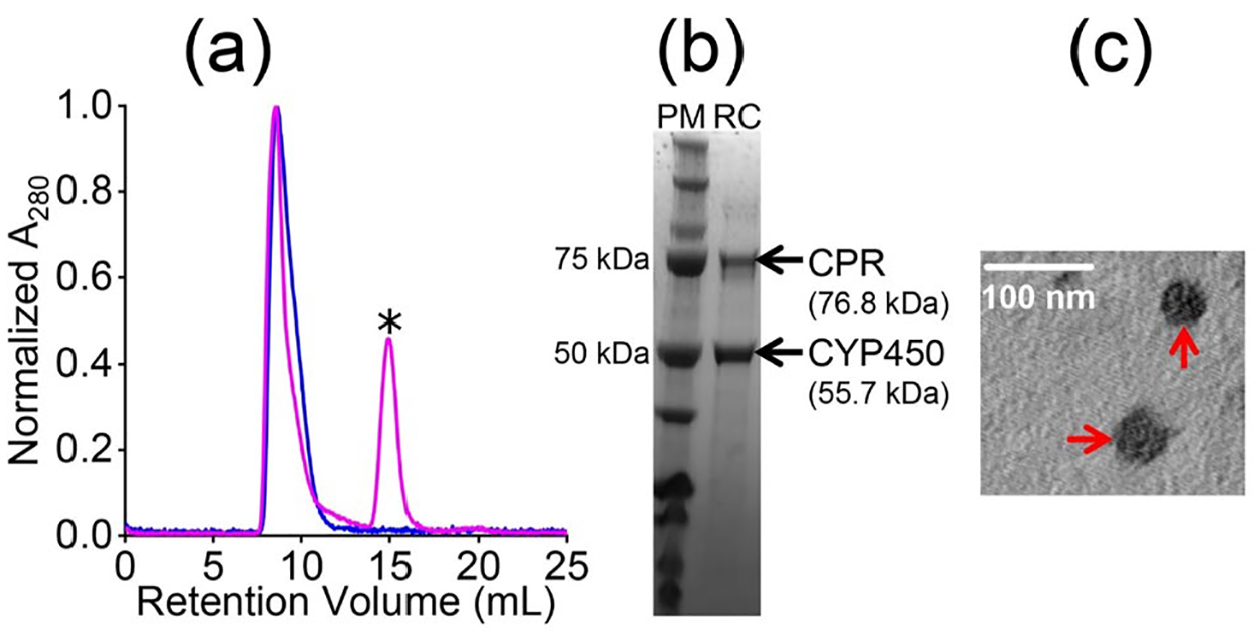
Reconstitution, purification, and characterization of CYP450 2B4-CPR redox complex in pentyl-inulin 7:3 w/w DMPC:DMPG nanodiscs. (a) SEC profiles of 7:3 w/w pentyl-inulin DMPC:DMPG nanodiscs with CYP450 2B4 (blue) or CYP450 2B4-CPR complex (magenta). The peak indicated with ‘*’ appeared from CPR (See Figure S2). The sample was purified using 10 mM Tris buffer (pH 7.4) containing 50 mM NaCl. (b) SDS-PAGE analysis of the purity of the CYP450-CPR complex. PM: protein marker and RC; redox complex. (c) TEM image of the CYP450 2B4-CPR complex. The nanodiscs are indicated with red arrows; scale bar: 100 nm.
Figure 5.
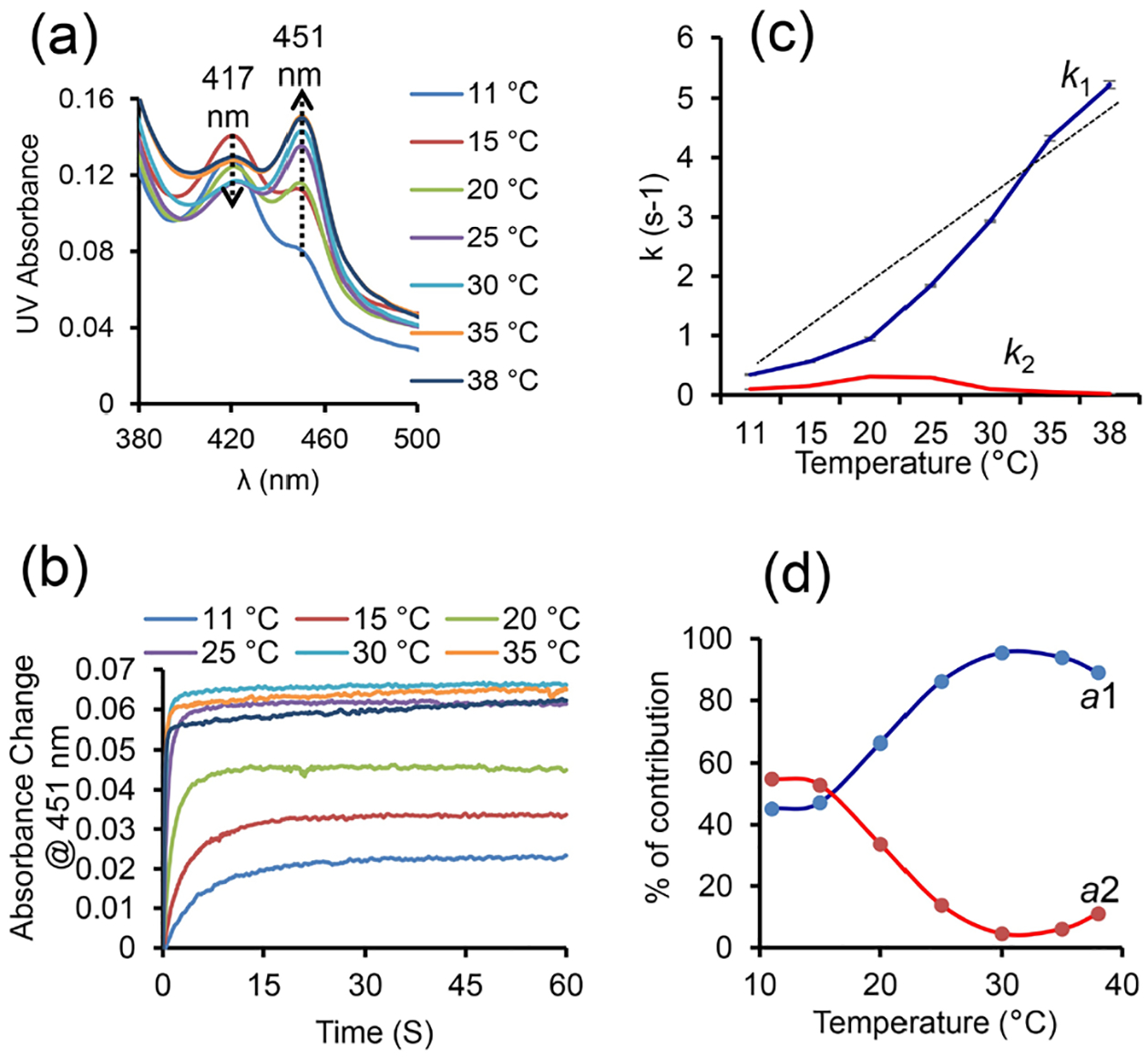
The kinetics of reduction of CO-bound CYP450 by CPR in the presence of benzphetamine at different temperatures (11 – 38 °C). (a) Soret-band region in the UV spectra of CO-bound CYP450 2B4 showing a decrease in the intensity of the peak at 417 nm and the appearance of a new peak at 451.25 nm (indicated with arrows). (b) Temperature-dependent kinetic traces (@451.25 nm) from the reduced (Fe2+) CO-bound CYP450 2B4 in the presence of CPR and benzphetamine. (c) Redox reaction rate constants at different temperatures. The straight-line shows a non-linear change of k1 with temperature. (d) Contribution of the slow and the rapid phases of reactions calculated using their amplitudes (a1 and a2) at different temperatures. The samples used in stopped-flow experiments were prepared in a 10 mM Tris buffer (pH 7.4) containing 50 mM NaCl. After mixing, the sample consisted of 1.5 μM proteins, as well as 10 and 20 molar equivalents of benzphetamine and NADPH, respectively.
The absorption peak intensity (and the rate of rise) increased with temperature, suggesting more efficient electron transfer at higher temperatures (Figures 5(a, b), and S4). The protein concentration (~1.6 μM) measured from the peak at 451 nm after the redox reaction in the presence of CPR and NADPH at a higher temperature (~30 °C) was similar to that measured using sodium dithionate before the redox reaction, indicating a complete conversion of CYP450 2B4 to its reduced CO-bound form (Figures 5(a, b), and S4, S5). The 451 nm absorbance of FBD of CPR is negligible due to its smaller extinction coefficient (ε=12.2 mM−1 cm−1) as compared to that of CYP450 (ε=91 mM−1 cm−1); hence it does not interfere with the measurements. Above the main phase-transition temperature (Tm) of DMPC:DMPG (7:3 w/w) in polymer-nanodiscs, lipids present in a more dynamic fluid-lamellar-phase would likely to enhance the structural interactions between CYP450 2B4 and CPR to form a functional redox complex, and subsequently an efficient electron transfer. The kinetics data were best-fitted to a double-exponential equation (eq. 1), indicating that the electron transfer occurred in a biphasic fashion,55 resulting in two rate constants, k1 and k2, with amplitudes a1 and a2 (Figures 5c and S4). The biphasic fashion of the redox reaction may be due to the conformational heterogeneity of proteins.37 Among the two phases, the rapid phase of the redox reaction is likely to have occurred from those conformations of CYP450 and CPR that favor their interaction more efficiently in a lipid bilayer. However, it is difficult to capture them experimentally as the structure of the CYP450-CPR complex is stabilized by transient interactions.56 The measured k1, calculated from the physiologically relevant rapid phase of the redox reaction, increased slowly/linearly with temperature from 11 to 15 °C, and 15 to 20 °C (ripple phase of lipids), and then rapidly near and above Tm (Figure 5c and Table S2). In contrast, only a slight change was observed for k2 (corresponding to the slow-phase of the reaction) around Tm of lipids (Figure 5c and Table S2). DLS measurements showed an increase in the hydrodynamic radius of polymer-nanodiscs with temperature suggesting an increase in lipid flexibility when they are in the liquid-crystalline phase (above Tm) as compared to the gel phase (below Tm) (Figures 6 and S3). Thus, the efficient electron transfer occurring at elevated temperatures could be due to the dynamic lamellar nature of the lipid bilayer that may assist productive structural interactions between CYP450 and CPR and lipids. The observed electron transfer rates in polymer-nanodiscs are similar to that reported by other investigators in a mixture of DLPC lipids and glycerol.57 The amplitudes a1 (for k1) and a2 (for k2) were substantially different at near and above Tm (Figure S6 and Table S2). The calculated percentages of the two different electron transfer processes (from the amplitudes) indicated only a small difference in their contribution to the redox reaction below Tm. Whereas at near and above Tm, the reduction of CYP450 2B4 was mainly through the rapid phase of the redox reaction (Figure 5d).
Figure 6.
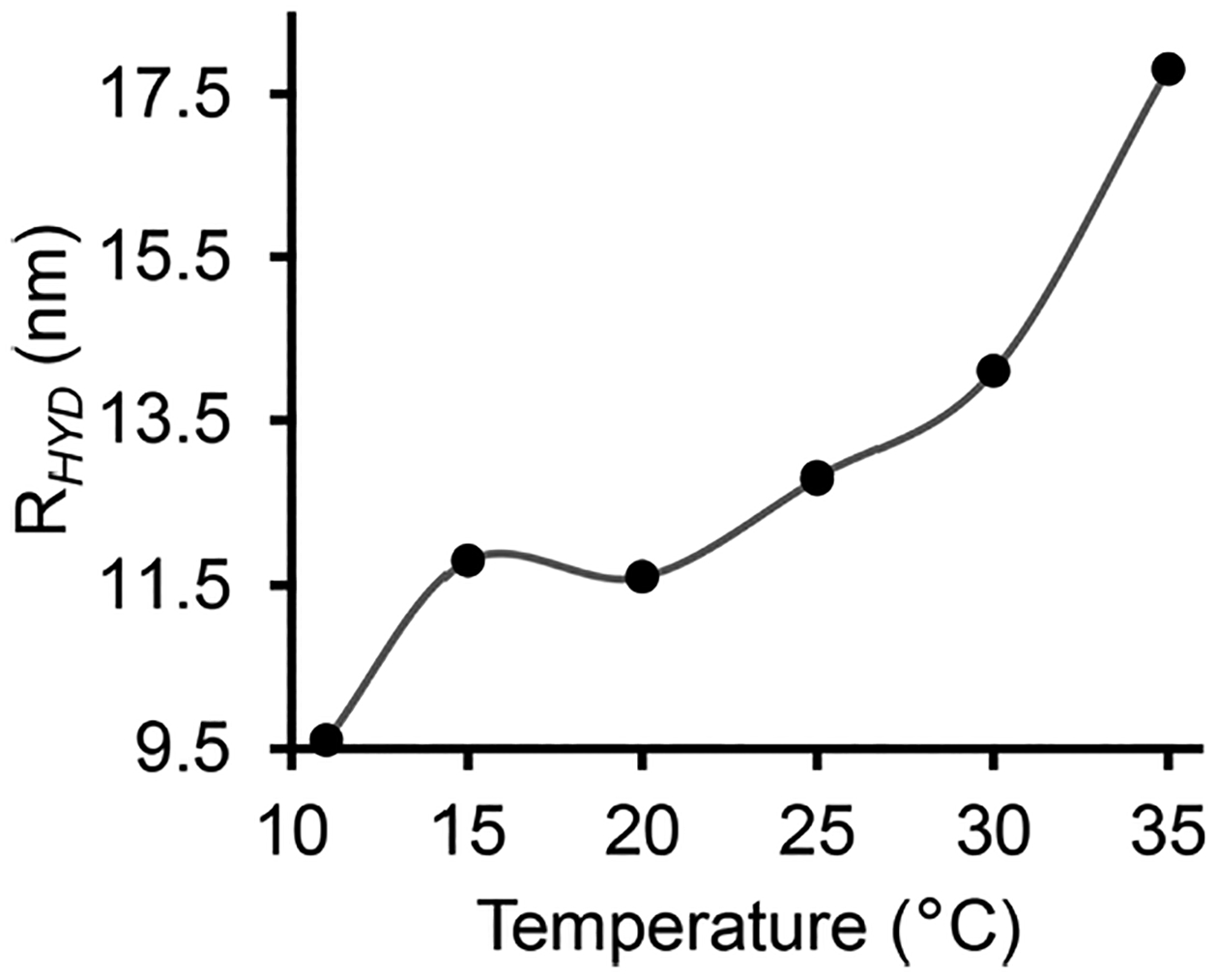
The hydrodynamic radius of polymer-nanodiscs with redox complex measured by DLS at different temperatures (11 – 35 °C). DLS profiles are shown in Figure S3.
The above-reported experimental results successfully demonstrate the feasibility of reconstituting a functional redox complex composed of oppositely charged proteins in non-ionic inulin-based nanodiscs. It would be fruitful to carry out structural studies using NMR spectroscopy to better understand the structural interactions between CYP450 and CPR and lipids. The ability to align the nanodiscs in an external magnetic field could be utilized in solid-state NMR experiments to probe the topology and structural interactions between the transmembrane domains of CYP450 and CPR.
CONCLUSIONS
To the best of our knowledge, this is the first study demonstrating the feasibility of overcoming the difficulties posed by the highly charged nanodiscs’ belts to reconstitute membrane proteins with opposite net charges. The reported results on the oppositely charged CYP450-CPR complex suggest that the non-ionic inulin-based polymer-nanodiscs can be used to study the structure, dynamics, and function of differently charged membrane protein complexes. Our results show an efficient electron transfer at a higher temperature (>25 °C) where the lipids are in the liquid-crystalline-phase.31, 35 The observed electron transfer rates are similar to the reported values,55, 57 demonstrating the suitability of non-ionic polymer-nanodiscs to study the CYP450-redox complex and to screen substrates/drugs metabolized by the P450 enzyme. Future studies focusing on the characterization of the CYP450-CPR redox complex with and without the transmembrane domains in the presence of various substrates would provide insights into the significance of the lipid bilayer on the enzymatic function of CYP450.34, 49, 58–61 Since pentyl-inulin forms nanodiscs at a wide range of pH conditions,26 membrane proteins functioning at different pH conditions could be directly isolated along with local native lipids from cells.62 Additionally, due to the large-size (~30 nm), pentyl-inulin-nanodiscs can accommodate high molecular weight proteins/complexes for high-resolution cryo-EM and solid-state NMR studies to characterize the structure and dynamics of transmembrane domains.26, 63, 64
Supplementary Material
ACKNOWLEDGMENT
This study was supported by the National Institutes of Health (NIH) (R35 GM139572 to A.R.).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Protein sequences, size-exclusion chromatography, SDS-PAGE analysis, DLS and UV spectra, and electron-transfer kinetic data (PDF).
The synthetic polymers used in this study are designed and produced by the Ramamoorthy laboratory, and are US patented.
REFERENCES
- (1).Dürr UHN; Gildenberg M; Ramamoorthy A The magic of bicelles lights up membrane protein structure. Chem. Rev 2012, 112, 6054–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Denisov IG; Sligar SG Nanodiscs in membrane biochemistry and biophysics. Chem. Rev 2017, 117, 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Denisov IG; Sligar SG Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol 2016, 23, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sligar SG; Denisov IG Nanodiscs: A toolkit for membrane protein science. Protein Sci 2021, 30, 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Günsel U; Hagn F Lipid nanodiscs for high-resolution NMR studies of membrane proteins. Chem. Rev 2021, 122, 9395–9421. [DOI] [PubMed] [Google Scholar]
- (6).Brown CJ; Trieber C; Overduin M Structural biology of endogenous membrane protein assemblies in native nanodiscs. Curr. Opin. Struct 2021, 69, 70–77. [DOI] [PubMed] [Google Scholar]
- (7).Orekhov PS; Bozdaganyan ME; Voskoboynikova N; Mulkidjanian AY; Karlova MG; Yudenko A; Remeeva A; Ryzhykau YL; Gushchin I; Gordeliy VI; et al. Mechanisms of formation, structure, and dynamics of lipoprotein discs stabilized by amphiphilic copolymers: A comprehensive review. Nanomaterials 2022, 12, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hagn F; Nasr ML; Wagner G Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR. Nat. Protoc 2018, 13, 79–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Denisov IG; Grinkova YV; Lazarides AA; Sligar SG Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc 2004, 126, 3477–3487. [DOI] [PubMed] [Google Scholar]
- (10).Epand RM; Epand RF; Sayer BG; Melacini G; Palgulachari MN; Segrest JP; Anantharamaiah GM An apolipoprotein AI mimetic peptide: Membrane interactions and the role of cholesterol. Biochemistry 2004, 43, 5073–5083. [DOI] [PubMed] [Google Scholar]
- (11).Knowles TJ; Finka R; Smith C; Lin Y-P; Dafforn T; Overduin M Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc 2009, 131, 7484–7485. [DOI] [PubMed] [Google Scholar]
- (12).Oluwole AO; Danielczak B; Meister A; Babalola JO; Vargas C; Keller S Solubilization of membrane proteins into functional lipid-bilayer nanodiscs using a diisobutylene/maleic acid copolymer. Angew. Chem. Int. Ed 2017, 56, 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Bayburt TH; Sligar SG Membrane protein assembly into nanodiscs. FEBS Lett 2010, 584, 1721–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ritchie TK; Grinkova YV; Bayburt TH; Denisov IG; Zolnerciks JK; Atkins WM; Sligar SG Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol 2009, 464, 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Barnaba C; Sahoo BR; Ravula T; Medina-Meza IG; Im S-C; Anantharamaiah GM; Waskell L; Ramamoorthy A Cytochrome-P450-induced ordering of microsomal membranes modulates affinity for drugs. Angew. Chem. Int. Ed 2018, 57, 3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Barnaba C; Ravula T; Medina-Meza IG; Im S-C; Anantharamaiah GM; Waskell L; Ramamoorthy A Lipid-exchange in nanodiscs discloses membrane boundaries of cytochrome-P450 reductase. ChemComm 2018, 54, 6336–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gentry KA; Prade E; Barnaba C; Zhang M; Mahajan M; Im S-C; Anantharamaiah GM; Nagao S; Waskell L; Ramamoorthy A Kinetic and structural characterization of the effects of membrane on the complex of cytochrome b 5 and cytochrome c. Sci. Rep 2017, 7, 7793–7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Krishnarjuna B; Yamazaki T; Anantharamaiah GM; Ramamoorthy A Nanodisc reconstitution of flavin mononucleotide binding domain of cytochrome-P450-reductase enables high-resolution NMR probing. ChemComm 2021, 57, 4819–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Krishnarjuna B; Ramamoorthy A Detergent-free isolation of membrane proteins and strategies to study them in a near-native membrane environment. Biomolecules 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Dörr JM; Koorengevel MC; Schäfer M; Prokofyev AV; Scheidelaar S; van der Cruijsen EAW; Dafforn TR; Baldus M; Killian JA Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. USA 2014, 111, 18607–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Pollock NL; Lee SC; Patel JH; Gulamhussein AA; Rothnie AJ Structure and function of membrane proteins encapsulated in a polymer-bound lipid bilayer. BBA-Biomembranes 2018, 1860, 809–817. [DOI] [PubMed] [Google Scholar]
- (22).Dörr JM; Scheidelaar S; Koorengevel MC; Dominguez JJ; Schäfer M; van Walree CA; Killian JA The styrene–maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys 2016, 45, 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Fiori MC; Zheng W; Kamilar E; Simiyu G; Altenberg GA; Liang H Extraction and reconstitution of membrane proteins into lipid nanodiscs encased by zwitterionic styrene-maleic amide copolymers. Sci. Rep 2020, 10, 9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ravula T; Hardin NZ; Bai J; Im SC; Waskell L; Ramamoorthy A Effect of polymer charge on functional reconstitution of membrane proteins in polymer nanodiscs. ChemComm 2018, 54, 9615–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Krishnarjuna B; Ravula T; Ramamoorthy A Detergent-free extraction, reconstitution and characterization of membrane-anchored cytochrome-b5 in native lipids. ChemComm 2020, 56, 6511–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ravula T; Ramamoorthy A Synthesis, characterization, and nanodisc formation of non-ionic polymers. Angew. Chem. Int. Ed 2021, 60, 16885–16888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lee SC; Knowles TJ; Postis VL; Jamshad M; Parslow RA; Lin YP; Goldman A; Sridhar P; Overduin M; Muench SP; et al. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc 2016, 11, 1149–1162. [DOI] [PubMed] [Google Scholar]
- (28).Gulamhussein AA; Uddin R; Tighe BJ; Poyner DR; Rothnie AJ A comparison of SMA (styrene maleic acid) and DIBMA (di-isobutylene maleic acid) for membrane protein purification. BBA-Biomembranes 2020, 1862, 183281. [DOI] [PubMed] [Google Scholar]
- (29).Guengerich FP Cytochrome P450 research and The Journal of Biological Chemistry. J. Biol. Chem 2019, 294, 1671–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Rendic S; Guengerich FP Metabolism and interactions of chloroquine and hydroxychloroquine with human cytochrome P450 enzymes and drug transporters. Curr. Drug Metab 2020, 21, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Nath A; Grinkova YV; Sligar SG; Atkins WM Ligand binding to cytochrome P450 3A4 in phospholipid bilayer nanodiscs: the effect of model membranes. J. Biol. Chem 2007, 282, 28309–28320. [DOI] [PubMed] [Google Scholar]
- (32).Baylon JL; Lenov IL; Sligar SG; Tajkhorshid E Characterizing the membrane-bound state of cytochrome P450 3A4: structure, depth of insertion, and orientation. J. Am. Chem. Soc 2013, 135, 8542–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Brignac-Huber LM; Park JW; Reed JR; Backes WL Cytochrome P450 organization and function are modulated by endoplasmic reticulum phospholipid heterogeneity. Drug Metab. Dispos 2016, 44, 1859–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Šrejber M; Navrátilová V; Paloncýová M; Bazgier V; Berka K; Anzenbacher P; Otyepka M Membrane-attached mammalian cytochromes P450: An overview of the membrane’s effects on structure, drug binding, and interactions with redox partners. J. Inorg. Biochem 2018, 183, 117–136. [DOI] [PubMed] [Google Scholar]
- (35).Laursen T; Lam HYM; Sørensen KK; Tian P; Hansen CC; Groves JT; Jensen KJ; Christensen SM Membrane anchoring facilitates colocalization of enzymes in plant cytochrome P450 redox systems. Commun. Biol 2021, 4, 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Mustafa G; Nandekar PP; Camp TJ; Bruce NJ; Gregory MC; Sligar SG; Wade RC Influence of transmembrane helix mutations on cytochrome P450-membrane interactions and function. Biophys. J 2019, 116, 419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Meling DD; McDougle DR; Das A CYP2J2 epoxygenase membrane anchor plays an important role in facilitating electron transfer from CPR. J. Inorg. Biochem 2015, 142, 47–53. [DOI] [PubMed] [Google Scholar]
- (38).Luthra A; Gregory M; Grinkova YV; Denisov IG; Sligar SG Nanodiscs in the studies of membrane-bound cytochrome P450 enzymes. Methods Mol. Biol 2013, 987, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Jumper J; Evans R; Pritzel A; Green T; Figurnov M; Ronneberger O; Tunyasuvunakool K; Bates R; Žídek A; Potapenko A; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Saribas AS; Gruenke L; Waskell L Overexpression and purification of the membrane-bound cytochrome P450 2B4. Protein Expr. Purif 2001, 21, 303–309. [DOI] [PubMed] [Google Scholar]
- (41).Hamdane D; Xia C; Im SC; Zhang H; Kim JJ; Waskell L Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem 2009, 284, 11374–11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhang M; Huang R; Im S-C; Waskell L; Ramamoorthy A Effects of membrane mimetics on cytochrome P450-cytochrome b5 interactions characterized by NMR spectroscopy. J. Biol. Chem 2015, 290, 12705–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ahuja S; Jahr N; Im SC; Vivekanandan S; Popovych N; Le Clair SV; Huang R; Soong R; Xu J; Yamamoto K; et al. A model of the membrane-bound cytochrome b5-cytochrome P450 complex from NMR and mutagenesis data. J. Biol. Chem 2013, 288, 22080–22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Dürr UH; Yamamoto K; Im S-C; Waskell L; Ramamoorthy A. J. J. o. t. A. C. S. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b 5. J. Am. Chem. Soc 2007, 129, 6670–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Clarke TA; Im S-C; Bidwai A; Waskell LJJ o. B. C. The role of the length and sequence of the linker domain of cytochrome b5 in stimulating cytochrome P450 2B4 catalysis. J. Biol. Chem 2004, 279, 36809–36818. [DOI] [PubMed] [Google Scholar]
- (46).Bennett AE; Rienstra CM; Auger M; Lakshmi KV; Griffin RG Heteronuclear decoupling in rotating solids. J. Chem. Phys 1995, 103, 6951–6958. [Google Scholar]
- (47).Guengerich FP; Martin MV; Sohl CD; Cheng Q Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat. Protoc 2009, 4, 1245–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Otey CR High-throughput carbon monoxide binding assay for cytochromes P450. Methods Mol. Biol 2003, 230, 137–139. [DOI] [PubMed] [Google Scholar]
- (49).McClary WD; Sumida JP; Scian M; Paço L; Atkins WM Membrane fluidity modulates thermal stability and ligand binding of cytochrome P4503A4 in lipid nanodiscs. Biochemistry 2016, 55, 6258–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zhang H; Myshkin E; Waskell L Role of cytochrome b5 in catalysis by cytochrome P450 2B4. Biochem. Biophys. Res. Commun 2005, 338, 499–506. [DOI] [PubMed] [Google Scholar]
- (51).Louka S; Barry SM; Heyes DJ; Mubarak MQE; Ali HS; Alkhalaf LM; Munro AW; Scrutton NS; Challis GL; de Visser SP Catalytic mechanism of aromatic nitration by cytochrome P450 TxtE: involvement of a ferric-peroxynitrite intermediate. J. Am. Chem. Soc 2020, 142, 15764–15779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Pallan PS; Wang C; Lei L; Yoshimoto FK; Auchus RJ; Waterman MR; Guengerich FP; Egli M Human cytochrome P450 21A2, the major steroid 21-hydroxylase. J. Biol. Chem 2015, 290, 13128–13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Davydov DR; Fernando H; Baas BJ; Sligar SG; Halpert JR Kinetics of dithionite-dependent reduction of cytochrome P450 3A4: heterogeneity of the enzyme caused by its oligomerization. Biochemistry 2005, 44, 13902–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Stepien P; Augustyn B; Poojari C; Galan W; Polit A; Vattulainen I; Wisnieska-Becker A; Rog T Complexity of seemingly simple lipid nanodiscs. BBA-Biomembranes 2020, 1862, 183420. [DOI] [PubMed] [Google Scholar]
- (55).Im S-C; Waskell L The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5). Arch. Biochem. Biophys 2011, 507, 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Mukherjee G; Nandekar PP; Wade RC An electron transfer competent structural ensemble of membrane-bound cytochrome P450 1A1 and cytochrome P450 oxidoreductase. Commun. Biol 2021, 4, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Zhang H; Hamdane D; Im SC; Waskell L Cytochrome b5 inhibits electron transfer from NADPH-cytochrome P450 reductase to ferric cytochrome P450 2B4. J. Biol. Chem 2008, 283, 5217–5225. [DOI] [PubMed] [Google Scholar]
- (58).Liu K-C; Hughes JMX; Hay S; Scrutton NS Liver microsomal lipid enhances the activity and redox coupling of colocalized cytochrome P450 reductase-cytochrome P450 3A4 in nanodiscs. FEBS J 2017, 284, 2302–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Navrátilová V; Paloncýová M; Berka K; Otyepka M Effect of lipid charge on membrane immersion of cytochrome P450 3A4. J. Phys. Chem. B 2016, 120, 11205–11213. [DOI] [PubMed] [Google Scholar]
- (60).Laursen T; Jensen K; Møller BL Conformational changes of the NADPH-dependent cytochrome P450 reductase in the course of electron transfer to cytochromes P450. BBA-Proteins Proteom 2011, 1814, 132–138. [DOI] [PubMed] [Google Scholar]
- (61).Barnaba C; Gentry K; Sumangala N; Ramamoorthy A The catalytic function of cytochrome P450 is entwined with its membrane-bound nature. F1000Res 2017, 6, 662–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Krishnarjuna B; Ravula T; Ramamoorthy A Detergent-free isolation of CYP450-reductase’s FMN-binding domain in E.coli lipid-nanodiscs using a charge-free polymer. ChemComm 2022, 58, 4913–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Huang R; Yamamoto K; Zhang M; Popovych N; Hung I; Im S-C; Gan Z; Waskell L; Ramamoorthy A Probing the transmembrane structure and dynamics of microsomal NADPH-cytochrome P450 oxidoreductase by solid-state NMR. Biophys. J 2014, 106, 2126–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Yamamoto K; Caporini MA; Im S-C; Waskell L; Ramamoorthy A Transmembrane interactions of full-length mammalian bitopic cytochrome-P450-cytochrome-b5 complex in lipid bilayers revealed by sensitivity-enhanced dynamic nuclear polarization solid-state NMR spectroscopy. Sci. Rep 2017, 7, 4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


