Abstract
Mutants of the Arabidopsis thaliana genes, EDM2 (Enhanced Downy Mildew 2), EDM3 (Enhanced Downy Mildew 3) and IBM2 (Increase in Bonsai Methylation 2) are known to show defects in a diverse set of defense and developmental processes. For example, they jointly exhibit enhanced levels of basal defense and stunted growth. Here we show that these two phenotypes are functionally connected by their dependency on the salicylic acid biosynthesis gene SID2 and the basal defense regulatory gene PAD4. Stunted growth of edm2, edm3 and ibm2 plants is a consequence of up-regulated basal defense. Constitutively enhanced activity of reactive oxygen species-generating peroxidases, we observed in these mutants, appears also to contribute to both, their enhanced basal defense and their growth retardation phenotypes. Furthermore, we found the histone H3 demethylase gene IBM1, a direct regulatory target of EDM2, EDM3 and IBM2, to be at least partially required for the basal defense and growth-related effects observed in these mutants. We recently reported that EDM2, EDM3 and IBM2 coordinate basal immunity with the timing of the floral transition by gradually reducing the extent of this defense mechanism prior to flowering. Together with these observations, data presented here show that at least some of the diverse phenotypic effects in edm2, edm3 and ibm2 mutants are genetically interlinked and functionally connected. Our new results show that repression of basal immunity by EDM2, EDM3 and IBM2 limits negative impact on growth and development.
Introduction
Sustained or constitutive activation of immune responses in plants is often associated with reduced growth and other developmental abnormalities, a phenomenon related to the concept of defense-growth trade-off. It has long been believed that such effects are due to competition of immunity-related and developmental processes for limited metabolic resources [1]. When plants are attacked by microbial pathogens, metabolic resources are preferentially allocated to defense, resulting in enhanced immunity, but at the expense of reduced growth [2, 3].
The Arabidopsis thaliana (Arabidopsis) Plant Homeodomain (PHD) finger protein-encoding EDM2 gene and the RNA Recognition Motif (RRM) domain protein-encoding genes EDM3/AIPP1 (hereafter EDM3) and IBM2/ASI1/SG1 (hereafter IBM2) are known to jointly affect multiple immunity-related and developmental processes [4–12]. They cooperate in a strong race-specific pathogen defense mechanism mediated by the disease resistance gene RPP7, that encodes an NLR-type immune receptor [13–15]. They also work together in an unspecific immune response, effective against a wide range of pathogens, termed basal immunity or basal resistance [16, 17]. While they promote RPP7-mediated immunity, EDM2, EDM3 and IBM2 suppress basal defense responses. The latter process is coordinated by these three genes with the floral transition [17]. Besides this and other developmental roles, EDM2, EDM3 and IBM2 promote overall growth, as their mutants exhibit reduced fresh weight and smaller rosette leaves [5, 11].
Mircoarray and RNA-seq studies with their mutants have shown EDM2, EDM3 and IBM2 to affect transcript levels of large overlapping sets of genes and transposable elements, while epigenome profiling revealed joint effects on methylation of the nucleobase cytosine and/or the repressive histone mark H3K9me2 at numerous chromatin sites [5, 8–10, 12, 16]. By chromatin-immunoprecipitation (ChIP) the EDM2, EDM3 and IBM2 proteins were found to co-localize at some of these loci [7–10, 12, 13, 16, 18]. RPP7 and the histone H3K9 demethylase gene IBM1 are among the direct target sites of EDM2, EDM3 and IBM2. Upon co-recruitment to heterochromatic stretches at each of these two (and several other) target loci, they prevent premature transcript termination at alternative polyadenylation signals, thereby promoting the synthesis of full-length mRNAs [8–10, 12, 13, 15, 16]. Consistent with their co-localization at chromatin sites, physical interactions among these three proteins were observed. While both EDM2 and IBM2 appear to directly interact with EDM3, interactions between EDM2 and IBM2 seem indirect and require EDM3 as a molecular bridge [12]. Recent results strongly suggested that EDM3/IBM2 interactions can be isoform specific [17]. Due to alternative splicing, two distinct protein isoforms are expressed for each of these two genes; a shorter one (EDM3S; IBM2S) and a longer one (EDM3L; IBM2L). Regarding their role in the suppression of basal immunity and the promotion of the floral transition, only the longer isoforms (EDM3L and IBM2L) are involved and not the shorter ones i.e. EDM3S and IBM2S [17].
Consistent with their role in suppressing the basal defense response, constitutive up-regulation in the transcripts of large overlapping sets of defense-related genes and immune receptor genes has been observed in the mutants of EDM2, EDM3 and IBM2 [15–17]. Thus, edm2, edm3 and ibm2 mutants exhibit three typical effects common to many other Arabidopsis mutants with constitutively elevated levels of basal immunity [19–23]: (1) constitutively elevated expression of defense genes, (2) enhanced basal defense and (3) retarded growth. However, it has been unclear, weather these effects are independent or causally connected.
Here we show that enhanced basal defense and growth retardation in edm2, edm3 and ibm2 mutants are genetically coupled by their dependency on the salicylic acid-associated defense genes SID2 and PAD4, implying that the latter effect is likely a consequence of the former one. We further present data suggesting that increased peroxidase activity, as a part of the constitutive basal defense response, is a partial cause of reduced edm2, edm3 or ibm2 growth rates. In addition we found that the direct EDM2, EDM3 and IBM2 target gene IBM1, is partially required for the basal defense and growth-related phenotypes. Each of these effects is mediated by the longer isoforms of EDM3 and IBM2 but not the shorter ones. Together with our previous report on the coordination of basal defense with the timing of the floral transition by EDM2, EDM3 and IBM2, our new results show that these three chromatin-associated regulators link various defense and developmental processes. This is likely contributing to a balanced defense-growth trade-off that enables efficient developmental transitions and limits growth penalties due to energetically costly basal immune responses.
Results
EDM2, EDM3L and IBM2L positively affect plant growth
Previously, we showed that edm2 mutants exhibit decreased fresh weight and rosette leaf expansion compared to their wild type parental lines [5]. Both fresh weight of aerial plant parts and rosette leaf expansion are also decreased in mutants of EDM3 and IBM2 (Fig 1A and 1B and S1 Fig). We used the previously described sg1-3 mutant allele of IBM2 [11], edm2-2 allele of EDM2 [13] and edm3-2 of EDM3 [17] for all experiments in this study. We further found that edm2, edm3, and ibm2 mutants have shorter primary roots (Fig 1C and S1 Fig). Moreover, only the longer isoforms of EDM3 and IBM2 we previously described, EDM3L and IBM2L [17], can rescue each of these two growth-related defects in edm3 or ibm2 mutants, respectively. Functional complementation by the shorter isoforms EDM3S and IBM2S failed as the respective transgenic lines exhibit similar phenotypes as their parental mutants (S1 Fig). Overall, these data show that EDM3L and IBM2L, like EDM2, positively affect overall plant growth.
Fig 1. EDM2, EDM3 and IBM2 positively affect overall plant growth.
A. Representative images of 25-day-old Arabidopsis plants of the indicated genotypes grown in soil. B. Average fresh weight of aerial parts from 25-day-old whole plants of the indicated genotypes. Error bars represent standard errors from three independent experiments, each with eighteen plants. C. Primary root length of 7-day-old plants of the indicated genotypes grown on agar plates. Primary root lengths of more than twelve plants of each genotype were measured using ImageJ. Data information: Asterisks indicate significant differences compared to Col-0 based on Student’s t-test. (**, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
Enhanced basal immunity in edm2, edm3 and ibm2 suppresses plant growth
To examine if growth defects in edm2, edm3 and ibm2 mutants are caused by their constitutively enhanced basal immunity, we used Arabidopsis lines with defects in defense responses controlled by salicylic acid (SA). The sid2-2 mutant is compromised in the defense-associated biosynthesis of this phytohormone [24], while the transgenic NahG line cannot accumulate SA, due to the expression of a bacterial SA hydroxylase gene [25]. The pad4-1 mutant is deficient in a SA-responsive signaling step [26]. We crossed edm3 and ibm2 mutants to each of these three lines and performed experiments with lines homozygous for each altered component (mutation or transgene). In addition, we generated edm2-2;sid2-2 and edm2-2;pad4-1 double mutants. We first examined if enhanced immunity in edm2, edm3 and ibm2 is reduced by blockage of SA-mediated defense. We tested Arabidopsis seedlings one week after infection with the Noco2 isolate of the pathogenic oomycete Hyaloperonospora arabidopsidis (Hpa) infection, which is virulent on wild type Col-0 plants and triggers basal defense in this accession [27]. As shown previously [17], edm2, edm3 and ibm2 plants show reduced susceptibility against HpaNoco2 compared to Col-0, indicating that basal defense in these single mutants is enhanced (Fig 2A–2D). However, we observed complete loss of this enhanced basal defense phenotype in all double mutants as well as the edm3-2;NahG and sg1-3/ibm2;NahG lines.
Fig 2. Enhanced basal immunity in mutants of EDM2, EDM3 and IBM2 requires salicylic acid, SID2, and PAD4.
A-D. Enhanced basal immunity in edm2, edm3 and ibm2 mutants was disrupted by introducing the sid2-2 or pad4 mutations or the NahG transgene. Two-week-old seedlings were sprayed-inoculated with 3 x 104/ml HpaNoco2 spores. HpaNoco2 spores were counted one week post infection, which were divided by the corresponding fresh weight and shown as percentage relative to wild type. E-G. Fresh weight of aerial parts from 24-day-old whole plants (E), 23-day-old whole plants (F) or 25-day-old whole plants (G) of the indicated genotypes, which were grown on soil. Data information: Data shown in each separate panel were generated simultaneously. Error bars represent standard errors from three independent experiments. Asterisks indicate significant differences compared to Col-0 based on Student’s t-test (A-D) or by one-way ANOVA with Brown-Forsythe and Welch’s test (E) or by ordinary one-way ANOVA (F & G). (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, no significance).
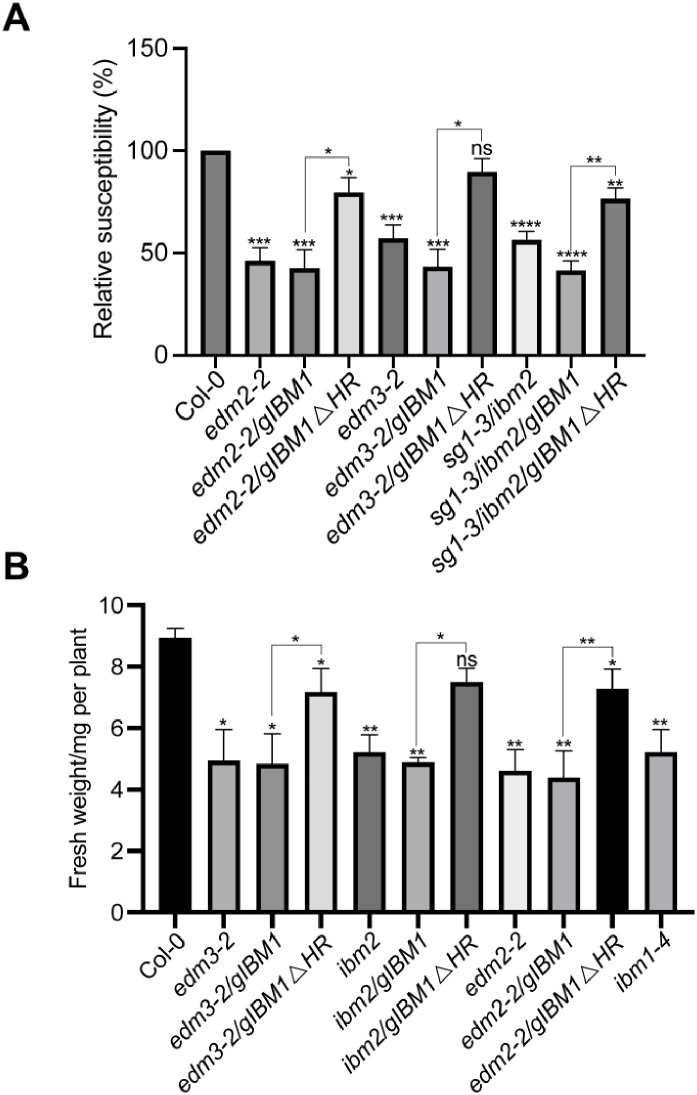
We further observed the growth-related effects on the fresh weight of aerial plant parts and primary root length of edm2, edm3 and ibm2 mutants to be partly or fully reversed to wild type levels in the respective double mutants with sid2-2, or pad4 (Fig 2E–2G and S2 Fig). We made the same observation with edm3;NahG and ibm2;NahG lines (Fig 2E–2G and S2 Fig). One deviation are the fresh weight measurements of ibm2 plants in Fig 2G, which seem lower than Col-0, but are not significantly different from this wild type reference line. The Student’s t-test p value for this is slightly above the significance cutoff. However, compared to Col-0 we observed fresh weight of aerial plant parts and overall growth of ibm2 plants to be significantly reduced with very high consistency in all other experiments of this study (Figs 1, 2E, 6B; S1B, S1E & S1F, S2A, S2D & S2E and S3). Still the fresh weight of aerial parts of ibm2;pad4 double mutants in this set of experiments is significantly higher than that of the ibm2 single mutant (Fig 2G).
Overall, these findings show that enhanced immunity in mutants of EDM2, EDM3 and IBM2 relies on the SA-dependent basal defense pathway. As this pathway is further required for the reduction of aerial part fresh weight and primary root length in edm2, edm3 and ibm2 plants, we conclude that the growth-related effects are a consequence of the enhanced basal defense phenotype of these mutants.
EDM2, EDM3 and IBM2 suppress ROS-generating peroxidase activity by down-regulation of type III peroxidase genes
A reactive oxygen species (ROS)-generating oxidative burst is an early event among plant immune responses [19–23, 28–30]. In Arabidopsis, NADPH oxidases and cell wall-associated type III peroxidases are two main sources of ROS in the apoplast [30, 31]. ROS are not only involved in immune response but also affect plant growth. ROS accumulation can stiffen cell walls by cross-linking its components, thereby inhibiting cell expansion [32]. Due to these dual effects, ROS have been linked to the defense-growth trade-off as a shared component of both processes [33].
Re-inspecting our previous RNA-seq analyses on EDM2-mediated transcriptome changes [16] we found 27 type III peroxidase genes and two NADPH oxidase genes, RbohD and RbohF, to be significantly up-regulated in edm2 plants compared to their wild type parental line, while the gene encoding the H2O2 scavenging enzyme, CAT1, is significantly down-regulated in this mutant. Since over-accumulation of the ROS H2O2 can reduce cell wall extensibility [32], the joint upregulation of type III peroxidase and NADPH-oxidase genes may contribute to growth-related effects in edm2 plants.
We selected 11 of these 27 type-III peroxidase genes, which show either particularly strongly increased transcript levels in edm2 plants or are possible direct target genes of EDM2 and IBM2 based on previous ChIP-seq analyses [18]. For nine of these 11 genes (except PRX31 and PRX62) we consistently observed in qRT-PCRs constitutively elevated transcript levels in the tested edm2-2, edm3-2 and sg1-3/ibm2 mutants compared to Col-0 (Fig 3A). While we could not confirm the up-regulation of RbohD and RbohF (Fig 3B), we observed by qRT-PCR a significant up-regulation of RbohB, another member of this family of NADPH oxidase genes in all three tested mutants (Fig 3B).
Fig 3. EDM2, EDM3 and IBM2 suppress expression levels of genes involved in the generation of ROS as well as activity levels of peroxidases.
A. The transcript levels of selected peroxidase genes in mutants of EDM2, EDM3 and IBM2 in the aerial parts measured by qRT-PCR. Log2-Fold Change (FD) relative to transcript levels in wild type was used to create this heatmap. The brightest red of the above scale represents the highest increase relative to wild type plants. Numbers above the scale bar represent log2 fold change values of transcript level increases relative to Col-0. B. Transcript levels of NADPH oxidase genes in mutants of EDM2, EDM3 and IBM2 in the aerial parts measured by qRT-PCR. C & D. Relative peroxidase activity in mutants of EDM2, EDM3 and IBM2. Peroxidase activity was measured in the aerial parts of two-week-old plants, divided by fresh weight and shown relative to the wild type plants. Data information: Error bars represent standard errors from three independent experiments. Asterisks indicate significant differences analyzed by student’s t-test. (*, p < 0.05; **, p < 0.01; ****, p < 0.0001; ns, no significance).
We further tested if peroxidase enzyme activity is also increased in these mutants. Type III peroxidases are known to be localized to the cell wall. Thus, the transcriptional upregulation of these enzymes we observed in mutants of EDM2, EDM3 and IBM2 should lead to increased peroxidase activity in their ionically bound protein fractions. Indeed, we found that compared to Col-0, the edm2, edm3 and ibm2 mutants jointly exhibit a noticeable increase in constitutive peroxidase activity in the ionically bound fraction (Fig 3C), but not the soluble fraction (Fig 3D).
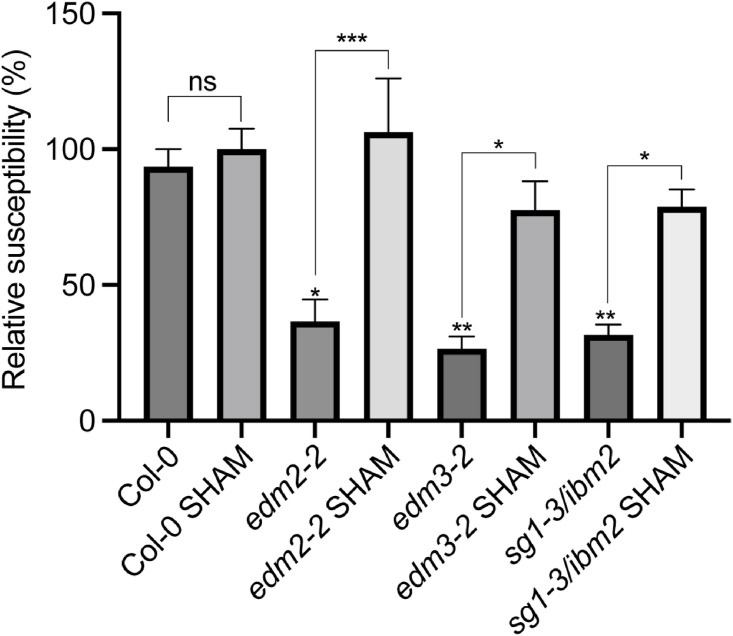
To test if the increased peroxidase activity contributes to the growth defects observed in edm2, edm3 and ibm2 plants, we treated them with 20μM of the peroxidase inhibitor salicylhydroxamic acid (SHAM), at day 14 and day 16 and compared their rosette areas at day 25. The concentration and timing of SHAM treatment we applied has been previously reported as suitable [34]. We observed a significant increase in rosette areas in all tested mutants after SHAM treatment compared to untreated plants, while there is no difference in wild type after treatment with this inhibitor (Fig 4). Overall, these data suggest that increased peroxidase activity contributes to growth defects in edm2, edm3 and ibm2 mutants.
Fig 4. Growth defects in mutants of EDM2, EDM3 and IBM2 are partly rescued by inhibiting peroxidase activity.
Rosette areas of 25-day-old plants of the indicated genotypes. 20μM SHAM was applied to soil grown plants at day 14 and day 16. Rosette area (n ≥ 16) was measured using ImageJ. Data information: Asterisks indicate significant differences between SHAM and non-SHAM treated plants of the same genotype based on Student’s t-tests. (*, p < 0.05; **, p < 0.01; ****, p < 0.0001; ns, no significance).
Basal HpaNoco2 resistance in mutants of EDM2, EDM3 and IBM2 is decreased by inhibiting peroxidase activity
To examine if enhanced basal resistance in edm2, edm3 and ibm2 mutants is partially due to the increased type III peroxidase activity, we pre-treated all three mutants with SHAM 24 hours before infection with HpaNoco2. Compared to untreated plants we observed a larger number of HpaNoco2 spores in all three mutant plants after treatment with SHAM and a mild increase in Col-0 wild type plants with SHAM treatment (Fig 5), suggesting increased peroxidase activity contributes to enhanced immunity in all three mutants.
Fig 5. Increased type III peroxidase activity is likely to contribute to enhanced basal defense in mutants of EDM2, EDM3 and IBM2.
Two-week-old plants of the indicated genotypes were pre-treated with 20μM SHAM 24 hours before sprayed-inoculated with 1 x 104/ml HpaNoco2 spores. HpaNoco2 spores numbers were determined one week after infection. Spore numbers were normalized to the corresponding fresh weights. Data information: Error bars represent standard errors from three independent experiments. Asterisks indicate significant differences between plants of the same genotype based one-way ANOVA. (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, no significance).
IBM1L partly rescues growth defects and disease susceptibility in mutants of EDM2, EDM3 and IBM2
At the epigenome and transcriptome levels mutations in the histone H3 demethylase gene IBM1 phenocopy many effects of edm2, edm3 and ibm2 mutations. Mutants of IBM1 are also growth-retarded. EDM2, EDM3 and IBM2 are known to promote the expression of the functional full-length IBM1 isoform (IBM1L) by suppressing proximal transcript polyadenylation at an alternative polyadenylation signal in a heterochromatic repeat region in the seventh IBM1 intron [8, 9]. To examine dependency of EDM2, EDM3 and IBM2-mediated basal defense and growth-related effects on IBM1, we transformed into edm2, edm3 and ibm2 mutants a genomic clone containing the entire IBM1 gene (gIBM1) or a mutated version of this clone with a deletion of the heterochromatic repeats in its seventh intron (gIBM1ΔHR). While gIBM1 was shown before to rescue various phenotypes of ibm1 mutants, gIBM1ΔHR, but not gIBM1, rescues low expression levels of the functional full-length IBM1L isoform and leaf abnormalities in edm2 and ibm2 mutants [8, 9]. The gIBM1ΔHR clone lacks the proximal alternative polyadenylation signal in intron 7 and, therefore, does not require EDM2, EDM3 and IBM2 for proper expression of IBM1L. We observed that HpaNoco2 susceptibility, aerial part fresh weight and primary root length were at least partially restored to wild type levels in edm2, edm3 and ibm2 mutants containing gIBM1ΔHR, but not gIBM1 (Fig 6A and 6B and S3 Fig). Compared to Col-0 in most cases HpaNoco2 susceptibility levels, aerial part fresh weight and primary root length were still significantly reduced in the mutants containing gIBM1ΔHR. However, in all cases these values were also significantly increased compared to their respective gIBM1-containing counterparts. Thus, the histone H3 demethylase gene IBM1, is at least partially responsible for EDM2, EDM3 and IBM2-mediated suppression of basal defense and the resulting promoting effects on growth.
Fig 6. Growth defects and HpaNoco2 susceptibility were partly rescued by IBM1L.
A. HpaNoco2 susceptibility is rescued in mutants of EDM2, EDM3 and IBM2 by a genomic IBM1 clone with a deletion of its heterochromatic repeats in intron 7 (gIBM1ΔHR), but not by the complete genomic region lacking the deletion (gIBM1). Two-week old plants were spray-inoculated with 3 x 104/ml HpaNoco2 spores. HpaNoco2 spores were counted one week post infection. The resulting spore numbers were divided by the respective fresh weight and shown as percentage relative to wild type. B. Fresh weight of aerial parts from twelve-day-old whole plants of the indicated genotypes. Data information: Error bars represent standard errors from three independent experiments. Asterisks indicate significant difference compared to Col-0 based on Student’s t-test (*, p < 0.05; **, p < 0.01; ***, p< 0.001; ****, p < 0.0001, ns, no significance).
Discussion
Numerous Arabidopsis mutants with constitutively activated immunity have been described before, such as cpr (constitutive expresser of PR genes) [22], cim (constitutive immunity) [23], lsd (lesion-simulating disease resistance) [20], acd (accelerated cell death) [35], dnd (defense no death) [36] and edr (enhanced disease resistance) [37] mutants. Common to mutants of these classes are constitutively enhanced expression levels of defense-related genes, enhanced basal defense and retarded growth or other developmental defects. These effects are typically dependent on SA-mediated defense signaling. Mutants of EDM2, EDM3 and IBM2 exhibit the same set of phenotypic effects. Using double mutants we showed enhanced basal defense and retarded rosette and root growth of edm2, edm3 and ibm2 plants to depend on the SA biosynthesis gene SID2 and the SA signaling gene PAD4. Expression of the bacterial SA hydroxylase gene NahG leads to the same effects in edm3 and ibm2 plants. Thus, both the enhanced basal defense and retarded growth phenotypes of the edm2, edm3 and ibm2 mutants are genetically interlinked. As blockage of SA-mediated defense responses rescues the growth defects of these mutants, their growth retardation must be a consequence of the enhanced basal defense. Consequently EDM2, EDM3 and IBM2 contribute to the defense-growth trade of by limiting the extent of energetically costly basal defense responses, thereby, prioritizing overall growth (Fig 7). We recently showed the coordination of basal defense with the timing of the floral transition to be mediated by the respective longer isoforms of EDM3 and IBM2. This also applies to the growth promoting effects, which is consistent with the view that these processes are interlinked.
Fig 7. Model illustrating joint effects of EDM2, EDM3 and IBM2 on basal immunity and growth.
A. In wild type Arabidopsis, EDM2, EDM3 and IBM2 suppress SA-dependent basal immunity. This effect is at least partially mediated by their promoting effect on expression of the full-length IBM1 isoform IBM1L. Reduced basal immunity has a promoting effect on plant growth. B. In single mutants of EDM2, EDM3 or IBM2, SA-dependent basal defense responses are de-repressed resulting in reduced plant growth. The increase if SA-dependent basal immunity is at least partially due to reduced levels of IBM1L.
While EDM2, EDM3 and IBM2 suppress basal defense, they have the opposite effect on immunity mediated by the NLR-type immune receptor genes RPP7 and RPP4, the expression of which they promote [4, 12, 14–17]. In particular RPP7-mediated immunity may be energetically costly to plants, as this defense mechanism is unusually strong leading to very tight disease resistance [4, 38]. In addition, mutants of RPP7 exhibit enhanced overall growth, suggesting that even in a resting state this immune receptor consumes energy-related resources (Yan Lai, Tokuji Tsuchiya; Jianqiang. Wang, and Thomas. Eulgem, unpublished). Thus, the contribution to the defense-growth trade-off of EDM2, EDM3 and IBM2 seems complex. Rather than serving as general suppressors immune responses, EDM2, EDM3 and IBM2 seem to mediate a certain balance of defense-related processes, by having counter-directional effects and promoting some defense mechanisms at the expense of others. In addition, they coordinate the extent of basal defense with at least one important developmental transition, the one from vegetative growth to flowering.
As a likely joint contributor of the elevated basal immunity in edm2, edm3 and ibm2 plants we identified constitutively increased activity of type III peroxidases, a class of enzymes known to contribute to the defense-associated oxidative burst. The oxidative burst is a very early immune response, occurring within minutes when plants are invaded by pathogens [39]. Plant defenses require ROS generated in the oxidative burst, which serve as defensive antimicrobial compounds, cross-linkers of cell wall macromolecules as well as defense signaling molecules [40, 41]. Expression of a set of type III peroxidase-encoding genes is constitutively upregulated in edm2, edm3 and ibm2 mutants. Consistent with the known localization of type III peroxidases to plant cell walls [31], we observed increased peroxidase activity in anionic protein fractions and not in soluble protein fractions of edm2, edm3 and ibm2 plants. Blockage of peroxidase activity by the inhibitor SHAM partially reversed the basal defense and growth levels of these mutants to those observed in their wild type parents. Thus, increased type III-mediated accumulation of ROS may contribute to the enhanced basal defense phenotype observed in edm2, edm3 and ibm2 plants. Apoplastic ROS accumulation may also reduce cell wall extensibility possibly due to lignin formation and cross-linking of other macromolecular components [32, 42], thereby suppressing plant cell growth. However, using standard H2O2 detection assays, we were unable to detect any significant increase of this reactive oxygen species in edm2, edm3 and ibm2 plants. Perhaps the actual increase of ROS is only incremental and too weak to be detectable by the assays we applied.
Many epigenomic and transcriptomic effects caused by EDM2, EDM3, IBM2 are known to be mediated by their direct target gene IBM1. By suppressing proximal polyadenylation of IBM1 transcripts EDM2, EDM3 and IBM2 jointly promote expression of the functional full-length IBM1L isoform. Here we found suppression of basal defense as well as the promotion of rosette and root growth by EDM2, EDM3 and IBM2 to depend at least partially on IBM1L. We already reported overlapping effects on the expression of defense-related and immune receptor genes by EDM2 and IBM1 [16]. However, we also observed that each of these regulators affects transcript levels of separate sets of defense-related and immune receptor genes. IBM1, in contrast to EDM2, EDM3 and IBM2, does not affect RPP7 expression and immunity-mediated by this NLR gene.
Collectively our results show joint effects of EDM2, EDM3 and IBM2 on basal immunity and overall plant growth to be genetically interlinked and functionally connected by their dependency on SID2, PAD4 and IBM1. This view is further supported by effects we observed using the type III peroxidase inhibitor SHAM. The promotion of plant root and rosette growth by EDM2, EDM3 and IBM2 must be a consequence of their suppressive effects on basal immunity. Generally these three chromatin-associated regulators seem to contribute to a balanced defense-growth trade off by prioritizing overall plant growth over basal defense responses. Results of this study capture only a fraction of the roles EDM2, EDM3 and IBM2 have in coordinating immune mechanisms and developmental/growth-related processes. As outlined above, they also align the extent of basal defense with the timing of the floral transition and prioritize RPP7-mediated race-specific immunity over basal defense. Additional research is needed to uncover the mechanics of these coordinative processes and to understand the full scope and significance their have in mediating a balanced defense-growth trade off.
Materials and methods
Plant material and growth conditions
All the genotypes except specified are in Col-0 background. The following single mutants used in this study: edm2-2 [4], edm3-2 [17], sg1-3/ibm2 [11], sid2-2 [43], pad4 [26], and ibm1-4 [16]. The following double mutants used in this study: edm2-2;edm3-2, edm2-2;sg1-3/ibm2, edm3-2;sg1-3/ibm2 were described previously [17]. The following transgenic lines used in this study: EDM3Snp-1, EDM3Snp-2, EDM3Lnp-1, EDM3Lnp-2, IBM2S-1 and IBM2L-1 were described previously [17], NahG [44]. The genomic IBM1 region (gIBM1) or gIBM1 with a deletion of the heterochromatic repeats in its 7th intron (gIBM1ΔHR) were cloned as described previously [9] and transformed into edm2-2, edm3-2 and sg1-3/ibm2, respectively, to generate the gIBM1 and gIBM1ΔHR transgenic lines. Double mutants were made by crossing and F2 seeds were screened by genotyping PCR to select homozygotes. Plants are grown either on ½ MS medium [45] or in soil under long day (16h/8h light-dark cycles).
Primary root length measurement
Plants were grown vertically on ½ MS medium for five or seven days. Primary root length of the indicated genotypes was measured using ImageJ.
Rosette area measurements
Seedlings were untreated or treated with 20μM SHAM (S607-5G, Sigma) at day 14 and day16. Rosette areas were pictured and measured using ImageJ at day 25.
RNA extraction and qRT-PCR
Total RNA of the aerial parts of two weeks old plants was extracted using Trizol reagent (Life technologies) according to the instructions of the manufacturer. 1μg of RNA was reversed transcribed into cDNA using Maxima Reverse Transcriptase (Fisher scientific). The cDNA products were used for Real-time PCR with CFX CONNECT detection system (Bio-Rad). All primers used are listed in S1 Table.
Peroxidase activity measurement and inhibitor experiments
Two weeks old seedlings were used for peroxidase activity measurement as described previously [46]. Effects of peroxidase inhibition were tested using salicylhydroxamic acid (SHAM).
Hyaloperonospora arabidopsidis infection assay
HpaNoco2 infection assays were performed as previously described [47]. Briefly, two week-old soil-grown Arabidopsis seedlings were infected by foliar spray with a water-based suspension of 1–3 x 104 spores/ml of asexual HpaNoco2 spores, then covered by saran wrap and kept at 18°C under short day conditions (8h-day/16h-night cycle) for 7 days. The average numbers of spores per 12 seedlings were determined using a hemocytometer and normalized to the fresh weight of the aerial plant parts.
Supporting information
A Fresh weight of aerial parts from 15-day-old Col-0, edm3-2 or EDM3 isoform-specific complementation lines (EDM3Snp-1, EDM3Lnp-1) grown in soil. EDM3Snp-1 and EDM3Lnp-1 express in the edm3-2 mutant background either the short or long EDM3 isoform, respectively, driven by the native EDM3 promoter (np). B. Fresh weight of aerial parts from 12-day old Col-0, ibm2 and IBM2 isoform-specific complementation lines (IBMS-1, IBM2L-1) grown in soil. IBMS-1, IBM2L-1 express in the ibm2 mutant background either the short or long IBM2 isoform, respectively, driven by the native IBM2 promoter. The sg1-3 mutant allele of IBM2 was used for all experiments. C & E. Primary root length of 5-day-old plants of Col-0, edm3-2 and ibm2 plants as well as EDM3-and IBM2-isoform specific complementation lines grown on agar plates. D & F. Representative images of plants used in panels C and E. Data information: Error bars represent standard errors from three independent experiments. Asterisks indicate significant differences compared to Col-0 based on Student’s t-test. (*, p < 0.05; **, p < 0.01; ****, p < 0.0001; ns, no significance). n ≥ 28 (C) and n ≥ 20 (E).
(TIF)
Primary root length of the indicated genotypes was measured using ImageJ. Data information: Data shown in each separate panel were generated simultaneously. Asterisks indicate significant differences compared to Col-0 based on one-way ANOVA (A-E). (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, no significance). n ≥ 18 (A), n ≥ 24 (B), n ≥ 33 (C), n ≥ 17 (D), n ≥ 21 (E).
(TIF)
Primary root lengths of five day-old plants of the indicated genotypes were measured using ImageJ. Error bars represent standard errors. Asterisks indicate significant differences compared to Col-0 based on one-way ANOVA. (**, p < 0.01; ****, p < 0.0001). n ≥ 20.
(TIF)
(DOCX)
Acknowledgments
We thank Drs. Xuemei Chen and Meng Chen (both University of California, Riverside, USA) for advice on this project.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
This work was supported by National Science Foundation (NSF) grant IOS-1457329 to TE. URL of funders web site: https://www.nsf.gov/div/index.jsp?div=IOS The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Karasov TL, Chae E, Herman JJ, Bergelson J. Mechanisms to Mitigate the Trade-Off between Growth and Defense. Plant Cell. 2017;29: 666–680. doi: 10.1105/tpc.16.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kempel A, Schädler M, Chrobock T, Fischer M, van Kleunen M. Tradeoffs associated with constitutive and induced plant resistance against herbivory. Proc Natl Acad Sci U S A. 2011;108: 5685–5689. doi: 10.1073/pnas.1016508108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Züst T, Rasmann S, Agrawal AA. Growth–defense tradeoffs for two major anti‐herbivore traits of the common milkweed Asclepias syriaca. Oikos. 2015. Available: https://onlinelibrary.wiley.com/doi/abs/10.1111/oik.02075 [Google Scholar]
- 4.Eulgem T, Tsuchiya T, Wang X-J, Beasley B, Cuzick A, Tör M, et al. EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 2007;49: 829–839. doi: 10.1111/j.1365-313X.2006.02999.x [DOI] [PubMed] [Google Scholar]
- 5.Tsuchiya T, Eulgem T. Co-option of EDM2 to distinct regulatory modules in Arabidopsis thaliana development. BMC Plant Biology. 2010. p. 203. doi: 10.1186/1471-2229-10-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsuchiya T, Eulgem T. The Arabidopsis defense component EDM2 affects the floral transition in an FLC-dependent manner. Plant J. 2010;62: 518–528. doi: 10.1111/j.1365-313X.2010.04169.x [DOI] [PubMed] [Google Scholar]
- 7.Tsuchiya T, Eulgem T. Mutations in EDM2 selectively affect silencing states of transposons and induce plant developmental plasticity. Sci Rep. 2013;3: 1701. doi: 10.1038/srep01701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei M, La H, Lu K, Wang P, Miki D, Ren Z, et al. Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns. Proc Natl Acad Sci U S A. 2014;111: 527–532. doi: 10.1073/pnas.1320106110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saze H, Kitayama J, Takashima K, Miura S, Harukawa Y, Ito T, et al. Mechanism for full-length RNA processing of Arabidopsis genes containing intragenic heterochromatin. Nat Commun. 2013;4: 2301. doi: 10.1038/ncomms3301 [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Duan C-G, Tang K, Wang B, Zhang H, Lei M, et al. RNA-binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin-containing gene IBM1. Proc Natl Acad Sci U S A. 2013;110: 15467–15472. doi: 10.1073/pnas.1315399110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coustham V, Vlad D, Deremetz A, Gy I, Cubillos FA, Kerdaffrec E, et al. SHOOT GROWTH1 maintains Arabidopsis epigenomes by regulating IBM1. PLoS One. 2014;9: e84687. doi: 10.1371/journal.pone.0084687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan C-G, Wang X, Zhang L, Xiong X, Zhang Z, Tang K, et al. A protein complex regulates RNA processing of intronic heterochromatin-containing genes in Arabidopsis. Proc Natl Acad Sci U S A. 2017;114: E7377–E7384. doi: 10.1073/pnas.1710683114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuchiya T, Eulgem T. An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc Natl Acad Sci U S A. 2013;110: E3535–43. doi: 10.1073/pnas.1312545110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deremetz A, Le Roux C, Idir Y, Brousse C, Agorio A, Gy I, et al. Antagonistic Actions of FPA and IBM2 Regulate Transcript Processing from Genes Containing Heterochromatin. Plant Physiol. 2019;180: 392–403. doi: 10.1104/pp.18.01106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai Y, Cuzick A, Lu XM, Wang J, Katiyar N, Tsuchiya T, et al. The Arabidopsis RRM domain protein EDM3 mediates race-specific disease resistance by controlling H3K9me2-dependent alternative polyadenylation of RPP7 immune receptor transcripts. Plant J. 2019;97: 646–660. doi: 10.1111/tpj.14148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai Y, Lu XM, Daron J, Pan S, Wang J, Wang W, et al. The Arabidopsis PHD-finger protein EDM2 has multiple roles in balancing NLR immune receptor gene expression. PLoS Genet. 2020;16: e1008993. doi: 10.1371/journal.pgen.1008993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Eulgem T. The Arabidopsis RRM domain proteins EDM3 and IBM2 coordinate the floral transition and basal immune responses. Plant J. 2023. doi: 10.1111/tpj.16364 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y-Z, Lin J, Ren Z, Chen C-X, Miki D, Xie S-S, et al. Genome-wide distribution and functions of the AAE complex in epigenetic regulation in Arabidopsis. J Integr Plant Biol. 2021;63: 707–722. doi: 10.1111/jipb.13068 [DOI] [PubMed] [Google Scholar]
- 19.Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6: 1845–1857. doi: 10.1105/tpc.6.12.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL. Arabidopsis mutants simulating disease resistance response. Cell. 1994;77: 565–577. doi: 10.1016/0092-8674(94)90218-6 [DOI] [PubMed] [Google Scholar]
- 21.Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, et al. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103: 1111–1120. doi: 10.1016/s0092-8674(00)00213-0 [DOI] [PubMed] [Google Scholar]
- 22.Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in arabidopsis. Plant Cell. 2000;12: 2175–2190. doi: 10.1105/tpc.12.11.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maleck K, Neuenschwander U, Cade RM, Dietrich RA, Dangl JL, Ryals JA. Isolation and characterization of broad-spectrum disease-resistant Arabidopsis mutants. Genetics. 2002;160: 1661–1671. doi: 10.1093/genetics/160.4.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414: 562–565. doi: 10.1038/35107108 [DOI] [PubMed] [Google Scholar]
- 25.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, et al. Requirement of salicylic Acid for the induction of systemic acquired resistance. Science. 1993;261: 754–756. doi: 10.1126/science.261.5122.754 [DOI] [PubMed] [Google Scholar]
- 26.Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, et al. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci U S A. 1999;96: 13583–13588. doi: 10.1073/pnas.96.23.13583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell. 1996;8: 2033–2046. doi: 10.1105/tpc.8.11.2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, et al. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 2012;24: 275–287. doi: 10.1105/tpc.111.093039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torres MA, Dangl JL, Jones JDG. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci U S A. 2002;99: 517–522. doi: 10.1073/pnas.012452499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolwell GP, Wojtaszek P. Mechanisms for the generation of reactive oxygen species in plant defence–a broad perspective. Physiological and Molecular Plant Pathology. 1997. pp. 347–366. doi: 10.1006/pmpp.1997.0129 [DOI] [Google Scholar]
- 31.Smirnoff N, Arnaud D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019;221: 1197–1214. doi: 10.1111/nph.15488 [DOI] [PubMed] [Google Scholar]
- 32.Schopfer P. Hydrogen peroxide-mediated cell-wall stiffening in vitro in maize coleoptiles. Planta. 1996;199: 43–49. [Google Scholar]
- 33.Neuser J, Metzen CC, Dreyer BH, Feulner C, van Dongen JT, Schmidt RR, et al. HBI1 Mediates the Trade-off between Growth and Immunity through Its Impact on Apoplastic ROS Homeostasis. Cell Rep. 2019;28: 1670–1678.e3. doi: 10.1016/j.celrep.2019.07.029 [DOI] [PubMed] [Google Scholar]
- 34.Lu D, Wang T, Persson S, Mueller-Roeber B, Schippers JHM. Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nat Commun. 2014;5: 3767. doi: 10.1038/ncomms4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenberg JT, Ausubel FM. Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J. 1993;4: 327–341. doi: 10.1046/j.1365-313x.1993.04020327.x [DOI] [PubMed] [Google Scholar]
- 36.Yu IC, Parker J, Bent AF. Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci U S A. 1998;95: 7819–7824. doi: 10.1073/pnas.95.13.7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frye CA, Tang D, Innes RW. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci U S A. 2001;98: 373–378. doi: 10.1073/pnas.98.1.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDowell JM, Dangl JL. Signal transduction in the plant immune response. Trends Biochem Sci. 2000;25: 79–82. doi: 10.1016/s0968-0004(99)01532-7 [DOI] [PubMed] [Google Scholar]
- 39.Wojtaszek P. Oxidative burst: an early plant response to pathogen infection. Biochem J. 1997;322 (Pt 3): 681–692. doi: 10.1042/bj3220681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorrain S, Vailleau F, Balagué C, Roby D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8: 263–271. doi: 10.1016/S1360-1385(03)00108-0 [DOI] [PubMed] [Google Scholar]
- 41.Moeder W, Yoshioka K. Lesion mimic mutants: A classical, yet still fundamental approach to study programmed cell death. Plant Signal Behav. 2008;3: 764–767. doi: 10.4161/psb.3.10.6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee Y, Rubio MC, Alassimone J, Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153: 402–412. doi: 10.1016/j.cell.2013.02.045 [DOI] [PubMed] [Google Scholar]
- 43.Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, et al. Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 2000;24: 205–218. doi: 10.1046/j.1365-313x.2000.00870.x [DOI] [PubMed] [Google Scholar]
- 44.Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, et al. A central role of salicylic Acid in plant disease resistance. Science. 1994;266: 1247–1250. doi: 10.1126/science.266.5188.1247 [DOI] [PubMed] [Google Scholar]
- 45.Classic Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962. Available: http://wolfe91.free.fr/MMBI/milieu%20MS/Murashige.pdf [Google Scholar]
- 46.Bindschedler LV, Dewdney J, Blee KA, Stone JM, Asai T, Plotnikov J, et al. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 2006;47: 851–863. doi: 10.1111/j.1365-313X.2006.02837.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDowell JM, Cuzick A, Can C, Beynon J, Dangl JL, Holub EB. Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 2000;22: 523–529. doi: 10.1046/j.1365-313x.2000.00771.x [DOI] [PubMed] [Google Scholar]