Abstract
Background
The chemotherapy resistance often leads to chemotherapy failure. This study aims to explore the molecular mechanism by which MUC1 regulates paclitaxel resistance in lung adenocarcinoma (LUAD), providing scientific basis for future target selection.
Methods
The bioinformatics method was used to analyse the mRNA and protein expression characteristics of MUC1 in LUAD. RT-qPCR and ELISA were used to detect the mRNA and protein expression, flow cytometry was used to detect CD133+ cells, and cell viability was detected by CCK-8 assay. The mRNA-seq was performed to analyse the changes in expression profile, GO and KEGG analysis were used to explore the potential biological functions.
Results
MUC1 is highly expressed in LUAD patients and is associated with a higher tumour infiltration. In paclitaxel resistance LUAD cells (A549/TAX cells), the expression of MUC1, EGFR/p-EGFR and IL-6 were higher than that of A549 cells, the proportion of CD133+ cells was significantly increased, and the expression of cancer stem cell (CSCs) transcription factors (NANOG, OCT4 and SOX2) were significantly up-regulated. After knocking down MUC1 in A549/Tax cells, the activity of A549/Tax cells was significantly decreased. Correspondingly, the expression of EGFR, IL-6, OCT4, NANOG, and SOX2 were significantly down-regulated. The mRNA-seq showed that knocking down MUC1 affected the gene expression, DEGs mainly enriched in NF-κB and MAPK signalling pathway.
Conclusion
MUC1 was highly expressed in A549/TAX cells, and MUC1-EGFR crosstalk with IL-6 may be due to the activation of NF-κB and MAPK pathways, which promote the enrichment of CSCs and lead to paclitaxel resistance.
Keywords: MUC1, lung adenocarcinoma, cancer cell stemness, paclitaxel resistance, NF-κB, MAPK
Introduction
Lung cancer remains the most common cause of cancer death worldwide. Most patients were in the advanced stage at the time of diagnosis, with a high rate of recurrence and metastasis [1,2]. Lung adenocarcinoma (LUAD) was the most common lung cancer, accounting for approximately 40% of all lung cancer cases [3]. The treatment for lung cancer mainly includes surgery, chemotherapy, radiotherapy, immunotherapy [4]. Paclitaxel is one of the commonly used drugs for chemotherapy, but its resistance often leads to chemotherapy failure [5]. Recently, targeted therapy and its combination with other treatments have shown promising results. However, similar to traditional chemotherapy, these new targeted drugs also tend to fail due to the development of drug resistance [6]. Therefore, overcoming drug resistance is a challenging issue in the treatment of LUAD.
Some mechanisms have been proposed to explain the emergence of paclitaxel resistance, one of which is the therapeutic tolerance mediated by cancer stem cells (CSCs) [7]. The current chemotherapy and radio technology kill most of cancer cells, but due to the specific drug resistance mechanism of CSCs, they cannot be eliminated [8–10]. CSCs are a subpopulation of cancer cells with the ability to self-renew and differentiate. Researches have reported that CSCs are involved in cancer resistance, metastasis, and recurrence, and significantly affect tumour treatment [9]. During the treatment of cancer, CSCs exert their drug resistance by maintaining dormant state, increasing DNA repair capacity, turning off apoptotic pathways, managing reactive oxygen species and reactive nitrogen species in a highly efficient manner, and by manipulating the tumour microenvironment (TME) [7,9]. It has been proposed that targeting CSC subpopulations can lead to tumour regression and reduce the possibility of tumour recurrence after treatment. [10]. Erlotinib, a tyrosine kinase inhibitor (TIK) of the epidermal growth factor receptor (EGFR), can inhibit the enrichment of cervical CSCs induced by the MUC1-EGFR-CREB/GRβ-IL-6 axis in cervical cancer [11], but there are no relevant reports on this pathway in LUAD.
Mucin1 (MUC1) is a type I transmembrane protein with a highly glycosylated extracellular domain [12]. In healthy tissues, MUC1 exists on the surface of all epithelial tissues and serves as a protective barrier for cells through its extracellular domain [13,14]. Nevertheless, in cancer cells, MUC1 has intracellular signalling function and is closely related to the treatment and progression of different epithelial cancers [15,16]. MUC1 can also up-regulate the expression of ATP-binding cassette transporter B1 (ABCB1) in EGFR-dependent manner to induce chemotherapy resistance [17]. Previous studies have shown that MUC1-C plays a crucial role in inducing stemness and paclitaxel resistance in human non-small cell lung cancer (NSCLC) A549 cells, but the molecular mechanism by which MUC1 induces paclitaxel resistance in LUAD is still unclear [18].
This study analysed the expression characteristics of MUC1 in LUAD patients through bioinformatics, and compared the expression of MUC1, EGFR and IL-6 in human lung cancer A549 cells and paclitaxel resistance LUAD A549/TAX cells and detection of cancer cell stemness in two types of cells. Furthermore, we knocked down MUC1 in A549/TAX cells and performed mRNA sequencing (mRNA-seq) to further analyse the molecular mechanism of MUC1 regulating the stemness of paclitaxel-resistant LUAD cells, providing scientific basis for future target selection.
Materials and methods
Expression analysis of MUC1 in LUAD patients
The gene expression data and clinical data of MUC1 in LUAD were downloaded from The Cancer Genome Atlas (TCGA) database and the expression of MUC1 in LUAD samples and matched normal tissues were compared. Moreover, we also analysed the expression of MUC1 in different clinical phenotypes. The protein expression of MUC1 in LUAD tissues and normal tissues were retrieved in the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/). The expression of MUC1 in different tumour tissues and matched normal tissues were compared by the Tumour Immune Estimation Resource (TIMER) database (http://timer.cistrome.org/). The study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of the 363 Hospital.
Immune infiltrate analysis of MUC1
CIBERSORT was used to evaluate the infiltration status of 22 immune cells in the high and low MUC1 groups in the TCGA-LUAD dataset. The ‘Pearson’ method was used to calculate the correlation between MUC1 expression and differential immune cells, an R-value of −1 indicates complete negative correlation, +1 indicates complete positive correlation, and 0 indicates no correlation. p < 0.05 was considered to be significant. Through the TIMER database (https://cistrome.shinyapps.io/timer/), the influence of copy number variation of MUC1 on immune cell infiltration was evaluated. The ESTIMATE algorithm was used to calculate the immune score, matrix score, tumour purity, and ESTIMATE score for each LUAD patient.
Cell culture
Human lung cancer A549 cells were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China), and paclitaxel resistance LUAD A549/TAX cells were purchased from Shanghai MEIXUAN biological science and technology LTD (Shanghai, China). A549 cells were cultured in DMEM medium supplemented with 10% foetal bovine serum (FBS) and penicillin/streptomycin (100 U/mL), while A549/TAX cells were additionally added with 200 ng/mL paclitaxel in the above medium to maintain their drug resistance. Then place the culture bottle in a 5% CO2 incubator at 37 °C for static cultivation.
RNA extraction and real-time qPCR (RT-qPCR)
TRIzol (TIANGEN, Beijing, China) was used to extract total RNA from cells and the FastKing cDNA first strand synthesis kit (TIANGEN, Beijing, China) was used for reverse transcribed to synthesize cDNA. The SuperReal PreMix Plus (TIANGEN, Beijing, China) was used to amplify the target genes in the ABI StepOne Plus fluorescence quantitative PCR instrument, 2-ΔΔCT method was used to perform relative quantitative analysis. The primers were listed in Supplementary Table 1.
ELISA
According to the instructions of manufacturers, the protein expression of MUC1, EGFR, and IL-6 were measured using protein ELISA kit (Kexing, Shanghai, China). The absorbance (optical density value) of each well in sequence was measured at 450 nm, repeats each result 3 times, and determined the protein concentration by comparing the relative absorbance between the sample and the standard.
Flow cytometry assay
Flow cytometry was used to detect the proportion of CD133+ cells. Firstly, cells were treated with 6 well plates and cell suspensions were collected and rinsed with phosphate-buffered saline (PBS) buffer. Subsequently, count 1 × 106 cells were resuspended in 100 µL staining buffer and added 5 µL antibodies (APC anti-human CD133), incubated at room temperature in dark for 30 min. After incubation, 1 mL PBS was added to each sample for resuspension centrifugation and discarded the supernatant. Finally, 500 µL PBS was added to each sample for measurement the proportion of CD133+ cells using flow cytometry (NovoCell, Agilent, USA) and analysed using FlowJo 7.6 software (Tree Star, Inc, CA, USA).
siRNA transfection
All siRNAs were purchased from hippo bio (Huzhou, China). The transfection reagent (Lipo8000) was mixed with siRNA to transfect A549 and A549/TAX cells. The cell culture plate was placed at 37 °C in a 5% CO2 incubator for 24 h, and fresh culture medium could be changed after 4-6 h of transfection. The siRNA sequences used for transfection were listed in Supplementary Table 2.
Cell counting kit 8 (CCK 8) assay
The cell counting kit 8 was used to detect the viability of negative control A549/TAX-NC cells and MUC1 knockdown A549/TAX siRNA-MUC1 cells. The cells were normally cultured to 90%, digested with trypsin, and after counting, the cell concentration was adjusted to 1 × 104 cells/mL. Add 100 µL of adjusted cell suspension to each well in 96 well plates, with 3 wells in each group, and incubate in a 5% CO2 incubator at 37 °C for subsequent testing. After reaching the cultivation time, remove the culture plate and add 10 µL of CCK-8 solution from the reagent kit to the 96-well cell culture plate, and then incubate in a 37 °C incubator for another 4 h. The absorbance at 450 nm was measured using a microplate reader (Molecular Devices, LLC).
mRNA-sequencing and differential expression analysis
The illumina high-throughput sequencing platform was used to perform mRNA sequencing of A549/TAX-NC cells and A549/TAX siRNA-MUC1 cells, and generate a large amount of raw data. The fastp software was used to filter the original sequencing data and obtained high-quality clean data. Salmon v0.13.0 was used for mRNA gene expression analysis. The differentially expressed genes (DEGs) was obtained by DESeq2, with screening parameters of P adj <0.05 and |log2 fold change (FC)| >1. The GO functional enrichment and KEGG pathway analysis of DEGs was performed by used David database (https://david.ncifcrf.gov/tools.jsp).
Statistical analysis
GraphPad Prism 5.0 was used for statistical analysis. The data was represented by mean ± standard deviation (SD), with at least 3 replicates for each group of data. The difference analysis of variables was evaluated using a student’s t-test. p < 0.05 was considered statistically significant for the difference.
Results
The expression characteristics of MUC1 in LUAD
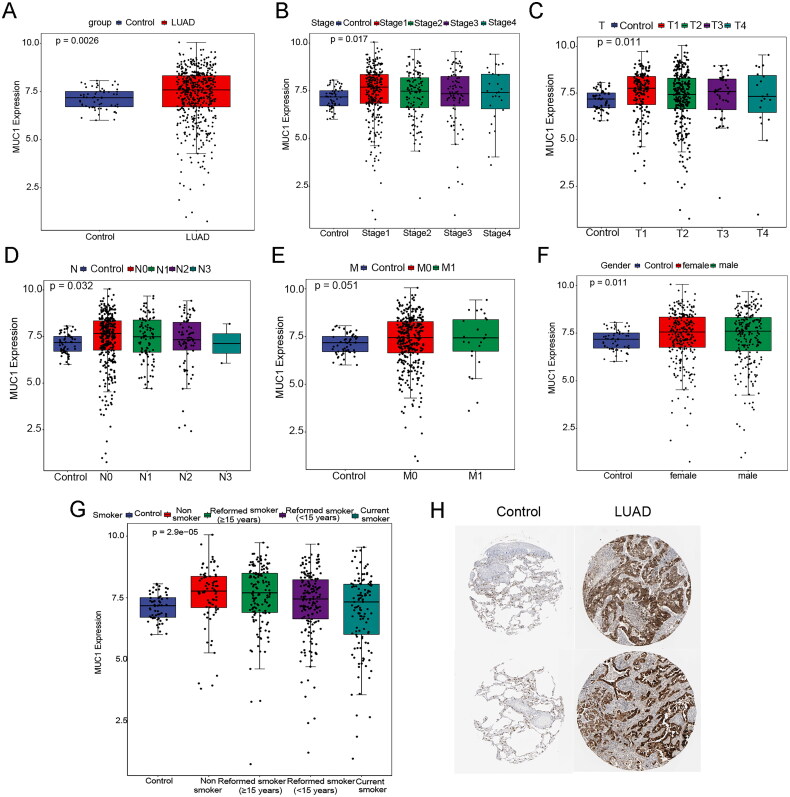
To explore the expression characteristics of MUC1 in LUAD, the mRNA expression of MUC1 in LUAD samples in the TCGA data sets were first mined. It was found that the mRNA level of MUC1 was significantly higher in tumour tissues than in normal tissues (Figure 1(A)) and MUCI also exhibited the same expression pattern in different clinical features (Figure 1(B–G)). Consistently, the protein expression of MUC1 in LUAD tissues was higher than that in normal tissues (Figure 1(H)). Furthermore, we investigated the expression of MUC1 in different types of tumour tissues and matched normal tissues and found that the expression of MUC1 was significantly up-regulated in LUAD, while significantly down-regulated in lung squamous cell carcinoma (LUSC), indicating that MUC1 has different effects on different types of cancer (Supplementary Figure 1).
Figure 1.
The expression characteristics of MUC1 in lung adenocarcinoma (LUAD). (A) the expression of MUC1 in normal and LUAD tissues; (B) the expression of MUC1 in different stages; (C) the expression of MUC1 in different T stages; (D) the expression of MUC1 in different N stages; (E) the expression of MUC1 in different M stages; (F) the expression of MUC1 in different genders; (G) the expression of MUC1 in different smoking history; (H) the protein expression of MUC1 in normal and LUAD tissues.
The immune infiltration analysis of MUC1 expression
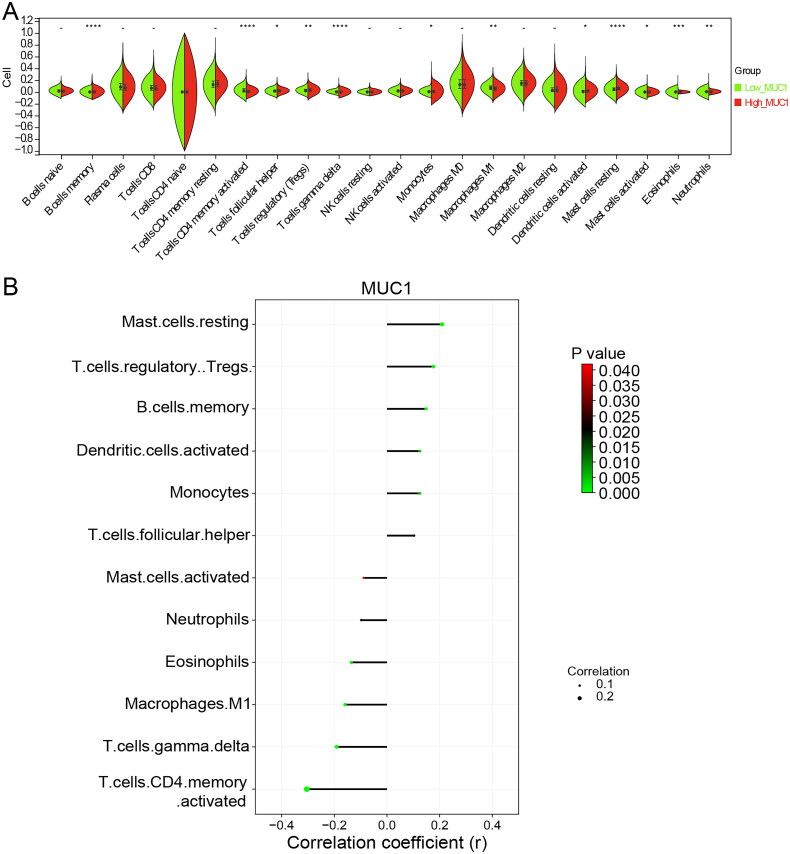
According to the median expression of MUC1, LUAD patients in TCGA were divided into high-MUC1 group and low-MUC1 group, and the DEGs between the two groups were analysed. A total of 585 DEGs were obtained with a standard of p < 0.05 and |log2 FC| >0.5, of which 363 DEGs were up-regulated and 222 DEGs were down-regulated in high-MUC1 group (Supplementary Figure 2). Studies have shown that MUC1 is closely related to tumour immune microenvironment of NSCLC [19]. To explore the relationship between MUC1 and tumour immune microenvironment, the immune cells abundance of high-MUC1 group and low-MUC1 group was analysed. As depicted in Figure 2(A), the high-MUC1 group and low-MUC1 group showed a statistical difference in the abundance of 12 immune cells. Among them, T cells follicular helper, T cells regulatory, Dendritic cells, Mast cells resting, Monocytes, and B cells were positive correlated with MUC1 expression, while T cells CD4+ memory activated, T cells gamma delta, Eosinophils, Macrophages M1, Master cell activated, and Neutrophils were negative correlated with MUC1 expression (p < 0.05) (Figure 2(B)).
Figure 2.
The immune infiltration analysis of MUC1 expression. (A) The immune cell infiltration in high-MUC1 group and low-MUC1 group; (B) the correlation between MUC1 expression and differential immune cells, the length of the line represents the correlation, the direction of the line represents the positive or negative correlation, and the colour and size of the end point represents p-value.
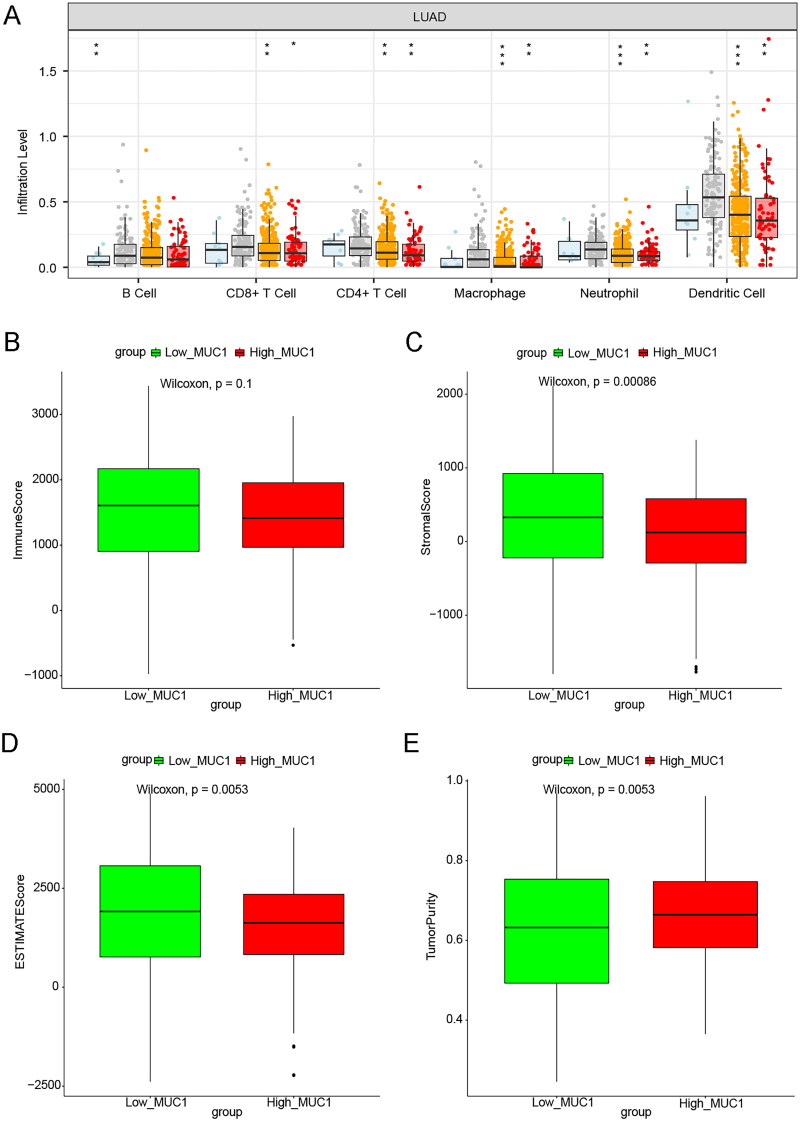
Further, the TIMER database was used to investigate the effect of MUC1 copy number variation on immune cell infiltration, and we found that the copy number variation of MUC1 was related to the infiltration degree of B cells, CD8+ cells, CD4+ cells, macrophages, Neutrophil, and Dendritic cells (Figure 3(A)). Meanwhile, ESTIMATE algorithm was used to calculate the immune score, stromal score, tumour purity, and ESTIMATE score. It was found that the stromal score and ESTIMATE score in the high-MUC1 group were lower than that in the low-MUC1 group, while the tumour purity was higher than that in the low-MUC1 group (Figure 3(B)).
Figure 3.
The immune correlation analysis between high-MUC1 group and low-MUC1 group. (A) The effect of MUC1 copy number variation on the immune cells infiltration in LUAD; B-E) the immune score M (B), stromal score (C), tumour purity (D), and ESTIMATE score (E) in high-MUC1 group and low-MUC1 group. *p < 0.05; **p < 0.01; *** p < 0.001; **** p < 0.0001.
MUC1 was up-regulated in paclitaxel-resistant lung cancer cells, and increased cancer cell stemness
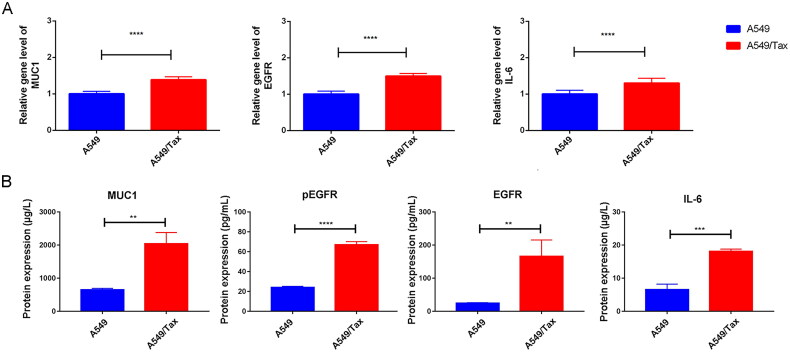
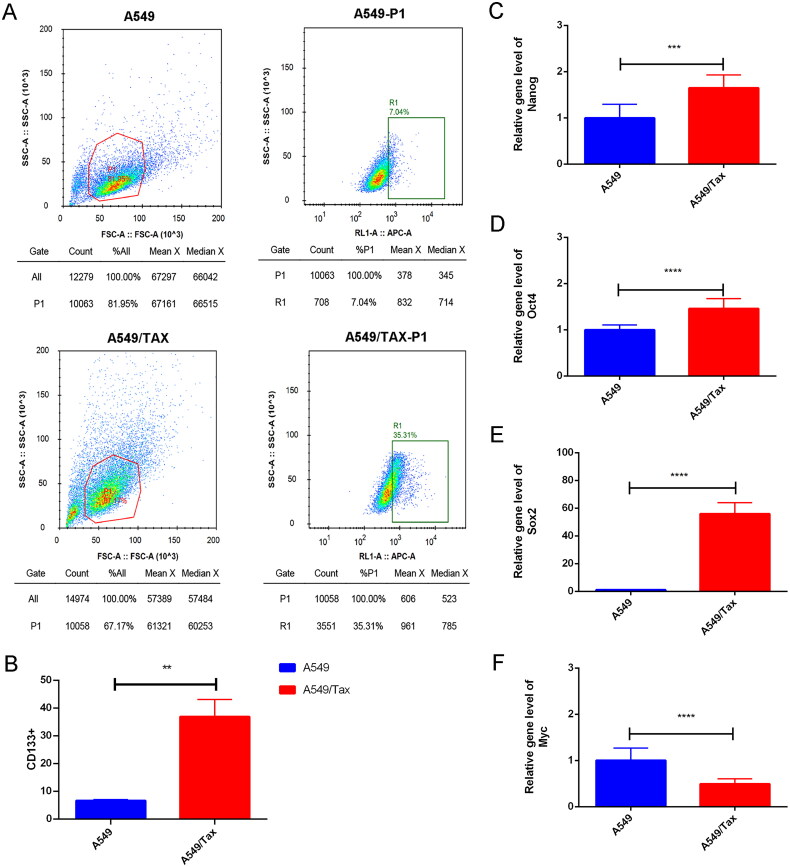
To explore the correlation between MUC1 and paclitaxel resistance LUAD, the expression of MUC1 in A549 cells and A549/TAX cells were detected by RT-qPCR, and the results showed that the expression of MUC1 in A549/TAX cells was significantly higher than that in A549 cells (Figure 4(A)). Previous studies have shown that MUC1 may through MUC1-EGFR-CREB/GRβ axis stimulate IL-6 expression to induce cervical CSCs enrichment, and thus we detected the expression of EGFR and IL-6 in both types of cells. The mRNA and protein expression levels of EGFR/p-EGFR and IL-6 in A549/TAX cells was significantly increased compared with A549 cells (Figure 4(A,B)). Meanwhile, the proportion of CD133+ cells was also significantly increased in A549/TAX cells (Figure 5(A,B)). For cancer cells, the acquisition of stem cell stemness means that the cancer cells are more aggressive and lead to a poor clinical prognosis. Therefore, we detected the expression of stem cell pluripotency factors. The mRNA expression of NANOG, OCT4, and SOX2 in A549/Tax cells were significantly increased, while the expression of MYC was significantly decreased (Figure 5(C–F)). These results indicated that the cancer cell stemness was increased in paclitaxel resistance LUAD A549/TAX cells.
Figure 4.
The expression of MUC1, EGFR, and IL-6 in A549 and A549/TAX cells. (A) The expression of MUC1, EGFR, and IL-6 in LUAD cells (A549) and paclitaxel-resistant LUAD cells (A549/TAX); (B) the protein expression of MUC1, EGFR/p-EGFR, and IL-6 in A549 cells and A549/TAX cells; **p < 0.01; *** p < 0.001; **** p < 0.0001.
Figure 5.
Cancer cell stemness increases in paclitaxel resistant LUAD cells. (A) The flow cytometry (FCM) assay for the expression of CD133; (B) the quantitative analysis showed that CD133 was significantly increased in A549/TAX cells; (C–F) the expression of nanog (C), Oct4 (D), Sox2 (E), and myc (F) in A549 cells and A549/TAX cells. **p < 0.01; *** p < 0.001; **** p < 0.0001.
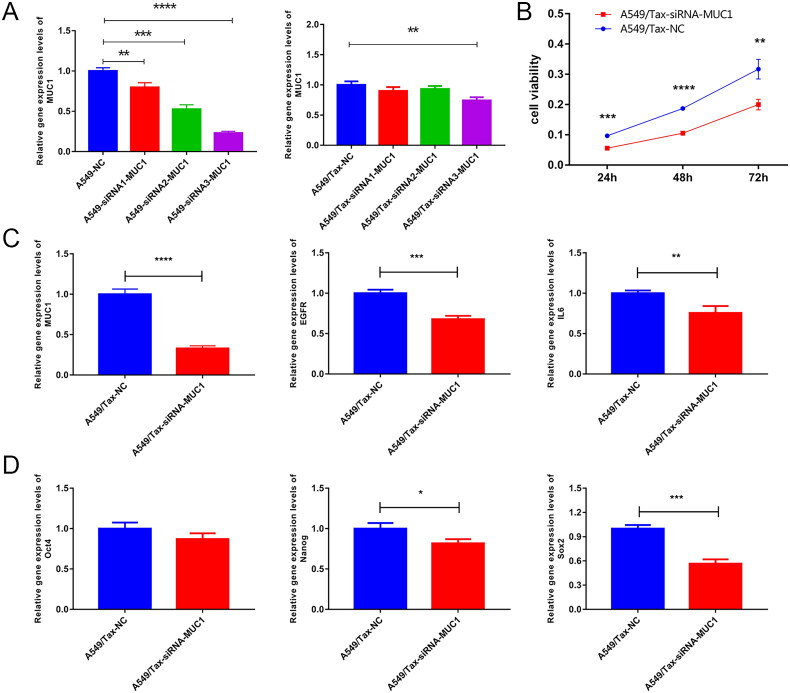
MUC1 knockdown decreased the stemness of A549/TAX cells
To further investigate the effect of MUC1 on paclitaxel resistance LUAD, we knocked down the MUC1 gene in A549/TAX cells and selected siRNA3 as an effective interference target for subsequent experiments based on RT-qPCR results (Figure 6(A)). The CCK-8 data showed that knocked down the MUC1 gene significantly reduced A549/TAX activity (Figure 6(B)). Compared with paclitaxel-resistant LUAD, knocked down MUC1 significantly decreased the expression of EGFR, IL-6, OCT4, NANOG, and SOX2. Therefore, we speculated that knocked down MUC1 may affect the stemness of paclitaxel resistance LUAD A549/TAX cells (Figure 6(C,D)).
Figure 6.
Knock down MUC1 reduces the expression of stem cell pluripotency factor in paclitaxel resistant LUAD cells. (A) screening of effective interference targets for knock down MUC1 in A549 cells and A549/tax cells; (B) cell Counting kit-8 assay (CCK-8) detects the cellular activity of negative control cells (A549/TAX-NC) and MUC1 interfering cells (A549/TAX-siRNA-MUC1); (C) the expression of MUC1, EGFR, and IL-6 in A549/TAX-NC and A549/TAX-siRNA-MUC1 cells; (D) the expression of Oct4, Sox2, and myc in A549/TAX-NC and A549/TAX-siRNA-MUC1 cells. *p < 0.05; **p < 0.01; *** p < 0.001; **** p < 0.0001.
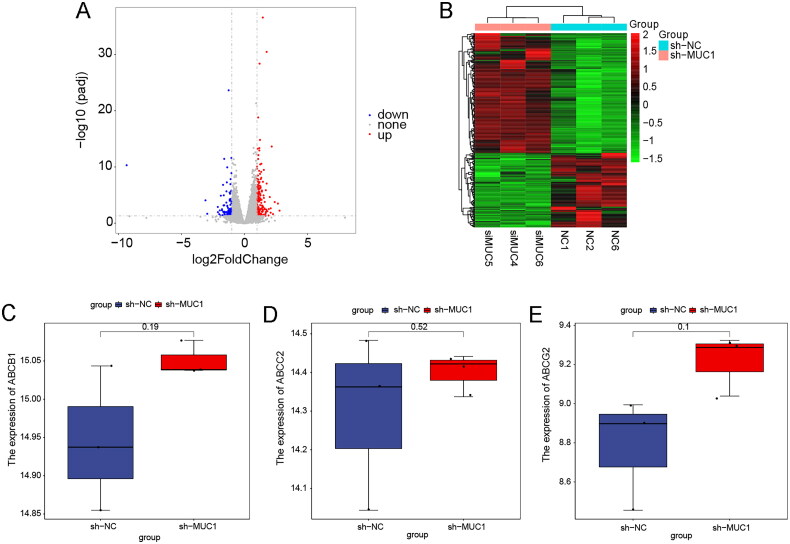
Identification of DEGs and functional enrichment between A549/Tax-NC and A549/TAX-siRNA-MUC1
To explore the mechanism by which MUC1 affects the stemness of paclitaxel resistance LUAD A549/TAX cells, the mRNA-sequencing on the negative control A549/TAX-NC cells and MUC1 knockdown A549/TAX-siRNA-MUC1 cells was performed. A total of 282 DEGs were screened out with a standard of p < 0.05 and |Log2FC| >1, of which 174 genes were up-regulated, and 108 genes were down-regulated. The volcano plot and heat map of DEGs were shown in Figure 7(A,B), and the top 20 DEGs were listed in Table 1. As is well known, the ABCB1 encoded by p-glycoprotein, the ABCC2 and ABCG2 genes encoded by transport proteins MRP2 and BCRP, respectively, play a role in the development of paclitaxel-resistance. To further explore the mechanism of paclitaxel-resistance, we analysed the gene expression of ABCB1, ABCC2, and ABCG2 in the mRNA-seq dataset. There was no significant difference in the expression of ABCB1, ABCC2, and ABCG2 between A549/TAX-NC cells and A549/TAX siRNA MUC1, indicating that MUC1 may regulate paclitaxel resistance not by p-glycoprotein encodes the ABCB1 (Figure 7(C–E)).
Figure 7.
The differentially expressed genes analysis of between A549/Tax-NC and A549/TAX-siRNA-MUC1. (A–B) The volcano plot (A) and heat map (B) of DEGs between A549/Tax-NC and A549/TAX-siRNA-MUC1; (C–E) the expression of ABCB1 (C), ABCC2 (D), and ABCG2 (E) between A549/Tax-NC and A549/TAX-siRNA-MUC1.
Table 1.
The top 20 DEGs between A549/tax and A549/TAX-siRNA-MUC1.
| Gene | Log2FoldChange | P value | Up/down |
|---|---|---|---|
| CXCL8 | 1.461740183 | 1.88E-41 | up |
| PTGS2 | 1.762389853 | 4.82E-35 | up |
| CXCL5 | 1.206154353 | 9.03E-33 | up |
| ATF3 | 1.094625077 | 6.78E-23 | up |
| TNFAIP3 | 1.239737232 | 7.66E-19 | up |
| DIO2 | 2.169883634 | 1.33E-17 | up |
| KLF6 | 1.185910112 | 3.12E-17 | up |
| PTP4A1 | 1.047409875 | 4.50E-17 | up |
| SLC4A4 | 1.182930912 | 9.92E-17 | up |
| CXCL2 | 1.107211101 | 9.04E-16 | up |
| PGAM1 | −1.251114978 | 6.64E-28 | down |
| CPNE7 | −1.036274022 | 3.06E-15 | down |
| TMEM35B | −1.601671145 | 4.69E-15 | down |
| AC011511.4 | −9.345851797 | 9.85E-14 | down |
| SHC2 | −1.365144437 | 2.53E-13 | down |
| IER5L | −1.032185661 | 3.74E-12 | down |
| EMP3 | −1.092783207 | 6.64E-11 | down |
| SAMD11 | −1.394955248 | 3.00E-10 | down |
| COL16A1 | −1.112313271 | 6.77E-10 | down |
| RPL21P16 | −1.694509565 | 9.73E-10 | down |
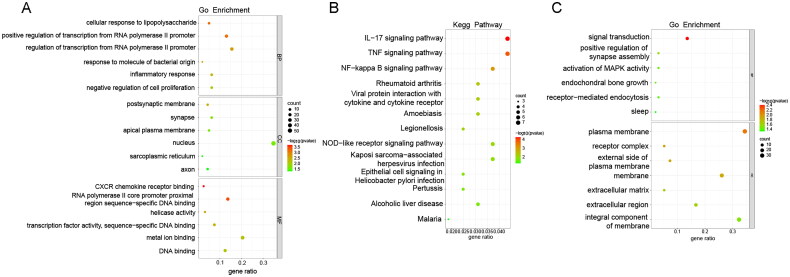
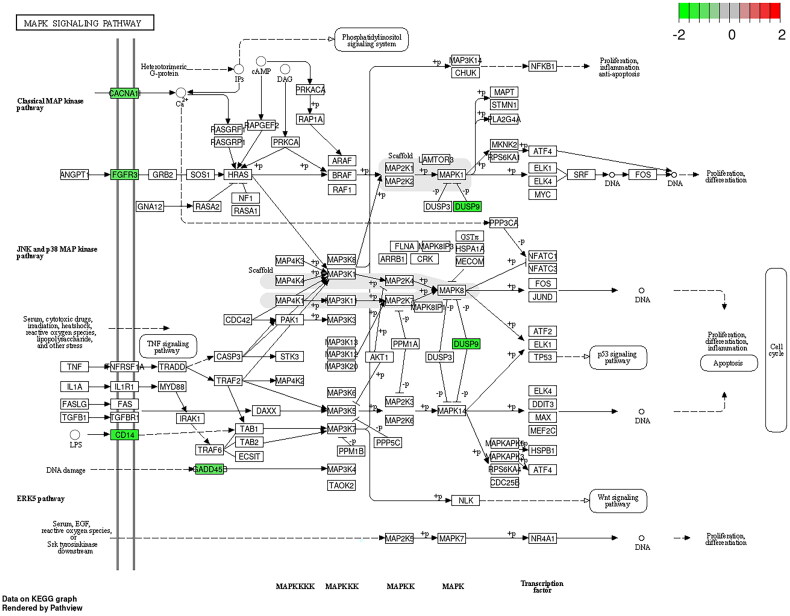
GO functional enrichment and KEGG pathway analysis of up-regulated and down-regulated genes were performed by the David database, respectively. In the up-regulated genes, GO enriched to the top terms were ‘cellular response to lipopolysaccharide’ in the biological process (BP), ‘postsynaptic membrane’ in the cell component (CC), and ‘CXCR chemokine receptor binding’ in the molecular function (MF), respectively (Figure 8(A)). KEGG pathway analysis showed that the up-regulated DEGs were mainly enriched in IL-17 signalling pathway, TNF signalling pathway, and NF-kappa B signalling pathway (Figure 8(B)). In the down-regulated genes, GO enriched to the top terms were ‘signal transduction’ in the BP and ‘plasma membrane’ in the CC, respectively (Figure 8(C)). KEGG pathway analysis showed that the down-regulated DEGs were enriched in MAPK signalling pathway (Figure 9). MUC1 may regulate the stemness of paclitaxel resistance LUAD cells through these key pathways.
Figure 8.
GO and KEGG analysis of DEGs between A549/Tax-NC and A549/TAX-siRNA-MUC1. (A) The GO enrichment of up-regulated DEGs; (B) The KEGG pathway of up-regulated DEGs; (C) The GO enrichment of down-regulated DEGs.
Figure 9.
The KEGG pathway of down-regulated DEGs.
Discussion
To date, chemotherapy is still the main treatment for patients with LUAD [4]. However, the application of chemotherapy is limited by intrinsic or acquired drug resistance [20]. Chemotherapy resistance involves multiple genes and related pathways, the role of MUC1 in resistance has been reported, but the molecular mechanism in LUAD is still unclear [9,21]. This study is the first to explore the molecular mechanism of MUC1 regulating paclitaxel resistance in LUAD cells. MUC1 is highly expressed in LUAD and with extensive tumour infiltration. In vitro experiments have confirmed that MUC1 affects cancer cell stemness through the MUC1-EGFR-IL-6 pathway, characterized by a significant decrease in the expression of IL-6 and stem cell transcription factors after knocking down MUC1 in A549/TAX cells. Furthermore, we explored the impact of knocking out MUC1 on the expression profile in A549/TAX cells, and speculated that the crosstalk between MUC1-EGFR and IL-6 may be due to NF-κB and MAPK pathways.
MUC1, as a well-known oncogenic gene, is usually overexpressed in various epithelial adenocarcinomas [22–25]. MUC1 regulate the growth, proliferation, metastasis, apoptosis, and development processes of cancer cells by participating in different signalling pathways [26,27]. The overexpression of MUC1 in NSCLC is associated with poor disease-free survival and overall survival [28]. This study found that the expression of MUC1 in tumour tissues was significantly higher than that in normal tissues of LUAD patients by mining the TCGA database. In addition, the pan cancer analysis showed that the expression of MUC1 was significantly up-regulated in LUAD, and significantly down-regulated in LUSC, indicated that MUC1 has different effects on different cancer types. A bioinformatics analysis of NSCLC shows that MUC1 has a higher expression frequency in adenocarcinoma and other non-small cell lung cancer compared to squamous cell carcinoma, which is consistent with our study [28]. MUC1-C promotes the suppressive immune microenvironment in non-small cell lung cancer, which is a potential target for reprogramming of the tumour microenvironment. In this study, it is also showed extensive infiltration of tumour immune cells in the high expression MUC1 group, confirming previous studies [19]. Moreover, MUC1 has been confirmed to be related to the formation of drug resistance [21]. A recent study showed that the extracellular domain of MUC1 acts as a hydrophilic barrier and regulates cellular permeability to paclitaxel, thereby inhibiting its uptake [29]. Therefore, it is necessary for us to study the correlation between MUC1 and paclitaxel-resistance in LUAD alone.
MUC1 was over-expressed in NSCLC tissues and paclitaxel resistance NSCLC A549/PR cells [18]. Consistently, in the present study, the expression of MUC1 was significantly increased in A549/TAX cells compared with A549 cells. A study in NSCLC has shown that MUC1 plays a central role in the integration of PD-L1, and the down-regulation of MUC1 inhibits the progression disease of NSCLC [30]. Besides, co-administration of the MUC1–EGFR–ABCB1 inhibitors with paclitaxel significantly blocked tumour growth and relapse in xenograft mouse model [17]. Similarly, in vitro and in vivo analyses have consistently shown that MUC1 cytoplasmic domain (MUC1-C) is involved in the stemness and paclitaxel-resistance of cancer cells, and the inhibition of MUC1-C expression combined with paclitaxel treatment was sufficient to reduce the sphere-forming ability and survival of paclitaxel-resistance LUAD A549/PTX cells [31]. The above results consistently indicate that MUC1 is the dominant factor for paclitaxel resistance, and regulating the expression of MUC1 may help overcome chemotherapy-induced paclitaxel resistance.
In cervical cancer, MUC1 may pass through MUC1-EGFR-CREB/GRβ axis stimulation of IL-6 expression induces CSCs enrichment, leading to drug resistance [11]. Hence, we compared the expression of related genes and proteins in A549 cells and A549/TAX cells, and found that the expression of EGFR/p-EGFR and IL-6 in A549/TAX cells were significantly increased. It was reported that the CD133 and MUC1 expression were associated with an aggressive tumour phenotype, and MUC1 is highly expressed in pancreatic CD133+ cells [32]. CD133 serves as a stemness biomarker for CD133+ CSCs, which have been found in lung cancer tissues [33,34]. Given that, the proportion of CD133+ cells was detected using flow cytometry. Compared with A549 cells, the proportion of CD133+ cells and the expression of MUC1 were significantly increased in A549/Tax cells, indicating that the high expression of MUC1 and the stemness of cancer cells may be involved in paclitaxel resistance. NANOG, OCT4, SOX2, and MYC are stem cell transcription factors that are believed to promote the acquisition of stemness in cancer cells and are overexpressed in CSCs [35–37]. Here, the expression of NANOG, OCT4, and SOX2 were significantly increased in A549/Tax cells, indicating that cancer cell stemness was stronger in the A549/Tax cells, and the development of paclitaxel resistance is related to the stemness of cancer cells.
To further investigate the correlation between MUC1 and paclitaxel resistance and stemness in LUAD, MUC1 was knocked down in A549/TAX cells, and the expressions of EGFR, IL-6, OCT4, NANOG, and SOX2 were significantly down-regulated, suggesting that knocking down MUC1 affected the stemness of paclitaxel-resistance LUAD cells. The CCK8 assay showed that the cell activity in the knocked down MUC1 cells was significantly lower than that in the negative control, indicating that knocking down MUC1 affected the cell activity of paclitaxel-resistance LUAD cells. To further investigate its molecular mechanism, mRNA-seq was conducted to analyze the changes in the RNA expression profile after MUC1 knockdown in A549/Tax cells. The first mechanism of the paclitaxel-resistance reported that p-glycoprotein encodes the ABCB1 gene, and can actively pump paclitaxel out of cells to induce paclitaxel-resistance. Paclitaxel is also transported by MRP2 and BCRP, which encoding ABCC2 and ABCG2, respectively [38]. However, in our mRNA-sequencing data, the expression of ABCB1, ABCC2, and ABCG2 were no significant difference between A549/TAX-NC cells and A549/TAX siRNA MUC1 cells. Similarly, a study in human lung cancer cell lines has shown that no correlation between the expression of ABCB1 in paclitaxel sensitive and resistant cell lines at the mRNA level, and there may be more than one mechanism for the development of paclitaxel-resistance [39]. Notably, some DEGs were enriched in NF-κB and MAPK pathway, which are involved in the regulation of cancer stem cells [40]. Therefore, MUC1 may regulate paclitaxel resistance not by p-glycoprotein encodes the ABCB1 but by NF-κB and MAPK pathway-mediated cancer stem cells.
The activation of NF-κB signaling pathway is related to the initiation and progression of many human cancers, inhibiting NF-κB can prevent EMT in lung cancer cells and induce apoptosis in CSCs [41,42]. Previous studies have shown that MUC1 may help cancer cells self-renew through NF-κB pathway, while the silence of MUC1 reduces the activity of NF-κB [27,43,44]. In inflammatory breast cancer, Syndecan-1/Notch/EGFR activates Akt-mediated NF-κB signaling pathway crosstalk regulates the expression of Interleukin 6 (IL-6) and other inflammatory factors to promote the formation of CSCs colony [45]. Consistently, the KEGG pathway analysis in this study indicates that some DEGs were enriched in the NF-κB pathway, indicating that MUC1-EGFR crosstalk with IL-6 may be through activation of the NF-κB pathway. Besides, the overexpression of MUC1 enhances the activation of the MAPK pathway and reduces cell adhesion through its cytoplasmic domain [46]. The p38 MAPK signalling pathway has been proven to regulate NF-κB pathway [47,48], while the DEGs in this study were also enriched in the MAPK pathway. Therefore, based on previous studies and the mRNA seq of this study, we speculate that MUC1-EGFR may lead to the activation of NF-κB and MAPK pathways, and their crosstalk with IL-6 promote the enrichment of CSCs, thereby inducing the development of paclitaxel resistance. Therefore, targeting NF- κB and MAPK pathways may provide multiple benefits in response to paclitaxel resistance, and finding reasonable drugs to regulate paclitaxel resistance combined with chemotherapy is a promising treatment strategy for LUAD. In the future, we will conduct further experiments to identify key targets for chemotherapy-induced paclitaxel resistance of LUAD.
In conclusion, our study confirms the key role of MUC1 in the regulation of paclitaxel resistance. MUC1 is highly expressed in paclitaxel resistance LUAD cells, and MUC1-EGFR crosstalk with IL-6 may be due to the activation of NF-κB and MAPK pathways, which promote the enrichment of CSCs and lead to paclitaxel resistance. In the treatment of LUAD, regulating the expression of MUC1 and the involved molecules or pathways in combination with chemotherapy may help overcome chemotherapy-induced paclitaxel resistance of LUAD.
Supplementary Material
Acknowledgments
Not applicable.
Funding Statement
This work was supported by the Chengdu Health Commission Medical Foundation (Grant: 2020067), General Medical research Fund project (Grant: TYYLKYJJ-2022-044), and Hospital-level incubation project of 363 Hospital (Grant: 2021FH001).
Author contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; Hongyu Xu and Zedong Du conceived and designed the study; Hongyu Xu provided administrative support; Hongyu Xu, Zhihui Li, Xianguo Liu, and Zedong Du provided materials and samples for this study; Hongyu Xu, Xianguo Liu, Xuan Zhang and Xueting Li collected and assembled the data; Hongyu Xu and Xuan Zhang analyzed and interpreted the data.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated or analysed during this study are included in this published article.
References
- 1.Nasim F, Sabath BF, Eapen GA.. Lung cancer. Med Clin North Am. 2019;103(3):1–15. doi: 10.1016/j.mcna.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Cufer T, Ovcaricek T, O’Brien ME.. Systemic therapy of advanced non-small cell lung cancer: major-developments of the last 5-years. Eur J Cancer. 2013;49(6):1216–1225. doi: 10.1016/j.ejca.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 3.Nagy-Mignotte H, Guillem P, Vesin A, et al. Primary lung adenocarcinoma: characteristics by smoking habit and sex. Eur Respir J. 2011;38(6):1412–1419. doi: 10.1183/09031936.00191710. [DOI] [PubMed] [Google Scholar]
- 4.Zappa C, Mousa SA.. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5(3):288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orr GA, Verdier-Pinard P, McDaid H, et al. Mechanisms of taxol resistance related to microtubules. Oncogene. 2003;22(47):7280–7295. doi: 10.1038/sj.onc.1206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denisenko TV, Budkevich IN, Zhivotovsky B.. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018;9(2):117. doi: 10.1038/s41419-017-0063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinbichler TB, Dudás J, Skvortsov S, et al. Therapy resistance mediated by cancer stem cells. Semin Cancer Biol. 2018;53:156–167. doi: 10.1016/j.semcancer.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Vinogradov S, Wei X.. Cancer stem cells and drug resistance: the potential of nanomedicine. Nanomedicine (Lond). 2012;7(4):597–615. doi: 10.2217/nnm.12.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Wang Z, Ajani JA, et al. Drug resistance and cancer stem cells. Cell Commun Signal. 2021;19(1):19. doi: 10.1186/s12964-020-00627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Dong Q, Li J, et al. Targeting cancer stem cells and their niche: perspectives for future therapeutic targets and strategies. Semin Cancer Biol. 2018;53:139–155. doi: 10.1016/j.semcancer.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Lv Y, Cang W, Li Q, et al. Erlotinib overcomes paclitaxel-resistant cancer stem cells by blocking the EGFR-CREB/GRβ-IL-6 axis in MUC1-positive cervical cancer. Oncogenesis. 2019;8(12):70. doi: 10.1038/s41389-019-0179-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W, Zhang Z, Zhang S, et al. MUC1: structure, function, and clinic application in epithelial cancers. Int J Mol Sci. 2021;22(12):6567. doi: 10.3390/ijms22126567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kashyap B, Kullaa AM.. Regulation of mucin 1 expression and its relationship with oral diseases. Arch Oral Biol. 2020. seq;117:104791. doi: 10.1016/j.archoralbio.2020.104791. [DOI] [PubMed] [Google Scholar]
- 14.Gao T, Cen Q, Lei H.. A review on development of MUC1-based cancer vaccine. Biomed Pharmacother. 2020. Dec;132:110888. doi: 10.1016/j.biopha.2020.110888. [DOI] [PubMed] [Google Scholar]
- 15.Johansson ME, Sjövall H, Hansson GC.. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013;10(6):352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cascio S, Finn OJ.. Intra- and extra-cellular events related to altered glycosylation of MUC1 promote chronic inflammation, tumor progression, invasion, and metastasis. Biomolecules. 2016;6(4):39. doi: 10.3390/biom6040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin W, Liao X, Lv Y, et al. MUC1 induces acquired chemoresistance by upregulating ABCB1 in EGFR-dependent manner. Cell Death Dis. 2017;8(8):e2980–e2980. doi: 10.1038/cddis.2017.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Gao H, Li H, et al. Downregulated mucin 1 alleviates paclitaxel resistance in non‑small cell lung cancer cells. Mol Med Rep. 2020;22(4):2966–2972. doi: 10.3892/mmr.2020.11349. [DOI] [PubMed] [Google Scholar]
- 19.Bouillez A, Adeegbe D, Jin C, et al. MUC1-C promotes the suppressive immune microenvironment in non-small cell lung cancer. Oncoimmunology. 2017;6(9):e1338998. doi: 10.1080/2162402x.2017.1338998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith AG, Macleod KF.. Autophagy, cancer stem cells and drug resistance. J Pathol. 2019;247(5):708–718. doi: 10.1002/path.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin W, Zhang M, Dong C, et al. The multifaceted role of MUC1 in tumor therapy resistance. Clin Exp Med. 2022;23(5):1441–1474. doi: 10.1007/s10238-022-00978-y. [DOI] [PubMed] [Google Scholar]
- 22.Joshi S, Kumar S, Bafna S, et al. Genetically engineered mucin mouse models for inflammation and cancer. Cancer Metastasis Rev. 2015;34(4):593–609. doi: 10.1007/s10555-015-9549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroeder JA, Thompson MC, Gardner MM, et al. Transgenic MUC1 interacts with epidermal growth factor receptor and correlates with mitogen-activated protein kinase activation in the mouse mammary gland. J Biol Chem. 2001;276(16):13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- 24.Gebregiworgis T, Purohit V, Shukla SK, et al. Glucose limitation alters glutamine metabolism in MUC1-Overexpressing pancreatic cancer cells. J Proteome Res. 2017;16(10):3536–3546. doi: 10.1021/acs.jproteome.7b00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inagaki Y, Gao J, Song P, et al. Clinicopathological utility of sialoglycoconjugates in diagnosing and treating colorectal cancer. World J Gastroenterol. 2014;20(20):6123–6132. doi: 10.3748/wjg.v20.i20.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duraisamy S, Ramasamy S, Kharbanda S, et al. Distinct evolution of the human carcinoma-associated transmembrane mucins, MUC1, MUC4 and MUC16. Gene. 2006;373:28–34. doi: 10.1016/j.gene.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 27.Ahmad R, Raina D, Trivedi V, et al. MUC1 oncoprotein activates the IkappaB kinase beta complex and constitutive NF-kappaB signalling. Nat Cell Biol. 2007;9(12):1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Situ D, Wang J, Ma Y, et al. Expression and prognostic relevance of MUC1 in stage IB non-small cell lung cancer. Med Oncol. 2011;28 Suppl 1: s596–604. doi: 10.1007/s12032-010-9752-4. [DOI] [PubMed] [Google Scholar]
- 29.Miyazaki K, Kishimoto H, Kobayashi H, et al. The glycosylated N-Terminal domain of MUC1 is involved in chemoresistance by modulating drug permeation across the plasma membrane. Mol Pharmacol. 2023;103(3):166–175. doi: 10.1124/molpharm.122.000597. [DOI] [PubMed] [Google Scholar]
- 30.Bouillez A, Rajabi H, Jin C, et al. MUC1-C integrates PD-L1 induction with repression of immune effectors in non-small-cell lung cancer. Oncogene. 2017;36(28):4037–4046. doi: 10.1038/onc.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ham SY, Kwon T, Bak Y, et al. Mucin 1-mediated chemo-resistance in lung cancer cells. Oncogenesis. 2016;5(1):e185–e185. doi: 10.1038/oncsis.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sousa AM, Rei M, Freitas R, et al. Effect of MUC1/β-catenin interaction on the tumorigenic capacity of pancreatic CD133(+) cells. Oncol Lett. 2016;12(3):1811–1817. doi: 10.3892/ol.2016.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Xu ZY, Wang LF, et al. CD133+ cancer stem cells in lung cancer. Front Biosci (Landmark Ed). 2013;18(2):447–453. doi: 10.2741/4113. [DOI] [PubMed] [Google Scholar]
- 34.Korkaya H, Kim GI, Davis A, et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol Cell. 2012;47(4):570–584. doi: 10.1016/j.molcel.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grubelnik G, Boštjančič E, Pavlič A, et al. NANOG expression in human development and cancerogenesis. Exp Biol Med (Maywood). 2020;245(5):456–464. doi: 10.1177/1535370220905560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohiuddin IS, Wei SJ, Kang MH.. Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim Biophys Acta Mol Basis Dis. 2020;1866(4):165432. doi: 10.1016/j.bbadis.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirzaei S, Paskeh MDA, Entezari M, et al. SOX2 function in cancers: association with growth, invasion, stemness and therapy response. Biomed Pharmacother. 2022;156:113860. doi: 10.1016/j.biopha.2022.113860. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y, Sadée W.. Membrane transporters and channels in chemoresistance and -sensitivity of tumor cells. Cancer Lett. 2006;239(2):168–182. doi: 10.1016/j.canlet.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 39.Shimomura M, Yaoi T, Itoh K, et al. Drug resistance to paclitaxel is not only associated with ABCB1 mRNA expression but also with drug accumulation in intracellular compartments in human lung cancer cell lines. Int J Oncol. 2012;40(4):995–1004. doi: 10.3892/ijo.2011.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butti R, Das S, Gunasekaran VP, et al. Receptor tyrosine kinases (RTKs) in breast cancer: signaling, therapeutic implications and challenges. Mol Cancer. 2018;17(1):34. doi: 10.1186/s12943-018-0797-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1(5):a000141–a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zakaria N, Mohd Yusoff N, Zakaria Z, et al. Inhibition of NF-κB signaling reduces the stemness characteristics of lung cancer stem cells. Front Oncol. 2018;8:166. doi: 10.3389/fonc.2018.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alam M, Rajabi H, Ahmad R, et al. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget. 2014;5(9):2622–2634. doi: 10.18632/oncotarget.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmad R, Raina D, Joshi MD, et al. MUC1-C oncoprotein functions as a direct activator of the nuclear factor-kappaB p65 transcription factor. Cancer Res. 2009;69(17):7013–7021. doi: 10.1158/0008-5472.Can-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ibrahim SA, Gadalla R, El-Ghonaimy EA, et al. Syndecan-1 is a novel molecular marker for triple negative inflammatory breast cancer and modulates the cancer stem cell phenotype via the IL-6/STAT3, notch and EGFR signaling pathways. Mol Cancer. 2017;16(1):57. doi: 10.1186/s12943-017-0621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosseinzadeh A, Merikhian P, Naseri N, et al. MUC1 is a potential target to overcome trastuzumab resistance in breast cancer therapy. Cancer Cell Int. 2022;22(1):110. doi: 10.1186/s12935-022-02523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seumois G, Ramírez-Suástegui C, Schmiedel BJ, et al. Single-cell transcriptomic analysis of allergen-specific T cells in allergy and asthma. Sci Immunol. 2020;5(48):eaba6087. doi: 10.1126/sciimmunol.aba6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li S, Wang H, Ma R, et al. Schisandrin B inhibits epithelial‑mesenchymal transition and stemness of large‑cell lung cancer cells and tumorigenesis in xenografts via inhibiting the NF‑κB and p38 MAPK signaling pathways. Oncol Rep. 2021;45(6):115. doi: 10.3892/or.2021.8066. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article.