Abstract
Background
The lung microbiome is an inflammatory stimulus whose role in COPD pathogenesis is incompletely understood. We hypothesised that the frequent exacerbator phenotype is associated with decreased α-diversity and increased lung inflammation. Our objective was to assess correlations between the frequent exacerbator phenotype, the microbiome and inflammation longitudinally during exacerbation-free periods.
Methods
We conducted a case–control longitudinal observational study of the frequent exacerbator phenotype and characteristics of the airway microbiome. 81 subjects (41 frequent and 40 infrequent exacerbators) provided nasal, oral and sputum microbiome samples at two visits over 2–4 months. Exacerbation phenotype, relevant clinical factors and sputum cytokine values were associated with microbiome findings.
Results
The frequent exacerbator phenotype was associated with lower sputum microbiome α-diversity (p=0.0031). This decrease in α-diversity among frequent exacerbators was enhanced when the sputum bacterial culture was positive (p<0.001). Older age was associated with decreased sputum microbiome α-diversity (p=0.0030). Between-visit β-diversity was increased among frequent exacerbators and those who experienced a COPD exacerbation between visits (p=0.025 and p=0.014, respectively). Sputum cytokine values did not differ based on exacerbation phenotype or other clinical characteristics. Interleukin (IL)-17A was negatively associated with α-diversity, while IL-6 and IL-8 were positively associated with α-diversity (p=0.012, p=0.012 and p=0.0496, respectively). IL-22, IL-17A and IL-5 levels were positively associated with Moraxella abundance (p=0.027, p=0.0014 and p=0.0020, respectively).
Conclusions
Even during exacerbation-free intervals, the COPD frequent exacerbator phenotype is associated with decreased sputum microbiome α-diversity and increased β-diversity. Decreased sputum microbiome α-diversity and Moraxella abundance are associated with lung inflammation.
Tweetable abstract
Low sputum microbiome α-diversity and decreased compositional stability (increased β-diversity) are associated with the COPD frequent exacerbator phenotype. Low sputum microbiome α-diversity is also associated with lung inflammation. https://bit.ly/3SEA8M5
Introduction
COPD is a leading cause of death; however, the mechanisms driving its progression remain incompletely understood. One recently recognised mechanism is inflammation triggered by the lung microbiome. Cycles of recurrent lung infection, inflammation and antibiotic use may disrupt the microbiome with downstream consequences for lung function [1]. Even in the absence of clinical infection, COPD patients with pathogenic bacteria in their airways have higher levels of sputum and systemic inflammatory markers and increased pulmonary symptoms [2, 3].
The COPD frequent exacerbator phenotype identifies a subset of patients at high risk of recurrent COPD exacerbation. Frequent exacerbators suffer increased morbidity and mortality compared to those who experience exacerbations less often [4–7]. Approximately half of COPD exacerbations are attributed to bacterial infection, and pathogenic bacteria such as Haemophilus influenzae, Moraxella catarrhalis and Streptococcus pneumoniae are often identified in the lung microbiome of COPD patients even during periods of stable lung disease [3].
Inflammatory markers are increased in the sputum of frequent exacerbators [8–10] and COPD patients colonised with potentially pathogenic bacteria [9, 11–15]. The COPD lung microbiome provides an inflammatory stimulus, even in the absence of overt lung infection. In particular, interleukin (IL)-17A, IL-8, IL-6, IL-1β, IL-22, IL-5 and leukotriene B4 (LTB4) levels in sputum have been associated with various components of the COPD lung microbiome during exacerbation-free intervals [2, 8, 11–19]. It remains unclear which particular components of the microbiome (bacterial biomass, α-diversity, the particular taxa present, etc.) are most closely associated with sputum inflammation or the frequent exacerbator phenotype.
We and others have shown that the sputum microbiome of frequent exacerbators has lower α-diversity compared to nonfrequent exacerbators [18, 20–26]. However, few of these studies evaluate the COPD lung microbiome solely during periods of clinical stability (when findings are less influenced by exacerbation treatments), longitudinally and including an analysis of concurrent lung inflammation. We undertook the present case–control longitudinal observational study of COPD exacerbation phenotype, the upper airway and sputum microbiome and lung inflammation to address these gaps.
Methods
Study design and recruitment
We conducted a case–control longitudinal observational study of exacerbation phenotype and characteristics of the upper airway and sputum microbiome. All participants were recruited from a single site and were aged ≥40 years with COPD. Frequent exacerbators had at least one severe exacerbation (an exacerbation requiring hospital admission or emergency department visit) in the past 12 months, in accordance with descriptions found in Global Initiative for Chronic Obstructive Lung Disease (GOLD) guidelines [27]. Infrequent exacerbators must have had no exacerbations in the prior 24 months. Recruitment and all visits/samples were deferred until participants had recovered for ≥1 month from the most recent exacerbation. The protocol was approved by the Minneapolis VA IRB (#4541-B). Additional details on recruitment and other methods can be found in the supplementary material. Sequence reads from 22 participants have been published previously (National Center for Biotechnology Information (NCBI) Sequence Read Archive Accession PRJNA543785) [20]; however, all data analysed here were re-sequenced for this analysis (NCBI Sequence Read Archive Accession PRJNA944199).
Study procedures
At visit 1, participants provided their medical history, underwent spirometry, completed the St George's Respiratory Questionnaire (SGRQ), and provided oral wash, nasal swab and induced sputum samples. All participants returned for a second study visit ∼2 months following visit 1, where they provided information on interim COPD exacerbations, repeated the SGRQ, and provided oral, nasal and sputum samples. Visit 2 was deferred (for up to 4 months following visit 1) if the participant reported any COPD exacerbations or antibiotic use in the 1 month prior to the visit. Exacerbation phenotype was determined at visit 1 and was not revised based on exacerbations observed during the study.
Sample processing, 16S rRNA gene quantification and MiSeq sequencing
All samples and negative controls were extracted using the MO BIO PowerSoil DNA Isolation Kit (QIAGEN, Germantown, MD, USA). Extracted DNA underwent 16S rRNA gene quantification using droplet digital PCR (ddPCR; supplementary table E1) and 16S rRNA gene V4 MiSeq sequencing. 16S rRNA V4 sequences were processed as described in the supplementary material.
Sputum culture results
The clinical microbiology laboratory performed Gram stain and aerobic culture on all sputum samples. Any organism identified in culture was considered a pathogen.
Cytokine analyses
Sputum samples were submitted to the University of Minnesota Cytokine Reference Laboratory for determination of LTB4, granulocyte–macrophage colony-stimulating factor (GM-CSF), IL-8, IL-6, IL-1β, IL-17A, IL-22, IL-5 and tumour necrosis factor (TNF)-α (R&D Systems, Minneapolis, MN, USA).
Statistical analyses
Data presented are from visit 1 only (V1) or both visits (BV). Linear regression, including random-effects censored regression models, linear mixed models (LMM) and generalised estimating equations (GEE), were used to test for associations between variables. Analyses of α-diversity metrics were adjusted for age, forced expiratory volume in 1 s % predicted (FEV1pp), body mass index (BMI), current tobacco use and current alcohol use. β-diversity was assessed via the Bray–Curtis dissimilarity metric and illustrated via principal coordinates analysis (PCoA). Univariate PERMANOVA analyses of β-diversity were performed for each anatomical site. Permutation tests were used to determine if within-subject visit 1–visit 2 similarity differed by permuting levels of categorical clinical factors. After eliminating genera present in <10% of samples, tests of association between taxa and clinical characteristics used Holm's procedure to control the family-wise error rate across all taxa at 5%. GM-CSF and TNF-α were not analysed and IL-22 results were dichotomised (detected versus not detected), as too few samples contained quantifiable results. Random-effects censored regression models were used to test for associations between clinical or microbiome characteristics and cytokine levels (modelled as the response variable). All analyses were conducted in R version 3.6.0.
Results
Cohort
81 subjects consisting of 40 infrequent exacerbators and 41 frequent exacerbators provided data and samples during two visits over 2–4 months (supplementary table E2). Subjects were balanced with respect to age, gender, race, inhaled corticosteroid (ICS) use and dental care habits (table 1). Consistent with the Department of Veterans Affairs patient population, most subjects were male. Frequent exacerbator phenotype was associated with lower BMI (median 27.9 kg·m−2 versus 30.2 kg·m−2, p=0.022), lower FEV1pp (44.0% pred versus 52.5% pred, p<0.001) and a higher number of COPD exacerbations in the past 12 months (median two versus 0, mean 2.39 versus 0). Frequent exacerbators were less likely than infrequent exacerbators to be current tobacco users (19.5% frequent exacerbators versus 42.5% infrequent exacerbators, p=0.032). Six subjects (five frequent exacerbators, one infrequent exacerbator) reported a COPD exacerbation at visit 2 and six subjects (four frequent exacerbators, two infrequent exacerbators) reported antibiotic use at visit 2. Relationships between microbiome measures at each site and clinical factors (exacerbation phenotype, age, FEV1pp, ICS use, pack-years of tobacco use, current tobacco use, toothbrushing frequency, SGRQ score, pathogen detection in sputum samples, and experiencing a COPD exacerbation or use of an antibiotic between study visits) were examined; unless mentioned, these analyses did not reveal an association.
TABLE 1.
Subject baseline characteristics
| Infrequent exacerbator | Frequent exacerbator | Overall | p-value # | |
| Subjects | 40 | 41 | 81 | |
| Male | 40 (100) | 40 (97.6) | 80 (98.8) | 1.00 |
| Age years, median (IQR) | 69 (5) | 69 (8) | 69 (7) | 0.632 |
| Caucasian white | 37 (92.5) | 40 (97.6) | 77 (95.1) | 0.359 |
| BMI kg·m−2, median (IQR) | 30.2 (7.79) | 27.93 (9.26) | 29.57 (8.46) | 0.022 |
| COPD severity | ||||
| Moderate | 24 (60) | 14 (34.1) | 38 (46.9) | |
| Severe | 14 (35) | 18 (43.9) | 32 (39.5) | |
| Very severe | 2 (5) | 9 (22) | 11 (13.6) | |
| FEV1 % predicted, median (IQR) | 52.5 (19) | 44 (17) | 48 (19) | <0.001 |
| COPD exacerbations in the past 12 months, median (IQR) | 0 (0) | 2 (1) | 1 (2) | <0.001 |
| COPD hospitalisations in the past 12 months, median (IQR) | 0 (0) | 0 (1) | 0 (0) | <0.001 |
| Inhaled corticosteroids (yes) | 11 (27.5) | 14 (34.1) | 25 (30.9) | 0.632 |
| Smoking pack-years, median (IQR) | 41 (23.12) | 50 (23) | 50 (24.5) | 0.059 |
| Current tobacco use (yes) | 17 (42.5) | 8 (19.5) | 25 (30.9) | 0.032 |
| Current alcohol use (yes) | 24 (60) | 32 (78) | 56 (69.1) | 0.096 |
| Brush teeth at least once daily¶ | 30 (75) | 30 (75)¶ | 60 (75)¶ | 1.00 |
| SGRQ score, median (IQR) | 43.62 (10.49) | 52.4 (20.6) | 47.29 (17.84) | 0.074 |
Data are presented as n or n (%), unless otherwise stated. IQR: interquartile range; BMI: body mass index; FEV1: forced expiratory volume in 1 s; SGRQ: St George's Respiratory Questionnaire. #: a two-sample t-test was conducted for all continuous variables and a Fisher exact test for all categorical variables; ¶: one subject did not provide frequency of brushing teeth.
α-Diversity
α-Diversity was assessed using Shannon diversity, Simpson diversity and Chao1 metrics. For simplicity, Simpson diversity findings are discussed in detail here (supplementary figure E1), as they are largely consistent with the Shannon and Chao1 diversity findings.
Nasal samples
No significant relationships between nasal sample Simpson diversity and any of the evaluated clinical factors were observed.
Oral samples
In multiple models of oral wash Simpson diversity, FEV1pp (a model covariate) was consistently associated with increased oral wash Simpson diversity (frequent exacerbator phenotype V1 linear regression, FEV1pp coefficient estimate (CE) 0.0008, 95% CI 0.00002 to 0.0016; p=0.027 (figure 1a); BV GEE, FEV1pp CE 0.0007, 95% CI 0.0001 to 0.0013; p=0.023). In light of this association, we further evaluated potential relationships between frequent exacerbator phenotype, FEV1pp and Simpson diversity. In a model of frequent exacerbator phenotype, FEV1pp and their interaction, frequent exacerbator phenotype and the interaction of frequent exacerbator phenotype and FEV1pp were associated with oral wash Simpson diversity (interaction BV GEE CE 0.0010, 95% CI 0.00002 to 0.0020; p=0.048). Higher SGRQ scores (indicating worse quality of life) were associated with lower oral wash Simpson diversity (V1 linear regression, CE −0.0007, 95% CI −0.0013 to −0.00011; p=0.018 (figure 1b); and BV GEE, CE −0.0005, 95% CI −0.0009 to −0.0001; p=0.020).
FIGURE 1.
Oral wash α-diversity is associated with forced expiratory volume in 1 s % predicted (FEV1pp) and COPD-related quality of life. a) Oral wash Simpson diversity at visit 1 is associated with FEV1pp (a model covariate) in a model of frequent exacerbator phenotype (frequent exacerbator phenotype V1 linear regression (LR), FEV1pp coefficient estimate (CE) 0.0008, 95% CI 0.00002 to 0.0016; p=0.027. FEV1pp is associated with low Simpson diversity in the adjusted model. b) Oral wash Simpson diversity at visit 1 is associated with St George's Respiratory Questionnaire (SGRQ) score at visit 1 (LR, CE −0.0007, 95% CI −0.0013 to −0.00011; p=0.018). Higher SGRQ scores indicate worse COPD-related quality of life.
Sputum samples
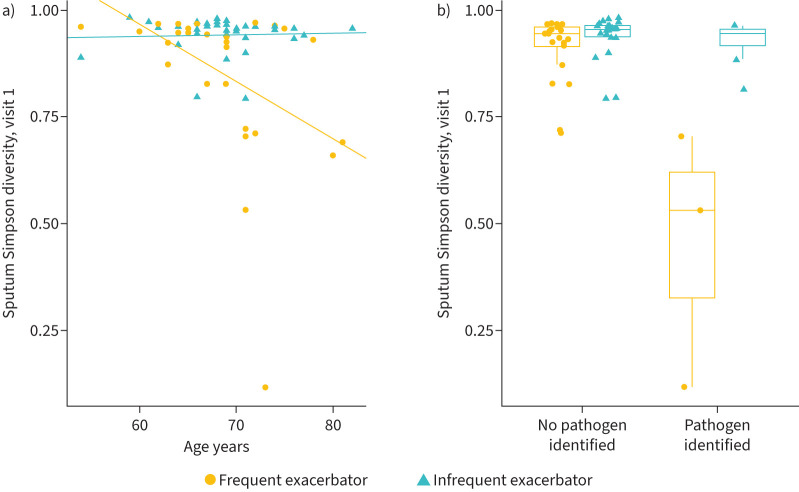
The frequent exacerbator phenotype was associated with lower Simpson diversity in sputum at visit 1 and both visits (V1 linear regression, CE −0.077, 95% CI −0.15 to −0.0048; p=0.041 (supplementary figure E2); BV GEE, CE −0.075, 95% CI −0.12 to −0.025; p=0.0031). Older age (another model covariate) was also associated with lower sputum Simpson diversity in a model of exacerbation phenotype (V1 linear regression, age CE −0.0090, 95% CI −0.015 to −0.0025; p=0.0080 (figure 2a and supplementary figure E3); BV GEE, age CE −0.0074, 95% CI −0.012 to −0.0025; p=0.0030).
FIGURE 2.
Sputum α-diversity is associated with exacerbation phenotype, age and culture results. a) Both the frequent exacerbator phenotype and older age were significantly associated with lower Simpson diversity at visit 1 (linear regression (LR), coefficient estimate (CE) −0.077, 95% CI −0.15 to −0.0048; p=0.041 and CE −0.0090, 95% CI −0.015 to −0.0025; p=0.0080, respectively). When the interaction of exacerbation phenotype and age was added to the model, only the interaction of age and exacerbation phenotype was significant (LR, CE −0.013, 95% CI −0.024 to −0.0024; p=0.020). The regression lines represent the association between phenotype, age and Simpson diversity. The significant interaction between age and phenotype indicates that older frequent exacerbators have lower sputum α-diversity than younger frequent exacerbators or older infrequent exacerbators. b) Presence of a pathogen in sputum culture was associated with lower sputum Simpson diversity at visit 1 (LR, CE −0.19, 95% CI −0.31 to −0.074; p=0.0028). When frequent exacerbator phenotype, pathogen and their interaction were included in the model, only their interaction was significant (visit 1 LR, CE −0.46, 95% CI −0.63 to −0.30; p<0.001). The association between sputum culture positivity during clinically stable periods and Simpson diversity differs based on exacerbation phenotype.
When exacerbation phenotype, age, and their interaction were included in a model of sputum α-diversity, their interaction was significantly associated with lower Simpson diversity (V1 linear regression, interaction CE −0.013, 95% CI −0.024 to −0.0024; p=0.020 (figure 2a); BV GEE, interaction CE −0.0099, 95% CI −0.018 to −0.0017; p=0.017). This shows that the association between exacerbation phenotype and sputum Simpson diversity differs based on age.
Sputum samples from six subjects (three frequent exacerbators and three infrequent exacerbators) were positive for clinically relevant respiratory pathogens (M. catarrhalis, H. influenzae, methicillin-resistant S. aureus and Klebsiella aerogenes) at visit 1. Presence of a pathogen was associated with lower sputum Simpson diversity at visit 1 and both visits (V1 linear regression, CE −0.19, 95% CI −0.31 to −0.074; p=0.0028; and BV LMM, CE −0.20, 95% CI −0.26 to −0.14; p<0.001). When frequent exacerbator phenotype, pathogen and their interaction were included in the model, only their interaction was significant at visit 1 and both visits (V1 linear regression, CE −0.46, 95% CI −0.63 to −0.30; p<0.001 (figure 2b); BV LMM, CE −0.27, 95% CI −0.38 to −0.15; p<0.001). This shows that the association between sputum culture positivity during clinically stable periods and Simpson diversity differs based on exacerbation phenotype.
In summary, sputum α-diversity is associated with frequent exacerbator phenotype, age and pathogen detection in sputum culture during clinically stable periods. The association between frequent exacerbator phenotype and decreased sputum α-diversity is also modified by older age or the identification of a pathogen from sputum culture.
α-Diversity over time
Visit 1 values were significantly associated with visit 2 values at all sites (data not shown). When visit 1 values were included in the model, self-reported COPD exacerbation or self-reported use of an antibiotic between study visits was not associated with α-diversity at visit 2. Exacerbation phenotype, age, FEV1pp, current tobacco use, pack-years of tobacco use and SGRQ score were not associated with a change in α-diversity between study visits.
β-Diversity
Environmental, equipment and regent control samples were distinct from subject samples (supplementary figure E4, supplementary table E3). After subsampling to include only subject samples from visit 1, PCoA revealed significant clustering by anatomic site (PERMANOVA, p=0.001 for all pairwise testing; figure 3).
FIGURE 3.

β-Diversity reveals clustering by anatomic site. Principal coordinates (PC) analysis using Bray–Curtis dissimilarity on visit 1 samples demonstrating significant clustering by anatomic site (PERMANOVA, p=0.001 for all pairwise testing). Centroids are illustrated by black dots, with lines connecting each sample to its corresponding centroid.
PERMANOVA analyses
Clustering on PCoA based on exacerbation phenotype and other clinical factors was investigated using PERMANOVA analyses. The analyses were conducted at each anatomic site separately and using visit 1 data, unless noted below. At visit 1, nasal samples clustered based on FEV1pp (PERMANOVA, R2=0.029, p=0.033). Several other PERMANOVA results with p<0.10 are provided in supplementary table E4. When visit 2 data were analysed, sputum samples from participants who reported between-visit antibiotic use for any indication clustered separately from participants who did not report between-visit antibiotic use (PERMANOVA, R2=0.029, p=0.049).
β-Diversity over time
Visit 1 samples were compared to corresponding visit 2 samples for all subjects and sites to assess the stability of microbiome composition over time. Among nasal samples, the frequent exacerbator phenotype and antibiotic use between study visits was associated with decreased similarity between paired samples (permutation testing (P), p=0.044 and p=0.032, respectively). There were no associations with oral wash similarity between visits. Among sputum samples, the frequent exacerbator phenotype (versus infrequent exacerbator phenotype) and experiencing a COPD exacerbation (versus no COPD exacerbation) between visits were associated with lower similarity between paired samples (P, p=0.025 and p=0.014, respectively; figure 4 and supplementary figure E5).
FIGURE 4.
Within-subject microbiome composition stability. Self-report of a COPD exacerbation between study visits corresponded with an increase in microbiome compositional changes (increased β-diversity, or a decrease in sample similarity) compared with subjects who did not experience a COPD exacerbation between study visits. Please note that one frequent exacerbator who reported an exacerbation between study visits is not represented here, as this subject did not provide both sputum samples. The frequent exacerbator phenotype also corresponded with an increase in microbiome compositional changes (increased β-diversity, or a decrease in sample similarity) between study visits compared with the infrequent exacerbator phenotype (permutation analyses, p=0.014 and p=0.025, respectively).
Bacterial taxa
We investigated the bacterial taxa present in each sample to determine potential associations with relevant clinical factors (97 tests). Many taxa were associated with clinical site, in accordance with clinical findings and the human microbiome literature. Nasal samples were enriched with Corynebacterium, Staphylococcus, Cutibacterium and Moraxella (among others) compared with oral and sputum samples (linear regression, all p<0.05 following Holm correction; supplementary table E5). Oral and sputum samples were enriched with Veillonella, Rothia, Prevotella, Streptococcus and Haemophilus (among others) when compared with nasal samples (linear regression, all p<0.05 following Holm correction; supplementary table E5). Across all anatomic sites, Mannheimia abundance was positively associated with age (linear regression, p=0.0014); Mogibacterium abundance was lower among frequent exacerbators compared to infrequent exacerbators (p=0.029); Leuconostoc abundance was negatively associated with FEV1pp (p=0.020); Bulleidia abundance was higher among current tobacco users (p=0.013); and Pseudomonas abundance was positively associated with pack-years of tobacco use (p<0.0001; supplementary table E6).
Sputum cytokine analyses
Sputum sample cytokine levels were tested for an association with clinical factors and sputum microbiome characteristics. Cytokines were chosen for analysis based on prior reported associations with culture or microbiome results, and analyses of the seven evaluable cytokines are provided here. Samples from all available visits were analysed using GEE or random-effects censored regression models, as appropriate, accounting for visit.
Cytokines associated with clinical characteristics
None of the clinical factors (exacerbation phenotype, age, FEV1pp, pack-years of tobacco exposure, current tobacco use or SGRQ score) were associated with cytokine levels in a random-effects censored regression model.
Cytokines associated with α-diversity (Simpson)
Three sputum cytokines were associated with sputum sample α-diversity on univariate analysis. IL-17A levels were negatively associated with Simpson diversity, while IL-6 and IL-8 were positively associated with Simpson diversity (random-effects censored regression model, IL-17A CE −1.3, 95% CI −2.2 to −0.50; p=0.012; IL-6 CE 6.0, 95% CI 2.2 to 9.8; p=0.012; and IL-8 CE 4.2, 95% CI 1.0 to 7.4; p=0.0496, respectively; figure 5). IL-22, IL-5, IL-1β and LTB4 were not associated with Simpson diversity.
FIGURE 5.
Multiple sputum cytokines are associated with α-diversity. Increased sputum concentrations of interkeukin (IL)-17A were associated with decreased Simpson diversity in the sputum, while increased sputum concentrations of IL-6 and IL-8 were associated with increased Simpson diversity (random effects censored regression model, IL-17A coefficient estimate (CE) −1.3, 95% CI −2.2 to −0.50; p=0.012; IL-6 CE 6.0, 95% CI 2.2 to 9.8; p=0.012; and IL-8 CE 4.2, 95% CI 1.0 to 7.4; p=0.0496, respectively).
Cytokines associated with pathogen abundance
Presence of typical COPD pathogens in sputum samples, such as Streptococcus, Moraxella or Haemophilus, have been associated with exacerbation phenotype, increased inflammation and decreased α-diversity. We found that IL-22, IL-17A and IL-5 levels were positively associated with Moraxella abundance (GEE with Holm correction, IL-22 CE 10.26, 95% CI 3.96 to 16.55; p=0.027; random-effects censored regression model with Holm correction, IL-17A CE 2.01, 95% CI 1.02 to 2.99; p=0.0014; and IL-5 CE 1.87, 95% CI 0. 93 to 2.81, p=0.002, respectively; figure 6). There were no significant associations with other cytokines or the genera Streptococcus or Haemophilus.
FIGURE 6.
Sputum cytokines are associated with Moraxella abundance. Increased sputum concentrations of interleukin (IL)-22, IL-17A and IL-5 were correlated with increased abundance of Moraxella in sputum samples (generalised estimating equations with Holm correction, IL-22 coefficient estimate (CE) 10.26, 95% CI 3.96 to 16.55; p=0.027; random-effects censored regression model with Holm correction, IL-17A CE 2.01, 95% CI 1.02 to 2.99; p=0.0014; and IL-5 CE 1.87, 95% CI 0.93 to 2.81; p=0.002, respectively).
Discussion
Our case–control longitudinal observational study of the upper airway and sputum microbiome during periods of clinical stability identified low sputum microbiome α-diversity as a key feature of the COPD frequent exacerbator phenotype. In turn, low sputum microbiome α-diversity was associated with airway bacterial colonisation and lung inflammation, two other characteristics consistent with the increased morbidity and mortality associated with the frequent exacerbator phenotype. Additionally, the frequent exacerbator phenotype was associated with decreased microbiome compositional stability (increased β-diversity) on longitudinal sputum sampling. These findings suggest that low α-diversity and an unstable sputum microbiome are key features of the frequent exacerbator phenotype.
In addition to our findings on the frequent exacerbator phenotype, we also detected associations between the sputum, oral or nasal microbiome and age and COPD-related symptom severity. Also, among the small number of subjects who experienced an exacerbation between study visits, we observed compositional changes (increased β-diversity) in the microbiome following the occurrence of a COPD exacerbation. Our study determined that the frequent exacerbator phenotype is associated with low sputum microbiome α-diversity. Low α-diversity among frequent exacerbators has been found by many, but not all, investigators, possibly related to differences in how exacerbation frequency was analysed [20, 22–26, 28, 29]. In the case of the sputum microbiome, this homogeneity was enhanced among older participants and among participants with airway bacterial colonisation. This phenotype also correlated with increased Mogibacterium, an oral taxon associated with oral inflammation and stable COPD [30, 31].
In this study and others, older age was associated with decreased α-diversity among sputum samples [23, 28]. Here, we also determined that older age is associated with a further decrease in α-diversity among frequent exacerbators. Although ageing is associated with an increased likelihood of COPD diagnosis, as well as increased susceptibility to lung infections, declines in lung function, and additional courses of antibiotics, the associations between ageing and the microbiome are relatively understudied. The ageing COPD lung microbiome and the gut–lung axis may exist at a juncture between declining lung function, immunosenescence, nutritional changes and increased antibiotic exposure [32]. Our work suggests that age itself may influence the COPD lung microbiome, possibly via mechanisms independent of typical factors (such as COPD exacerbations) that are known to influence the lung microbiome.
Our analysis of associations between the microbiome and tobacco use encompassed both pack-years of tobacco use as well as current (versus former) tobacco use status. Consistent with many of the previous studies [33–35], which identified more tobacco-associated microbiome changes of the upper versus lower airway, we identified that greater pack-years of tobacco exposure were associated with increased Pseudomonas abundance, while current (versus former) tobacco use was associated with increased Bulleidia abundance, primarily in the oropharynx. Bulleidia has previously been associated with tobacco use and lung cancer [36, 37].
The present study also evaluated the longitudinal stability of the upper and lower airway microbiome. Although microbiome findings were generally similar on repeated sampling (low β-diversity), we identified several scenarios in which β-diversity was increased. The sputum microbiome of frequent exacerbators exhibited decreased similarity (increased β-diversity) compared with infrequent exacerbators. Subjects who experienced a COPD exacerbation between study visits also exhibited decreased compositional similarity versus those who did not report interim exacerbations.
We also found significant differences in sputum cytokine levels which associated with sputum microbiome α-diversity, but not related to exacerbation phenotype, COPD severity, age, tobacco use or COPD-related quality of life. Low sputum α-diversity was associated with increased concentrations of sputum IL-17A and decreased concentrations of IL-6 and IL-8. Moraxella abundance in the sputum microbiome was also associated with increased concentrations of IL-22, IL-17A and IL-5. IL-17A, IL-22 and IL-6 are key mediators of a T-helper (Th)17 response, often at mucosal sites [38]. IL-8 is a neutrophil chemoattractant. IL-5 is involved in Th2 responses and eosinophil recruitment. In prior COPD studies, elevated sputum IL-6 and IL-8 levels have been associated with tobacco use, lower FEV1pp, frequent exacerbations and acute exacerbation (versus clinical stability) [8, 39–42]. Our finding of a positive association between IL-6 and IL-8 levels and α-diversity is somewhat unexpected, but it is possible that the acute rise in IL-6 and IL-8 observed during exacerbations is not reflected in our samples, which were collected during exacerbation-free intervals. Furthermore, few investigators have assessed the sputum microbiome (specifically low α-diversity) in relation to inflammatory cytokines.
Our manuscript has several strengths. We used well-defined frequent exacerbator and infrequent exacerbator phenotypes consistent with GOLD guidelines [27], allowing us to address associations between the microbiome and exacerbation phenotype. We deferred all study visits for 1 month following a COPD exacerbation or antibiotic use for any reason, in order to focus on the microbiome during periods of clinical stability. This approach minimises the influence of recent antibiotic or systemic steroid use on our microbiome findings, to the extent possible in an observational study of COPD. Lastly, our longitudinal approach allowed us to assess the stability of microbiome composition in relationship to exacerbation phenotype and recent COPD exacerbations. Our use of sputum inflammatory cytokines, in the context of the microbiome findings, identified clinical correlates of our microbiome findings.
Despite these strengths, our study had several relative weaknesses. We are unable to assess the influence of sex on the microbiome, as our single-centre study was conducted at a Veterans Affairs hospital with a limited female population. Sputum samples may be contaminated by saliva during expectoration and therefore may not reflect only the lower airway microbiome. Despite this potential limitation, we note that most of our key microbiome findings were identified only in the sputum microbiome and not identified in the oral microbiome. This suggests that sputum analysis can identify microbiome associations unique to the lower airways, despite potential upper airway contamination.
In conclusion, we found that frequent exacerbators exhibit lower sputum microbiome α-diversity, which is enhanced by older age or bacterial colonisation of the airways. Sputum microbiome α-diversity is a significant correlate of lung inflammation. The sputum microbiome composition of frequent exacerbators changes more over time when compared to the compositional stability of infrequent exacerbators.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00595-2023.SUPPLEMENT (322.8KB, pdf)
Figure E1 00595-2023.supplement_fig_E1 (224.8KB, jpg)
Figure E2 00595-2023.supplement_fig_E2 (277.7KB, jpg)
Figure E3 00595-2023.supplement_fig_E3 (244.7KB, jpg)
Figure E4 00595-2023.supplement_fig_E4 (314.1KB, jpg)
Figure E5 00595-2023.supplement_fig_E5 (298.7KB, jpg)
Table E3 00595-2023.TABLEE3 (74.4KB, pdf)
Acknowledgements
The authors wish to thank Susan Johnson (Minneapolis VA Medical Center, Minneapolis, MN) for assistance with study recruitment. This material is based upon work supported by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development (Clinical Sciences Research and Development). The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the US government.
Provenance: Submitted article, peer reviewed.
Ethics statement: The protocol was approved by the Minneapolis VA IRB (#4541-B).
Author contributions: Conception and design: A.A. Pragman, C.S. Reilly and C.H. Wendt; data acquisition: A.A. Pragman, S.W. Hodgson and A. Zank; data analysis: A.A. Pragman, T. Wu, C.S. Reilly and C.H. Wendt; data interpretation: A.A. Pragman, T. Wu, C.S. Reilly and C.H. Wu; drafting the manuscript: A.A. Pragman; revision and final approval of the version to be published: all authors.
Conflict of interest: A. Pragman reports support for attending meetings and/or travel from the US Department of Veterans Affairs outside the submitted work.
Conflict of interest: S.W. Hodgson has nothing to disclose.
Conflict of interest: T. Wu has nothing to disclose.
Conflict of interest: A. Zank has nothing to disclose.
Conflict of interest: C.S. Reilly reports grants or contracts from NIH, outside the submitted work; and participation on a data safety monitoring or advisory board for the Mayo Clinic and Washington University, outside the submitted work.
Conflict of interest: C.H. Wendt has nothing to disclose.
Support statement: This publication was supported by grant number 1UL1RR033183 from the National Center for Research Resources and by grant number 8UL1TR000114-02 from the National Center for Advancing Translational Sciences of the National Institutes of Health (NIH) to the University of Minnesota Clinical and Translational Science Institute (CTSI). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the CTSI or the NIH. The University of Minnesota CTSI is part of a national Clinical and Translational Science Award consortium created to accelerate laboratory discoveries into treatments for patients. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Sethi S, Maloney J, Grove L, et al. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006; 173: 991–998. doi: 10.1164/rccm.200509-1525OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desai H, Eschberger K, Wrona C, et al. Bacterial colonization increases daily symptoms in patients with chronic obstructive pulmonary disease. Ann Am Thorac Soc 2014; 11: 303–309. doi: 10.1513/AnnalsATS.201310-350OC [DOI] [PubMed] [Google Scholar]
- 3.Sethi S, Murphy TF. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin Microbiol Rev 2001; 14: 336–363. doi: 10.1128/CMR.14.2.336-363.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carolan BJ, Sutherland ER. Clinical phenotypes of chronic obstructive pulmonary disease and asthma: recent advances. J Allergy Clin Immunol 2013; 131: 627–634. doi: 10.1016/j.jaci.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 5.Hurst JR, Vestbo J, Anzueto A, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med 2010; 363: 1128–1138. doi: 10.1056/NEJMoa0909883 [DOI] [PubMed] [Google Scholar]
- 6.Soler-Cataluña JJ, Martínez-García MA, Román Sánchez P, et al. Severe acute exacerbations and mortality in patients with chronic obstructive pulmonary disease. Thorax 2005; 60: 925–931. doi: 10.1136/thx.2005.040527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spencer S, Jones PW. Time course of recovery of health status following an infective exacerbation of chronic bronchitis. Thorax 2003; 58: 589–593. doi: 10.1136/thorax.58.7.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhowmik A, Seemungal TA, Sapsford RJ, et al. Relation of sputum inflammatory markers to symptoms and lung function changes in COPD exacerbations. Thorax 2000; 55: 114–120. doi: 10.1136/thorax.55.2.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcha DS, Thurston SJ, Patel AR, et al. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax 2012; 67: 1075–1080. doi: 10.1136/thoraxjnl-2012-201924 [DOI] [PubMed] [Google Scholar]
- 10.Thomsen M, Ingebrigtsen TS, Marott JL, et al. Inflammatory biomarkers and exacerbations in chronic obstructive pulmonary disease. JAMA 2013; 309: 2353–2361. doi: 10.1001/jama.2013.5732 [DOI] [PubMed] [Google Scholar]
- 11.Barker BL, Haldar K, Patel H, et al. Association between pathogens detected using quantitative polymerase chain reaction with airway inflammation in COPD at stable state and exacerbations. Chest 2014; 147: 46–55. doi: 10.1378/chest.14-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill AT, Campbell EJ, Hill SL, et al. Association between airway bacterial load and markers of airway inflammation in patients with stable chronic bronchitis. Am J Med 2000; 109: 288–295. doi: 10.1016/S0002-9343(00)00507-6 [DOI] [PubMed] [Google Scholar]
- 13.Marin A, Garcia-Aymerich J, Sauleda J, et al. Effect of bronchial colonisation on airway and systemic inflammation in stable COPD. COPD 2012; 9: 121–130. doi: 10.3109/15412555.2011.636407 [DOI] [PubMed] [Google Scholar]
- 14.Patel IS, Seemungal TA, Wilks M, et al. Relationship between bacterial colonisation and the frequency, character, and severity of COPD exacerbations. Thorax 2002; 57: 759–764. doi: 10.1136/thorax.57.9.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stockley RA, Hill AT, Hill SL, et al. Bronchial inflammation: its relationship to colonizing microbial load and α1-antitrypsin deficiency. Chest 2000; 117: 291S–293S. doi: 10.1378/chest.117.5_suppl_1.291S [DOI] [PubMed] [Google Scholar]
- 16.Singh R, Mackay AJ, Patel AR, et al. Inflammatory thresholds and the species-specific effects of colonising bacteria in stable chronic obstructive pulmonary disease. Respir Res 2014; 15: 114. doi: 10.1186/s12931-014-0114-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramsheh MY, Haldar K, Esteve-Codina A, et al. Lung microbiome composition and bronchial epithelial gene expression in patients with COPD versus healthy individuals: a bacterial 16S rRNA gene sequencing and host transcriptomic analysis. Lancet Microbe 2021; 2: e300–e310. doi: 10.1016/S2666-5247(21)00035-5 [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Locantore N, Haldar K, et al. Inflammatory endotype-associated airway microbiome in chronic obstructive pulmonary disease clinical stability and exacerbations: a multicohort longitudinal analysis. Am J Respir Crit Care Med 2021; 203: 1488–1502. doi: 10.1164/rccm.202009-3448OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tufvesson E, Bjermer L, Ekberg M. Patients with chronic obstructive pulmonary disease and chronically colonized with Haemophilus influenzae during stable disease phase have increased airway inflammation. Int J Chron Obstruct Pulmon Dis 2015; 10: 881–889. doi: 10.2147/COPD.S78748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pragman AA, Knutson KA, Gould TJ, et al. Chronic obstructive pulmonary disease upper airway microbiota alpha diversity is associated with exacerbation phenotype: a case–control observational study. Respir Res 2019; 20: 114. doi: 10.1186/s12931-019-1080-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Singh R, Miller BE, et al. Sputum microbiome temporal variability and dysbiosis in chronic obstructive pulmonary disease exacerbations: an analysis of the COPDMAP study. Thorax 2017; 73: 331–338. doi: 10.1136/thoraxjnl-2017-210741 [DOI] [PubMed] [Google Scholar]
- 22.Mayhew D, Devos N, Lambert C, et al. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax 2018; 73: 422–430. doi: 10.1136/thoraxjnl-2017-210408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, Wang B, Tan M, et al. Analysis of sputum microbial metagenome in COPD based on exacerbation frequency and lung function: a case control study. Respir Res 2022; 23: 321. doi: 10.1186/s12931-022-02246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dicker AJ, Huang JTJ, Lonergan M, et al. The sputum microbiome, airway inflammation, and mortality in chronic obstructive pulmonary disease. J Allergy Clin Immunol 2021; 147: 158–167. doi: 10.1016/j.jaci.2020.02.040 [DOI] [PubMed] [Google Scholar]
- 25.Yang CY, Li SW, Chin CY, et al. Association of exacerbation phenotype with the sputum microbiome in chronic obstructive pulmonary disease patients during the clinically stable state. J Transl Med 2021; 19: 121. doi: 10.1186/s12967-021-02788-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su L, Qiao Y, Luo J, et al. Exome and sputum microbiota as predictive markers of frequent exacerbations in chronic obstructive pulmonary disease. Biomolecules 2022; 12: 1481. doi: 10.3390/biom12101481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agustí A, Celli BR, Criner GJ, et al. Global Initiative for Chronic Obstructive Lung Disease 2023 report: GOLD executive summary. Eur Respir J 2023; 61: 2300239. doi: 10.1183/13993003.00239-2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millares L, Pascual S, Montón C, et al. Relationship between the respiratory microbiome and the severity of airflow limitation, history of exacerbations and circulating eosinophils in COPD patients. BMC Pulm Med 2019; 19: 112. doi: 10.1186/s12890-019-0867-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Maschera B, Lea S, et al. Airway host–microbiome interactions in chronic obstructive pulmonary disease. Respir Res 2019; 20: 113. doi: 10.1186/s12931-019-1085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q, Pu Y, Lu H, et al. Porphyromonas, Treponema, and Mogibacterium promote IL8/IFNγ/TNFα-based pro-inflammation in patients with medication-related osteonecrosis of the jaw. J Oral Microbiol 2020; 13: 1851112. doi: 10.1080/20002297.2020.1851112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang YJ, Sethi S, Murphy T, et al. Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J Clin Microbiol 2014; 52: 2813–2823. doi: 10.1128/JCM.00035-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saint-Criq V, Lugo-Villarino G, Thomas M. Dysbiosis, malnutrition and enhanced gut–lung axis contribute to age-related respiratory diseases. Ageing Res Rev 2021; 66: 101235. doi: 10.1016/j.arr.2020.101235 [DOI] [PubMed] [Google Scholar]
- 33.Morris A, Beck JM, Schloss PD, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med 2013; 187: 1067–1075. doi: 10.1164/rccm.201210-1913OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeiffer S, Herzmann C, Gaede KI, et al. Different responses of the oral, nasal and lung microbiomes to cigarette smoke. Thorax 2022; 77: 191–195. doi: 10.1136/thoraxjnl-2020-216153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ying KL, Brasky TM, Freudenheim JL, et al. Saliva and lung microbiome associations with electronic cigarette use and smoking. Cancer Prev Res 2022; 15: 435–446. doi: 10.1158/1940-6207.CAPR-21-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camelo-Castillo AJ, Mira A, Pico A, et al. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front Microbiol 2015; 6: 119. doi: 10.3389/fmicb.2015.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsay JJ, Wu BG, Badri MH, et al. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am J Respir Crit Care Med 2018; 198: 1188–1198. doi: 10.1164/rccm.201710-2118OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnell A, Littman DR, Kuchroo VK. TH17 cell heterogeneity and its role in tissue inflammation. Nat Immunol 2023; 24: 19–29. doi: 10.1038/s41590-022-01387-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hacievliyagil SS, Gunen H, Mutlu LC, et al. Association between cytokines in induced sputum and severity of chronic obstructive pulmonary disease. Respir Med 2006; 100: 846–854. doi: 10.1016/j.rmed.2005.08.022 [DOI] [PubMed] [Google Scholar]
- 40.Wedzicha JA, Seemungal TA, MacCallum PK, et al. Acute exacerbations of chronic obstructive pulmonary disease are accompanied by elevations of plasma fibrinogen and serum IL-6 levels. Thromb Haemost 2000; 84: 210–215. doi: 10.1055/s-0037-1613998 [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson TM, Hurst JR, Perera WR, et al. Effect of interactions between lower airway bacterial and rhinoviral infection in exacerbations of COPD. Chest 2006; 129: 317–324. doi: 10.1378/chest.129.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aaron SD, Angel JB, Lunau M, et al. Granulocyte inflammatory markers and airway infection during acute exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2001; 163: 349–355. doi: 10.1164/ajrccm.163.2.2003122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00595-2023.SUPPLEMENT (322.8KB, pdf)
Figure E1 00595-2023.supplement_fig_E1 (224.8KB, jpg)
Figure E2 00595-2023.supplement_fig_E2 (277.7KB, jpg)
Figure E3 00595-2023.supplement_fig_E3 (244.7KB, jpg)
Figure E4 00595-2023.supplement_fig_E4 (314.1KB, jpg)
Figure E5 00595-2023.supplement_fig_E5 (298.7KB, jpg)
Table E3 00595-2023.TABLEE3 (74.4KB, pdf)