Abstract
Our previous work identified a cosmid clone containing a 43-kb insert from Enterococcus faecalis OG1RF that produced a nonprotein antigen in Escherichia coli. In the present work, we studied this clone in detail. Periodate treatment of lysates of the clone confirmed that the antigen was carbohydrate in nature. Analysis of DNA sequences and transposon insertion mutants suggested that the insert contained a multicistronic gene cluster. Database comparison showed that the cluster contained genes similar to genes involved in the biosynthesis of dTDP-rhamnose, glycosyltransferases, and ABC transporters involved in the export of sugar polymers from both gram-positive and gram-negative organisms. Insertions in several genes within the cluster abolished the immunoreactivity of the clone. This is the first report on a gene cluster of E. faecalis involved in the biosynthesis of an antigenic polysaccharide.
Many studies and reviews have discussed the important roles of polysaccharides in bacterial physiology and pathogenesis (2, 15, 46). As components of bacterial cell walls, capsules, lipopolysaccharides of gram-negative organisms, and accessory polymers of gram-positive cell walls, they not only offer structural rigidity for maintaining the shape of cells but also affect surface permeability, charge, and hydrophobicity and consequently affect the way bacteria interact with the environment. One important aspect of polysaccharides relates to their immunological and biological properties in tissues of the host, such as inhibition of phagocytosis and stimulation of cytokine production (17). These properties are critical in determining the outcome of the intricate interactions between bacteria and the host immune system. Many polysaccharides are also the immunodeterminants of serotype-specific antigens, including the O-antigen side chains of lipopolysaccharide and group and type antigens of some streptococci. Thus, it is clear that knowledge of cell surface architecture is important in understanding bacterial behavior inside the host, as well as in designing new therapeutics and serodiagnostic tools.
Biosynthesis of polysaccharides generally involves synthesis of sugar precursors in the cytoplasm, formation and polymerization of repeating units, and export to the cell surface. The formation of repeating units usually is initiated by the transfer of a sugar precursor onto a lipid carrier, such as undecaprenol-phosphate, to which subsequent sugar residues can be added. The sugar residue attached to the lipid carrier is often the first sugar in the repeating units (1, 44, 56), although there are exceptions as in Escherichia coli O8 and O9 serotype-specific O antigens in which the sugar residues attached to the lipid carrier do not appear in the final polysaccharides (41). Genes for the biosynthesis of polysaccharides are generally arranged in clusters of one or several transcriptional units (13, 32, 44, 56).
Enterococci (formerly group D streptococci) are among the leading causes of nosocomial infections in the United States, with the majority of clinical isolates being classified as Enterococcus faecalis (34). Besides their low-level intrinsic resistance to many antibiotics, enterococci have also developed high-level resistance to other antibiotics, including vancomycin and β-lactams (34). Very little information is available on cell surface-associated polysaccharides of enterococci. Early studies on the type-specific antigens in enterococci indicated that they were polysaccharides in the cell walls (6, 19, 45, 47). Type 1 antigen seems to be the most frequently identified type among E. faecalis isolates, at least in some studies (45, 47). Crude cell wall extracts from E. faecalis type 1 strains were found to contain rhamnose, glucose, galactose, N-acetylglucosamine (GlcNAc), and N-acetylgalactosamine (6). Pazur et al. demonstrated that enzymatic activities to synthesize thymidine diphosphate glucose (TDP-Glc) and to convert TDP-Glc to thymidine diphosphate rhamnose (TDP-Rha) were present in the cell extracts of E. faecalis N (40). TDP-Glc and TDP-Rha are the precursors from which glucose and rhamnose can be transferred to synthesize polysaccharides. An immunogenic glycan was also found in the cell walls of E. faecalis N (38, 39). The glycan was shown to be a diheteroglycan of glucose and galactose, but no rhamnose was present. No genetic information on polysaccharide biosynthesis in enterococci is available.
In a previous study, we reported a cosmid clone carrying a DNA fragment from E. faecalis OG1RF that produced a nonprotein antigen in E. coli (57). We hypothesized that the antigen could be a polysaccharide of OG1RF. To test this hypothesis, we have carried out further studies including DNA sequence analysis and characterization of transposon mutants. The results showed that the DNA fragment contains a gene cluster with homology to genes important in the assembly and export of polysaccharide from various organisms. Transposon insertions in three open reading frames (ORFs) within the cluster caused the loss of antigen expression in E. coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Strains and plasmids used in this study are listed in Table 1. E. coli cells were grown in Luria-Bertani (LB) broth or on LB agar with appropriate antibiotics overnight at 37°C. Enterococci were grown in brain heart infusion (BHI) broth or on BHI agar (Difco) overnight at 37°C for routine purposes. Antibiotics were used at the following concentrations: chloramphenicol at 25 μg/ml, kanamycin at 50 μg/ml, and ampicillin at 50 μg/ml. Isopropylthio-β-d-galactoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-galactoside (X-Gal) were used at 0.5 mM and 80 μg/ml, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain, plasmid, or cosmid | Characteristics | Reference and/ or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− 80dlacZΔ(lacZYA-argF)U169 endA1 recA1 hsdR17 (rK− mK+) deoR thi-1 supE44 λ− gyrA96 relA1 | |
| Sφ874 | F−trp lacZ strA thi upp ΔsbcB-rfb | C. Whitfield (36) |
| AB1133 | K-12; thr-1 leuB6(Δgpt-proA)66 hisG4 argE3 thi-1 rfbD1 lacY ara-14 galK2 xyl-5 mtl-1 mgl-51 rpsL31 kdgK51 supE44 | P. Rick (42) |
| 21548 | AB1133; rfe::Tn10 Tcr | P. Rick (42) |
| LO6E6II | E. coli library clone of OG1RF | 57 |
| TX5159 | DH5α(pTEX5159) | Formerly called BO4B6I (57) |
| TX5169 | Sφ874(pTEX5159) | This study |
| E. faecalis | ||
| OG1RF | 35 | |
| TX52 | Clinical isolate from a patient with endocarditis | 57 |
| Plasmids and cosmids | ||
| pBeloBAC11 | Cosmid vector, F′ replicon, Cmr | H. Shizuya (57) |
| pBluescript SK (−) | High-copy-number cloning vector derived from pUC19, Apr | Stratagene |
| pTEX5159 | pBeloBAC11 with a 43-kb insert from OG1RF which contains a gene cluster for polysaccharide biosynthesis | 57 |
| pTEX5159A-U | pTEX5159::Tn7 Cmr Kmr | This study |
| pTEX5175 | 3,195-bp EcoRI-HindIII fragment of pTEX5159 subcloned in pBluescript SK (−), Apr | This study |
Patient sera were collected from patients with E. faecalis endocarditis infections and had high titers against E. faecalis strains (57).
In vitro transposition.
Cosmid DNA from clone TX5159 (formerly BO4B6I) (57) was prepared by the alkaline-sodium dodecyl sulfate (SDS) method and purified in a CsCl-ethidium bromide gradient as described by Sambrook et al. (43). In vitro tranposition using a Tn7-based system (3, 4, 48) was performed by Matthew C. Biery and Nancy L. Craig, Department of Molecular Biology and Genetics, Howard Hughes Medical Institute, Johns Hopkins University.
DNA manipulations and transformation of E. coli.
DNA preparation, purification, restriction digestion, agarose gel electrophoresis, and ligation were performed by using standard methods (43) unless otherwise stated. Routine preparation of E. coli competent cells and transformation of DNA into E. coli were performed by the one-step procedure (9). Transformation of cosmid DNA that had been subjected to in vitro transposition was carried out by using Subcloning Efficiency DH5α competent cells (GIBCO BRL, Life Technologies, Gaithersburg, Md.); transformants were randomly picked and stored in 96-well microtiter dishes in LB broth containing 15% glycerol.
Proteinase K, periodate treatment, SDS-polyacrylamide gel electrophoresis (PAGE), and Western blot analysis.
Proteinase K treatment of the cell lysates of E. coli clones was based on a method described previously (57), with slight modifications. Briefly, 20 ml of an overnight culture of E. coli clones was centrifuged and resuspended in 700 μl of phosphate-buffer saline. The suspension was split into two aliquots; 50 μl of proteinase K (50 mg/ml); Fisher Scientific, Fair Lawn, N.J.) was added to the first aliquot, and 4 μl of 100 mM phenylmethylsulfonyl fluoride and 50 μl of H2O were added to the second. The aliquots were incubated at 50°C overnight and stored at −20°C. Treatment by periodate followed part of the procedure for the ECL Glycoprotein Detection Module (Amersham Life Science, Little Chalfont, Engalnd). Twenty-five microliters of each of the above aliquots (proteinase K treated and untreated) were mixed with 25 μl of 200 mM acetate buffer (pH 5.5.), then 25 μl of 30 mM sodium metaperiodate (freshly prepared) was added, and the mixture was incubated at room temperature for 1 h in the dark. The reaction was stopped by addition of 25 μl of 20 mM sodium metabisulfite and stored at −20°C in the dark. For controls, 25 μl of 200 mM acetate buffer instead of 30 mM sodium metaperiodate was used.
For SDS-PAGE and Western blot analysis, twofold serial dilutions of the samples were made in PBS phosphate-buffered saline; 15 μl from each dilution was mixed with 15 μl of 2× loading buffer (25 mM Tris Tris-Cl [pH 6.8], 9% [vol/vol] glycerol, 2.5% [vol/vol] β-mercaptoethanol, 0.0025% bromophenol blue, 4% SDS), boiled for 5 min, and loaded onto the gel. The conditions for SDS-PAGE and Western blotting were based on the method described previously (57).
Immunoblot analysis of bacterial colonies.
Colonies were inoculated onto NitroBind nitrocellulose transfer membranes (Micron Separations, Inc., Westborough, Mass.) placed on LB agar plates containing proper antibiotics and were then incubated at 37°C overnight. The membranes were lifted, placed in a chloroform chamber for 15 min, and incubated in lysis buffer. Subsequent steps were as described previously (57).
PCR amplification from the ends of the transposon and purification of PCR products.
NLC94 and NLC272 are primers that allow DNA synthesis to proceed outward from the ends of the Tn7 transposon. Primers GW248, GW250, GW251, GW254, GW273, GW280, and GW283 are primers hybridizing to regions in the insert of the cosmid. The sequence and location of each primer are summarized in Table 2 (see also Fig. 3).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) | Position (5′-3′) |
|---|---|---|
| GW248 | GAAGACATTGGTTATTTGAC | 8342–8323 in contig C, complement strand |
| GW250 | TCCTACATGCTAAAACGGTC | 3152–3171 in contig C |
| GW251 | AAAATCCACAATAAAGTTAG | 2989–2970 in contig C, complement strand |
| GW254 | TGGGATGGGAAATCGTTCTG | 17646–17665 in contig C |
| GW273 | ATAAAATGAAATGTTCCACCC | 501–482 bp from the end of contig B (proximal to contig C) |
| GW278 | GGAAACGGTGGGAGTATCAG | 16324–16343 in contig C |
| GW280 | CTTTAGCAGCATTTTGGAGT | 1882–1901 in contig C |
| GW283 | TTTGTTTGGTTCCATCTGTC | 9916–9897 in contig C, complement strand |
| NLC94 | AAAGTCCAGTATGCTTTTTCACAGCTAAC | End of Tn7 |
| NLC272 | ATTTTCGTATTAGCTTACGACGCTACACCC | End of Tn7 |
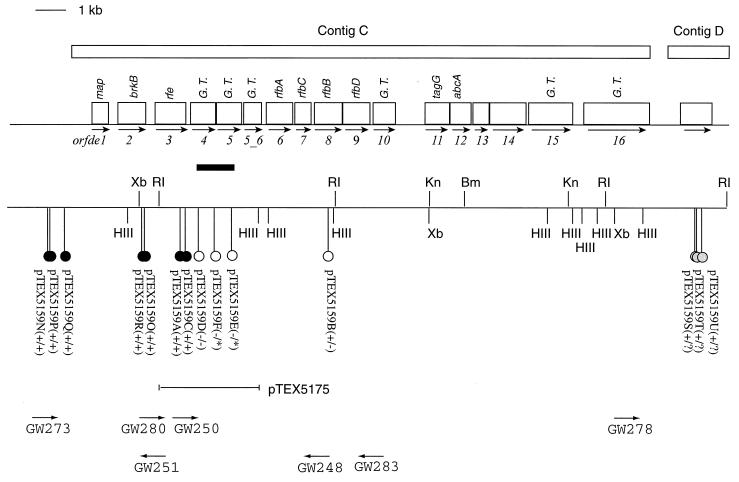
FIG. 3.
The polysaccharide biosynthesis gene cluster and flanking regions. The ORFs are shown as open boxes, the direction of the arrows underneath the boxes indicates the direction of transcription, and the best-gapped BLASTx hits of some of the ORFs are indicated above the boxes. The six ORFs that showed some similarities to glycosyltransferases are labeled G. T. Tn7 transposon insertions are shown as lollipops. The name of each cosmid carrying the corresponding insertion is given underneath followed by pluses and/or minuses in parentheses to indicate the immunoreactivity. The sign before the slash indicates the immunoreactivity in DH5α, and the sign after the slash indicates the immunoreactivity in Sφ874. Open-circle lollipops represent transposon insertions that rendered the clones immunonegative, filled-circle lollipops represent those that did not affect the immunoreactivity of the clones, and shaded-circle lollipops in contig D are those that had undergone DNA rearrangements in Sφ874. The black bar below orfde4 and orfde5 indicates the region where the initial 10 immunonegative clones were mapped. Three of these 10 clones, pTEX5159D to pTEX5159F, are shown as open-circle lollipops below the black bar. Primers used in characterizing transposon insertions are shown at the bottom, with the direction of arrows indicating the direction of DNA synthesis. Primers are not drawn in scale. Bm, BamHI; HIII, HindIII; Kn, KpnI; RI, EcoRI; Xb, XbaI.
Miniprep DNA from cosmid clones with transposon insertions was used as templates. The GeneAmp XL PCR kit (Perkin-Elmer, Branchburg, N.J.) was used to amplify DNA by using combinations of one of the NLC primers and one of the GW primers as instructed by the manufacturer. The reactions were performed in a model 9600 GeneAmp PCR system (Perkin-Elmer) with an initial denaturation at 96°C for 1 min, a regular three-step cycling reaction with 94°C denaturation for 20 s, 50°C annealing for 20 s, and 72°C extension for 7 min for 16 cycles, followed by a second three-step cycling reaction using the same temperatures as in the previous one but with an increase in extension time at 72°C for 15 s per cycle for 16 cycles, and finally an incubation at 72°C for 10 min.
Agarose gel electrophoresis and ethidium bromide staining were performed to examine the PCR products. The desired products were excised from the gels and purified by using a QIAquick gel extraction kit (Qiagen Inc., Santa Clarita, Calif.). The DNA concentration of each purified product was measured in a model 7625 Microplate Fluorometer (Cambridge Technology).
Sublconing of restriction fragments into pBluescript SK (−).
Subcloning was based on the method described previously (57), with sight modifications. Cosmid DNA was digested with EcoRI, BamHI, or DraI and purified by phenol-chloroform extraction followed by ethanol precipitation. Vector pBluescript SK (−) DNA was digested with EcoRI, BamHI, or EcoRV (to ligate with DraI) and dephosphorylated with shrimp alkaline phosphatase (United States Biochemical, Cleveland, Ohio). After ligation, the mixture was transformed into E. coli DH5α and plated on LB plates with ampicillin, IPTG, and X-Gal. White colonies were picked and analyzed by restriction digestion of miniprep DNA.
DNA sequencing and sequence analysis.
Three types of templates were prepared for sequencing: DNA from pBluescript SK (−) clones prepared by using a Qiagen Plasmid Mini kit (Qiagen Inc., Chatsworth, Calif.); purified PCR products; and TX5159 cosmid DNA purified in a CsCl-ethidium bromide gradient. Primers hybridizing to the T3 and T7 regions in pBluescript SK (−) and to the ends of the Tn7 transposon, as well as primers hybridizing to the ends of the sequences from the previous rounds, were used as sequencing primers. Sequencing reactions were performed by the Taq Dye-deoxy Terminator method and a model 377 DNA sequencing system (Applied Biosystems, Foster City, Calif.). The GelAssemble program in the Genetics Computer Group (GCG; Madison, Wis.) software package was used to assemble the nucleotide sequences. Gapped BLASTx at the National Center for Biotechnology Information (NCBI) was used to search for homologous sequences in the database. ORF Finder at NCBI was used to identify potential ORFs in the sequences. The terminator program in the GCG package was used to search for terminator-like sequences. The ProtParam program at ExPASy was used to calculate the theoretical parameters including molecular weight, isoelectric point, and hydrophobicity of each deduced amino acid sequence. Hydropathy plots were obtained by using the ProtScale program at ExPASy according to the method of Kyte and Doolittle (24) with a window size of 9. The DAS (dense alignment surface method) (10) transmembrane segment prediction program was also used to predict transmembrane domains. Potential signal peptide cleavage sites were analyzed by using SignalP (37). Overall amino acid sequence comparison was done with the Bestfit program in the GCG package, using the default parameters. Potential ribosome-binding sites (RBSs) and promoter sequences were examined manually.
Elution of antibodies specific to the polysaccharide.
Cell lysates of TX5159 were treated with proteinase K, subjected to SDS-PAGE, transferred to nitrocellulose membranes, and incubated with one of the patient sera as described above. The membranes were then incubated with 10 ml of 100 mM glycine (pH 2.5) for 30 min at room temperature. After neutralization with 1 ml of 1 M Tris (pH 8.0), the solution was transferred to a clean tube and stored at −20°C until ready to use (16).
Nucleotide sequence accession number.
The nucleotide sequence reported here was submitted to GenBank under accession no. AF071085.
RESULTS
Periodate treatment of E. coli(pTEX5159) lysates.
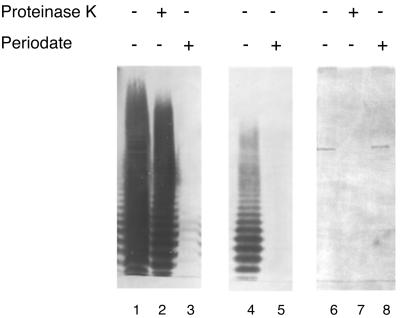
In our previous study, TX5159 (formerly BO4B6I) was found to react with four patient sera but not with a rabbit serum prepared against surface protein extracts from an E. faecalis strain isolated from a patient with endocarditis (57). The supernatant from an overnight culture of TX5159 also reacted with one of the patient sera in a dot blot (data not shown). The antigenic material in the TX5159 cell lysates was resistant to extensive proteinase K degradation, suggesting the material was not a protein. To further examine the nature of this antigen, the lysate was subjected to treatment with sodium metaperiodate, which preferentially oxidizes carbohydrates, and then subjected to Western blot analysis using one of the patient sera (Fig. 1). The immunostaining disappeared completely after periodate treatment, suggesting that the antigenic material is a carbohydrate. The material was produced in E. coli in sufficient amounts so that after a 1:32 dilution of the sample (equivalent to 3 to 4 μl of the overnight culture), there was still a strong reaction on the Western blot. The appearance of the diluted material in the Western blot resembled the ladder pattern seen with repeating units of polysaccharides; 10 bands could be clearly observed. A cosmid clone previously found to encode a protein antigen, the autolysin of E. faecalis (57), was used as a control. This antigen band disappeared after proteinase K treatment but remained intact after incubation with periodate.
FIG. 1.
Effects of proteinase K and periodate on cell lysates of TX5159. Cell lysates of an overnight culture were treated with either proteinase K or periodate. The samples were loaded for SDS-PAGE and transferred to a membrane after electrophoresis. The membrane was blotted with patient serum at 1:500 dilution in 1% skim milk. Lane 1, TX5159; lane 2, TX5159 plus proteinase K; lane 3, TX5159 plus periodate; lane 4, TX5159, diluted 32-fold; lane 5, diluted TX5159 lysates plus periodate; LO6E6II was (used as a control for the treatments); lane 6, LO6E6II lane 7, LO6E6II plus proteinase K; lane 8, LO6E6II plus periodate.
Cosmid pTEX5159 was transformed into E. coli Sφ874, which lacks the entire rfb region for O-antigen biosynthesis. The cell lysates of TX5159 was subjected to proteinase K treatment and Western blotting to examine whether any E. coli rfb genes were involved in generating the immunoreactive ladder pattern. The pattern of TX5169 was the same pattern as that of TX5159 (data not shown.
DNA sequencing of pTEX5159 and sequence analysis of contig C.
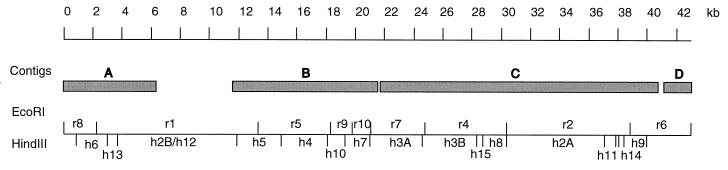
DNA sequencing resulted in four cotigs (A to D) along the insert of pTEX5159 (Fig. 2). The total number of bases sequenced is 94,434, and the total unique sequence is 34,170 bases long. The average coverage of the four contigs is 2.76×, and the coverage of contig C, is 3×, slightly higher than for the other contigs. About 17.5% of the total unique sequence are single pass, most at the ends of each contig. A restriction map of the 43-kb insert of TX5159 was generated by using the information from DNA sequencing and restriction enzyme digestion (RED) analysis of transposon insertion clones made by in vitro tranposition (described below) (Fig. 2). Based on the restriction map, gaps between contigs B and C and between contigs C and D should be less than 500 bp, while a gap of about 4.5 kb is present between contigs A and B. Database comparison of the sequences from the four contigs showed that contig C contains ORFs with similarity to genes involved in polysaccharide biosynthesis.
FIG. 2.
Restriction map of the insert in pTEX5159. The top line shows the scale. The shaded boxes represent contigs A to D. Below the boxes, the horizontal line represents the insert, the short vertical lines above the horizontal line represent the EcoRI sites, and the vertical lines below show the HindIII sites. The EcoRI fragments were designated r1 to r10, with r1 being the largest in size and r10 the smallest, while the HindIII fragments were designated h1 to h15. Fragment h2A had the same mobility in agarose gel electrophoresis as h2B; the same was true of h3A and h3B.
Contig C contains 18,817 bases with an overall G+C content of 36.7%. Fifteen ORFs (orfde1 to orfde15), all of which had the same transcriptional orientation (Table 3; Fig. 3), were identified by ORF Finder at NCBI. A partial coding region was found in the 3′ flanking region of orfde15 and was oriented for transcription in the same orientation as the others. This was designated orfde16. Manual examination of the region between orfde5 and orfde6 revealed that there could be an ORF of 585 bp in the same transcriptional orientation as the other ORFs in the contig, with TTG as the start codon. This was designated orfde5_6. There was a large (1,011-bp) region between orfde10 and orfde11. Numerous stop codons were found in this intergenic region, and database searches did not find any sequence similarities, suggesting this may be noncoding region.
TABLE 3.
Positions of ORFs in contig C and features of the predicted gene products
| ORF | Putative RBS and start sitea | Potential coding region
|
Predicted gene productb
|
||||
|---|---|---|---|---|---|---|---|
| Location in nucleo- tide sequence | % G+C | Length (amino acids) | MW (103) | pI | GRAVY | ||
| orfde1 | TTTAGGAGAGATAGTATGATT | 778–1314 | 40.6 | 178 | 19.8 | 5.10 | −1.292 |
| orfde2 | AATAAGGAGAGTGTGTGATGAAG | 1558–2466 | 35.9 | 302 | 34.0 | 9.82 | 6.762 |
| orfde3 | ACCAGGAGAGCGACGAATCAATACCAAAAATATGCCT | 2718–3710 | 36.3 | 330 | 36.3 | 9.85 | 9.497 |
| orfde4 | AGTAAGGAGAAATGAGCATGCAA | 3831–4670 | 34.2 | 279 | 32.3 | 5.65 | −2.620 |
| orfde5 | AATGGGGAAATTTTTTAACAAATGGGG | 4597–5445 | 37.2 | 282 | 32.7 | 5.63 | −1.770 |
| orfde5_6 | AGTGAAGAAGATGAACTAATTATTTCAGATGATGGTT CTACTGATCTACCTTGGAA | 5577–6161 | 39.0 | 194 | 22.2 | 9.78 | −1.789 |
| orfde6 | TTTAAGGAGATTGTTCATGAAA | 6283–7149 | 38.5 | 288 | 32.1 | 4.82 | −1.719 |
| orfde7 | AATAGGAGCGAGGAAACGTGAAA | 7162–7734 | 38.2 | 190 | 21.3 | 5.30 | −2.679 |
| orfde8 | AGTAGGAGAGTTTCAATTATGAAA | 7759–8787 | 38.3 | 342 | 38.4 | 4.99 | −5.047 |
| orfde9 | TGTGAGATAACAGTCGAAAGGCGATGGCA | 8823–9722 | 38.3 | 299 | 33.8 | 4.68 | −3.632 |
| orfde10 | AGTGGAAAGGTGTTTCTCTTTATGAAG | 9781–10506 | 37.7 | 211 | 26.9 | 6.23 | 0.357 |
| orfde11 | TACGGAAGGAATAAGTATATGTTT | 11518–12312 | 32.6 | 264 | 30.7 | 9.48 | 6.549 |
| orfde12 | TATAAGGAGAGAACGACATGTCA | 12325–13005 | 37.7 | 226 | 25.4 | 5.18 | −1.195 |
| orfde13 | TATGGAGATACGATGGAC | 13019–13543 | 36.0 | 174 | 20.2 | 4.89 | −3.937 |
| orfde14 | AATGGAAGGATACGACAAAAGATGAAG | 13533–14714 | 36.5 | 393 | 45.1 | 4.56 | −3.176 |
| orfde15 | TTTGGAAGGCGATCAGGTTTATGGTT | 14936–16543 | 39.1 | 536 | 61.0 | 5.39 | −4.153 |
| orfde16 | TATAAGGAGAATGCACATGAAT | 16700–18817 (3′ partial) | |||||
Putative RBSs are underlined, and start codons are in boldface.
Molecular weight (MW), isoelectric point (pI), and grand average hydropathicity (GRAVY) were calculated by using ProtParam at ExPASy. GRAVY is based on the Kyte-Doolittle formulation (24); the more negative the value is, the more hydrophilic the protein is.
Putative RBSs were found upstream of each coding region within 11 bp of the start codon except for orfde3 (21 bp) and orfde5_6 (39 bp) (Table 2). However, no obvious start sites were found 5′ to the ATG at nucleotide position 2718 of orfde3 and the TTG at 5577 of orfde5_6. Therefore, since the start sites for orfde3 and orfde5_6 were not certain, the coding region from nucleotide positions 2718 to 3710 identified by ORF Finder was used for orfde3 and the coding region from nucleotide positions 5577 to 6161 identified by manual examination was used for orfde5_6 in our analysis.
Putative promoter-like (−35 and −10 like) sequences were found upstream of orfde2, orfde3, orfde4, orfde5_6, orfde6, orfde11, and orfde16. However, since enterococcal DNA has high A+T content, the apparent promoter-like sequences may not be real promoters, and functional analysis such as primer extension or mutagenesis will be needed to verify the true promoter regions. The 3′ end of orfde4 overlapped the 5′ end of orfde5 for 74 bp; thus, these two ORFs could be transcriptionally and translationally coupled. The intergenic regions from orfde6 to orfde10 are 12, 24, 35, and 58 bp, respectively; these could form a transcriptional unit. The intergenic regions from orfde11 to orfde13 are 12 and 13 bp, respectively. The 3′ end of orfde13 overlapped the 5′ end of orfde14 for 11 bp; therefore, orfde11 to orfde14 could be a transcriptional unit. There is a 221-bp intergenic region between orfde14 and orfde15, but no promoter-like sequences were identified. Analysis of terminator-like sequences using the terminator program in the package GCG revealed many candidate sequences, including some within genes, and so the significant of these is not clear.
Database similarities of the genes in contig C and characteristics of the deduced gene products.
BLASTx was performed to search for sequence similarities in the databases (Table 4). Orfde1 showed strong similarity with a putative methionine aminopeptidase A (map) from Synechocystis sp. (21) and various other organisms. Orde2 showed some similarity to the serum resistance locus BrkB protein of Bordetella pertussis (11), which is required for the resistance of B. pertussis to complement-dependent killing by normal human serum and contains domains with homology to some transporters. Hydrophobicity analysis indicated that the orfde2 gene product is a hydrophobic protein with four to six potential transmembrane regions. A putative signal peptide cleavage site was found between amino acids 50 and 51 (LTA-VG).
TABLE 4.
Database homologies of the ORFs in contig C
| ORF | BLASTx hit | Accession no. | Organism | Expect value | Reference(s) |
|---|---|---|---|---|---|
| orfde1 | Methionine aminopeptidase A (map) | P53579 | Synechocystis sp. | 6e-59 | 21 |
| orfde2 | Serum resistance locus brkB | D64004 | Synechocystis sp. | 2e-07 | |
| Serum resistance locus brkB | I40328 | Bordetella pertussis | 2e-04 | 11 | |
| orfde3 | Putative undecaprenyl-phosphate N-acetylglucosaminyltransferase (tagO) | gn1|PID|e11 | Bacillus subtilis | 1e-57 | Unpublished data |
| 68896 | |||||
| Lipophilic protein which affects bacterial lysis rate and methicillin resistance level (llm) | D21131 | Staphylococcus aureus | 4e-57 | 29 | |
| Undecaprenyl-phosphate N-acetylglucosaminyltransferase (rfe) | S30678 | Escherichia coli | 4e-19 | 31 | |
| orfde4 | EpsX | X81320 | Acinetobacter calcoaceticus | 1e-25 | Unpublished data |
| Hypothetical 31.1-kDa protein in gnd-rfc intergenic region (orf264) | P36667 | E. coli | 4e-12 | 27, 59 | |
| orfde5 | RfbC protein | S28579 | Yersinia enterocolitica | 0.14 | |
| dTDP-rhamnosyltransferase (rfbF) | P37782 | Shigella flexneri | 0.14 | 33 | |
| orfde5_6 | Unnamed protein product | AB002668 | Actinobacillus actinomycetemcomitans | 5e-18 | 60 |
| MigA | U70729 | Pseudomonas aeruginosa | 5e-11 | 54 | |
| PssF | AF028810 | Rhizobium leguminosarum bv. viciae | 3e-09 | 18 | |
| orfde6 | Glucose-1-phosphate thymidyltransferase (cps19fL) | U09239 | Streptococcus pneumoniae | e-131 | 32 |
| orfde7 | dTDP-4-dehydrorhamnose 3,5-epimerase (rfbC) | P26394 | Salmonella enterica | 1e-44 | 20 |
| orfde8 | dTDP-glucose-4,6-dehydratase (rmlB) | D78182 | Streptococcus mutans | e-150 | 52 |
| orfde9 | 32.3-kDa cps19fO gene product | U09239 | S. pneumoniae | 2e-97 | 32 |
| dTDP-4-keto-l-rhamnose reductase (rmlD) | AB000631 | S. mutans | 1e-94 | 51 | |
| orfde10 | Unnamed protein product | AB002668 | A. actinomycetemcomitans | 1e-30 | 60 |
| ss-1,3-N-Acetylglucosaminyltransferase | X85787 | S. pneumoniae | 0.001 | 22, 23 | |
| orfde11 | Teichoic acid translocation permease protein TagG (tagG) | P42953 | B. subtilis | 2e-48 | 25 |
| O-antigen export system permease protein RfbA (rfbA) | Q50862 | Myxococcus xanthus | 3e-23 | 14 | |
| orfde12 | ATP-binding cassette membrane transporter AbcA (abcA) | Q07698 | Aeromonas salmonicida | 5e-58 | 8 |
| O-antigen export system ATP-binding protein RfbB (rfbB) | Q50803 | M. xanthus | 8e-56 | 14 | |
| Teichoic acid translocation ATP-binding protein TagH (tagH) | P42954 | B. subtilis | 1e-51 | 25 | |
| orfde13 | No good match | ||||
| orfde14 | No good match | ||||
| orfde15 | Hypothetical 102.8-kDa protein Y4gI | P55465 | Rhizobium sp. strain NGR 234 | 4e-68 | 12 |
| O-antigen biosynthesis protein RfbC (rfbC) | U36795 | M. xanthus | 5e-52 | 14 | |
| EpsI | U40830 | Streptococcus thermophilus | 6e-14 | 50 | |
| EspG | U93364 | Lactococcus lactis cremoris | 2e-12 | 53 | |
| β-1,4-Galactosyltransferase | X85787 | S. pneumoniae | 1e-11 | 22, 23 | |
| orfde16 | Hypothetical 102.8-kDa protein Y4gI O-antigen biosynthesis protein RfbC (rfbC) | P55465 | Rhizobium sp. strain NGR234 | e-128 | 12 |
| Q50864 | M. xanthus | 6e-16 | 14 |
Orfde3 showed similarities to the undecaprenyl-phosphate α-N-acetylglucosaminyltransferase encoded by the rfe genes from various organisms, including E. coli and Bacillus subtilis, and a lipophilic protein which affects bacterial lysis rate and methicillin resistance level (llm) of Staphylococcus aureus (29). A potential signal peptide cleavage site was found between amino acids 26 and 27 of Orfde3 (IIP-QD). Hydropathy plot indicated that Orfde3 is highly hydrophobic with 10 to 11 potential transmembrane domains, similar to the E. coli Rfe protein (31). This E. coli enzyme catalyzes the transfer of GlcNAc onto undecaprenyl phosphate to synthesize GlcNAc-pyrophosphorylundecaprenol, the first lipid-linked intermediate in the biosynthesis of enterobacterial common antigens (31), and was shown to be required for the initial step in the formation of repeating units of several O antigens (1, 41).
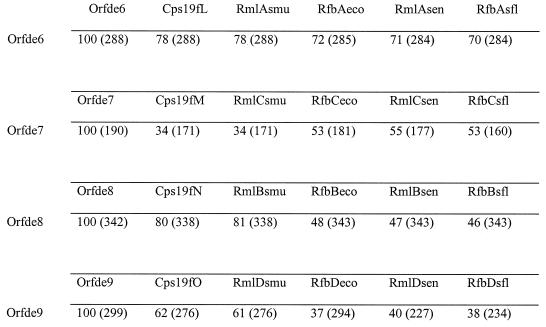
BLASTx results showed that Orfde6 to Orfde9 had extremely strong sequence homology with gene products glucose-1-phosphate thymidyltransferse, dTDP-4-dehydrorhamnose 3,5-epimerase, dTDP-glucose-4,6-dehydratase, and dTDP-4-keto-l-rhamnose reductase, respectively, of various bacteria (Table 4). These four enzymes catalyze the biosynthesis of dTDP-rhamnose from glucose-1-phosphate. The overall amino acid sequences of Orfde6 to Orfde9 were compared with the corresponding translated sequences from two gram-positive and three gram-negative organisms: cps19fLMNO from the Streptococcus pneumoniae type 19F capsular polysaccharide gene cluster (13, 32), rmlACB and rmlD of the Streptococcus mutans dTDP-rhamnose biosynthesis pathway (51, 52), rfbBDAC from the E. coli O7 rfb gene cluster (30), rmlBDAC from the Salmonella typhimurium LT2 rfb gene cluster (20), and rfbBDAC from the Shigella flexneri type 2a rhamnose biosynthesis operon (28) (Fig. 4). Orfde6 showed high similarities with the glucose-1-phosphate thymidyltransferase from all five organisms, being from 70 to 78% identical. Orfde8 showed 80 and 81% identity to the cps19fN and rmlC gene products from the two gram-positive organisms, while its identity to the gram-negative proteins was lower (46 to 48%) but at the same level as the identity between the streptococal and gram-negative proteins (47 to 50%). This was also observed with Orfde9, which showed higher identities to the two streptoccal proteins (62 and 61%) than to the gram-negative ones (37 to 40%). However, Orfde7 showed stronger identities to the three gram-negative proteins (53 to 55%) than to the gram-positive ones (both 34%). Orfde6 to Orfde9 are generally hydrophilic, and the hydropathy plots of Orfde6 to Orfde9 exhibited remarkable similarities to those of the corresponding sequences from these other organisms (data not shown).
FIG. 4.
Comparison of amino acid sequences of dTDP-rhamnose biosynthesis genes from various bacteria. Percentage identity was obtained by using Bestfit; each number in parentheses indicates the length of the segment that gave the percentage identity in amino acids. Default parameters were used for comparisons. Orfde6 to Orfde9 are from E. faecalis OG1RF. Cps19fL to -O, Streptococcus pneumoniae type 19F; smu, Streptococcus mutans; eco, Escherichia coli; sen, Salmonella enterica; sf1, Shigella flexneri. Cps19fL, Rm1Asmu, RfbAeco, Rm1Asen, and RfbAsf1, glucose-1-phosphate thymidyltransferases; Cps19fM, Rm1Csmu, RfbCeco, and RfbCsf1, dTDP-4-dehydrorhamnose 3,5-epimerases; Cps19fN, Rm1Bsmu, RfbBeco, RmlBsen, and RfbBsf1, dTDP-glucose-4,6-dehydratases; Cps19fO, Rm1Dsmu, RfbDeco, RmlDsen, and RfbDsf1, dTDP-4-keto-l-rhamnose reductases.
Orfde11 showed strong similarity with the integral membrane protein component of ATP-binding cassette (ABC) transport systems involved in sugar polymer export. The highest similarity was with the teichoic acid translocation permease TagG or B. subtilis (25). Hydrophobicity analysis showed that the orfde11 gene product is highly hydrophobic with five to six transmembrane domains, consistent with being an integral membrane protein. Orfde12 was similar to the ATP-binding protein component of several sugar polymer export systems, including the O-antigen export systems of Myxococcus xanthus (14) and Klebsiella pneumoniae (7). It contains the characteristic ABC consensus sequence of the ATP-binding proteins in the transport system. It seems likely that Orfde11 and Orfde12 comprise an ABC transport system for exporting the polysaccharide.
Orfde4, Orfde5, Orfde5_6, Orfde10, Orfde15, and Orfde16 (3′ partial) showed various degrees of similarity to glycosyltransferases. No signal cleavage site was identified in any of these sequences. Except for Orfde10, they were not predicted to be membrane proteins. Orfde10 was predicted to contain two transmembrane domains, one near the N terminus and the other near the C terminus. The two proteins that showed the highest similarity to Orfde4, the epsX gene product of Acinetobacter calcoaceticus and a hypothetical 31.1-kDa protein encoded by orf264 in the gnd-rfc intergenic region of E. coli (27, 59), have no clearly documented function. However, it has been shown that when orf264 was introduced into E. coli K-12 strains EMG2 and WE110, whose orf264 genes are defective, the two strains produced an O antigen. This led to the conclusion that orf264 was essential for the biosynthesis of O antigen in E. coli K-12 strains and was thought likely to be rhamnosyltransferase (27, 59). The third-best BLASTx expect value was with a glycosyltransferase encoded by spsQ of Sphingomonas strain S88 (58). The gene spsQ was identified in a gene cluster essential for the synthesis of a capsular polysaccharide in Sphingomonas strain S88 and is thought to be a glycosyltransferase, suggesting that orfde4 may also encode a glycosyltransferase. Orfde5 showed only very low similarity to the dTDP-rhamnosyltransferase (encoded by rfbF) of Shigella flexneri (33). Bestfit comparison of the amino acid sequences of RfbF and Orfde5 showed a 121-amino-acid stretch with 34% similarity and 24% identity. It has been noted that the sequence similarities among glycosyltransferases are generally low (26), and thus the homology between RfbF and Orfde5 may be significant. Both Orfde5_6 and Orfde10 were found to be similar to several putative glycosyltransferases and proteins of unknown function. Orfde15 and Orfde16 shared some sequence similarity between themselves and were homologous to a similar set of proteins, most of which were glycosyltransferases. The two best matches were the hypothetical 102.8-kDa protein Y4gI of Rhizobium sp. strain NGR234 (12) and the O-antigen biosynthesis protein RfbC of M. xanthus (14).
Orfde13 and Orfde14 showed no significant similarity to any sequences in the databases.
Sequence analysis of contig D.
Contig D has 2,943 bases. One ORF and 1,029 bp was identified in the middle of contig D and did not reveal any database similarities. The end of contig D was from vector pBeloBAC11. The rest of the sequence in the contig did not show any similarities to sequences in the database.
Immunoscreening of TX5159 with transposon insertions.
We screened 282 transposon insertion clones, generated by in vitro transposition of a Tn7 derivative, by using a patient serum and subjected immunonegative clones to a second screen. Ten clones, DH5α(pTEX5159D) to DH5α(pTEX5159M), were found to be immunonegative. Five of the 10 cones had single insertions in EcoRI fragment r4, while the other five had double insertions, with one in r4 and the other in either r1 or r5. Clones with single insertions in only r1 or r5 were still immunopositive, suggesting that the insertions in r4 were responsible for the loss of the immunoreactivity of these clones. Cosmid DNA from three clones, DH5α(pTEX5159D), DH5α(pTEX5159E), and DH5α(pTEX5159F), were transformed into Sφ874, and the transformants were tested by colony immunoblotting. Sφ874(pTEX5159D) was negative, while transformants with pTEX5159E and pTEX5159F were not able to regrow after the initial appearance on the primary transformation plates.
Characterization of transposon insertion mutants.
To determine the sites of the insertions, cosmid DNA from the 10 clones that were immunonegative was prepared and subjected to PCR amplification (Table 5). All insertions in EcoRI fragment r4 were localized to a small region, shown as a black bar in Fig. 3. PCR products amplified by using primer pairs GW248-NLC94 and GW250-NLC272 from three immunonegative clones, DH5α(pTEX5159D), DH5α(pTEX5159E), and DH5α(pTEX5159F), were gel purified and subjected to DNA sequencing. The sequences showed that the insertions were in orfde4 and orfde5, which encode potential glycosyltransferases (Fig. 3). The insertion in DH5α(pTEX5159D) was at nucleotide position 4191 in contig C, 361 bp downstream of the start site of orfde4 (equivalent to amino acid 119 from the N terminus of Orfde4); the insertion in DH5α(pTEX5159E) was at 5238, in the middle of orfde5 (equivalent to amino acid 214 from the N terminus of Orfde5); and the insertion in DH5α(pTEX5159F) was at 4701, 105 bp downstream of the start of orfde5 (equivalent to amino acid 35 from the N terminus of Orfde5). The sites of insertion in the other seven clones were estimated based on their PCR product sizes in relation to those from the three clones that had been sequenced. It appeared that all were in orfde4 or orfde5. The insertion in DH5α(pTEX5159L) could be either at the end of orfde4 or at the beginning of orfde5.
TABLE 5.
Characterization of transposon mutants
| Cosmid | Site of insertiona | Immuno- reactivityb
|
|
|---|---|---|---|
| DH5α | Sφ874 | ||
| pTEX5159G | orfde4 | − | |
| pTEX5159D | orfde4 | − | − |
| pTEX5159H | − | ||
| pTEX5159I | orfde5 | − | |
| pTEX5159E | orfde5 | − | DNGe |
| pTEX5159J | orfde5 | − | |
| pTEX5159K | orfde5 | − | |
| pTEX5159F | orfde5 | − | DNG |
| pTEX5159L | orfde4/5 | − | |
| pTEX5159M | − | ||
| pTEX5159A | orfde3 | + | + |
| pTEX5159B | orfde8 | + | − |
| pTEX5159C | orfde3-orfde4 | + | + |
| pTEX5159Nc | r7, 0.7 kb upstream of contig C | + | + |
| pTEX5159Oc | orfde2 | + | + |
| pTEX5159Qc | r7, 0.2 kb upstream of contig C | + | + |
| pTEX5159Pc | r7, 0.7 kb upstream of contig C | + | + |
| pTEX5159Rc | orfde2 | + | + |
| pTEX5159Sc | r6, 1.8 kb downstream of contig C | + | −?d |
| pTEX5159Tc | r6, 1.8 kb downstream of contig C | + | −d |
| pTEX5159Uc | r6, 1.9 kb downstream of contig C | + | −d |
ORF where the transposon had inserted. Sites of insertion of those that were not sequenced were estimated based on their PCR product sizes in relation to the ones that were sequenced.
Tested by using colonies of each clone in immunoblot analyses with patient serum.
Not been subjected to PCR amplification. The insertion site was determined by RED analysis.
The clone was transformed into Sφ874, resulted in large deletions in the insert, and was immunonegative.
Sφ874 transformants of pTEX5159E and pTEX5159F did not grow (DNG) when streaked from the primary transformation plates.
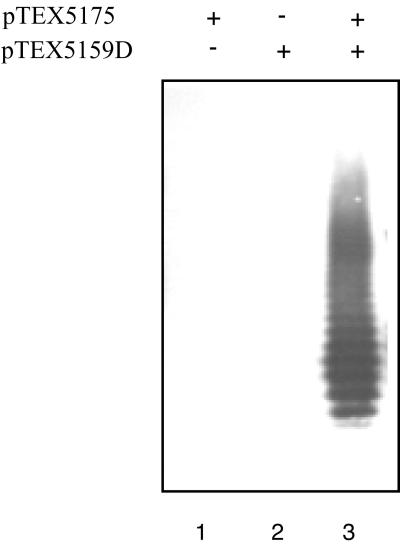
To prove the orfde4-5 region was responsible for the mutant phenotype, a complementation test was performed. An EcoRI-HindIII fragment which contains the 3′ half of orfde3, all of orfde4 and orfde5, and almost all of orfde5_6, was cloned in pBluescript SK (−), and the resulting construct was designated pTEX5175. The plasmid was transformed into the immunonegative insertion mutant DH5α(pTEX5159D) as well as into DH5α. Cell lysates of the transformants were treated with proteinase K and subjected to Western blot analysis. Plasmid pTEX5175 restored the immunoreactivity of DH5α(pTEX5159D), while DH5α(pTEX5175) was immunonegative (Fig. 5).
FIG. 5.
Complementation of DH5α(pTEX5159D) with pTEX5175. Samples were treated with proteinase K and analyzed as described in the text. Lane 1, DH5α(pTEX5175); lane 2, DH5α(pTEX5159D); lane 3, DH5α(pTEX5159D + pTEX5175).
Clones with insertions in r7 or r6, at the ends of contig C, were found through RED analysis of random clones (Table 5). Five clones were found to have insertions in r7; PCR results showed that insertion in pTEX5159N, pTEX5159Q, and pTEX5159P were upstream of contig C, while insertions in pTEX5159O and pTEX5159R were in orfde2. Three clones, pTEX5159S, pTEX5159T, and pTEX5159U, had insertions 1.8 to 1.9 kb downstream of contig C. These clones were all immunopositive in colony blots in DH5α. When transformed into Sφ874, the five clones with insertions in r7 remained positive. However, Sφ874 transformants of pTEX5159S, pTEX5159T, and pTEX5159U seemed to have undergone deletions or rearrangements in Sφ874, based on RED analysis, and were immunonegative.
Complementation by E. coli O-antigen biosynthesis genes.
After RED analysis of random clones, three clones, DH5α(pTEX5159A), DH5α(pTEX5159B), and DH5α(pTEX5159C) were found to have insertions in r4 but still reacted in colony immunoblots. PCR products amplified by using primer pairs GW283-NLC94 and GW280-NC272 from these three clones were subjected to DNA sequencing (Table 5). The insertion in DH5α(pTEX5159C) was at nucleotide position 3728 in contig C, which is in the region between orfde3 and orfde4 (Fig. 3). This finding suggested that orfde4 was transcribed independently.
The insertion in DH5α(pTEX5159B) was at nucleotide position 8388 in the middle of orfde8 (equivalent to amino acid 210 from the N terminus of Orfde8), which is highly similar to the gene for dTDP-glucose-4,6-dehydratase, the second enzyme in the dTDP-rhamnose biosynthesis pathway of various bacteria. Rhamnose is a major component in the O antigens of gram-negative bacteria and in capsules of some gram-positive bacteria. Thus, it was surprising to find that DH5α(pTEX5159B) was still immunopositive. Since most E. coli K-12 strains should carry genes for dTDP-rhamnose synthesis (27, 59), however, it was possible that DH5α(pTEX5159B) was complemented by the E. coli genes. To test this, cosmid DNA from DH5α(pTEX5159B) was transformed into E. coli Sφ874, which lacks the entire rfb region. The transformant was found to be immunonegative in colony blots (Table 5), indicating that complementation of the insertion mutation occurred in DH5α but not in Sφ874.
DH5α(pTEX5159A) had an insertion at nucleotide position 3525, which is 187 bp upstream of the 3′ end of orfde3 (equivalent to 61 amino acids from the C terminus of Orfde3). Orfde3 is similar to the undecaprenyl-phosphate α-N-acetylglucosaminyltransferase that catalyzes the transfer of GlcNAc to the lipid carrier, the first step in the synthesis of the repeating units of several O antigens (1, 41). Since the E. coli rfe gene is outside the rfb region, both DH5α and Sφ874 possess this gene. To test the possibility that this clone was being complemented by the rfe gene of E. coli, cosmids pTEX5159A and pTEX5159 were transformed into an E. coli rfe strain, 21548, as well as into the parent strain AB1133 (rfe+). The cell lysates of the transformants were subjected to proteinase to proteinase K treatment and Western blot analysis as described above. All four clones showed the characteristic ladder pattern as with TX5159 (data not shown).
Detection of the expression of this polysaccharide in E. faecalis.
Antibody was eluted from nitrocellulose membranes following Western blotting of proteinase K-treated extracts from TX5159 with a patient serum. The eluted antibody reacted with TX5159 but not with the cloning host DH5α. The eluted antibody was used to examine E. faecalis OG1RF and TX52 (57) under different growth conditions, including exposure to horse and rabbit sera, at different temperatures, and in minimal growth media. Several clinical from patients with E. faecalis endocarditis were also examined after growth in BHI broth. These clinical strains include isolates from patients from whom the tested sera were collected. No positive reaction was detected (data not shown).
DISCUSSION
In a previous study, we described a cosmid clone of E. faecalis (TX5159) that produced proteinase K-resistant antigenic material in E. coli (7). In this report, we provide evidence that the antigenic material was carbohydrate in nature and that its production did not involve the E. coli rfb genes. Furthermore, the antigen appears as a ladder after PAGE, suggesting a structure of repeating units, such as those of polysaccharides. Since the clone contains a number of genes with sequence similarity to polysaccharide biosynthesis functions, we conclude that genes for the biosynthesis of an enterococcal antigenic polysaccharide had been cloned and expressed in E. coli.
Early studies indicated that antigenic polysaccharides could be detected in the cell walls of enterococci and, moreover, that many of these were rhamnose-containing polysaccharides (6, 19, 45, 47). However, no studies to our knowledge suggested the presence of capsules in enterococci. In our previous report, TX5159 was found to react with four patient sera but not with a rabbit serum prepared against surface protein extracts from an E. faecalis strain grown in BHI (57). The rabbit serum was made from rabbits immunized with zwittergent surface extracts of E. faecalis, and it is possible that the polysaccharide was not present. In this study, we failed to detect the antigen in E. faecalis OG1RF and TX52 (57) under different growth conditions or in several E. faecalis clinical isolates. This raised the possibility that the antigen is due to E. coli and not enterococcus. PCR amplification of OG1RF genomic DNA by using intragenic primers to orfde2 to orfde16 resulted in PCR products of the expected sizes (data not shown). Southern blot analysis using pTEX5159 DNA as a probe against genomic DNA of OG1RF, TX52, and DH5α indicated that the probe hybridized to OG1RF and TX52 but not to DH5α (data not shown), suggesting that they are enterococcal genes. Therefore, in this model, the patients would also be making antibody against the E. coli polysaccharide, and the enterococcal genes would be complementing E. coli genes that were defective in the laboratory strains. We note that in our screens for immunonegative transposon mutants in DH5α, where all the mutants carried insertions in orfde4 and orfde5, orfde4 shows some sequence similarity to orf264 in the gnd-rfc intergenic region of E. coli (27, 59). In most of the E. coli K-12 strains, there is an IS5 insertion in orfde264, which is responsible for the loss of O-antigen production in these K-12 strains. This finding raised the possibility that orfde4 complemented the defective orf264 of DH5α and thus restored its ability to make an O antigen. This possibility can be ruled out because of the following observations. Firstly, unlike DH5α, which still maintains most of the rfb genes, Sφ874 lacks the entire rfb region for O-antigen biosynthesis. However, TX5169 had the same reaction pattern with the patient serum as TX5159. Second, plasmid pTEX5175, which contains the complete sequence of orfde4, orfde5, and their flanking regions, alone did not make DH5α immunoreactive. Thus, the production of the antigenic polysaccharide required the presence of pTEX5159 but not the E. coli rfb genes.
The four patient sera used were collected from endocarditis patients infected with E. faecalis in different regions in the United States. The fact that TX5159 reacted strongly with all four sera suggested that the polysaccharide was produced in all of these patients and was not due to one particular strain or one particular patient. It has long been known that the biosynthesis of many polysaccharides (e.g., E. coli group IA and group II K antigens, and colanic acid [56]) is regulated, and many regulatory strategies have been used to achieve control of expression in response to different changes in the environment. Thus, expression of the polysaccharide in E. faecalis may be controlled so that it is only expressed under certain conditions, such as those found in infection. The use of more sensitive detection methods with a wider range of growth conditions is under way. Preliminary results from reverse transcription-PCR using primers to orfde4 and orfde6 suggest that the mRNA ttanscripts to those two genes could be detected in RNA extracts from OG1RF in exponential phase but not in stationary phase (data not shown). The other ORFs in contigs C and D are now being tested.
Comparison of the ORFs in contig C to database sequences revealed similarities to genes involved in polysaccharide biosynthesis and export from both gram-negative and gram-positive organisms. Orfde3 showed similarities to the undecaprenyl-phosphate α-N-acetylglucosaminyltransferase encoded by the rfe genes from various organisms and Llm of S. aureus (29). The activity of the lipophilic protein encoded by llm is not known, although it has been proposed to be involved in the metabolism of cell surface components such as peptidoglycan (29). Rfe is required for the synthesis of the first lipid-linked intermediate in the biosynthesis of enterobacterial common antigen and several O antigens (1, 41, 42). Hydropathy plots of the enterococcal Orfde3 and Rfe of E. coli showed remarkable similarities between the two, indicating similar overall distribution of hydrophobic residues. However, a Tn7 insertion near the 3′end of orfde3 (pTEX5159A) did not affect the expression of the polysaccharide in E. coli 21548 (rfe), Sφ874 (rfb), AB1133 (rfe+), or DH5α. Several possibilities may explain this result: (i) the C-terminal 60 or so amino acids may not be required for enzymatic activity, (ii) the gene is not required for the synthesis of the polysaccharide in E. coli, and (iii) orfde3 codes for some other function(s) instead of the undecaprenyl-phosphate α-N-acetylglucosaminyltransferase and is complemented by similar functions from E. coli. Further mutational studies of orfde3 may help answer some of these questions. One conclusion that can be drawn from results with pTEX5159A is that the insertion did not affect the expression of downstream genes, consistent with the result for DH5α(pTEX5159C), which had an insertion between orfde3 and orfde4 and was immunopositive, as well as the observation that orfde4 appeared to have its own promoter, based on DNA sequence.
orfde6 to orfde9 showed high sequence similarities to genes in the dTDP-rhamnose biosynthesis pathway of various organisms. As noted above, rhamnose has been found as a component in enterococcal polysaccharides; the transposon insertion in orfde8 (pTEX5159B) resulted in loss of immunoreactivity in Sφ874 (rfb) but not in DH5α. This finding indicated that the E. coli rfbBDAC genes could provide this function and provides further support for the conclusion the orfde6 to orfde9 encode a dTDP-rhamnose biosynthesis pathway.
The G+C contents of orfde6 to orfde9 are very close to each other, from 38.2 to 38.5%, while those of the other ORFs in the cluster vary from 32.6 to 40.6%, suggesting that orfde6 to orfde9 may have originated from one source while the other ORFs in the cluster may have been assembled from several sources during evolution. This is consistent with observations in other bacteria such as E. coli and Salmonella spp. (20, 30, 49, 55, 59).
Several reports have shown the involvement of ABC transport systems in the translocation of sugar polymers across the cytoplasmic membrane in both gram-negative and gram-positive bacteria )14, 25, 56, 61) or the presence of ABC transporter genes within polysaccharide biosynthesis clusters (5, 61). It seems that the use of ABC transporters could be one of the common mechanisms for exporting sugar polymers. The strong sequence similarities of orfde11 and orfde12 to components in the ABC transport system, in particular to the ABC tranporters for teichoic acids of B. subtilis (25) and the O antigen of M. xanthus (14), suggest that these two ORFs may play similar roles (14) in the export of the E. faecalis polysaccharide to the cell surface.
The products of six ORFs, orfde4, orfde5, orfde5_6, orfde10, orfde15, and orfde16 (3′ partial), showed various degrees of similarities to glycosyltransferases. One feature of these ORFs is that, except for orfde10, they encode hydrophilic proteins, consistent with the findings for other glycosyltransferases and the notion that the assembly of repeat units occurs on the cytoplasmic side of the membrane. The sequence homology of these ORF products to glycosyltransferases was not very strong, as has been recognized in general among glycosyltransferases. Thus, it would be difficult to propose, based on sequence comparison, the substrate specificity encoded by these ORFs. Further analysis, such as enzyme activity assays, is needed to elucidate their functions. Transposon insertions in orfde4 and orfde5 abolished the immunoreactivity in both DH5α and Sφ874, suggesting that these two genes are required for the production of the polysaccharide and that there are not equivalent functions in the E. coli host strains. The introduction of pTEX5175 into DH5α(pTEX5159D) restored its immunoreactivity, suggesting that its transposon insertion did not affect the expression of orfde6 and downstream genes. This is consistent with the results from sequence analysis that orfde4 and orfde6 may be transcribed from their own promoters and are in two individual transcriptional units.
Analysis of Tn7 transposon insertion mutants also provided evidence that orfde1 and orfde2 are not essential for antigen production. The involvement of orfde3 is not certain and requires further investigation. No Tn7 insertions in the other ORFs in contig C were identified. However, based on sequence analysis, it is likely that the other ORFs are also involved in the synthesis and export pathway. Tn7 insertions in contig D resulted in large deletions or rearrangement in the cosmid, suggesting that disruption in this region may be toxic to the host. Whether contig D is required in the biosynthesis pathway remains unclear.
In conclusion, a gene cluster of OG1RF was cloned and produced an antigenic polysaccharide in E. coli. Analysis of DNA sequences and transposon mutants indicated that it is a multicistronic region containing genes for dTDP-rhamnose synthesis, assembly of repeat units, and export of the polysaccharide. This is the first report of an enterococcal gene cluster involved in the biosynthesis of an antigenic polysaccharide. Further study of this cluster and the effects of such a polysaccharide on bacterial cell surface permeability, response to antimicrobial agents, and the host immune system may have important implications for research in enterococci as well as for polysaccharides in other organisms.
ACKNOWLEDGMENTS
We are grateful to Matthew C. Biery and Nancy L. Craig, Department of Molecular Biology and Genetics, Johns Hopkins University, for performing the in vitro transposition reactions, and as well as Huy Phan, University of Texas Medical School at Houston, for mapping the transposon insertion clones. We thank Chris Whitfield, Department of Microbiology, University of Guelph, Guelph, Ontario, Canada, and Paul Rick, Department of Microbiology, Uniformed Services of the Health Sciences, for providing strains and instructive advice. We also thank Miguel Valvano, Department of Microbiology and Immunology, University of Western Ontario, London, Ontario, Canada, for helpful advice.
This work was supported by the NIH grant AI 33516 to Barbara E. Murray.
REFERENCES
- 1.Alexander D C, Valvano M A. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J Bacteriol. 1994;176:7079–7084. doi: 10.1128/jb.176.22.7079-7084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archibald A R, Hancock I C, Harwood C R. Cell wall structure, synthesis, and turnover. In: Sonenshein A L, Hock J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 381–410. [Google Scholar]
- 3.Bainton R, Games P, Craig N. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991;65:805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- 4.Bainton R J, Kubo K M, Feng J-N, Craig N L. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- 5.Becker A, Ruberg S, Kuster H, Roxlau A A, Keller M, Ivashina T, Cheng H-P, Walker G C, Puhler A. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J Bacteriol. 1997;179:1375–1384. doi: 10.1128/jb.179.4.1375-1384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleiweis A S, Krause R M. The cell walls of group D streptococci. I. The immunochemistry of the type 1 carbohydrate. J Exp Med. 1965;122:237–249. doi: 10.1084/jem.122.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bronner D, Clarke B R, Whitfield C. Identification of an ATP-binding cassette transporter system required for translocation of lipopolysaccharide O-antigen side-chains across the cytoplasmic membrane of Klebsiella pneumoniae serotype O1. Mol Microbiol. 1994;14:505–519. doi: 10.1111/j.1365-2958.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 8.Chu S, Trust T J. An Aeromonas salmonicida gene which influences a-protein expression in Escherichia coli encodes a protein containing an ATP-binding cassette and maps beside the surface array protein gene. J Bacteriol. 1993;175:3105–3114. doi: 10.1128/jb.175.10.3105-3114.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung C T, Niemela S Z, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in procariotic membrane protein, the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez R C, Weiss A A. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect Immun. 1994;62:4727–4738. doi: 10.1128/iai.62.11.4727-4738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 13.Guidolin A, Morona J K, Morona R, Hansman D, Paton J C. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect Immun. 1994;62:5384–5396. doi: 10.1128/iai.62.12.5384-5396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo D, Bowden M G, Pershad R, Kaplan H B. The Myxococcus xanthus rfbABC operon encodes an ATP-binding cassette transporter homolog required for O-antigen biosynthesis and multicellular development. J Bacteriol. 1996;178:1631–1639. doi: 10.1128/jb.178.6.1631-1639.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock I C. Microbial cell surface architecture. In: Mozes N, Handley P S, Busscher H J, Rouxhet P G, editors. Microbial cell surface analysis: structure and physicochemical methods. New York, N.Y: VCH Publishers, Inc.; 1991. pp. 21–59. [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. p. 498. [Google Scholar]
- 17.Henderson B, Poole S, Wilson M. Bacterial modulins: a novel class of virulence factors which cause host tissue pathology by inducing cytokine synthesis. Microbiol Rev. 1996;60:316–341. doi: 10.1128/mr.60.2.316-341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivashina, T. V., M. R. Sadykov, A. A. Kanapin, and V. N. Ksenzenko. Unpublished data.
- 19.Jelinkova J, Rotta J. Identification and typing of enterococci. Methods Microbiol. 1978;12:199–220. [Google Scholar]
- 20.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko T, Tanaka A, Sato S, Kotani H, Sazuka T, Miyajima N, Sugiura M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 1995;2:153–166. doi: 10.1093/dnares/2.4.153. [DOI] [PubMed] [Google Scholar]
- 22.Kolkman M A, Morrison D A, van der Zeijst B A, Nuijten P J. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of glycosyl transferase gene cps14E. J Bacteriol. 1996;178:3736–3741. doi: 10.1128/jb.178.13.3736-3741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 24.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 25.Lazarevic V, Karamata D. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol. 1995;16:345–355. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 26.Liu D, Haase A M, Lindqvist L, Lindberg A A, Reeves P R. Glycosyl transferases of O-antigen biosynthesis in Salmonella enterica: identification and characterization of transferase genes of groups B, C2, and E1. J Bacteriol. 1993;175:3408–3413. doi: 10.1128/jb.175.11.3408-3413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu D, Reeves P R. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140(Pt. 1):49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- 28.Macpherson D F, Manning P A, Morona R. Characterization of the TDP-rhamnose biosynthesis genes encoded in the rfb locus of Shigella flexneri. Mol Microbiol. 1994;11:281–292. doi: 10.1111/j.1365-2958.1994.tb00308.x. [DOI] [PubMed] [Google Scholar]
- 29.Maki H, Yamaguchi T, Murakami K. Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J Bacteriol. 1994;176:4993–5000. doi: 10.1128/jb.176.16.4993-5000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marolda C L, Valvano M A. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J Bacteriol. 1995;177:5539–5546. doi: 10.1128/jb.177.19.5539-5546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier-Dieter U, Barr K, Starman R, Hatch L, Rick P D. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. J Biol Chem. 1992;267:746–753. [PubMed] [Google Scholar]
- 32.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthesis pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 33.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray B E. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray B E, Singh K V, Ross R P, Heath J D, Dunny G M, Weinstock G M. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuhard J, Thomassen E. Altered deoxyribonucleotide pools in P2 eductants of Escherichia coli K-12 due to deletion of the dcd gene. J Bacteriol. 1976;26:999–1001. doi: 10.1128/jb.126.2.999-1001.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Pazur J H, Cepure A, Kane J A. Glycans from streptococcal cell walls. The molecular structure of an antigenic diheteroglycan of glucose and galactose from Streptococcus faecalis. J Biol Chem. 1973;248:279–284. [PubMed] [Google Scholar]
- 39.Pazur J H, Forsberg S L. Determination of the sugar sequences and the glycosidic-bond arrangements of immunogenic heteroglycans. Carbohydr Res. 1978;60:167–178. doi: 10.1016/s0008-6215(00)83474-6. [DOI] [PubMed] [Google Scholar]
- 40.Pazur J H, Shuey E W. The enzymatic synthesis of thymidine diphosphate glucose and its conversion to thymidine diphosphate rhamnose. J Biol Chem. 1960;236:1780–1785. [PubMed] [Google Scholar]
- 41.Rick P D, Hubbard G L, Barr K. Role of the rfe gene in the synthesis of the O8 antigen in Escherichia coli K-12. J Bacteriol. 1994;176:2877–2884. doi: 10.1128/jb.176.10.2877-2884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rick P D, Mayer H, Neumeyer B A, Wolski S, Bitter-Suermann D. Biosynthesis of enterobacterial common antigen. J Bacteriol. 1985;162:494–503. doi: 10.1128/jb.162.2.494-503.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharpe M E, Shattock P M F. The serological typing of group D streptococci associated with outbreaks of neonatal diarrhoea. J Gen Microbiol. 1952;6:150–165. doi: 10.1099/00221287-6-1-2-150. [DOI] [PubMed] [Google Scholar]
- 46.Shockman G D, Barrett J F. Structure, function and assembly of cell walls of gram positive bacteria. Annu Rev Microbiol. 1983;37:501–527. doi: 10.1146/annurev.mi.37.100183.002441. [DOI] [PubMed] [Google Scholar]
- 47.Smyth J, Matthews H, Halpenny M K, Brandis H, Colman G. Biotyping, serotyping and phage typing of Streptococcus faecalis isolated from dental plaque in the human mouth. J Med Microbiol. 1987;23:45–54. doi: 10.1099/00222615-23-1-45. [DOI] [PubMed] [Google Scholar]
- 48.Stellwagen A E, Craig N L. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein that activates the bacterial transposon Tn7. Genetics. 1997;145:573–585. doi: 10.1093/genetics/145.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stingele F, Neeser J-R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsukioka Y, Yamashita Y, Nakano Y, Oho T, Koga T. Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J Bacteriol. 1997;179:4411–4414. doi: 10.1128/jb.179.13.4411-4414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsukioka Y, Yamashita Y, Oho T, Nakano Y, Koga T. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J Bacteriol. 1997;179:1126–1134. doi: 10.1128/jb.179.4.1126-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Kranenburg R, Marugg J D, van Swam I I, Willem N J, de Vos W M. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Lory S, Ramphal R, Jin S. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol Microbiol. 1996;22:1005–1012. doi: 10.1046/j.1365-2958.1996.01533.x. [DOI] [PubMed] [Google Scholar]
- 55.Wang L, Romana L R, Reeves P R. Molecular analysis of a Salmonella enterica group E1 rfb gene cluster: O antigen and the genetic basis of the major polymorphism. Genetics. 1992;130:429–443. doi: 10.1093/genetics/130.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitfield C, Valvano M A. Biosynthesis and expression of cell-surface polysaccharides in gram-negative bacteria. Adv Microb Physiol. 1993;35:135–246. doi: 10.1016/s0065-2911(08)60099-5. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y, Jiang L, Murray B E, Weinstock G M. Enterococcus faecalis antigens in human infections. Infect Immun. 1997;65:4207–4215. doi: 10.1128/iai.65.10.4207-4215.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamazaki M, Thorne L, Mikolajczak M, Armentrout R W, Pollock T J. Linkage of genes essential for synthesis of a polysaccharide capsule in Sphingomonas strain S88. J Bacteriol. 1996;178:2676–2687. doi: 10.1128/jb.178.9.2676-2687.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao Z, Valvano M A. Genetic analysis of the O-specific lipopolysaccharide synthesis region (rfb) of Escherichia coli K-12 W3110: identification of genes that confer group 6 specificity to Shigella flexneri serotype Y and 4a. J Bacteriol. 1994;176:4133–4143. doi: 10.1128/jb.176.13.4133-4143.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida Y, Nakano Y, Yamashita Y, Koga T. Identification of a genetic locus essential for serotype b-specific antigen synthesis in Actinobacillus actinomycemcomitans. Infect Immun. 1998;66:107–114. doi: 10.1128/iai.66.1.107-114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L, Al-Hendy A, Toivanen P, Skurnik M. Genetic organization and sequence of the rfb gene cluster of Yersinia enterocolitica serotype O:3: similarities to the dTDP-l-rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Mol Microbiol. 1993;9:309–321. doi: 10.1111/j.1365-2958.1993.tb01692.x. [DOI] [PubMed] [Google Scholar]